Abstract
In China, few molecular epidemiological data on hepatitis C virus (HCV) are available and all previous studies were limited by small sample sizes or specific population characteristics. Here, we report characterization of the epidemic history and transmission dynamics of HCV strains in China. We included HCV sequences of individuals belonging to three HCV surveillance programs: 1) patients diagnosed with HIV infection at the Beijing HIV laboratory network, most of whom were people who inject drugs and former paid blood donors, 2) men who have sex with men, and 3) the general population. We also used publicly available HCV sequences sampled in China in our study. In total, we obtained 1,603 Ns5b and 865 C/E2 sequences from 1,811 individuals. The most common HCV strains were subtypes 1b (29.1%), 3b (25.5%) and 3a (15.1%). In transmission network analysis, factors independently associated with clustering included the region (OR: 0.37, 95% CI: 0.19–0.71), infection subtype (OR: 0.23, 95% CI: 0.1–0.52), and sampling period (OR: 0.43, 95% CI: 0.27–0.68). The history of the major HCV subtypes was complex, which coincided with some important sociomedical events in China. Of note, five of eight HCV subtype (1a, 1b, 2a, 3a, and 3b), which constituted 81.8% HCV strains genotyped in our study, showed a tendency towards decline in the effective population size during the past decade until present, which is a good omen for the goal of eliminating HCV by 2030 in China.
Introduction
Hepatitis C virus (HCV) infection is a major public health threat in China. The most recent estimate of the national prevalence of HCV infection is 0.7%, representing approximately 10 million people [1]. In 2020 alone, 194,066 individuals were newly diagnosed with HCV infection [2]. Moreover, an estimated 34,198 people died of cirrhosis attributed to HCV infection in 2017 [3]. Inspired by the exciting curable therapeutic effect of new all-oral antivirals with a short treatment duration, more manageable side effects, and improved sustained virologic response (SVR), in 2016, the WHO introduced ambitious global targets to eliminate HCV infection by 2030 [4,5]. To achieve these goals, China needs to develop national policies based on up-to-date and reliable epidemiological data. Previous national or quasi-national studies have determined HCV genotype distribution in China. However, all these studies are limited by small sample sizes, samples of a specific population (mostly from people who inject drugs [PWID] and former paid blood donors, whereas men who have sex with men [MSM] were seldom considered) and restricted geographic sampling [6–11]. Previous studies have also reconstructed the evolutionary history of HCV lineages in China and successfully linked the time scale of HCV evolution to unique historical events and past sociomedical conditions in China, such as the "Cultural Revolution" and "Encouraged Plasma Campaign" [12,13]. However, the outcomes of these theoretical studies have been limited by a relatively narrow span of sampling time.
Rapidly evolving RNA viruses, such as HIV and HCV, contain measurable footprints in their genome, which can be used for molecular transmission networks. Thus, by using nucleotide sequences, HIV transmission networks that link people who are infected with genetically similar isolates can be constructed, whereby linked people are presumed to have a direct or indirect epidemiologic connection and usually represent a “hotspot” of HIV transmission [14–16]. Over the last two decades, many clustering methods have been developed to define HIV transmission networks within a population. Broadly speaking, these methods can be grouped into two categories: methods that cluster directly on sequence variation via pairwise genetic distance measures, and methods that interpret this variation in the context of subtrees in a phylogeny. Phylogenetic analysis can be associated with high computational burden, especially for large sequence datasets. However, the genetic distance method can be computed rapidly. Therefore recent network analyses have favoured the generally faster and parameter-rich distanced-based methods [17,18].
These network analyses contribute significantly to our understanding of HIV epidemiology, for example, by providing information about HIV epidemics by identifying transmission linkages and by elucidating differences in transmission within and between populations [14–16]. HCV also evolves rapidly and shares the same routes of transmission as HIV; however, HCV transmission networks have never been characterized in China.
In this study, we aimed to update the genotype distribution, infer the molecular transmission networks and reconstruct the epidemic history of HCV in China using a substantially more comprehensive dataset and metadata than previous works.
Methods
Sampling strategy
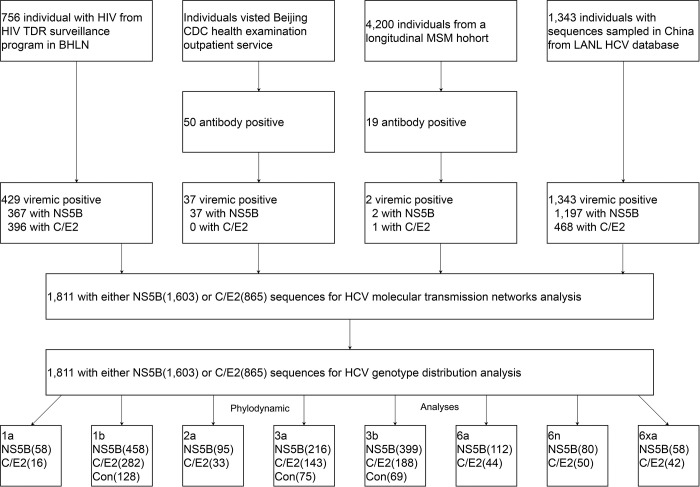
We designed a cross-sectional study to make full use of all available HCV genotyping data in China. The study population consisted of four separate groups of HCV-infected individuals (Fig 1 and S1 Table). The first group consisted of patients diagnosed with HIV infection at the Beijing HIV laboratory network (BHLN) from 1999 through 2017. The BHLN, established in 1986, is a collaborative network of laboratories involved in HIV diagnosis. It was authorized by the Beijing Municipal Commission of Health and includes a central reference laboratory in the Beijing Center for Disease Prevention and Control (CDC), four additional HIV reference laboratories (DiTan, YouAn, Peking Union Medical College, and People’s Liberation Army of China [PLA] General Hospital), and approximately 280 HIV screening laboratories [19,20].
Fig 1. Study profile.
BHLN = Beijing HIV laboratory network. LANL = Los Alamos National Laboratory. TDR = Transmitted drug resistance. MSM = Men who have sex with men. PWID = People who inject drugs.
