Abstract
Under low-Ca conditions, plants accumulate salicylic acid (SA) and induce SA-responsive genes. However, the relationship between SA and low-Ca tolerance remains unclear. Here, we demonstrated that the inhibition or suppression of nonexpressor of pathogenesis-related 1 (NPR1) activity, a major regulator of the SA signaling pathway in the defense response, improves shoot growth under low-Ca conditions. Furthermore, mutations in phytoalexin-deficient 4 (PAD4) or enhanced disease susceptibility 1 (EDS1), which are upstream regulators of NPR1, improved shoot growth under low-Ca conditions, suggesting that NPR1 suppressed growth under low-Ca conditions. In contrast, growth of SA induction–deficient 2-2 (sid2-2), which is an SA-deficient mutant, was sensitive to low Ca levels, suggesting that SA accumulation by SID2 was not related to growth inhibition under low-Ca conditions. Additionally, npr1-1 showed low-Ca tolerance, and the application of tenoxicam—an inhibitor of the NPR1-mediated activation of gene expression—also improved shoot growth under low Ca conditions. The low-Ca tolerance of double mutants pad4-1, npr1-1 and eds1-22 npr1-1 was similar to that of the single mutants, suggesting that PAD4 and EDS1 are involved in the same genetic pathway in suppressing growth under low-Ca conditions as NPR1. Cell death and low-Ca tolerance did not correlate among the mutants, suggesting that growth improvement in the mutants was not due to cell death inhibition. In conclusion, we revealed that NPR1 suppresses plant growth under low-Ca conditions and that the other SA-related genes influence plant growth and cell death.
Keywords: Arabidopsis, Calcium deficiency, Growth suppression, NPR1, Salicylic acid
Introduction
Ca is an important nutrient for plant growth. Insufficient uptake of Ca causes Ca deficiency symptoms in plants, which are characterized by necrosis in the tips of tissues and inhibition of growth (de Freitas and Mitcham 2012). For example, blossom-end-rot, a disorder characterized by a blackened area in tomato fruit, is induced by low Ca levels (Taylor and Locascio 2004).
Several studies have identified the genes that affect the symptoms of Ca deficiency. CAX1 is an H+/Ca2+ antiporter that transports Ca2+ from the cytosol to vacuoles (Hirschi et al. 1996). cax1 mutants show increased low-Ca tolerance (Bradshaw 2005), whereas the overexpression of CAX1 in crops results in severe Ca deficiency symptoms (Hirschi 1999, Zorrilla et al. 2019). Other genes involved in low-Ca tolerance are the callose synthase genes, Glucan Synthase–Like (GSL). Callose is synthesized and accumulated in the cell wall under low-Ca conditions, and gsl10 exhibits growth defects and severe cell death under low-Ca conditions (Shikanai et al. 2020). GSL1 and GSL8 are also involved in low-Ca tolerance (Shikanai et al. 2022a, 2022b), suggesting that supporting the cell wall structure with callose is important for low-Ca tolerance.
A recent study revealed that defense response genes, including salicylic acid (SA)–related genes, are upregulated under low-Ca conditions (Shikanai et al. 2020). Furthermore, a positive correlation between susceptibility to low-Ca conditions and accumulation of SA has been observed in Chinese cabbage (Brassica rapa ssp. pekinensis; Su et al. 2016). These findings suggest that SA may be involved in growth suppression and cell death development under low-Ca conditions.
Several genes are involved in SA biosynthesis and downstream signaling in defense responses, including phytoalexin-deficient 4 (PAD4) and enhanced disease susceptibility 1 (EDS1) (White 1979, Peng et al. 2021). PAD4 and EDS1 are major regulators of defense responses including resistance to Peronospora parasitica, an oomycete pathogen (Parker et al. 1996, Jirage et al. 1999, Lapin et al. 2020). PAD4 and EDS1 function as heterodimers that induce the expression of downstream genes including SA induction–deficient 2/isochorismate synthase 1 (SID2/ICS1; Zhou et al. 1998, Feys et al. 2001, Wagner et al. 2013). In Arabidopsis, SID2 is responsible for biosynthesizing a majority of SA upon pathogen infection (Wildermuth et al. 2001). Biosynthesized SA binds to the nonexpressor of pathogenesis-related (PR) genes (NPRs), such as NPR1, to promote the expressions of PR genes (Moreau et al. 2012, Wang et al. 2020, Peng et al. 2021).
These SA-related genes contribute to the response to various environmental stresses. For example, SA-deficient mutant nahG and SA signaling mutant npr1-1 exhibit salt tolerance (Hao et al. 2012). Mutation in PAD4, EDS1 or ICS1 leads to a sensitive phenotype to low-Fe stresses (Shen et al. 2016). However, the involvement of SA-related genes in response to low-Ca conditions remains unclear. In the present study, we showed that NPR1 inhibition improves shoot growth under low-Ca conditions. In addition, mutations in PAD4 or EDS1 improve growth and their growth-related effects are genetically on the same pathway as NPR1. Although NPR1, PAD4 and EDS1 are involved in SA signaling in pathogen-induced defense responses, SA accumulation by SID2 does not seem to be the cause of growth suppression under low-Ca conditions. Furthermore, although npr1-1, pad4-1 and eds1-22 showed low-Ca tolerance, the appearance of symptoms of cell death varied among them, suggesting that growth improvement in the mutants was not due to the inhibition of cell death. Our study provides genetic evidence for the involvement of SA-related genes in growth suppression under low-Ca conditions.
Results
Mutation in PAD4 or EDS1 improved shoot growth under low-Ca conditions
In a previous study, mRNA accumulation of SA-related defense response genes, such as PAD4, EDS1 and SAG101, was observed under low-Ca conditions using RNA sequencing (Shikanai et al. 2020). We confirmed the induction of these genes by qRT-PCR in Col-0 (Fig. 1A). As these are the major regulators of defense responses (Lapin et al. 2020), we focused on the contribution of these three genes to growth under low-Ca conditions using pad4-1, eds1-22 and sag101-3 mutants. Of these, pad4-1 is an ethyl methanesulfonate (EMS) mutant (W359stop; Jirage et al. 1999), whereas eds1-22 and sag101-3 contain T-DNA insertions in exons EDS1 and SAG101 (Chen et al. 2015), respectively.
