Abstract
The nitrate (NO3-) reducing bacteria resident in the oral cavity have been implicated as key mediators of nitric oxide (NO) homeostasis and human health. NO3--reducing oral bacteria reduce inorganic dietary NO3- to nitrite (NO2-) via the NO3--NO2--NO pathway. Studies of oral NO3--reducing bacteria have typically sampled from either the tongue surface or saliva. The aim of this study was to assess whether other areas in the mouth could contain a physiologically relevant abundance of NO3- reducing bacteria, which may be important for sampling in clinical studies. The bacterial composition of seven oral sample types from 300 individuals were compared using a meta-analysis of the Human Microbiome Project data. This analysis revealed significant differences in the proportions of 20 well-established oral bacteria and highly abundant NO3--reducing bacteria across each oral site. The genera included Actinomyces, Brevibacillus, Campylobacter, Capnocytophaga, Corynebacterium, Eikenella, Fusobacterium, Granulicatella, Haemophilus, Leptotrichia, Microbacterium, Neisseria, Porphyromonas, Prevotella, Propionibacterium, Rothia, Selenomonas, Staphylococcus, Streptococcus and Veillonella. The highest proportion of NO3--reducing bacteria was observed in saliva, where eight of the bacterial genera were found in higher proportion than on the tongue dorsum, whilst the lowest proportions were found in the hard oral surfaces. Saliva also demonstrated higher intra-individual variability and bacterial diversity. This study provides new information on where samples should be taken in the oral cavity to assess the abundance of NO3--reducing bacteria. Taking saliva samples may benefit physiological studies, as saliva contained the highest abundance of NO3- reducing bacteria and is less invasive than other sampling methods. These results inform future studies coupling oral NO3--reducing bacteria research with physiological outcomes affecting human health.
Introduction
Advances in sequencing technology have allowed characterisation and in-depth analysis of the bacteria of the human oral cavity, with over 700 bacterial species having been identified [1, 2]. It has been possible to elucidate the communities of bacteria involved in oral nitrate (NO3-) and nitrite (NO2-) reduction. In the human NO3--NO2—nitric oxide (NO) pathway, the ingested dietary NO3- is absorbed and enters the circulatory system [3]. The NO3- is actively taken up by the salivary glands [4] and NO3- is concentrated in the saliva [5]. Commensal bacteria reduce the NO3- to NO2- which is subsequently swallowed, enters the bloodstream, and may be reduced to NO under anoxic or hypoxic conditions [5]. NO produced via the NO3--NO2--NO pathway is important for a wide array of physiological processes [3]. Better understanding of the process of bacteria-mediated dietary NO3--reduction may aid the optimisation of NO3- administration for improving blood pressure regulation, cognition, protection against ischaemia-reperfusion injury and exercise performance [5–9].
The Human Microbiome Project (HMP) under National Institutes of Health (NIH) contains bacterial samples taken from many human sites, ranging from the oral cavity to the gut and skin microflora [10, 11]. Through use of the HMP, bacterial composition and abundance can be compared between body sites on a larger scale, which may not be practical in conventional clinical or physiological study settings. Therefore, analysis of the data generated by the HMP can be used to inform and direct smaller-scale studies.
The oral cavity comprises various bacterial niches, such as the teeth, hard and soft palates, gingival sulcus, cheek, lip and tongue dorsum [12]. Previous work has suggested that the highest reduction of NO3- occurs on the tongue surface, which has been associated with the presence of NO3--reducing oral bacteria [13]. However, whilst previous studies have determined some oral niche areas of bacterial colonisation, human sample sizes are typically low, with less than 20 study participants being common [1, 13–16].
The new availability of data from samples taken on a larger scale can enable a wider perspective on where oral bacteria colonise particular oral niches. Methods for measuring oral NO3--reducing bacteria composition conventionally involve sampling from either the saliva or tongue surface [16–19]. The genera indicated by previous research to include potent NO3- reducing species included Actinomyces, Brevibacillus, Campylobacter, Capnocytophaga, Corynebacterium, Eikenella, Fusobacterium, Granulicatella, Haemophilus, Leptotrichia, Microbacterium, Neisseria, Porphyromonas, Prevotella, Propionibacterium, Rothia, Selenomonas, Staphylococcus, Streptococcus and Veillonella [13, 16]. Of these, Doel et al. identified species within Actinomyces, Rothia and Veillonella as the most potent NO3- reducers [13].
Saliva samples may be collected due to ease of sampling, whereas samples taken from the tongue dorsum may have more physiological relevance because NO3--reduction is known to occur on the tongue surface [13]. However, samples taken from the tongue dorsum using a buccal brush may not provide a high yield of NO3--reducing bacteria [authors’ personal observations]. The aim of this study was, therefore, to determine the site(s) in the mouth where the highest abundance of NO3--reducing bacteria reside in a larger human population, such as those found in the HMP, in order to inform methodology for future studies investigating the relationships between oral NO3--reducing bacteria and human health.
Materials and methods
Data mining
Complete 16S operational taxonomic unit (OTU) tabulated data and sample mapping files were mined from the publicly available online NIH Human Microbiome Project Metagenomic 16S Sequence QIIME community profiling database [10, 11]. The HMP is a community resource project that provides free access to use the human microbiome data by the scientific community. The data are available to use under the original HMP ethical approval and informed consent (https://www.hmpdacc.org/hmp/resources/tools_protocols.php). The original data was obtained by the HMP Microbiome Project [10, 11], and are publicly accessible for download at: https://www.hmpdacc.org/HMQCP/. The authors did not have any special access of request privileges. Authors did not have access to participant identifying information and all samples were anonymous. The participants were healthy 18- to 40-yr-old adults and the exclusion criteria included the presence of systemic diseases including hypertension, cancer, immunodeficiency or autoimmune disorders, use of potential immunomodulators, and recent use of antibiotics or probiotics [20]. The OTU data contained participant sample identifiers, bacteria names, class and number of observations (OTUs). Sample identifiers and body sampling site were extracted from the HMP mapping file, where saliva and six oral sites were further extracted. Anonymised sample identifiers were used to match bacterial observations and oral sampling site. Data from the HMP included samples taken following completion of informed consent procedure from a cohort of 300 participants, where samples were taken at multiple sites [10, 11]. Original methods have been described by the HMP Consortium [11]. The 2012 HMP data were downloaded for the purposes of the present study on 15th of November 2016.