To ensure that as many sequences as possible were obtained, we adopted a cost-effective sampling strategy, which is one that obtains many sequences at a low cost. We mainly considered the two groups of populations most seriously affected by HCV infection in the BHLN: PWID and former paid blood donors. We acknowledge that we obtained HCV RNA first and designed a sampling strategy later. We obtained RNA remnants from China’s national HIV transmitted drug resistance surveillance programme conducted by the BHLN. This programme randomly selected approximately 40% of individuals with newly diagnosed HIV infection between 1999 and 2017 from the national HIV epidemic database maintained by the BHLN. In total, we obtained 9,059 RNA samples from heterosexual individuals (2,190), MSM (6,136), PWID (539), and former paid blood donors (194) [19,20]. Unfortunately, for most of these samples, HCV antibody test records were not available (except for four heterosexuals and two volunteer blood donors). Therefore, we devised cost-effective inclusion criteria. According to the literature, the national prevalence of HCV co-infection in people living with HIV is approximately 4.0%, 6.4%, 82.4%, and 92.9% for heterosexuals, MSM, PWID, and former paid blood donors [6]. There is an unduly high HCV prevalence among the PWID and former paid blood donors. In other words, if we were concerned with these two groups of populations, we could obtain more HCV sequences at a lower cost. Moreover, or most importantly, the plasma samples were not sufficient for performing another HCV antibody test. Therefore, we included all samples from PWID and former paid blood donors in the BHLN in our analysis. Four heterosexual and two volunteer blood donors who were HCV antibody positive were also included.
The second group consisted of individuals who visited the health examination outpatient service of the Beijing CDC in 1999 and had HCV antibody-positive records. We roughly deemed these individuals to be from the general population.
The third group consisted of individuals with HCV antibody-positive records from an MSM cohort. In this cohort, we conducted seven serial consecutive cross-sectional surveys of MSM from 2015 through 2021. The purpose of this survey was to track trends in the prevalence of HIV, HCV and syphilis in this population [21,22].
The fourth group consisted of publicly available sequences from the Los Alamos National Laboratory (LANL) HCV sequence database. We retrieved all sequences sampled in China with information on the province of origin and sample year and covering the same genomic region from the databases (data available as of Dec 1, 2021). The sampling year and locations were confirmed by reference to the original literature.
Patient inclusion and data collection
We extracted baseline data on individuals from the national HIV epidemic database or LANL HCV database, including demographic and population characteristics and CD4 cell count. For geographic location, we grouped individuals into 25 provinces according to Hukou, a basic system of household registration in China. It officially identifies a person as a resident of an area and includes identifying information such as name, parents, spouse, and date of birth.
Genotypic analysis
We performed population-based sequencing of the HCV Ns5b and C/E2 regions in all specimens using in-house methods [8]. These sequences correspond to nucleotides 8,400 to 9,100 and 927 to 2,040, respectively, in the H77 genome. We inferred HCV genotype by automated genotyping in context-based modelling for expeditious typing (COMET)-HCV [23], followed by maximum likelihood (ML) phylogenetic analysis of the sequences [24]. An ML tree was used to confirm the results of COMET.
Transmission network analysis
To construct the transmission network, we followed the protocol outlined by Wertheim et al. [14–16]. We aligned HCV sequences in a pairwise fashion and then evaluated Tamura-Nei 93 (TN93) distances for all sequences using the HyPhy package [25]. TN93 genetic distance was used because it can be computed rapidly and is the most complex genetic distances that can be represented by a closed-form solution [17,18].We performed stepwise transmission network analysis using a serial set of genetic threshold (0.005–0.045 subsitution/site, increment every 0.0025 subsitution/site) [26]. We selected 0.01 subsitution/site and 0.0325 substituion/site for Ns5b and C/E2 datasets, because this distance identifies the maximum number of clusters in the transmission network (S1 Fig). The degree (connectivity) of each individual was defined as the number of links (edges in the transmission network) to other individuals. Clusters were defined as connected components of the network comprising two or more nodes. We used Cytoscape (3.8.0) to visualize the networks.
Phylogenetic analysis
We aligned sequences by using the BioEdit tool and manually corrected the alignment according to the encoded reading frame. If several sequences from the same patient were available in the dataset, we retained only the oldest sequence. Long branch trees were reconfirmed regarding genotype, and those found to be misclassified were eliminated. All these efforts help to minimize the possibility of duplicate patient sampling. We reconstructed ML phylogenetic tree with the datasets using the GTR+CAT nucleotide substitution model in FastTree 2.1 [24]. Temporal signal was examined using root-to-tip regression in TempEst v1.5.3 [27]. The sequences whose sampling year is incongruent with genetic divergence were excluded for Bayesian analysis. We estimated time-calibrated phylogenies dated from time-stamped genome data using the Bayesian software package BEAST(version 1.10.4) [28]. We only did Bayesian evolutionary analysis for main eight HCV subtype (1a, 1b, 2a, 3a, 3b, 6a, 6n, and 6xa) because their datasets contain at least 10 dated sequences. We used the HKY nucleotide substitution model with codon partitions [29] and Bayesian SkyGrid tree prior [30] with an uncorrelated relaxed clock with a lognormal distribution [31,32].
For each dataset, at least three independent Markov chain Monte Carlo (MCMC) chains were run for 50 million generations with states sampled every 1,000 generations. Multiple MCMC chains were calculated to increase Effective Sample Size(ESS). Log files were combined using Logcombiner (v.1.10.4) to ensure sufficient convergence (ESS≧200) with 10% of posterior samples discarded as burn-in. MCMC mixing was diagnosed using visual trace inspection and calculation of ESS in Tracer (v.1.7.2) [33]. The ESS of a parameter sampled from an MCMC is the number of effectively independent draws from the posterior distribution that the Markov chain is equivalent to. Maximum clade credibility trees were summarized using TreeAnnotator after discarding 10% as burn-in (S1 File.The protocol for Bayesian estimation of past population dynamics using the Skygrid coalescent model.).
Ethical issues
All analyses were performed on de-identified datasets to protect the participants’ anonymity. The research ethics committee at the Beijing CDC approved this study, and all the methods in this study were performed in accordance with approved guidelines. By law (Law of the People’s Republic of China on the Prevention and Treatment of Infectious Diseases, and Regulations on AIDS Prevention and Treatment), consent was not required, as these data were collected and analysed in the course of routine public health surveillance.
Statistical analysis
Four sampling phases were established: 1994–2003, 2004–2008, 2009–2013, and 2014–2020. The most early (1994–2003) and recent (2014–2020) phases encompassed more years to account for the relatively fewer data available in these years. We compared categorical data with the x2 test and continuous data with one-way ANOVA, wherever appropriate. We analysed the variables for clustering using univariable and multivariable logistic regression. Variables considered were region, HCV subtype, population characteristics, and sampling phase. We analysed all variables separately and entered those associated (P<0.1) with the outcomes into the multivariable model. We present the results as odds ratios (ORs) with 95% confidence intervals (CIs). We performed all analyses using R (version 4.1.1; R Foundation, Vienna, Austria). We used listwise deletion to handle missing data.