Fig. 1.
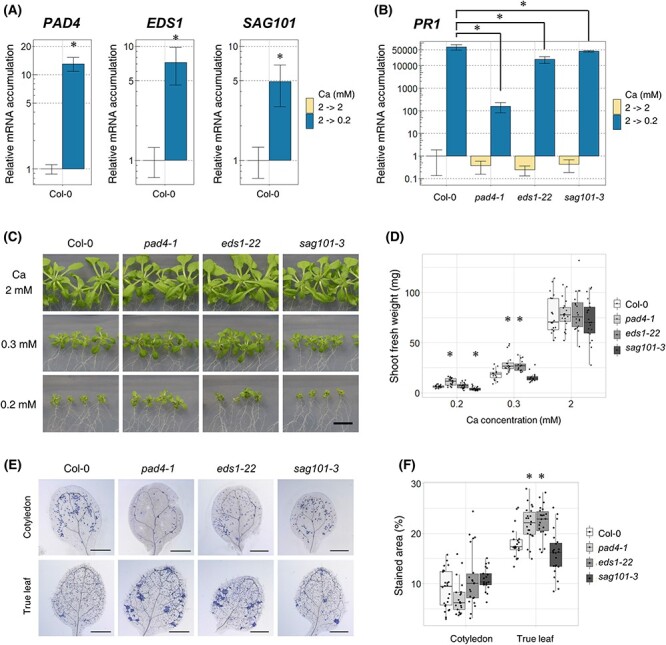
Mutation in PAD4 or EDS1 improved shoot growth under low-Ca conditions. (A) The accumulation of mRNA of PAD4, EDS1 and SAG101 in Col-0. Plants were grown on 2 mM Ca medium for 7 d, transferred to 2 mM Ca or 0.2 mM Ca medium and grown for 96 h. Relative mRNA accumulation was determined by setting the accumulation under 2 mM Ca to 1. Mean ± SD; n = 3–4; Welch’s t-test; *, P < 0.05. (B) Accumulation of PR1 mRNA in pad4-1, eds1-22 and sag101-3. The experiments were performed as described in (A). Mean ± SD; n = 3–4; Dunnett’s test; *, P < 0.05. (C and D) Shoot growth and fresh weight of pad4-1, eds1-22 and sag101-3 under normal- (2 mM) and low-Ca (0.3 and 0.2 mM) conditions. Plants were grown under the indicated Ca conditions for 18 d. Images of shoots (C) and shoot fresh weight (D). n = 16–21; Dunnett’s test compared with Col-0 in each condition; * P < 0.05; Bar = 1 cm. (E and F) The severity of cell death in pad4-1, eds1-22 and sag101-3. Plants were grown on 2 mM Ca medium for 7 d, transferred to 0.2 mM Ca medium and then grown for 96 h. Whole shoots were stained with trypan blue and their cotyledons and true leaves were photographed (E). The stained regions of the leaves were quantified (F). The stained area (%) was calculated by dividing the cell death area by the total leaf area. n = 21–24; Dunnett’s test compared Col-0 in each tissue; *, P < 0.05.
The effects of mutations on SA-related genes were detected by measuring the mRNA accumulation levels of PR1, a marker gene for SA-related defense responses (Linthorst and Van Loon 1991, Peng et al. 2021), in these mutants. The induction of PR1 under low-Ca conditions has been reported previously (Shikanai et al. 2020). PR1 expression under low-Ca conditions was lower in all mutants than in Col-0 plants (Fig. 1B), suggesting that the defense response was partly impaired in these mutants under low-Ca conditions.
Thereafter, the involvement of PAD4, EDS1 and SAG101 in growth under low-Ca conditions was evaluated by observing the growth of these mutants under normal- and low-Ca conditions. We found that pad4-1 and eds1-22 grew better than Col-0 under low-Ca (0.2 mM and/or 0.3 mM) conditions (Fig. 1C, D), whereas growth under the normal conditions was indistinguishable, indicating that PAD4 and EDS1 suppress shoot growth only under the low-Ca conditions.
In addition, cell death in newly emerging tissues was investigated as it is one of the symptoms of Ca deficiency disorders, together with growth suppression (de Freitas et al. 2016). Plants were grown in normal-Ca medium for 7 d. Thereafter, the tissues were transferred to low-Ca (0.2 mM) conditions and further incubated for 96 h. New true leaves emerged during the incubation period. We quantified cell death in the leaves using trypan blue staining and found that pad4-1 and eds1-22 showed severe cell death in true leaves, while sag101-3 did not (Fig. 1E, F). These results suggest that the improved growth of pad4-1 and eds1-22 was not due to the alleviation of cell death.
Next, we determined Ca concentrations in the shoots to observe the possible involvement of these genes in Ca distribution. The Ca concentration in pad4-1 was higher than that in Col-0 under both normal- and low-Ca conditions, whereas the Ca concentrations in eds1-22 and sag101-3 were not significantly different (Fig. S1A). Accordingly, there was no clear effect of mutations in SA-related genes on Ca distribution.
In conclusion, pad4-1 and eds1-22 mutants showed improved growth under low-Ca conditions, but it is unlikely that this was due to the alleviation of cell death or an increase in the Ca concentration. It is possible that PAD4 and EDS1 play roles in growth suppression under low-Ca conditions.
SA was induced by SID2 under low-Ca conditions, but SID2 mutants did not show low-Ca tolerance
PAD4 and EDS1 promote SA-dependent defense responses (Wiermer et al. 2005). Because SA can promote or inhibit plant growth in a dose-dependent manner (Kováčik et al. 2009, Nazar et al. 2015), we investigated how SA accumulation changes under low-Ca conditions. Under low-Ca conditions, SA accumulation in Col-0 plants was approximately 10-fold higher than that under normal conditions (Fig. 2A). This result is consistent with a previous report on Chinese cabbage in the viewpoint that SA is elevated under low-Ca conditions (Su et al. 2016).
Fig. 2.