Genus level classifications were mined from this dataset. Seven oral sample types and a total of 1288 samples were analysed from the oral cavity. The oral sites included attached keratinized gingiva (n = 183, 96 males, 87 females), buccal mucosa (n = 186, 96 males, 90 females), hard palate (n = 183, 95 males, 88 females), saliva (n = 166, 90 males, 76 females), subgingival plaque (n = 188, 97 males, 91 females), supragingival plaque (n = 192, 99 males, 93 females), and tongue dorsum (n = 190, 98 males, 92 females). Before proceeding with analysis, we compared the NO3- reducing bacteria between male and female subjects at each oral site. No significant differences were found between males and females within each oral site (P>0.05 for all comparisons). Consequently, we grouped the male and female samples together for comparison of NO3- reducing bacteria between oral sites.
Data analysis
Data downloaded from the HMP repository were processed and analysed using R statistical software [21]. Sample uniformity within each oral site, and between sites were visualised using multidimensional scaling (MDS) plots with the edgeR package [22]. Alpha diversity analyses were completed using vegan [23]. Median proportions were used to calculate Shannon Diversity Index and Simpson’s Diversity Index for visualization of diversity across the seven oral sample types, and the Chao 1 Index was employed to estimate species richness.
Using the HMP data, the Metacoder R package was used to generate an in-built taxmap object, which returned 985 phyla at 45383 observations [24]. For the detection of differences between bacterial abundances of each of the seven oral sample types specified, Metacoder was used to detect pairwise differences in log2-ratio of median proportions [24]. For each oral site sample, the differences in read proportions were extracted for analysis of 20 NO3--reducing bacteria of interest. For visualisation of bacterial differences in log2-ratio of median proportions across sites, heat trees were generated using Metacoder [24]. The percentage median relative abundance of genera and the corresponding standard error of the median were calculated to determine a list of the most relative abundant genera in each oral site. The most frequently occurring genera with the highest percentage relative abundance for each site were selected for direct comparison of the ten most abundant genera. The percentage median relative abundances of the remaining genera were aggregated to create the “Other” genera list.
Statistical analysis
Both Adonis and MRPP were used to identify any statistical differences in Shannon Diversity Index, and Simpson’s Diversity Index across the oral sites. The Wilcoxon signed-rank test was then applied find oral site-specific differences in diversity. For pairwise comparison between each oral site, statistical differences were computed using the Wilcoxon signed-rank test in R, due to the repeated sampling of each body site for each participant within the HMP database. Benjamini and Hochberg False discovery rate (FDR) correction was applied to control for multiple comparison testing and p < 0.05 was considered statistically significant.
Results
Analysis of alpha diversities
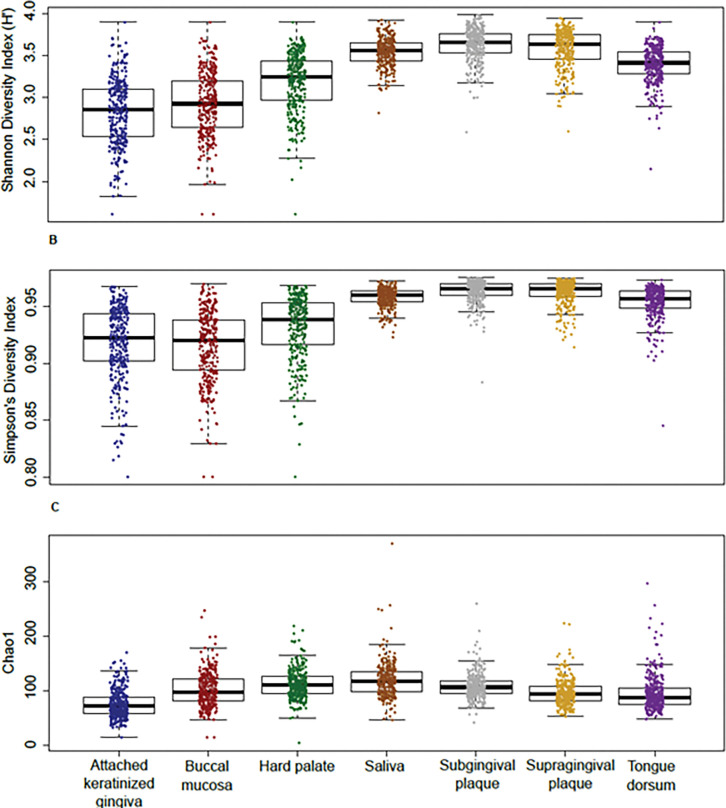
An overview of the bacterial composition of saliva and six oral sites revealed a high abundance of five major phyla: Firmicutes (44%), Actinobacteria (20%), Bacteroidetes (16%), Proteobacteria (15%) and Fusobacteria (4%). Shannon Diversity Index (H’) and Simpson’s Diversity Index showed that there were significant differences in microbial diversity across each oral site: Multiple Response Permutation Procedure (MRPP), p < 0.001; Permutational Multivariate Analysis of Variance Using Distance Matrices (ADONIS), p < 0.001. Chao1 indicated similar species richness (Fig 1). However, further testing showed that there were no significant differences in diversity between the saliva and subgingival plaque sites (p = 0.08), the saliva and supragingival plaque (p = 0.17) and the subgingival and supragingival plaque sites (p = 0.73).