Results
Study population
Our study population was four cohorts (Fig 1 and S1 Table). First, we included 756 individuals newly diagnosed with HIV from the national epidemiology database of China. The BHLN is authorized officially to participate in maintaining this database. Second, we included 50 individuals who visited the health examination outpatient service of the Beijing Center for Disease Prevention and Control (CDC) and had HCV antibody-positive records. Third, we included 19 individuals with HCV antibody-positive records from an MSM cohort that consist of 4,200 people recruited between 2015 and 2021.
From the above three cohorts, we included 825 individuals in our analysis. Amplification and sequencing of Ns5b and C/E2 fragments were successful for 342 (40.6%) individuals. For an additional 126 patients, sequences were obtained for either Ns5b alone (n = 64) or C/E2 fragment alone (n = 62). Hence, we were able to perform HCV genotyping and phylogenetic analysis for 468 (56.7%) individuals based on the availability of sequence data. The prevalence of viraemic HCV infection in PWID, former paid blood donors, and MSM was 60.7% (327 of 539), 40.7% (79 of 194), and 0.05% (2 of 4,200) respectively. The majority of the participants were men (80.1%). Han, Uygur and Yi ethnicities accounted for 43.0%, 38.4% and 10.3%, respectively. The median age was 32 years (interquartile range [IQR] 26–39). The CD4 was only available for individuals with HIV/HCV co-infection and the overall median baseline CD4 count was 336 cells per μL (IQR 240–461).
Fourth, we included all Ns5b and C/E2 sequences sampled in China with known sampling provinces and sampling years available in the LANL HCV sequence database. After rigorous phylogenetic analysis, we obtained both HCV Ns5b and C/E2 sequence fragments from 322 individuals and either of the fragments from 1,021(S2 File.Accession numbers).
Thus, we included 1,603 Ns5b and 865 C/E2 sequences from 1,811 individuals from 25 provinces of China in the final analysis (Fig 1 and S1 Table). The transmission risk group were predominantly PWID (77.1%), followed by general population (16.5%), former paid blood donor (5.7%), heterosexual (0.3%), MSM(0.1%), and volunteer blood donor (0.1%) (Table 1).
Table 1. Baseline characteristic by sampling phase.
| 1994–2003 | 2004–2008 | 2009–2013 | 2014–2020 | Total | |
|---|---|---|---|---|---|
| Sex | |||||
| Men | 123(85.4) | 271(80.7) | 69(78.4) | 48(68.6) | 511(80.1) |
| Women | 21(14.6) | 65(19.3) | 19(21.6) | 22(31.4) | 127(19.9) |
| Age at diagnosis(years)a | 27(23–31) | 32(26–37) | 34(28–39) | 44(33–72) | 32(26–39) |
| CD4 counts(cells per μL)b | 213(119–306) | 294(206–422) | 369(286–506) | 356(299–405) | 336(240–461) |
| Ethnicity | |||||
| Han | 71(75.5) | 86(36.1) | 47(53.4) | 9(12) | 213(43) |
| Uighur | 23(24.5) | 124(52.1) | 25(28.4) | 18(24) | 190(38.4) |
| Yi | 0(0) | 22(9.2) | 16(18.2) | 13(17.3) | 51(10.3) |
| Li | 0(0) | 0(0) | 0(0) | 34(45.3) | 34(6.9) |
| Other Minority | 0(0) | 6(2.5) | 0(0) | 1(1.3) | 7(1.4) |
| Population characteristicc | |||||
| Heterosexual | 2(1) | 0(0) | 2(0.3) | 0(0) | 4(0.3) |
| MSM | 0(0) | 0(0) | 0(0) | 2(0.7) | 2(0.1) |
| PWID | 153(77.3) | 180(75.9) | 442(67.7) | 288(99.3) | 1063(77.1) |
| Former paid blood donor | 4(2) | 57(24.1) | 18(2.8) | 0(0) | 79(5.7) |
| General population | 37(18.7) | 0(0) | 191(29.2) | 0(0) | 228(16.5) |
| Volunteer blood donor | 2(1) | 0(0) | 0(0) | 0(0) | 2(0.1) |
| Region | |||||
| North | 13(6) | 40(11.5) | 18(2) | 2(0.6) | 73(4) |
| Northeast | 2(0.9) | 7(2) | 2(0.2) | 0(0) | 11(0.6) |
| East | 0(0) | 5(1.4) | 367(39.8) | 0(0) | 372(20.5) |
| Central South | 2(0.9) | 37(10.7) | 281(30.4) | 43(13.3) | 363(20) |
| Southwest | 96(44.2) | 128(36.9) | 226(24.5) | 257(79.3) | 707(39) |
| Northwest | 54(24.9) | 128(36.9) | 27(2.9) | 20(6.2) | 229(12.6) |
| Unknown | 50(23) | 2(0.6) | 2(0.2) | 2(0.6) | 56(3.1) |
| Genotype and subtype | |||||
| 1a | 2(0.9) | 2(0.6) | 38(4.1) | 22(6.8) | 64(3.5) |
| 1b | 80(36.9) | 121(34.9) | 291(31.5) | 35(10.8) | 527(29.1) |
| 2a | 18(8.3) | 18(5.2) | 85(9.2) | 0(0) | 121(6.7) |
| 3a | 43(19.8) | 62(17.9) | 104(11.3) | 64(19.8) | 273(15.1) |
| 3b | 48(22.1) | 99(28.5) | 213(23.1) | 102(31.5) | 462(25.5) |
| 6a | 8(3.7) | 10(2.9) | 91(9.9) | 26(8) | 135(7.5) |
| 6n | 11(5.1) | 18(5.2) | 50(5.4) | 40(12.3) | 119(6.6) |
| 6xa | 7(3.2) | 12(3.5) | 36(3.9) | 13(4) | 68(3.8) |
| Other | 0(0) | 5(1.4) | 15(1.6) | 22(6.8) | 42(2.3) |
Data are n (%)
North = Beijing, Hebei, Shanxi, Inner Mongolia
Northeast = Liaoning, Heilongjiang
East = Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, Shandong
Central South = Henan, Hubei, Hunan, Guangdong,Guangxi, Hainan
Southwest = Chongqing, Sichuan, Guizhou, Yunnan
Northwest = Shannxi, Qinghai, Sinkiang
a Data for n = 429
b Data for n = 125
cData for n = 1378
dData for n = 1755
MSM = Men who have sex with men
PWID = People who inject drugs
Other = 6e, 6 g, 6 l, 6w, and 6v.