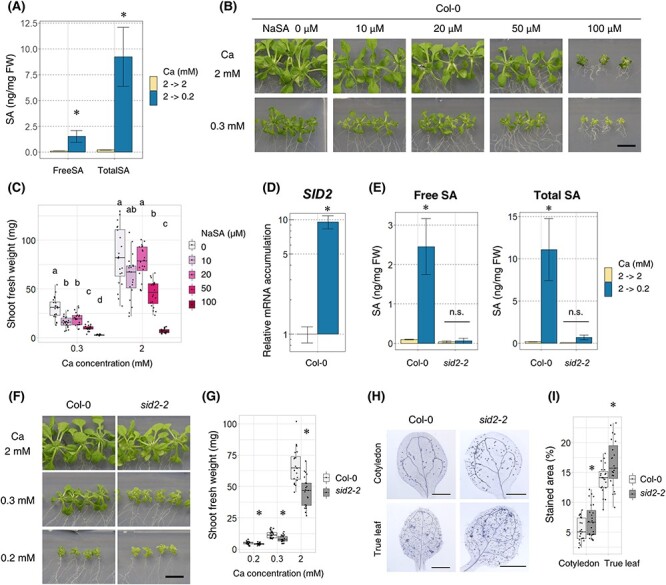
SA accumulation required SID2 under low-Ca conditions, but sid2 mutant did not show low-Ca tolerance. (A) SA accumulation in Col-0. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 2 mM Ca or 0.2 mM Ca medium and allowed to grow for 96 h. Entire shoots were collected, and SA concentration was measured using HPLC. Mean ± SD; n = 3; Welch’s t-test; *, P < 0.05. (B and C) Shoot growth and fresh weight of Col-0 under different concentrations of sodium salicylate (NaSA). Plants were grown with indicated Ca (0.3 and 2 mM) and NaSA (0, 10, 20, 50 and 100 µM) concentrations for 18 d. Shoots were photographed (B), and shoot fresh weight was measured (C). n = 18–21; Steel–Dwass test within samples of the same Ca concentration; P < 0.05. Different letters indicate significant differences. Bar = 1 cm. (D) The accumulation of SID2 mRNA in Col-0. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 2 mM Ca or 0.2 mM Ca medium and allowed to grow for 96 h. The mRNA accumulation levels under the 2 mM Ca condition were set as 1. Mean ± SD; n = 3–4; Welch’s t-test; *, P < 0.05. (E) SA accumulation in sid2-2. The experiments were performed as in (A). Mean ± SD; n = 3; Welch’s t-test; *, P < 0.05. n.s., not significant. (F and G) Shoot growth and fresh weight of sid2-2. The experiments were performed as mentioned in (B and C). n = 20–24; Dunnett’s test; *, P < 0.05; bar = 1 cm. (H and I) The severity of death in sid2-2. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 0.2 mM Ca medium and allowed to grow for 96 h. Entire shoot was stained with trypan blue, and true leaves and cotyledons were photographed (H). The stained regions in the leaves were quantified (I). The stained area (%) was calculated by dividing the cell death area by the whole leaf area for each leaf. n = 21–24; Welch’s t-test; *, P < 0.05. Abbreviation: n.s., not significant.
To evaluate the effect of SA on growth under low-Ca conditions, sodium salicylate (NaSA) was added to the medium and Col-0 plants were grown for 18 d. Under both normal- and low-Ca conditions, the growth of Col-0 was inhibited in an NaSA-dependent manner (Fig. 2B, C), indicating that high concentrations of exogenous SA can suppress shoot growth, irrespective of Ca conditions.
Next, we observed the growth phenotype of the low-SA mutant sid2-2 under low-Ca conditions. SID2 is responsible for the majority of SA biosynthesis in Arabidopsis (Wildermuth et al. 2001, Peng et al. 2021), and its mRNA expression was induced under low-Ca conditions (Fig. 2D). sid2-2 accumulated less than 10% of free and total SA under low-Ca conditions compared with Col-0 (Fig. 2E), indicating that SID2 contributes to SA accumulation under low-Ca conditions. However, sid2-2 was smaller than Col-0 under both normal- and low-Ca conditions, with increased cell death (Fig. 2F–I). These results suggest that the reduced accumulation of SA does not necessarily result in low-Ca tolerance.
Inhibition of NPR1 improved shoot growth under low-Ca conditions
Because sid2-2 did not show low-Ca tolerance, we hypothesized that another gene in the SA pathway is involved in growth inhibition independent of SA accumulation. One candidate gene is NPR1, which is involved in both SA-dependent and SA-independent defense responses (Lai et al. 2018, Yildiz et al. 2021). RNA sequence analysis has revealed mRNA accumulation of NPR1 under low-Ca conditions in a previous study (Shikanai et al. 2020), suggesting the possibility of NPR1 involvement in low-Ca tolerance. To assess this, we first confirmed the RNA sequencing results using qRT-PCR, and a 1.5-fold NPR1 induction was observed in Col-0 under low-Ca conditions (Fig. 3A). Next, to evaluate the effect of the NPR1 on low-Ca tolerance, we obtained an NPR1 mutant (npr1-1) and a partial loss-of-function mutant (EMS mutation, H334Y, Cao et al. 1997, Ding et al. 2020). npr1-1 showed improved growth under low-Ca conditions compared with Col-0 (Fig. 3B, C) and reduced cell death in true leaves (Fig. 3D, E). The Ca concentration in the shoots of npr1-1 was not significantly different from that in Col-0 (Fig. S1C). These results suggest that npr1-1 is low-Ca-tolerant and that this tolerance is not associated with an increase in Ca uptake.
Fig. 3.