Fig 1. Bacterial diversity in the oral cavity.
A Shannon Diversity (H’) Index of saliva and six oral sites. B Simpsons Diversity Index. C Chao1 species richness for saliva and six oral sites. For each panel, scatter plots show the individual samples. The box plots show the median and quartiles.
Pairwise comparison of NO3-reducing bacteria log2-ratio of median proportions at each oral site
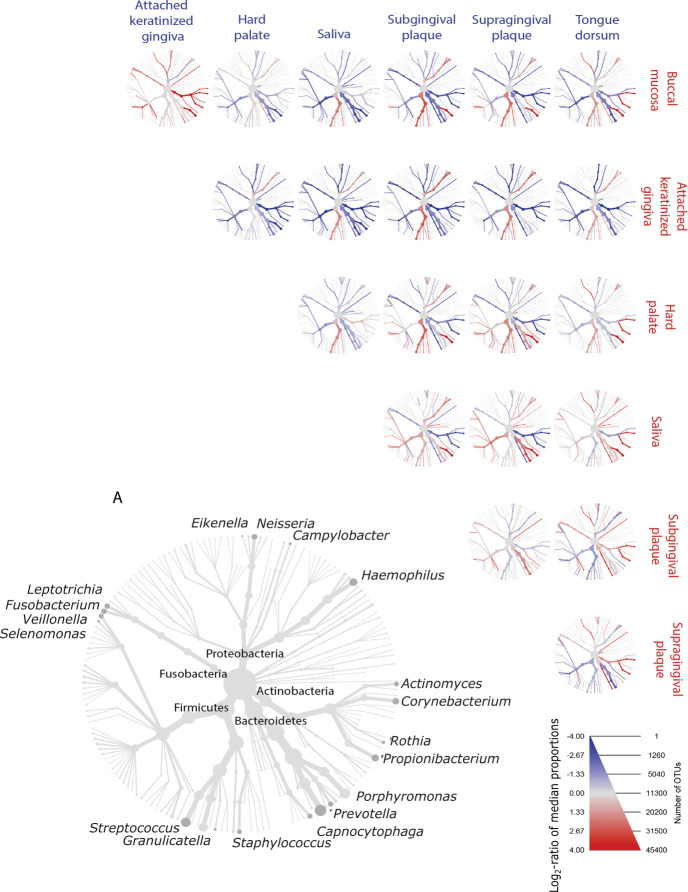
In saliva and six oral sites, the log2-ratio of median proportions of NO3--reducing bacteria were compared at different taxonomic levels. Significant differences were observed across phylum, class, order, family and genus (Fig 2).
Fig 2. Heat trees of pairwise comparisons showing the difference in median proportions of NO3--reducing bacteria between seven oral sample types.
The heat trees show statistically significant differences in log2-ratio of median proportions of NO3--reducing bacteria at seven oral sample types. Heat tree (A) is a labelled key of well-established NO3--reducing bacteria, where node size indicates the number of overall reads across all oral sites. The genera on the periphery of heat tree (A) correspond to the nearest branch ending with a dark grey node. Blue and red colours show the log2-ratio of median proportions observed at each oral site. Blue taxa correspond to the oral site label highlighted in blue, and red taxa correspond to the oral site label highlighted in red. Blue taxa are enriched in the blue oral sites labelled in the row, whilst red taxa are enriched in the red oral sites labelled in the column. The gradient of taxa colours corresponds to the difference in log2-ratio of bacterial median proportions, as shown in the legend.
The well-established NO3--reducing genera selected for comparison in the present study were Actinomyces, Brevibacillus, Campylobacter, Capnocytophaga, Corynebacterium, Eikenella, Fusobacterium, Granulicatella, Haemophilus, Leptotrichia, Microbacterium, Neisseria, Porphyromonas, Prevotella, Propionibacterium, Rothia, Selenomonas, Staphylococcus, Streptococcus and Veillonella.
Of the selected NO3--reducing bacteria, pairwise comparisons of the subgingival and supragingival plaque sites exhibited the least differences in proportions of NO3--reducing bacteria. The genera with the most significant differences in proportions observed in all oral sites included Corynebacterium, Propionibacterium and Eikenella. There were no significant differences in the proportions found in Brevibacillus, Staphylococcus, and Microbacterium in any of the pairwise comparisons.
Eight of the well-established genera including NO3--reducers were found in higher proportion in the saliva than on the tongue dorsum (p < 0.01). These included Campylobacter, Capnocytophaga, Corynebacterium, Eikenella, Porphyromonas, Prevotella, Propionibacterium, and Selenomonas. At the tongue dorsum site, there was a higher proportion of Actinomyces, Granulicatella, Neisseria, Rothia, and Streptococcus. No differences in proportions were found in Brevibacillus, Fusobacterium, Haemophilus, Leptotrichia, Microbacterium, Staphyloccocus, and Veillonella. In each of the site comparisons and each of the 20 selected genera comparisons, saliva had the highest significant proportions of NO3--reducing genera (Fig 2 and Table 1). Supragingival plaque had a significantly higher proportion of selected NO3--reducing genera compared to subgingival plaque, including Actinomyces, Capnocytophaga, Corynebacterium, Haemophilus, Neisseria, Rothia and Streptococcus (Table 1).