Phylogenetic analysis
We performed phylogenetic analysis using the merged Ns5b and C/E2 sequence dataset, which consisted of 1,603 and 865 sequences respectively. The phylogenetic tree confirmed the genotype assignment by COMET-HCV, and the genotype determinations between the Ns5b and C/E2 fragments were consistent (S2 Fig). All isolates in our study belong to four genotypes (1, 2, 3, and 6) and 13 subtypes (1a, 1b, 2a, 3a, 3b, 6a, 6e, 6g, 6l, 6n, 6v, 6w, and 6xa). The prevalence of genotypes 1, 2, 3, and 6 was 32.6%, 6.7%, 40.6%, and 20.1%, respectively. The most common HCV subtypes in order of decreasing frequency were 1b (29.1%), 3b (25.5%), 3a (15.1%), 6a (7.5%), 2a (6.7%), 6n (6.6%), 6xa (3.8%), and 1a (3.5%). Additional clades, including subtypes 6e, 6g, 6l, 6w, and 6v, were present in fewer than 1.0% of individuals. HCV genotype patterns differed between population groups. In most groups, subtype 1b was the most prevalent (Tables 1 and 2). Table 1 presents the temporal trends for these eight major subtypes. There was a decreasing trend for genotype 1b and a stable trend for 3a and 3b. Table 2 illustrates the geographical distribution of HCV subtypes in China.
Table 2. HCV genotype and subtype assignment by selected characteristics.
| 1a | 1b | 2a | 3a | 3b | 6a | 6n | 6xa | Other | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sexa | ||||||||||
| Men | 7(1.4) | 162(31.7) | 17(3.3) | 106(20.7) | 140(27.4) | 11(2.2) | 26(5.1) | 29(5.7) | 13(2.5) | 511(100) |
| Women | 1(0.8) | 45(35.4) | 11(8.7) | 17(13.4) | 22(17.3) | 15(11.8) | 3(2.4) | 1(0.8) | 12(9.4) | 127(100) |
| Ethnicityb | ||||||||||
| Han | 3(1.4) | 101(47.4) | 35(16.4) | 21(9.9) | 38(17.8) | 11(5.2) | 2(0.9) | 1(0.5) | 1(0.5) | 213(100) |
| Uyghur | 2(1.1) | 89(46.8) | 0(0) | 57(30.0) | 40(21.1) | 2(1.1) | 0(0) | 0(0) | 0(0) | 190(100) |
| Yi | 1(2) | 3(5.9) | 0(0) | 14(27.5) | 11(21.6) | 0(0) | 0(0) | 22(43.1) | 0(0) | 51(100) |
| Li | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 12(35.3) | 0(0) | 0(0) | 22(64.7) | 34(100) |
| Other minority | 0(0) | 3(42.9) | 0(0) | 2(28.6) | 2(28.6) | 0(0) | 0(0) | 0(0) | 0(0) | 7(100) |
| Population characteristicc | ||||||||||
| Heterosexual | 0(0) | 3(75) | 1(25) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 4(100) |
| MSM | 0(0) | 2(100) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 2(100) |
| PWID | 49(4.6) | 202(19) | 6(0.6) | 241(22.7) | 339(31.9) | 58(5.5) | 96(9) | 65(6.1) | 7(0.7) | 1063(100) |
| Former paid blood donor | 0(0) | 52(65.8) | 16(20.3) | 3(3.8) | 6(7.6) | 1(1.3) | 1(1.3) | 0(0) | 0(0) | 79(100) |
| General population | 0(0) | 169(74.1) | 37(16.2) | 5(2.2) | 8(3.5) | 7(3.1) | 2(0.9) | 0(0) | 0(0) | 228(100) |
| Volunteer blood donor | 0(0) | 1(50) | 0(0) | 0(0) | 1(50) | 0(0) | 0(0) | 0(0) | 0(0) | 2(100) |
| Regiond | ||||||||||
| North | 0(0) | 33(45.2) | 4(5.5) | 9(12.3) | 21(28.8) | 5(6.8) | 0(0) | 1(1.4) | 0(0) | 73(100) |
| Northeast | 0(0) | 4(36.4) | 1(9.1) | 1(9.1) | 4(36.4) | 1(9.1) | 0(0) | 0(0) | 0(0) | 11(100) |
| East | 3(0.8) | 187(50.3) | 30(8.1) | 49(13.2) | 46(12.4) | 37(9.9) | 13(3.5) | 1(0.3) | 6(1.6) | 372(100) |
| Central South | 15(4.1) | 105(28.9) | 65(17.9) | 14(3.9) | 53(14.6) | 68(18.7) | 10(2.8) | 3(0.8) | 30(8.3) | 363(100) |
| Southwest | 44(6.2) | 70(9.9) | 6(0.8) | 123(17.4) | 281(39.7) | 20(2.8) | 95(13.4) | 63(8.9) | 5(0.7) | 707(100) |
| Northwest | 2(0.9) | 98(42.8) | 1(0.4) | 74(32.3) | 51(22.3) | 2(0.9) | 1(0.4) | 0(0) | 0(0) | 229(100) |
| Co-infectione | ||||||||||
| HCV single infection | 0(0) | 25(34.2) | 13(17.8) | 0(0) | 0(0) | 13(17.8) | 0(0) | 0(0) | 22(30.1) | 73(100) |
| HCV/HIV co-infection | 34(4.2) | 219(26.9) | 22(2.7) | 177(21.7) | 227(27.9) | 51(6.3) | 43(5.3) | 37(4.5) | 5(0.6) | 815(100) |
Data are n (%)
aData for n = 638
bData for n = 493
cData for n = 1378
dData for n = 1755
eData for n = 886
North = Beijing, Hebei, Shanxi, Inner Mongolia
Northeast = Liaoning, Heilongjiang
East = Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, Shandong
Central South = Henan, Hubei, Hunan, Guangdong,Guangxi, Hainan
Southwest = Chongqing, Sichuan, Guizhou, Yunnan
Northwest = Shannxi, Qinghai, Sinkiang
MSM = Men who have sex with men
PWID = People who inject drugs
Other = 6e, 6 g, 6 l, 6w, and 6v.