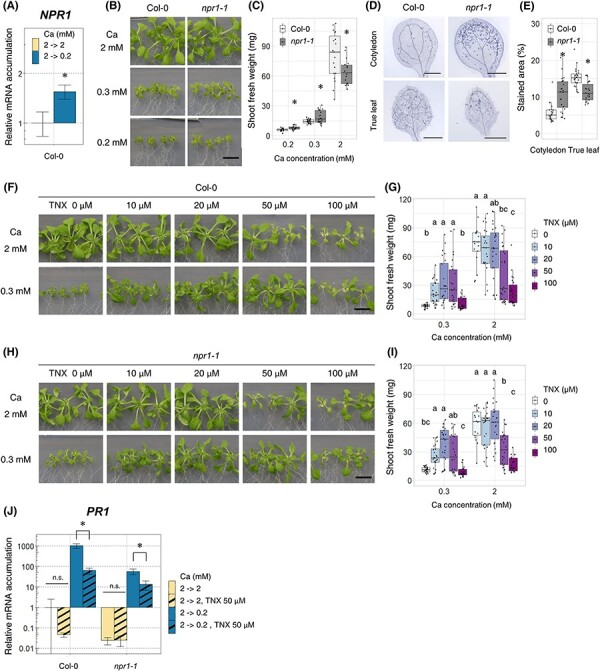
Inhibition of NPR1 improved the shoot growth under low-Ca conditions. (A) Accumulation of NPR1 mRNA in Col-0. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 2 mM Ca or 0.2 mM Ca medium and allowed to grow for 96 h. mRNA accumulation under 2 mM Ca conditions was set as 1. Mean ± SD; n = 3–4; Welch’s t-test; *, P < 0.05. (B and C) Shoot growth and fresh weight of npr1-1 under normal- (2 mM) and low-Ca (0.3 and 0.2 mM) conditions. Plants were grown with indicated Ca conditions for 18 d. Image of shoot was photographed (B), and shoot fresh weight was measured (C). n = 16–24; Welch t-test; *, P < 0.05; bar = 1 cm. (D and E) The severity of cell death of npr1-1. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 0.2 mM Ca medium and allowed to grow for 96 h. Entire shoot was stained with trypan blue, and true leaves and cotyledons were photographed (D). The stained regions in the leaves were quantified (E). The stained area (%) was calculated by division of the cell death area by the whole leaf area for each leaf. n = 22–24; Welch’s t-test; *, P < 0.05. (F and G) Shoot growth and fresh weight of Col-0 under different concentrations of TNX. Plants were grown with indicated Ca (0.3 and 2 mM) and TNX (0, 10, 20, 50 and 100 µM) concentrations for 18 d. The image of shoot was photographed (F), and the shoot fresh weight was measured (G). n = 15–25; Steel–Dwass test within samples of the same Ca concentration; P < 0.05. Different letters indicate significant differences. Bar = 1 cm. (H and I) Shoot growth and fresh weight of npr1-1 under different concentrations of TNX. The experiments were performed as in (F) and (G). n = 18–24; Steel–Dwass test within samples of the same Ca concentration; P < 0.05. Different letters indicate significant differences. Bar = 1 cm. (J) Accumulation of PR1 mRNA in the presence of TNX. Plants were first grown in 2 mM Ca, 0 µM TNX medium for 7 d and then transferred to 2 mM or 0.2 mM Ca and 0 µM or 50 µM TNX medium for 96 h. Average mRNA accumulation levels under 2 mM Ca with the 0 µM TNX condition was set as 1. Mean ± SD; n = 3–4; Welch’s t-test; *, P < 0.05. Abbreviation: n.s., not significant.
To further confirm the effect of NPR1 on the shoot growth using a different approach, we observed the growth of Col-0 and npr1-1 plants under different concentrations of tenoxicam (TNX). TNX is known as an anti-inflammatory drug in humans, and in Arabidopsis, TNX broadly suppresses SA-responsive genes by reducing NPR1 protein accumulation (Ishihama et al. 2021). In the presence of TNX, the shoot fresh weight of Col-0 under low-Ca conditions was 3–4 times higher than that under non-applied conditions (Fig. 3F, G), supporting the idea that inhibition of NPR1 activity can improve growth under low-Ca conditions. npr1-1 also showed growth recovery at 10 or 20 µM TNX (Fig. 3H, I). Considering that the npr1-1 mutation is not a null allele, the growth recovery of npr1-1 following TNX application can be explained by the inhibition of NPR1, which partially functions in this mutant. We further tested the effects of TNX on NPR1 activity via PR1 mRNA accumulation. PR1 mRNA accumulation was lower under TNX-treated conditions than in non-treated conditions for both Col-0 and npr1-1, indicating the inhibition of NPR1 activity by TNX under low-Ca conditions (Fig. 3J). Based on these data, we concluded that NPR1 suppresses the shoot growth under low-Ca conditions.
NPR1 is on the same genetic pathway as PAD4 and EDS1 for growth suppression under low-Ca conditions
As we revealed that PAD4, EDS1 and NPR1 inhibited shoot growth under low-Ca conditions, we investigated whether these genes were involved in the same genetic pathway in the suppression of growth under low-Ca conditions. In the defense response, PAD4 and EDS1 function together and indirectly activate NPR1 (Cui et al. 2018; Peng et al. 2021). To test whether they are involved in the same genetic pathway in the suppression of growth under low-Ca conditions, double mutants, pad4-1 npr1-1 and eds1-22 npr1-1 were established. Under 0.2 mM Ca, the growth of pad4-1 npr1-1 was not significantly different from that of pad4-1 and npr1-1 single mutants (Fig. 4A, B). Under 0.3 mM Ca, pad4-1 npr1-1 showed similar growth to pad4-1 but greater growth than npr1-1. eds1-22 npr1-1 showed similar growth to eds1-22 and npr1-1 under 0.2 mM and 0.3 mM Ca conditions (Fig. 4C, D). These results showed that growth improvement was not an additive effect between pad4-1 and npr1-1 or between eds1-22 and npr1-1, suggesting that PAD4 and NPR1 and EDS1 and NPR1 are in the same genetic pathway that suppresses growth under low-Ca conditions.
Fig. 4.