Table 1. Tables of pairwise comparison differences in log2-ratio median read proportions in selected NO3--reducing genera at six oral sites and saliva.
| Genera | Attached keratinized gingiva— Buccal mucosa |
Attached Keratinized gingiva— Hard palate |
Attached Keratinized gingiva— Saliva |
Attached Keratinized gingiva— Subgingival plaque |
Attached Keratinized gingiva— Supragingival plaque |
Attached Keratinized gingiva— Tongue dorsum |
|---|---|---|---|---|---|---|
| Actinomyces | -3.45 | -4.79 | -4.20 | -5.73 | -6.23 | -5.69 |
| Brevibacillus | 0 | 0 | 0 | 0 | 0 | 0 |
| Campylobacter | 0 | -2.58 | -5.36 | -4.60 | -3.56 | -3.81 |
| Capnocytophaga | -2.44 | -1.51 | -4.18 | -6.82 | -7.07 | -0.95 |
| Corynebacterium | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | 0 |
| Eikenella | 0 | 0 | -1.00E+25 | -1.00E+25 | -1.00E+25 | 0 |
| Fusobacterium | -1.41 | -1.74 | -3.31 | -4.91 | -3.23 | -3.02 |
| Granulicatella | 0 | -1.10 | 0 | 2.01 | 1.62 | -0.98 |
| Haemophilus | 0.82 | 1.37 | 0.95 | 3.13 | 2.30 | 0.94 |
| Leptotrichia | -3.34 | -3.81 | -4.49 | -5.78 | -6.29 | -4.88 |
| Microbacterium | 0 | 0 | 0 | 0 | 0 | 0 |
| Neisseria | -2.49 | -3.33 | -3.67 | -2.55 | -3.69 | -4.47 |
| Porphyromonas | 0 | -1.05 | -1.70 | -1.22 | 0 | 0 |
| Prevotella | 0 | -1.77 | -2.83 | -2.04 | 0 | -2.48 |
| Propionibacterium | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | 0 |
| Rothia | -4.37 | -5.33 | -4.53 | -5.83 | -6.82 | -5.36 |
| Selenomonas | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 | -1.00E+25 |
| Staphylococcus | 0 | 0 | 0 | 0 | 0 | 0 |
| Streptococcus | -0.19 | 0 | 1.78 | 2.57 | 1.94 | 1.26 |
| Veillonella | 0 | -1.15 | -2.10 | 0 | 0 | -2.17 |
| Genera |
Buccal mucosa—
Hard palate |
Buccal mucosa—
Saliva |
Buccal mucosa—
Subgingival plaque |
Buccal mucosa—
Supragingival plaque |
Buccal mucosa—
Tongue dorsum |
|
| Actinomyces | -1.34 | -0.75 | -2.28 | -2.79 | -2.25 | |
| Brevibacillus | 0 | 0 | 0 | 0 | 0 | |
| Campylobacter | -1.93 | -4.71 | -3.94 | -2.91 | -3.16 | |
| Capnocytophaga | 0.93 | -1.74 | -4.38 | -4.62 | 1.49 | |
| Corynebacterium | 0 | 0 | -5.92 | -6.79 | 1.00E+25 | |
| Eikenella | 0 | 0 | -1.00E+25 | -1.00E+25 | 0 | |
| Fusobacterium | 0 | -1.90 | -3.51 | -1.82 | -1.62 | |
| Granulicatella | -1.37 | 0 | 1.74 | 1.35 | -1.25 | |
| Haemophilus | 0.55 | 0 | 2.31 | 1.48 | 0 | |
| Leptotrichia | 0 | -1.15 | -2.44 | -2.95 | -1.54 | |
| Microbacterium | 0 | 0 | 0 | 0 | 0 | |
| Neisseria | -0.84 | -1.18 | 0 | -1.20 | -1.98 | |
| Porphyromonas | 0 | -1.22 | -0.74 | 0 | 0 | |
| Prevotella | -2.01 | -3.07 | -2.29 | 0 | -2.73 | |
| Propionibacterium | 0 | 0 | -4.27 | -4.39 | 1.00E+25 | |
| Rothia | 0 | 0 | -1.46 | -2.45 | 0 | |
| Selenomonas | -1.41 | -4.73 | -5.40 | -4.61 | 0 | |
| Staphylococcus | 0 | 0 | 0 | 0 | 0 | |
| Streptococcus | 0.28 | 1.97 | 2.76 | 2.13 | 1.45 | |
| Veillonella | -1.33 | -2.28 | 0 | 0 | -2.35 | |
| Genera |
Hard palate—
Saliva |
Hard palate—
Subgingival plaque |
Hard palate—
Supragingival plaque |
Hard palate—
Tongue dorsum |
Saliva—
Subgingival plaque |
|
| Actinomyces | 0.59 | -0.94 | -1.44 | -0.90 | -1.53 | |
| Brevibacillus | 0 | 0 | 0 | 0 | 0 | |