Network inference
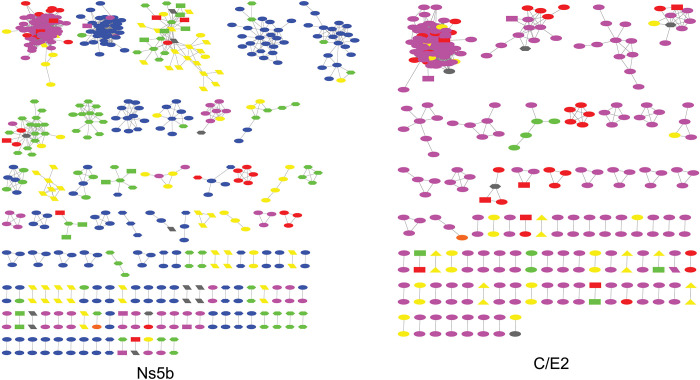
Using the Ns5b sequence (n = 1,603), we built an HCV transmission network representing 25 provinces of China. The network contains 111 connected components with ≥2 nodes (clusters) comprising 530 nodes (individual sequence) and 2,194 edges (undirected, potential links). The average degree (number of edges per node) was 4.1. The number of sequences per cluster ranged from 2–84 (median: 3, interquartile range:2–3) (Fig 2). In multivariable logistic analyses, being in a cluster was significantly associated with region (OR: 0.37, 95% CI: 0.19–0.71), subtype (OR: 0.23, 95% CI: 0.1–0.52), and sampling period(OR: 0.43, 95% CI: 0.27–0.68) (S2 Table).
Fig 2. HCV molecular transmission network in China.
Clusters with ≥2 cases (i.e., nodes) are depicted. Links (i.e., edges) indicate genetic distance≤0.01 substitutions/site for Ns5b and ≤0.0325 substitutions/site for C/E2. Shape indicates population groups: Diamond, heterosexual; ellipse, people inject drugs; rectangle, former blood donors; hexagon, unknown; triangle, general population; parallelogram, volunteer blood donors. Color indicate sampling region: Red, North; orange, Northeast; yellow, East; green, Central South; blue, Southwest; purple, Northwest. North = Beijing, Hebei, Shanxi, Inner Mongolia, Northeast = Liaoning, Heilongjiang, East = Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, Shandong, Central South = Henan, Hubei, Hunan, Guangdong,Guangxi, Hainan, Southwest = Chongqing, Sichuan, Guizhou, Yunnan, Northwest = Shannxi, Qinghai, Sinkiang.
We repeated the same network inference procedure for 865 C/E2 sequences. Although the available dataset is relatively smaller, we observed a similar pattern in the transmission network inferred using C/E2 sequences (Fig 2 and S3 Table).
Phylodynamic analyses and inference of divergence date
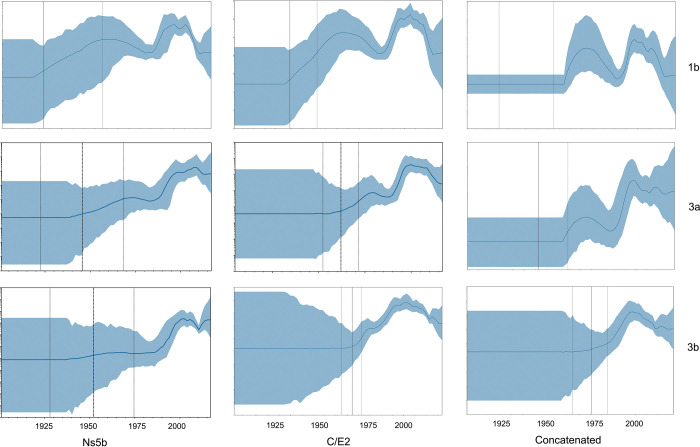
We performed a Bayesian SkyGrid Plots (BSP) analysis for 19 datasets: 1) six Ns5b datasets (1a, 1b, 2a, 3a, 3b, 6a, 6n, and 6xa), 2) six C/E2 datasets (1a, 1b, 2a, 3a, 3b, 6a, 6n, and 6xa), and 3) three Ns5b +C/E2 concatenated datasets (1b, 3a, and 3b). Table 3 and S3 Fig. show the date of the Time to the Most Recent Common Ancestor (TMRCA) for the eight major HCV subtypes. Among them, subtype 1a and 6n were the oldest, subtype 6xa was the youngest. The TMRCA dates for strains 1b, 2a, 3a, and 3b were in the same range, approximately 80 years ago. The BSP shown in Figs 3 and S4. depict the estimated change in the effective number of infected individuals over time. Of the eight major subtypes, the epidemic history of 1b was one of most complicated in our datasets: it showed an "M-shape" or "Roller Coaster" curve that consisted of two major epidemic waves. The first wave began circa 1910 and ended circa 1985, with a peak circa 1957. The increasing period of the wave coincides with the introduction of modern medicine in China (probably through the reuse and inadequate sterilization of glass and metal syringes). The decreasing period coincides with the two major social and political events in China: the "Great Leap Forward" (1958–1960) and "Cultural Revolution" (1966–1976). The second wave seemed to be sparked by the increase in PWID in the middle 1980s and was enhanced by the “Encouraged Plasma Campaign”(1993–2000) in the 1990s. This escalating trend was abruptly reversed in approximately 2000, when the Chinese government outlawed the use of paid blood donors. After that, despite a small rebound between 2005 and 2010, the 1b epidemic entered a downward trend from 2010 until the present. The other seven major subtypes have similar but relatively simple BSP curves. Of note, five major subtypes (1a, 1b, 2a, 3a, and 3b) exhibited a declining trend after 2010 until the present, whereas three subtypes (6a, 6n and 6xa) showed an increasing or stable trend. We repeated the same phylodynamic procedure using C/E2 sequences datasets, and TMRCA and BSP were roughly consistent with that of Ns5b except for subtype 1a (Figs 3 and S4). The BSP for concatenated datasets have smaller confidence limits but narrower time scale (Fig 3).
Table 3. The TMRCA of HCV in China.
| Ns5b | C/E2 | C/E2+ Ns5b | ||||
|---|---|---|---|---|---|---|
| Subtype | Numbers | Numbers | Numbers | |||
| 1a | 58 | 1899.6(1616.1–1981.5) | 16 | 2007.4(1960–2013.1) | ||
| 1b | 458 | 1915.2(1871.8–1942) | 282 | 1920.4(1897.4–1939.3) | 128 | 1897.3(1844.9–1935.4) |
| 2a | 95 | 1977.9(1963–1987.8) | 33 | 1800.2(190–1975.9) | ||
| 3a | 216 | 1948.9(1910.3–1974) | 143 | 1959.9(1947.9–1970.3) | 75 | 1955.4(1935.9–1971) |
| 3b | 399 | 1952.2(1918.8–1976.2) | 188 | 1962.9(1949.9–1974.7) | 69 | 1975.9(1961.7–1985.8) |
| 6a | 112 | 1964.6(1938.2–1982.2) | 44 | 1972.3(1954.1–1984.5) | ||
| 6n | 80 | 1967.8(1925–1993.5) | 50 | 1932.3(1836.5–1973.7) | ||
| 6xa | 58 | 1972.9(1935.5–1990.5) | 42 | 1976(1957.6–1988.5) |
TMRCA = Time to the Most Recent Common Ancestor.
aData are TMRCA (the 95% highest posterior density [HPD] interval).