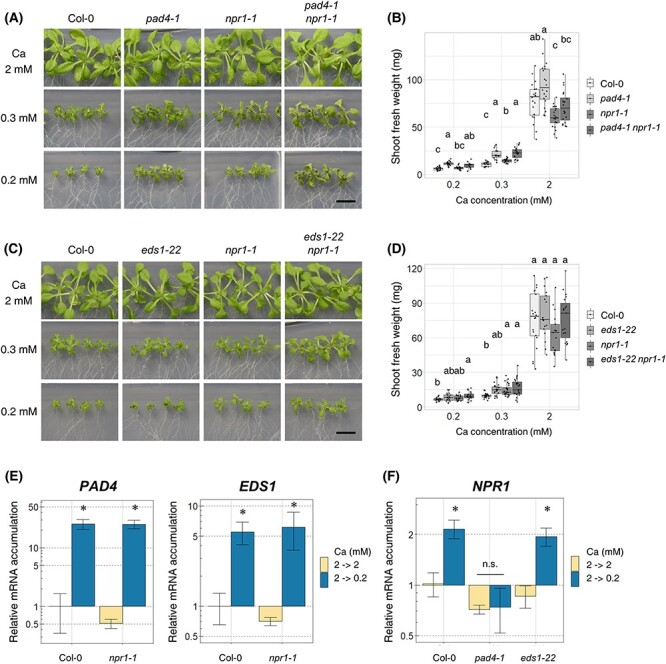
NPR1 is included in the same genetic pathway for growth regulation under low-Ca conditions as PAD4 or EDS1. (A and B) Shoot growth and fresh weight of pad4-1 npr1-1 under normal- (2 mM) and low-Ca (0.3 and 0.2 mM) conditions. Plants were grown with indicated Ca conditions for 18 d. Shoot was photographed (A), and the shoot fresh weight was measured (B). n = 14–21; Steel–Dwass test within samples of the same Ca concentration; P < 0.05. Different letters indicate significant differences. Bar = 1 cm. (C and D) Shoot growth and fresh weight of eds1-22 npr1-1. The experiments were performed as in (A) and (B). n = 17–24; Steel–Dwass test within samples of the same Ca concentration; P < 0.05. Different letters indicate significant differences. Bar = 1 cm. (E) The mRNA accumulation of PAD4 and EDS1 in npr1-1. First, plants were grown in 2 mM Ca medium for 7 d, then transferred to 2 mM Ca or 0.2 mM Ca medium and allowed to grow for 96 h. Average mRNA accumulation under 2 mM Ca conditions was set as 1. Mean ± SD. n = 3–4; Welch’s t-test; *, P < 0.05. (F) NPR1 mRNA accumulation in pad4-1 and eds1-22. The experiments were performed as in (E). Mean ± SD. n = 3-4. Welch’s t-test; *, P < 0.05. Abbreviation: n.s., not significant.
As shown in Figs. 1A and 3A, we confirmed that PAD4, EDS1 and NPR1 are all induced under low-Ca conditions. The induction of these genes has been reported in response to other biotic and abiotic stresses (Pst infection, Bartsch et al. 2006, Zhang et al. 2012, low temperature, Chen et al. 2015, Olate et al. 2018), and SA-induced NPR1 induction by PAD4 or EDS1 has been reported (Chen et al. 2021). To analyze whether the three genes were induced by each other under low-Ca conditions, we measured the mRNA accumulation of each gene in the other single mutants. npr1-1 showed similar upregulated levels of PAD4 and EDS1 as Col-0 in response to low Ca, suggesting that their transcript-level regulation is independent of NPR1 (Fig. 4E). In contrast, pad4-1 did not accumulate NPR1 under low-Ca conditions (Fig. 4F). These results suggest that PAD4 is upstream and regulates mRNA accumulation of NPR1.
Discussion
Growth inhibition by NPR1 under low Ca is possibly regulated by PAD4 and EDS1 independent of SA
Under several abiotic stresses, such as low temperatures (Scott et al. 2004), heavy metals (Tao et al. 2013) and salt (Hao et al. 2012), growth was recovered when SA accumulation was suppressed, suggesting that SA accumulation inhibits growth under these conditions. Under low-Ca conditions, the accumulation of SA or mRNA induction of defense response genes has also been reported under low-Ca conditions (Su et al. 2016, Shikanai et al. 2020, Fig. 2). However, in our study, we demonstrated that SA accumulation in sid2-2 was reduced, whereas growth inhibition under low-Ca conditions was maintained (Fig. 2), suggesting that SA accumulation is not likely responsible for growth inhibition under low-Ca conditions.
What could be the reason for the growth inhibition under low-Ca conditions? As shown in Fig. 3, a mutation in NPR1 and TNX application improved growth under low-Ca conditions. In addition to NPR1, mutations in either PAD4 or EDS1 also improved growth under low-Ca conditions (Fig. 1D). The double mutants, pad4-1 npr1-1 and eds1-22 npr1-1 showed growth recovery similar to the single mutants (Fig. 4). The upregulation of NPR1 was not observed in pad4-1 (Fig. 4F). Taken together, we propose that PAD4 and EDS1 activate NPR1, leading to growth inhibition under low-Ca conditions in an SA-independent manner (Fig. 5).
Fig. 5.
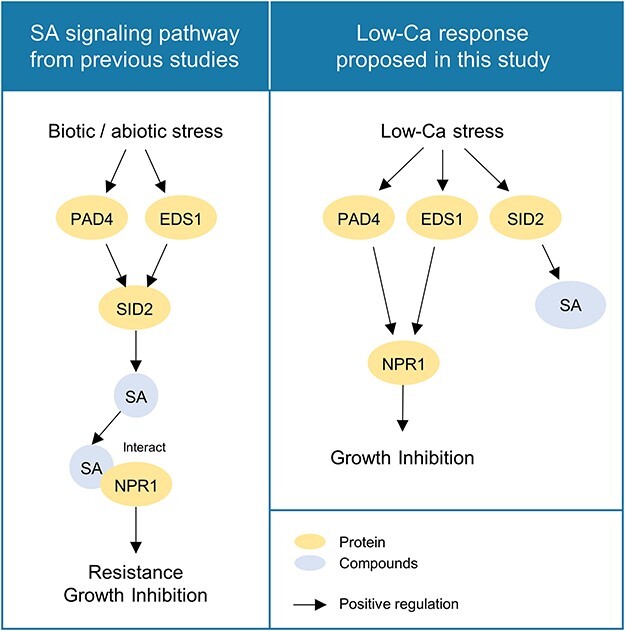
The proposed model for growth regulation under low-Ca conditions. Left: Reported SA signaling pathway. PAD4 and EDS1 induce mRNA accumulation of SID2, which is followed by SA biosynthesis (Zhou et al. 1998). The SA produced binds to NPR1, which induces gene expression, leading to stress resistance and growth inhibition. Right: Proposed SA signaling pathway under low-Ca conditions: mRNA accumulation of PAD4, EDS1 and SID2 was induced. SA is synthesized by SID2 induction but is not responsible for growth inhibition. PAD4 alone induces NPR1, and also, PAD4 and EDS1 together activates NPR1, leading to growth inhibition.