| Campylobacter | -2.78 | -2.01 | -0.98 | -1.23 | 0.77 | |
| Capnocytophaga | -2.67 | -5.31 | -5.56 | 0 | -2.64 | |
| Corynebacterium | 0 | -5.99 | -6.86 | 1.00E+25 | -5.36 | |
| Eikenella | -1.00E+25 | -1.00E+25 | -1.00E+25 | 0 | -5.76 | |
| Fusobacterium | -1.57 | -3.18 | -1.49 | -1.29 | -1.60 | |
| Granulicatella | 1.09 | 3.11 | 2.72 | 0 | 2.02 | |
| Haemophilus | -0.42 | 1.76 | 0.93 | -0.43 | 2.18 | |
| Leptotrichia | -0.68 | -1.97 | -2.48 | -1.07 | -1.29 | |
| Microbacterium | 0 | 0 | 0 | 0 | 0 | |
| Neisseria | 0 | 0 | -0.36 | -1.14 | 1.12 | |
| Porphyromonas | -0.66 | 0 | 0 | 0 | 0.48 | |
| Prevotella | -1.06 | 0 | 1.71 | -0.72 | 0.78 | |
| Propionibacterium | 0 | -3.80 | -3.92 | 1.00E+25 | -3.30 | |
| Rothia | 0.80 | 0 | -1.49 | 0 | -1.30 | |
| Selenomonas | -3.32 | -3.99 | -3.20 | 0 | 0 | |
| Staphylococcus | 0 | 0 | 0 | 0 | 0 | |
| Streptococcus | 1.69 | 2.48 | 1.85 | 1.17 | 0.80 | |
| Veillonella | -0.95 | 1.12 | 1.03 | -1.02 | 2.08 | |
| Genera |
Saliva—
Supragingival plaque |
Saliva—
Tongue dorsum |
Subgingival plaque—
Supragingival plaque |
Subgingival plaque—
Tongue dorsum |
Supragingival plaque—
Tongue dorsum |
|
| Actinomyces | -2.03 | -1.49 | -0.50 | 0 | 0.54 | |
| Brevibacillus | 0 | 0 | 0 | 0 | 0 | |
| Campylobacter | 1.81 | 1.56 | 1.04 | 0.79 | 0 | |
| Capnocytophaga | -2.88 | 3.23 | -0.24 | 5.87 | 6.11 | |
| Corynebacterium | -6.23 | 1.00E+25 | -0.87 | 1.00E+25 | 1.00E+25 | |
| Eikenella | -5.67 | 1.00E+25 | 0 | 1.00E+25 | 1.00E+25 | |
| Fusobacterium | 0 | 0 | 1.69 | 1.89 | 0 | |
| Granulicatella | 1.63 | -0.97 | 0 | -2.99 | -2.60 | |
| Haemophilus | 1.35 | 0 | -0.83 | -2.19 | -1.36 | |
| Leptotrichia | -1.80 | 0 | 0 | 0.90 | 1.41 | |
| Microbacterium | 0 | 0 | 0 | 0 | 0 | |
| Neisseria | 0 | -0.81 | -1.14 | -1.92 | -0.78 | |
| Porphyromonas | 0.87 | 0.96 | 0 | 0 | 0 | |
| Prevotella | 2.77 | 0.34 | 1.99 | 0 | -2.43 | |
| Propionibacterium | -3.43 | 1.00E+25 | 0 | 1.00E+25 | 1.00E+25 | |
| Rothia | -2.29 | -0.83 | -0.99 | 0 | 1.46 | |
| Selenomonas | 0.12 | 4.02 | 0.79 | 4.69 | 3.90 | |
| Staphylococcus | 0 | 0 | 0 | 0 | 0 | |
| Streptococcus | 0 | -0.52 | -0.63 | -1.31 | -0.69 | |
| Veillonella | 1.98 | 0 | 0 | -2.15 | -2.05 |
Values show the differences in log2-ratio of median proportions between each oral site. Each pairwise oral site comparison is shown, where the oral site first stated is compared to the second oral site stated. Positive values are the taxa which were enriched in the first oral site stated, whilst negative values are the taxa which were enriched in the second site stated. For example, in the attached keratinized gingiva and buccal mucosa pairwise comparison, Actinomyces was enriched in the buccal mucosa site, whilst Haemophilus was enriched in the attached keratinized gingiva site.
Determining genera in each oral site with the highest percentage median relative abundance
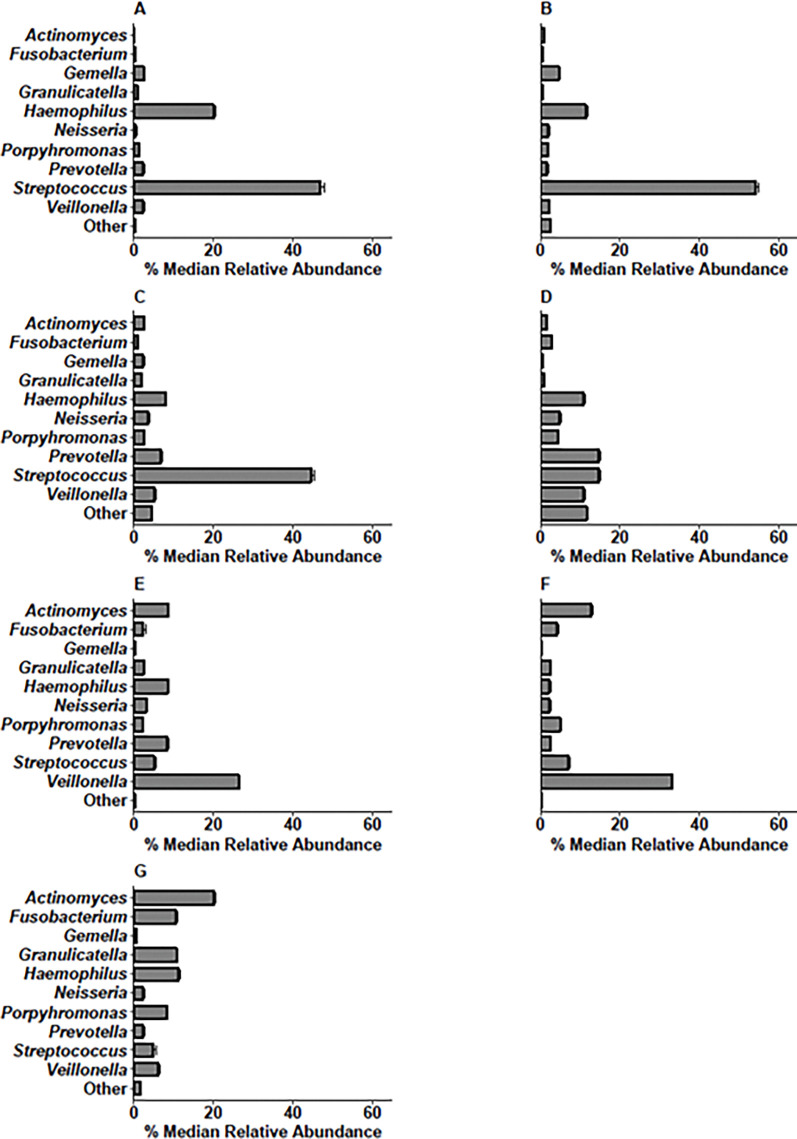
The percentage median relative abundance of genera was calculated for each oral site, and the ten genera found in the highest relative abundances were Actinomyces, Fusobacterium, Gemella, Granulicatella, Haemophilus, Neisseria, Porphyromonas, Prevotella, Streptococcus, and Veillonella (Fig 3).