Fig 3. The past population dynamics of HCV visualized using the Skygrid model.
The shaded portion is the 95% Bayesian credibility interval, and the solid line is the posterior median.
Discussion
Here, we report large amounts of data on HCV molecular epidemiology in China based on demographic and clinical data and HCV sequences from 1,811 patients of 25 provinces between 1994 and 2020. These data show that the HCV epidemic in China exhibits some degree of genetic diversity [34–41], consisting of four HCV subtypes and corresponding to 13 subtypes. Consistent with previous studies, the most prevalent HCV variant was subtype 1b, followed by 3b and 3a [6–11]. These subtypes are responsible for the majority of HCV cases globally [34–41]. Of note, five of eight major epidemic subtypes, together with 81.8% of HCV strains in our study, showed a declining tendency in effective population size during the past decade. In HCV transmission network analysis, 33.1% of patients grouped into 111 molecularly defined HCV transmission clusters.
Nakano, et al. [12] also reconstructed the population genetic history of HCV 1b in China and found that both groups of 1b grew at a rapid exponential rate during the "Cultural Revolution" of 1966–1976. They further attribute this rapid growth to the introduction of a million nonprofessional health-care providers ("barefoot doctors"). Barefoot doctors were healthcare providers who underwent basic medical training and worked in rural villages in China. The barefoot doctors system was developed and institutionalized in 1965 and broke down in the 1980s. Barefoot doctors included farmers, folk healers, rural healthcare providers and recent middle or secondary school graduates who received minimal basic medical and paramedical education.
Contrary to Nakano’s finding, we observed a declining trend for HCV 1b in the effective population size during the "Cultural Revolution", and we suggest that attributing the increasing trend only to the introduction of "barefoot doctors" during the "Cultural Revolution" is oversimplified. Indeed, the impacts of large historical events such as the "Cultural Revolution" on the epidemic dynamics of HCV are complicated. On the one hand, the closure of medical schools and specialist hospital departments led to the introduction of "barefoot doctors" into the medical system, which may have caused an increase in HCV infection. On the other hand, nearly all professional medical staff had to stop working and were dispersed across the countryside during that period, which led to a sharp decline in the total amount of medical activity, including unsafe injections. We believe that the latter was the real determinant for the declining trend in the "Cultural Revolution" period.
Pybus et al. [42] showed that genotype 6 infection worldwide descended from a common ancestor that existed approximately 1,100 to 1,350 years earlier. How stable endemic transmission of HCV genotype 6 could be maintained for such long a time period has always fascinated scientists. As introduction and transmission events of HCV genotype 6 occurred so many years ago, we can only speculate. We suggest that traditional tattooing, which once prevailed in some minor ethnic populations of Yunnan Province, is responsible [43]. We further suggest that Yunnan is the epidemic centre of HCV genotype 6 in China as well as that of HIV [44–46]. Yunnan is located in southwestern China, bordering Myanmar, Laos, and Vietnam. There are 16 ethnic minorities inhabiting the border, many of whom used to practice the custom of a traditional tattooing. The proximity and close cultural ties between populations in Yunnan and Southeast Asia countries have linked these groups for many years. It is plausible to speculate that HCV genotype 6 was introduced to China from Southeast Asian and maintained through traditional tattooing until the modern time, when this traditional custom was no longer popular.
To our knowledge, this is the largest study of its type thus far and involves the longest time period. Through this informative dataset, we conducted a national HCV molecular epidemiology study with broad representativeness and accurate phylogenetic reconstruction.
This analysis also has limitations. First, since approximately two-thirds of the sequences were from publicly available databases, most of the baseline characteristics of the patients were not available, which prevented us from including these variables in transmission cluster analysis and from making a more detailed investigation of the risk factors driving HCV epidemics in China. Second, because we used a cost-effective sampling method, participants with HIV/HCV co-infection or PWID were overrepresented in our study. Hence, the findings might not be fully representative of the whole population in China. Third, the number of the recently sampled sequences was relatively small (S5 Fig). Therefore, the small rebound observed between 2005 and 2010 in our study is more likely due to sampling biases (e.g., distribution of samples in time and, lack of convergence in chain) than a real trend in the data.
Fourth, we discarded 152(6.5%) sequences from the original dataset because we thought they had quality problems, which could reshape a dataset with no temporal signal into one that strongly supports phylogenetic molecular clock analysis. The original results without the filtering sequences are listed in S4 Table.
In summary, this national study of 1,811 patient HCV sequences describes the most recent data on HCV genotype distribution in China. The most common HCV strain was found to be 1b, followed by 3b and 3a. Phylodynamic analysis revealed a complex scenario that was most likely driven by a combination of social, demographic, and medical factors over both recent and historical timescales. Crucially, BSP analysis showed a declining trend up to the present for 81.8% of the HCV strains in our study, which is a good omen for the goal of eliminating HCV by 2030.
Supporting information
The threshold that was selected is highlighted in red.
(DOCX)
1,603 Ns5b and 865 C/E2 sequences from China were analyzed with HCV reference strains (NC_004102, D90208, AB047639, JN714194, JQ065709, HQ639936, DQ278894) as an out-group using Fasttree 2.1.
(DOCX)
TMRCA = Time to the Most Recent Common Ancestor. The solid line indicates the 95% highest posterior density [HPD] interval for TMRCA.
(DOCX)
The shaded portion is the 95% Bayesian credibility interval, and the solid line is the posterior median.
(DOCX)
(DOCX)
BHLN = Beijing HIV laboratory network; PWID = People who inject drugs; MSM = Men who have sex with men; NA = not available; LANL = Los Alamos National Laboratory.
(DOCX)
North = Beijing, Hebei, Shanxi, Inner Mongolia, Northeast = Liaoning, Heilongjiang, East = Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, Shandong, Central South = Henan, Hubei, Hunan, Guangdong,Guangxi, Hainan, Southwest = Chongqing, Sichuan, Guizhou, Yunnan, Northwest = Shannxi, Qinghai, Sinkiang; MSM = men who have sex with men, PWID = people who inject drugs; NA = not available; OR = odds ratio; aData are n (%); bUnivariable logistic regression analysis; cMultivariable logistic regression analysis; dData for n = 1552, eData for n = 1175, Other = 6e, 6g, 6l, 6w, and 6v.