NPR1 functions as a transcriptional co-activator (Lapin et al. 2020) and regulates gene mRNA accumulation in both SA-dependent and SA-independent manners (Lai et al. 2018, Yildiz et al. 2021). PAD4 and EDS1 form a heterodimer that functions as a major regulator of basal defense (Feys et al. 2001, Wagner et al. 2013). PAD4 and EDS1 induce the mRNA accumulation of genes such as AGD2-LIKE DEFENSE RESPONSE PROTEIN1, which activates NPR1 independent of SA (Cui et al. 2017, Sattely et al. 2018, Lapin et al. 2020, Yildiz et al. 2021). These previous studies support our hypothesis that NPR1 would be activated by PAD4 and EDS1 in an SA-independent manner under low-Ca conditions.
NPR1 mRNA accumulation was not observed in pad4-1 mutants under low-Ca conditions (Fig. 4F), whereas NPR1 mRNA was induced in the eds1-22 mutant, similar to that in Col-0 (Fig. 4F), suggesting that PAD4 could be responsible for NPR1 mRNA accumulation. A previous study has shown that EDS1 forms a heterodimer with PAD4 (Feys et al. 2001, Wagner et al. 2013), suggesting that PAD4 and EDS1 function together. These results suggest that PAD4 may contribute to both NPR1 mRNA accumulation and its protein activity, while EDS1 may contribute to NPR1 activity alone.
How does NPR1 activation inhibit the plant growth under low-Ca conditions? There is limited information available regarding the function of NPR1 in plant growth. Among the few studies conducted, Vanacker et al. (2001) showed that npr1 has a reduced leaf cell number and increased DNA content. This study suggests that NPR1 promotes cell division and/or suppresses endoreduplication.
Interestingly, under low-Ca conditions, npr1-1 showed better growth compared to wild type (Fig. 3C). This suggests that the activation of NPR1 could prevent growth under low Ca. This is in contradiction to the findings of Vanacker et al. (2001). Based on our study, NPR1 would inhibit growth under low Ca as further growth may exacerbate the Ca deficiency. Further study is needed to elucidate the function of NPR1 in plant growth and the molecular mechanism of NPR1 for growth inhibition under low-Ca conditions.
Possible function of SA in low-Ca tolerance
In this study, we quantified the severity of cell death in SA mutants, as cell death is a representative symptom of Ca deficiency disorder (de Freitas et al. 2016). The accumulation of SA causes a hypersensitive response (HR), a type of programmed cell death, in response to pathogen infection. The degree of HR severity varies depending on the mutant: disruption of PAD4 or EDS1 alleviates HR (Rustérucci et al. 2001, Wiermer et al. 2005), whereas npr1 displays a stronger HR compared to the wild type (Rate and Greenberg 2001). In contrast, under low-Ca conditions, the SA mutants used in this study showed the opposite cell death phenotype: pad4-1 and eds1-22 exhibited severe cell death compared to Col-0, whereas npr1-1 did not (Fig. 1F, 3E). Accordingly, the cell death symptom under low-Ca conditions seems to occur by a different process from HR. sid2-2, a mutant defective in SA production, exhibited more severe cell death than Col-0 (Fig. 2I). sid2-2, a mutant defective in SA production, exhibited more severe cell death than Col-0 (Fig. 2I). The result suggested that SA accumulation inhibits cell death under low-Ca conditions. SA contributes to cell wall modification under biotic stress by enhancing lignin and callose production (Li et al. 2017, van Butselaar and Van den Ackerveken 2020). Recent studies have shown that callose plays an important role in alleviating cell death under low-Ca conditions (Shikanai et al. 2020, 2022a, 2022b). Therefore, SA accumulation under low-Ca conditions may strengthen the cell wall structure, leading to the inhibition of cell death.
Conclusion
In this study, we found that SA-related genes including NPR1 regulate growth under low-Ca conditions. Our data demonstrated that PAD4, EDS1 and NPR1 inhibited growth under low-Ca conditions, whereas SA accumulation by SID2 was unlikely to be the cause of growth inhibition. Additionally, the growth improvement in the mutants are unlikely to be a result of the inhibition of cell death. These findings provide new insights into how plants regulate their growth in low-Ca environments. Our research is expected to be useful for understanding low-Ca responses in plants and their applications in agriculture.
Materials and Methods
Plant materials and growth conditions
Mutants pad4-1 (CS3806; Jirage et al. 1999), eds1-22 (SALK_071051; Chen et al. 2015), sag101-3 (SALK_022911; Chen et al. 2015), sid2-2 (CS16438; Wildermuth et al. 2001) and npr1-1 (CS3726; Cao et al. 1997) were obtained. Double mutants pad4-1, npr1-1 and eds1-22 npr1-1 were generated by crossing, and homozygous seeds were selected by genotyping. The primers used for genotyping are listed in Supplementary Table 1.
The composition of the medium was based on a previous study (Yamagami medium; Shikanai et al. 2015) with some modifications: 1% (w/v) sucrose (Sigma-Aldrich, Burlington, Massachusetts, code 84097-250G) and 1% (w/v) agar (Nacalai Tesque, Kyoto, Japan, code 01056-15). For application of NaSA or TNX, the chemicals were added after autoclaving the remaining medium.
All the wild type (ecotype Col-0) and mutant seeds were surface-sterilized with commercial chlorine bleach and sown on medium plates. The plates were kept at 4°C for 2 d for vernalization and then transferred to a growth chamber. Growth chamber was kept at 22°C, relative humidity 50%, 16 h light and 8 h dark. To measure the shoot fresh weight and Ca concentration in the shoots, plants were grown under indicated Ca conditions for 18 d. For RNA extraction, SA measurement or trypan blue staining, plants were first grown under normal-Ca (2 mM) conditions for 7 d and then transferred to normal-Ca (2 mM) or low-Ca (0.2 mM) conditions for 96 h.
Determination of Ca concentrations
The shoots of 2–6 plants (with dry weight of 5–10 mg) were collected for each sample and dried at 60°C overnight. The dried samples were weighed and transferred to Teflon tubes for nitric acid digestion. After adding 1 ml of nitric acid, the tubes were heated at 100°C until nitric acid evaporated. This step was repeated with 400 μl of nitric acid, followed by the addition of 400 μl of hydrogen peroxide and heating until evaporation. The residues were dissolved in 1 ml of 0.08 N nitric acid and diluted 10 times with 0.08 N nitric acid containing 1 ppb beryllium as an internal standard. Elemental concentrations were measured using inductively coupled plasma mass spectrometry (Agilent 7800).