Fig 3. Bar charts showing the percentage median relative abundance of bacterial genera in the attached keratinized gingiva, buccal mucosa, hard palate, saliva, subgingival plaque, supragingival plaque and tongue dorsum.
A attached keratinized gingiva, B buccal mucosa, C hard palate, D saliva, E subgingival plaque, F supragingival plaque, G tongue dorsum. Error bars show the standard error of median. The “Other” bacteria refer to the sum of the genera observed at a lower abundance.
Haemophilus and Streptococcus were found in all oral sites. In four of the seven oral sample types, Streptococcus was the most abundant (attached keratinized gingiva, 47%; buccal mucosa, 54%; hard palate, 45%). Prevotella and Streptococcus were found at the highest median relative abundance in saliva (15%). Saliva had a higher abundance of “Other” bacteria, which included the combined genera of lower relative abundances (12%). Veillonella were found in a higher median relative abundance in the saliva (11%), subgingival plaque (26%) and supragingival plaque (33%). In the tongue dorsum, subgingival and supragingival plaque sites, there was also a higher percentage median relative abundance of Actinomyces compared to other sites.
Visual analysis of clustering using multidimensional scaling plots
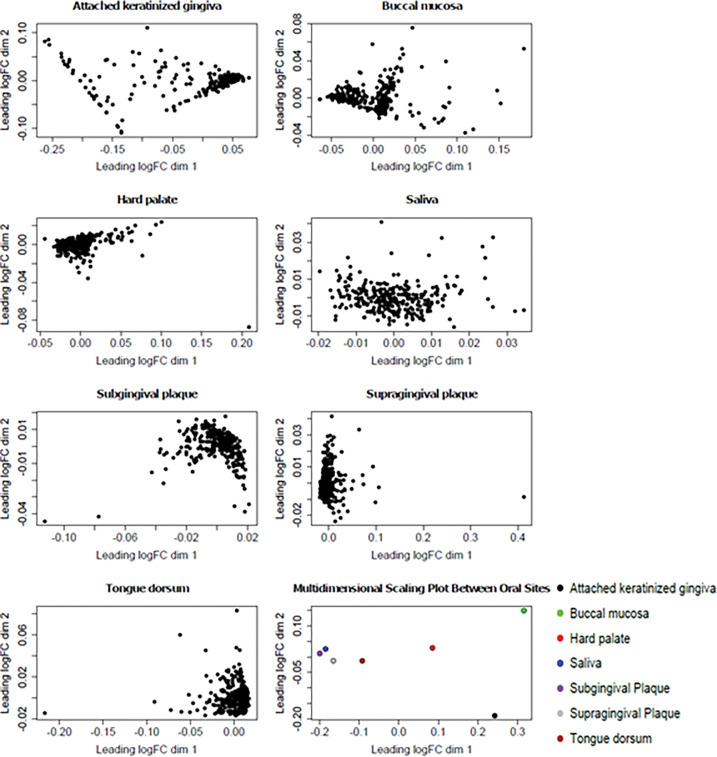
Multidimensional scaling plots of each of the seven oral sample types showed that there was low intra-site variability, and higher inter-site variability compared to intra-site variability (Fig 4). Distinct clustering was found in the hard palate, subgingival plaque, supragingival plaque, and tongue dorsum sites. The attached keratinized gingiva and buccal mucosa sites visually showed less pronounced clustering of individual samples. Saliva clustering of individual samples was the least apparent amongst the different oral sample types, with there being further distances between individual samples.
Fig 4. Multidimensional scaling plots (MDS) of individual samples in the attached keratinized gingiva, buccal mucosa, hard palate, saliva, subgingival plaque, supragingival plaque and tongue dorsum.
The multidimensional scaling plot of oral sites shows that the samples taken at each site share similarities. The attached keratinized gingiva shows some clustering, although there is higher intra-variability than the other sites, excluding saliva. Buccal mucosa samples appear to form two clusters which are not present within the other samples. The hard palate, subgingival plaque, supragingival plaque and tongue dorsum show clustering. Saliva samples appear to have the most variability between points.
Discussion
The relationships between the abundance of oral bacteria involved in the NO3--NO2--NO pathway, NO homeostasis and physiological processes, have been recognised as a potential target for improving blood pressure regulation, cognition, protection against ischaemia-reperfusion injury and exercise performance [3, 6–9]. Saliva and tongue swab samples are frequently used as measures of the oral microbial community and may be particularly useful for studies which aim to relate microbial communities to physiological or clinical outcomes. Saliva and tongue swab sampling may be used due to ease of sampling and high bacterial diversity and density [16–19]. However, due to discrepancies between sampling methods in previous reports, including whether the saliva or tongue dorsum was sampled, it is important to establish whether higher proportions of NO3--reducing bacteria were located elsewhere in the oral cavity. Our results provide an insight into which oral niches may be most appropriate to sample NO3--reducing bacteria involved in the NO3--NO2--NO pathway and highlight the benefits of saliva sampling in future physiological studies.
The overall oral bacterial composition was consistent with previous literature, where Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria and Fusobacteria were the most abundant phyla [25, 26]. This is further reflected at the family and genus levels, with Streptococcus, Prevotella, Neisseria and Haemophilus being most abundant genera [1, 15, 16, 26].