(DOCX)
North = Beijing, Hebei, Shanxi, Inner Mongolia, Northeast = Liaoning, Heilongjiang, East = Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, Shandong, Central South = Henan, Hubei, Hunan, Guangdong,Guangxi, Hainan, Southwest = Chongqing, Sichuan, Guizhou, Yunnan, Northwest = Shannxi, Qinghai, Sinkiang; MSM = men who have sex with men, PWID = people who inject drugs; NA = not available; OR = odds ratio; aData are n (%), bUnivariable logistic regression analysis, cMultivariable logistic regression analysis, dData for n = 849, eData for n = 846, Other = 6e, 6g, 6l, 6w, and 6v.
(DOCX)
TMRCA = Time to the Most Recent Common Ancestor. aData are TMRCA (the 95% highest posterior density [HPD] interval).
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the study participants and the staff at the collaborating clinical sites and laboratories. We thank the local health workers of the BHLN, who spent numerous hours and great effort in obtaining, verifying, and cleaning the data used in this study. We thank Dr. Xiang He from Guangdong Institute of Public Health for useful comments on drafts of the manuscript.
Data Availability
The minimal anonymized data sets necessary to replicate study findings have been uploaded in Open Science Framework (OSF) DOI: https://doi.org/10.17605/OSF.IO/NKD8Y. HCV sequences have been submitted to GenBank (See the Supporting Information file for accession numbers).
Funding Statement
This work was supported by China Capital's Funds for Health Improvement and Research (2022-1G-3011) to Jingrong Ye, Beijing Municipal Science & Technology Commission (D161100000416002), Beijing High-Level Public Health Doctor Cultivation Project (Academic Leader-01-04) to Hongyan Lu, Cultivation Fund of Beijing Center for Disease Prevention and Control (2019-BJYJ-13) to Yanming Sun. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.The Polaris Observatory HCV Collaborators. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modeling study. Lancet Gastroenterol Hepatol.2017; 2,161–176. [DOI] [PubMed] [Google Scholar]
- 2.National Health Commission of the People’s Republic of China. An overview of the epidemic situation of legal infectious diseases in China in 2020.2021;http://http://www.nhc.gov.cn/jkj/s3578/202103/f1a448b7df7d4760976fea6d55834966.shtml.
- 3.GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017.Lancet Gastroenterol Hepatol.2020; 5,245–266. doi: 10.1016/S2468-1253(19)30349-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assembly WHOS-NWH. Draft Global Health Sector Strategies Viral Hepatitis 2016–2021, 2016. http://www.who.int/hepatitis/strategy2016-2021/Draft_global_health_sector_strategy_viral_hepatitis_13nov.pdf (accessed Jun 1, 2020). [Google Scholar]
- 5.WHO. Combating Hepatitis B and C to Reach Elimination by 2030. Geneva: World Health Organization, 2016. https://www.who.int/hepatitis/publications/hep-elimination-by-2030-brief/en/ (accessed Jun 1, 2020). [Google Scholar]
- 6.Shang Hong, Zhong Ping, Liu Jing,et al.High Prevalence and Genetic Diversity of HCV among HIV-1 Infected People from Various High-Risk Groups in China. PLoS ONE.2010; 5,e10631. doi: 10.1371/journal.pone.0010631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Wang Y,Xia W,et al. New trends of HCV infection in China revealed by genetic analysis of viral sequences determined from first-time volunteer blood donors.J Viral Hepat.2011; 18,42–52. doi: 10.1111/j.1365-2893.2010.01280.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Chiyu, Wu Nana, Liu Jun,et al.HCV Subtype Characterization among Injection Drug Users: Implication for a Crucial Role of Zhenjiang in HCV Transmission in China. PLoS ONE.2011; 6,e16817. doi: 10.1371/journal.pone.0016817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Di, Li Lin, Liu Yongjian, Li Hanping, Xu Xiaoyuan, Li Jingyun. Different HCV Genotype Distributions of HIV-Infected Individuals in Henan and Guangxi, China. PLoS ONE.2012; 7,e50343. doi: 10.1371/journal.pone.0050343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Ling, Wang Min, Xia Wenjie, et al. Migration patterns of hepatitis C virus in China characterized for five major subtypes based on samples from 411 volunteer blood donors from 17 provinces and municipalities.J Virol.2014; 88,7120–9. doi: 10.1128/JVI.00414-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou S,Cella E,Zhou W,et al. Population dynamics of hepatitis C virus subtypes in injecting drug users on methadone maintenance treatment in China associated with economic and health reform. J Viral Hepat.2017; 24, 551–560. doi: 10.1111/jvh.12677 [DOI] [PubMed] [Google Scholar]
- 12.Nakano Tatsunori,Lu Ling,He Yunshao,Fu Yongshui,Robertson Betty H, Pybus Oliver G. Population genetic history of hepatitis C virus 1b infection in China.J Gen Virol.2006; 87,73–82. doi: 10.1099/vir.0.81360-0 [DOI] [PubMed] [Google Scholar]
- 13.Lu Ling, Tong Wangxia,Gu Lin,et al.The Current Hepatitis C Virus Prevalence in China May Have Resulted Mainly from an Officially Encouraged Plasma Campaign in the 1990s: a Coalescence Inference with Genetic Sequences.J Virol.2013; 87,12041–50. doi: 10.1128/JVI.01773-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertheim Joel O, Leigh Brown Andrew J, Hepler N Lance, et al. The Global Transmission Network of HIV-1. J Infect Dis.2014;209,1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacka Brendan,Applegate Tanya,Krajden Mel,et al.Phylogenetic Clustering of Hepatitis C Virus Among People Who Inject Drugs in Vancouver, Canada. Hepatology.2014; 60,1571–1580. doi: 10.1002/hep.27310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlet S R, Wertheim J O, Bull R A, et al. A molecular transmission network of recent hepatitis C infection in people with and without HIV: Implications for targeted treatment strategies.J Viral Hepat.2017; 24,404–411. doi: 10.1111/jvh.12652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan AS, Pybus OG, Sanders EJ, et al.Defining HIV-1 transmission clusters based on sequence data.AIDS.2017;31,1211–1222. doi: 10.1097/QAD.0000000000001470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon Art F. Y. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks.Virus Evol.2016;2,vew031. doi: 10.1093/ve/vew031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye Jingrong, Hao Mingqiang, Xing Hui, et al.Transmitted HIV drug resistance among individuals with newly diagnosed HIV infection: a multicenter observational study.AIDS.2020; 34,609–619. doi: 10.1097/QAD.0000000000002468 [DOI] [PubMed] [Google Scholar]