RNA extraction and qRT-PCR
Total RNA was extracted from frozen shoots harvested from 4–6 plants. Extraction was performed using the NucleoSpin RNA Plant (Macherey-Nagel, Düren, Germany) following the manufacturer’s protocol. About 300 ng of total RNA was reverse transcribed using PrimeScript RT Master Mix (Perfect Real Time) (Takara Bio) and prepared as 50 μl of cDNA solution. A total of 2 μl of the cDNA solution was used for real-time PCR. Thermal Cycler Dice Real-Time System III (Takara Bio, Kusatsu, Japan) and SYBR Premix Ex Taq II (Tli RNase H Plus) (Takara Bio) were used for real-time PCR. The expressions of ACT8 were used for normalizing mRNA accumulation of target genes. The primers used in this experiment are listed in Supplemental Table 2.
SA quantification
The shoot from 10–20 plants (80–160 mg) per sample was used for SA quantification. The samples were homogenized in 1 ml of 90% methanol and extracted using 1 ml of 100% methanol. The extracts were mixed and dried at 40°C, and the residues were dissolved in 4 ml of water at 80°C for 15 min. One milliliter of each extract was used for quantifying free and total SA. For total SA quantification, 1 ml of β-glucosidase (Merck, Darmstadt, Germany) solution (3 units/ml) was prepared with 0.1 M sodium acetate buffer, added to 1 ml of extract and incubated at 37°C for 4–6 h. For free SA quantification, 1 ml of 0.1 M sodium acetate buffer was added to 1 ml of extract. Subsequently, 2.5 ml of ethyl acetate–cyclohexane (1:1) and 50 µl of concentrated HCl were added and mixed intensely. The upper layer was dried at 35°C and dissolved in 1 ml of 20% methanol in 20 mM sodium acetate buffer. This solution was used for HPLC analysis (JASCO FP-1520S) using the CAPCELL PAK C18 MG column (5 µm, 150 mm × 2 mm) with 20% methanol in 20 mM sodium acetate buffer at a flow rate of 1 ml/min. Detection was performed at an excitation wavelength of 295 nm and an emission wavelength of 370 nm.
Trypan blue staining and quantification
Trypan blue staining was performed as previously described (Koch and Slusarenko 1990, Fukuda et al. 2016) with some modifications. Briefly, shoots were soaked in ethanol for 1 min and incubated in lactophenol-trypan blue solution (lactic acid:phenol:trypan blue:glycerol:ethanol:distilled water = 5 ml:5 g:5 mg:5 ml:20 ml:5 ml) at 70°C for 10 min. The samples were transferred to chloral hydrate [chloral hydrate:distilled water = 5:2 {w/w)], incubated at 22°C overnight and photographed.
Fiji software (Schindelin et al. 2012) and custom macros were used for the quantification of trypan blue staining results. The stained area (%) was calculated by dividing the cell death area by the leaf size. The cell death area was determined by increasing the color threshold until the leaf veins (xylem) were selected, as veins were stained in all leaves at a level similar to that of the cell death region. Leaf size was measured by selecting the edges of the leaves using the Polygon Sections function of the Fiji software.
Supplementary Material
Acknowledgments
We thank Atsushi Ishikawa for kindly providing pad4-1 and npr1-1.
Contributor Information
Yusuke Shikanai, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, 113-8657 Japan; Department of Agricultural Chemistry, Tokyo University of Agriculture, Setagaya-ku, Tokyo, 156-8502 Japan.
Miyuki Kusajima, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, 113-8657 Japan.
Hidemitsu Nakamura, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, 113-8657 Japan.
Toru Fujiwara, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, 113-8657 Japan.
Takehiro Kamiya, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, 113-8657 Japan.
Supplementary Data
Supplementary Data are available at PCP online.
Data Availability
The data underlying this article are available in the article and online supplementary material. Information on the mutants and primers is available in Supplementary Tables.
Funding
The Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Doctoral Course (DC) Research Fellowships (17J06965) to Y.S., Grant-in-Aid for Scientific Research (S) (19H05637) to T.F., and Grant-in-Aid for Scientific Research (B) to T.K. (21H02087).
Author Contributions
S.H., Y.S., T.F. and T.K. designed the study. S.H., Y.S., M.K. and H.N. performed the experiments. S.H. analyzed the data. S.H., Y.S., T.F. and T.K. wrote the manuscript.
Disclosures
The authors have no conflicts of interest to declare.