A limitation to the present study was that detailed participant clinical information was not available on the publicly accessible HMP database. Therefore, MDS plots were generated to qualitatively establish any outliers within each oral site. Further research is warranted to compare oral microbiomes related to specific host characteristics, including sex and health status. As clustering was generally observed in six of the seven oral sample types, samples were not immediately excluded, as the distances between each point may not reflect actual dissimilarity within each site. Therefore, the MDS plots overall showed low intra-individual variability in most oral sites. Saliva displayed higher intra-individual variability, represented by the spread between data points. However, because clinical information was not available, it is unclear whether this was due to differences between participants in the HMP cohort, or because of the high diversity of bacteria typically found in saliva [26, 27]. Furthermore, it is important to note that variation between samples of the same site may also occur due to differences in sampling techniques, for example, the pressure used to sample bacteria caught within the tongue crypt. Further understanding of how sampling techniques could influence the recording of bacterial abundance may be useful for future studies [28].
Comparison of alpha diversity at each oral site
The high diversity and species richness found in saliva and plaque sites compared to the other oral sites represents a larger mixture of microbial communities, which may also account for the higher intra-individual variability compared to the diversity and species richness found in the other oral sites [26, 27]. The high bacterial diversity, species richness and variability in saliva samples may be because saliva samples represent a composite of bacteria from all oral sites. Furthermore, variability in the salivary microbiome between humans may be a result of oral hygiene routine or physiological factors such as obesity and fitness [26, 27, 29].
Pairwise comparisons of predominant and NO3--reducing genera log2-ratio of median proportions at saliva and six oral sites
Pairwise comparisons between each oral site were used to determine which sites had the highest log2-ratio of median proportion of 20 predominant and highly abundant NO3--reducing bacteria. The well-established predominant and highly abundant NO3--reducing bacteria were selected based on previous studies [13, 16], and included Actinomyces, Brevibacillus, Campylobacter, Capnocytophaga, Corynebacterium, Eikenella, Fusobacterium, Granulicatella, Haemophilus, Leptotrichia, Microbacterium, Neisseria, Porphyromonas, Prevotella, Propionibacterium, Rothia, Selenomonas, Staphylococcus, Streptococcus, and Veillonella.
Saliva had significantly enriched proportions of eight NO3--reducing bacteria, whilst the tongue dorsum had a significantly higher proportion of Actinomyces, Granulicatella, Neisseria, Rothia, and Streptococcus, confirming the results of Aas et al. [1]. These results are important, as the tongue dorsum has been suggested as the site where most NO3--reduction occurs [13], but a higher abundance of NO3--reducing bacteria could be sampled from other oral sample types such as the saliva, which may be more appropriate for physiological studies that aim to collect samples of oral NO3--reducing bacteria.
In the attached keratinized gingiva site, the selected NO3--reducing genera had significantly reduced median proportions compared to the other oral sites. However, in all of the pairwise comparisons, Haemophilus was significantly enriched in the attached keratinized gingiva site, followed by Streptococcus which was significantly enriched when compared to the saliva, plaque sites and the tongue dorsum. The hard palate also had significantly enriched proportions of Streptococcus This may be a result of high bacterial cocci colonisation on the hard oral surfaces, such as Streptococcus and Haemophilus, where cocci bacteria can efficiently attach to epithelial cells and tooth enamel [30]. A similar result was found for the buccal mucosa site, consistent with previous studies [1, 16].
The subgingival and supragingival plaque sites exhibited less notable differences, consistent with previous research [31, 32]. Supragingival plaque had a significantly higher abundance of predominant NO3--reducing bacteria compared to subgingival plaque. The higher proportions of NO3--reducing bacteria in supragingival plaque are likely due to positioning within the oral cavity. Supragingival plaque is in frequent contact with saliva, and may also be more exposed to bacteria of the outside environment, such as in air or water, whereas subgingival plaque is located deeper in the oral cavity [31].
Percentage median relative abundances of genera in each oral site
The tongue dorsum had a low percentage median relative abundance of “Other” bacteria, which may be a result of the tongue scrapings reaching deeper oral niches in the tongue surface. The tongue dorsum also had a higher percentage median relative abundance of Actinomyces compared to the other oral sites. This may be because the tongue dorsum crypts are protected from the sheer force of salivary flow [27]. In saliva, the frequently recognised NO3--reducing Prevotella were found to be one of the genera with the highest percentage mean relative abundance. Consistent with the alpha diversity analyses, saliva had a high percentage median relative abundance of “Other” bacteria, compared to the specific oral sites. This may be because saliva represents NO3--reducing bacteria accumulated from all oral sites, as well as from food and fluid consumption. High proportions of Streptococcus, found within the oral sites are consistent with previous findings [14]. After undertaking routine oral hygiene procedures, Streptococcus, are typically some of the first to colonise oral sites, with these bacteria utilising facilitated attachment by salivary glycoproteins [30, 31]. High counts of Streptococcus may also account for a high abundance of Veillonella, and Actinomyces, as these bacteria form a symbiotic relationship within oral biofilms [32, 33].
Conclusions
Comparisons of the oral NO3--reducing genera in different oral sample types, using the HMP dataset, revealed the importance of selecting an appropriate oral site from which to sample bacteria likely to be involved in the NO3--NO2--NO pathway. Using only one method of sampling oral bacteria may lead to low abundances of NO3--reducing genera, particularly in small population physiological studies. Saliva samples are likely to provide a good representation of all NO3--reducing bacteria in the oral cavity, providing a high yield of NO3--reducing bacteria samples when compared to the tongue dorsum sample site. However, the measurement of bacterial abundance using saliva samples alone may lead to more variability between samples of the same type and may not capture bacteria deep within biofilms found in the plaque sites or crevices of the tongue dorsum. This comparison of NO3--reducing genera at each oral site provides a basis for where to sample NO3--reducing bacteria in future physiological studies investigating the relationship between the bacteria in the NO3--NO2--NO pathway and human health.