- 20.Ye Jingrong, Hao Mingqiang, Xing Hui, et al.Characterization of subtypes and transmitted drug resistance strains of HIV among Beijing residents between 2001-2016.PLoS One. 2020;26,e0230779. doi: 10.1371/journal.pone.0230779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Xiaoyan, Zhang Qiyun, He Xiong, et al. Trends in prevalence of HIV, syphilis, hepatitis C, hepatitis B, and sexual risk behavior among men who have sex with men. The results of 3 consecutive respondent-driven sampling surveys in Beijing, 2004 through 2006. J Acquir Immune Defic Syndr.2007; 45,581–7. [DOI] [PubMed] [Google Scholar]
- 22.Chen Qiang, Sun Yanming, Sun Weidong, et al. Trends of HIV incidence and prevalence among men who have sex with men in Beijing, China: Nine consecutive cross-sectional surveys, 2008–2016. PLoS One.2018;13, e0201953. doi: 10.1371/journal.pone.0201953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res.2014; 42,e144. doi: 10.1093/nar/gku739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Adam AP. Arkin. FastTree 2-approximatelymaximum-likelihood trees for large alignments. PLoS One.2010; 5, e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosakovsky Pond S.L., Frost S. D. W. and Muse S.V. HyPhy: hypothesis testing using phylogenies. Bioinformatics.2005; 21,676–679. doi: 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- 26.Wertheim Joel O., Pond Sergei L. Kosakovsky1, Forgione Lisa A., et al.Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathog.2017;13,e1006000. doi: 10.1371/journal.ppat.1006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut Andrew, Lam Tommy T, Carvalho Luiz Max, Pybus Oliver G .Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen).Virus Evol.2016; 2, vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marc A Suchard Philippe Lemey, Baele Guy, Ayres Daniel L, Drummond Alexei J, Andrew Rambaut. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4, vey016. doi: 10.1093/ve/vey016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro Beth, Rambaut Andrew, Drummond Alexei J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol.2006; 23, 7–9. doi: 10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- 30.Hill Verity,Baele Guy.Bayesian estimation of past population dynamics in BEAST 1.10 using the SkyGrid coalescent model.Mol Biol Evol.2019; 36,2620–2628. doi: 10.1093/molbev/msz172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drummond Alexei J, Ho Simon Y W,Phillips Matthew J,Rambaut Andrew. Relaxed phylogenetics and dating with confidence. PLoS Biol.2006;4, e88. doi: 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandev S Gill Philippe Lemey, Nuno R Faria Andrew Rambaut, Shapiro Beth, Suchard Marc A. Improving Bayesian population dynamics inference: a coalescent based model for multiple loci. Mol. Biol. Evol.2013;30, 713–724. doi: 10.1093/molbev/mss265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambaut Andrew, Alexei J Drummond Dong Xie, Baele Guy, Suchard Marc A .Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol.2018; 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Ngoc Chau, Thanh Thanh Tran Thi, Lan Phuong Tran Thi, et al. Differential prevalence and geographic distribution of hepatitis C virus genotypes in acute and chronic hepatitis C patients in Vietnam. PLoS ONE.2019; 14,e0212734. doi: 10.1371/journal.pone.0212734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouacida Lobna, Suin Vanessa, Hutse Veronik, et al. Distribution of HCV genotypes in Belgium from 2008 to 2015. PLoS ONE. 2018;13,e0207584. doi: 10.1371/journal.pone.0207584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palladino Claudia, Ezeonwumelu Ifeanyi Jude, Marcelino Rute, et al.Epidemic history of hepatitis C virus genotypes and subtypes in Portugal, Sci Rep.2018; 16,12266. doi: 10.1038/s41598-018-30528-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shier Medhat K, Iles James C, El-Wetidy Mohammad S, Ali Hebatallah H, Al Qattan Mohammad M. Molecular characterization and epidemic history of hepatitis C virus using core sequences of isolates from Central Province, Saudi Arabia. PLoS ONE. 2017;12,e0184163. doi: 10.1371/journal.pone.0184163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nouhin Janin, Iwamoto Momoko, sophearot prak,et al. Molecular epidemiology of hepatitis C virus in Cambodia during 2016–2017,Sci Rep.2019; 9, 7314. doi: 10.1038/s41598-019-43785-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petruzziello Arnolfo, Sabatino Rocco, Loquercio Giovanna,et al. (2019) Nine year distribution pattern of hepatitis C virus (HCV) genotypes in Southern Italy. PLoS ONE.2019; 14,e0212033. doi: 10.1371/journal.pone.0212033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNaughton Anna L, Cameron Iain Dugald,Wignall-Fleming Elizabeth B,et al.2015. Spatiotemporal reconstruction of the introduction of hepatitis C virus into Scotland and its subsequent regional transmission. J Virol.2015; 89:11223–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gower Erin, Estes Chris, Blach Sarah, Razavi-Shearer Kathryn, Razavi Homie.Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61, S45–S57. doi: 10.1016/j.jhep.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 42.Pybus Oliver G.,Barnes Eleanor,Taggart Rachel,et al.Genetic History of Hepatitis C Virus in East Asia.J Virol.2009; 83,1071–82. doi: 10.1128/JVI.01501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Jun,Survey on the status of the traditional tattoo and its static protection in Yunnan minorities, Proceedings of the 2011 annual meeting of the professional committee of ethnic museum of China association of museums, Xining Qinghai, 2011.8.1,329–344. [Google Scholar]
- 44.Meng Zhefeng, Xin Ruolei, Zhong Ping, et al.A new migration map of HIV-1 CRF07_BC in China: analysis of sequences from 12 provinces over a decade.PLoS One.2012;7,e52373. doi: 10.1371/journal.pone.0052373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Yi, He Xiang, Jenny H Hsi,et al.The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s.AIDS.2013; 27,1793–802. doi: 10.1097/QAD.0b013e328360db2d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Zhe, He Xiang, Wang Zhe, et al.Tracing the origin and history of HIV-1 subtype B’ epidemic by near full-length genome analyses.AIDS.2012;26,877–84. doi: 10.1097/QAD.0b013e328351430d [DOI] [PubMed] [Google Scholar]