References
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., et al. (2006) Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H.D. (2005) Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol. 167: 81–88. [DOI] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S. and Dong X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Chen H., Li M., Qi G., Zhao M., Liu L., Zhang J., et al. (2021) Two interacting transcriptional coactivators cooperatively control plant immune responses. Sci. Adv 7: eabl7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.F., Xu L., Tan W.J., Chen L., Qi H., Xie L.J., et al. (2015) Disruption of the Arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol. Plant. 8: 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Gobbato E., Kracher B., Qiu J., Bautor J. and Parker J.E. (2017) A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 213: 1802–1817. [DOI] [PubMed] [Google Scholar]
- Cui H., Qiu J., Zhou Y., Bhandari D.D., Zhao C., Bautor J. et al. (2018) Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in arabidopsis effector-triggered immunity. Molecular Plant. 11: 1053–1066. [DOI] [PubMed] [Google Scholar]
- de Freitas S.T., Amarante C.D. and Mitcham E.J. (2016) Calcium deficiency disorders in plants. In Postharvest Ripening Physiology of Crops. Edited by Pareek, S. pp. 477–502. CRC Press: Boca Raton, Florida. [Google Scholar]
- de Freitas S.T. and Mitcham E.I. (2012) 3 factors involved in fruit calcium deficiency disorders. Hortic Rev (Am Soc Hortic Sci) 40: 107–146. [Google Scholar]
- Ding Y., Dommel M.R., Wang C., Li Q., Zhao Q., Zhang X., et al. (2020) Differential quantitative requirements for NPR1 between basal immunity and systemic acquired resistance in Arabidopsis thaliana. Front. Plant Sci. 11: 570422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A. and Parker J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Segami S., Tomoyama T., Asaoka M., Nakanishi Y., Gunji S., et al. (2016) Lack of H+-pyrophosphatase prompts developmental damage in Arabidopsis leaves on ammonia-free culture medium. Front. Plant Sci. 7: 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Zhao Y., Jin D., Zhang L., Bi X., Chen H., et al. (2012) Salicylic acid-altering Arabidopsis mutants response to salt stress. Plant Soil. 354: 81–95. [Google Scholar]
- Hirschi K.D. (1999) Expression of Arabidopsis CAX1 in tobacco: altered calcium homeostasis and increased stress sensitivity. Plant Cell 11: 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K.D., Zhen R.G., Cunningham K.W., Rea P.A. and Fink G.R. (1996) CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93: 8782–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N., Choi S.W., Noutoshi Y., Saska I., Asai S., Takizawa K., et al. (2021) Oxicam-type non-steroidal anti-inflammatory drugs inhibit NPR1-mediated salicylic acid pathway. Nat. Commun. 12: 7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., et al. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E. and Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kováčik J., Grúz J., Bačkor M., Strnad M. and Repčák M. (2009) Salicylic acid-induced changes to growth and phenolic metabolism in Matricaria chamomilla plants. Plant Cell Rep. 28: 135–143. [DOI] [PubMed] [Google Scholar]
- Lai Y.S., Renna L., Yarema J., Ruberti C., He S.Y. and Brandizzi F. (2018) Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Aca. Sci. U.S.A. 115: E5203–E5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D., Bhandari D.D. and Parker J.E. (2020) Origins and immunity networking functions of EDS1 family proteins. Annu. Rev. Phytopathol. 58: 253–276. [DOI] [PubMed] [Google Scholar]
- Linthorst H.J. and Van Loon L.C. (1991) Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci 10: 123–150. [Google Scholar]
- Li J., Zhong R., Palva E.T. and Davis K.R. (2017) WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS One 12: e0183731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Tian M. and Klessig D.F. (2012) Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 22: 1631–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R., Umar S. and Khan N.A. (2015) Exogenous salicylic acid improves photosynthesis and growth through increase in ascorbate-glutathione metabolism and S assimilation in mustard under salt stress. Plant Signal Behav. 10: e1003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olate E., Jiménez-Gómez J.M., Holuigue L. and Salinas J. (2018) NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants 4: 811–823. [DOI] [PubMed] [Google Scholar]
- Parker J.E., Holub E.B., Frost L.N., Falk A., Gunn N.D. and Daniels M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Yang J., Li X. and Zhang Y. (2021) Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72: 761–791. [DOI] [PubMed] [Google Scholar]
- Rate D.N. and Greenberg J.T. (2001) The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J 27: 203–211. [DOI] [PubMed] [Google Scholar]
- Rustérucci C., Aviv D.H., Holt B.F. III, Dangl J.L. and Parker J.E. (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattely E.S., Holmes E.C., Rajniak J., Kim J.-G., Tang S. and Fischer C.R. (2018) N-Hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115: E4920–E4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E. and Mur L.A. (2004) Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Yang Y., Liu K., Zhang L., Guo H., Sun T., et al. (2016) Involvement of endogenous salicylic acid in iron-deficiency responses in Arabidopsis. J. Exp. Bot. 67: 4179–4193. [DOI] [PubMed] [Google Scholar]
- Shikanai Y., Asada M., Takafumi S., Enomoto Y., Yamagami M., Yamaguchi K., et al. (2022b) Role of GSL8 in low calcium tolerance in Arabidopsis thaliana. Plant Biotechnol. 39: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai Y., Takahashi S., Enomoto Y., Yamagami M., Yamaguchi K., Shigenobu S., et al. (2022a) Arabidopsis Glucan Synthase-Like1 (GSL1) is required for tolerance to low-calcium conditions and exhibits a function comparable to GSL10. Plant Cell Physiol. 63: 1474–1484. [DOI] [PubMed] [Google Scholar]
- Shikanai Y., Yamagami M., Shigenobu S., Yamaguchi K., Kamiya T. and Fujiwara T. (2015) Arabidopsis thaliana PRL1 is involved in low-calcium tolerance. Soil Sci Plant Nutr. 61: 951–956. [Google Scholar]
- Shikanai Y., Yoshida R., Hirano T., Enomoto Y., Li B., Asada M., et al. (2020) Callose synthesis suppresses cell death induced by low-calcium conditions in leaves. Plant Physiol. 182: 2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Yu S., Yu R., Zhang F., Yu Y., Zhang D., et al. (2016) Effects of endogenous salicylic acid during calcium deficiency-induced tipburn in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep 34: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S., Sun L., Ma C., Li L., Li G. and Hao L. (2013) Reducing basal salicylic acid enhances Arabidopsis tolerance to lead or cadmium. Plant Soil. 372: 309–318. [Google Scholar]
- Taylor M.D. and Locascio S.J. (2004) Blossom-end rot: a calcium deficiency. J Plant Nutr 27: 123–139. [Google Scholar]
- Vanacker H., Lu H., Rate D.N. and Greenberg J.T. (2001) A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28: 209–216. [DOI] [PubMed] [Google Scholar]
- van Butselaar T. and Van den Ackerveken G. (2020) Salicylic acid steers the growth–immunity tradeoff. Trends Plant Sci. 25: 566–576. [DOI] [PubMed] [Google Scholar]
- Wagner S., Stuttmann J., Rietz S., Guerois R., Brunstein E., Bautor J., et al. (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microb. 14: 619–630. [DOI] [PubMed] [Google Scholar]
- Wang W., Withers J., Li H., Zwack P.J., Rusnac D.V., Shi H., et al. (2020) Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature 586: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.F. (1979) Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99: 410–412. [DOI] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J. and Parker J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G. and Ausubel F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Yildiz I., Mantz M., Hartmann M., Zeier T., Kessel J., Thurow C., et al. (2021) The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming. Plant Physiol. 186: 1679–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y. and Mou Z. (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F. and Glazebrook J. (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla C., Schabow J.E., Chernov V. and Palta J.P. (2019) CAX1 vacuolar antiporter overexpression in potato results in calcium deficiency in leaves and tubers by sequestering calcium as calcium oxalate. Crop Sci. 59: 176–189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and online supplementary material. Information on the mutants and primers is available in Supplementary Tables.