Supporting information
(ZIP)
Acknowledgments
We thank Dr Ryan Ames for his valuable advice on data analysis during this project. We thank the Human Microbiome Project for the publicly available data used for analysis in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parahitiyawa NB, Scully C, Leung WK, Yam WC, Jin LJ, Samaranayake LP. Exploring the oral bacterial flora: current status and future directions. Oral Dis. 2010;16(2):136–45. doi: 10.1111/j.1601-0825.2009.01607.x [DOI] [PubMed] [Google Scholar]
- 3.Lundberg JO, Weitzberg E, Gladwin MT. The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nature Reviews Drug Discovery. 2008;7(2):156–67. doi: 10.1038/nrd2466 [DOI] [PubMed] [Google Scholar]
- 4.Qin L, Liu X, Sun Q, Fan Z, Xia D, Ding G, et al. Sialin (SLC17A5) functions as a nitrate transporter in the plasma membrane. Proc Natl Acad Sci U S A. 2012;109(33):13434–9. doi: 10.1073/pnas.1116633109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundberg JO, Gladwin MT, Ahluwalia A, Benjamin N, Bryan NS, Butler A, et al. Nitrate and nitrite in biology, nutrition and therapeutics. Nat Chem Biol. 2009;5(12):865–9. doi: 10.1038/nchembio.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Presley TD, Morgan AR, Bechtold E, Clodfelter W, Dove RW, Jennings JM, et al. Acute effect of a high nitrate diet on brain perfusion in older adults. Nitric Oxide. 2011;24(1):34–42. doi: 10.1016/j.niox.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate-reducing oral bacteria in blood pressure control. Free Radic Biol Med. 2013;55:93–100. doi: 10.1016/j.freeradbiomed.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashworth A, Mitchell K, Blackwell JR, Vanhatalo A, Jones AM. High-nitrate vegetable diet increases plasma nitrate and nitrite concentrations and reduces blood pressure in healthy women. Public Health Nutr. 2015;18(14):2669–78. doi: 10.1017/S1368980015000038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–23. doi: 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium. THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–17. doi: 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doel JJ, Benjamin N, Hector MP, Rogers M, Allaker RP. Evaluation of bacterial nitrate reduction in the human oral cavity. Eur J Oral Sci. 2005;113(1):14–9. doi: 10.1111/j.1600-0722.2004.00184.x [DOI] [PubMed] [Google Scholar]
- 14.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30(7):644–54. doi: 10.1034/j.1600-051x.2003.00376.x [DOI] [PubMed] [Google Scholar]
- 15.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. The ISME Journal. 2010;4(8):962–74. doi: 10.1038/ismej.2010.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, Parthasarathy DK, et al. Metagenomic analysis of nitrate-reducing bacteria in the oral cavity: implications for nitric oxide homeostasis. PLoS One. 2014;9(3):e88645. doi: 10.1371/journal.pone.0088645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro KB, Hotchkiss JH, Roe DA. Quantitative relationship between oral nitrate-reducing activity and the endogenous formation of N-nitrosoamino acids in humans. Food Chem Toxicol. 1991;29(11):751–5. doi: 10.1016/0278-6915(91)90183-8 [DOI] [PubMed] [Google Scholar]
- 18.Kanady JA, Aruni AW, Ninnis JR, Hopper AO, Blood JD, Byrd BL, et al. Nitrate reductase activity of bacteria in saliva of term and preterm infants. Nitric Oxide. 2012;27(4):193–200. doi: 10.1016/j.niox.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velmurugan S, Gan JM, Rathod KS, Khambata RS, Ghosh SM, Hartley A, et al. Dietary nitrate improves vascular function in patients with hypercholesterolemia: a randomized, double-blind, placebo-controlled study. Am J Clin Nutr. 2016;103(1):25–38. doi: 10.3945/ajcn.115.116244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. Faseb j. 2013;27(3):1012–22. doi: 10.1096/fj.12-220806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 22.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–7. doi: 10.1093/bioinformatics/btm453 [DOI] [PubMed] [Google Scholar]
- 23.Oksanen J SG, Blanchet F, Kindt R, Legendre P MP, O’Hara R, Solymos P, et al. vegan: Community Ecology Package. R package version 2.6–4 ed2022. [Google Scholar]
- 24.Foster ZSL, Sharpton TJ, Grünwald NJ. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLOS Computational Biology. 2017;13(2):e1005404. doi: 10.1371/journal.pcbi.1005404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith NBDAWDMTWM A. J. The Microbial Generation of Nitric Oxide in the Human Oral Cavity. Microbial Ecology in Health and Disease. 1999;11(1):23–7. [Google Scholar]
- 26.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19(4):636–43. doi: 10.1101/gr.084616.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall MW, Singh N, Ng KF, Lam DK, Goldberg MB, Tenenbaum HC, et al. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. npj Biofilms and Microbiomes. 2017;3(1):2. doi: 10.1038/s41522-016-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikutani T, Tamura F, Takahashi Y, Konishi K, Hamada R. A novel rapid oral bacteria detection apparatus for effective oral care to prevent pneumonia. Gerodontology. 2012;29(2):e560–5. doi: 10.1111/j.1741-2358.2011.00517.x [DOI] [PubMed] [Google Scholar]
- 29.Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Scientific Reports. 2016;6(1):22164. doi: 10.1038/srep22164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28(8):397–403. doi: 10.1089/dna.2009.0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neeser JR, Grafström RC, Woltz A, Brassart D, Fryder V, Guggenheim B. A 23 kDa membrane glycoprotein bearing NeuNAc alpha 2-3Gal beta 1-3GalNAc O-linked carbohydrate chains acts as a receptor for Streptococcus sanguis OMZ 9 on human buccal epithelial cells. Glycobiology. 1995;5(1):97–104. doi: 10.1093/glycob/5.1.97 [DOI] [PubMed] [Google Scholar]
- 32.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52(1):38–52. doi: 10.1111/j.1600-0757.2009.00311.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23(3):196–205. doi: 10.1111/j.1399-302X.2007.00411.x [DOI] [PubMed] [Google Scholar]