Abstract
Early life stress (ELS) can impact brain development and is a risk factor for neurodevelopmental disorders such as schizophrenia. Post-weaning social isolation (SI) is used to model ELS in animals, using isolation stress to disrupt a normal developmental trajectory. We aimed to investigate how SI affects the expression of genes in mouse hippocampus and to investigate how these changes related to the genetic basis of neurodevelopmental phenotypes. BL/6J mice were exposed to post-weaning SI (PD21-25) or treated as group-housed controls (n = 7–8 per group). RNA sequencing was performed on tissue samples from the hippocampus of adult male and female mice. Four hundred and 1,215 differentially-expressed genes (DEGs) at a false discovery rate of < 0.05 were detected between SI and control samples for males and females respectively. DEGS for both males and females were significantly overrepresented in gene ontologies related to synaptic structure and function, especially the post-synapse. DEGs were enriched for common variant (SNP) heritability in humans that contributes to risk of neuropsychiatric disorders (schizophrenia, bipolar disorder) and to cognitive function. DEGs were also enriched for genes harbouring rare de novo variants that contribute to autism spectrum disorder and other developmental disorders. Finally, cell type analysis revealed populations of hippocampal astrocytes that were enriched for DEGs, indicating effects in these cell types as well as neurons. Overall, these data suggest a convergence between genes dysregulated by the SI stressor in the mouse and genes associated with neurodevelopmental disorders and cognitive phenotypes in humans.
Introduction
Early life stress (ELS) includes childhood exposure to a range of adversities, and is associated with increased risk for neurodevelopmental disorders [1]. One such stressor is social isolation (SI), a situation in which an individual is deprived of typical and expected social interaction. These interactions are fundamental to normal social development [2] and exposure to SI during vulnerable periods of neurodevelopment can impact on both the behaviour and neurobiology of those affected [3, 4]. The detriment of SI-induced changes is clear, with evidence that exposure contributes to increased risk of anxiety and depression [5, 6], schizophrenia [7] and cognitive decline [8].
The use of rodent models of SI to investigate behaviour is common. Mice exposed to post-weaning SI tend to experience increased anxiety [9–16], increased depressive-like behaviour [11, 13], decreased cognitive ability [13, 14, 17–19], increased aggression [12, 20] and decreased sociability [10, 21] when compared to group-housed controls. Some of the neurobiology relevant to SI-induced behaviour in rodents has been mapped [22]. Targeted analyses have implicated neuronal growth factors such as BDNF [13, 16, 17, 20], hormones such as oxytocin [23], inflammatory mediators such as cytokines [24–26] and the function of most main neurotransmitter systems [22] as being involved in behaviours found in SI-exposed rodents. Rodents subjected to SI display dysregulation of genes crucial to the function of glutamatergic [14, 27, 28], GABAergic [28–30], dopaminergic [31] neurotransmission, all of which are systems thought to have a part to play in psychiatric disorders [32].
To our knowledge, no mouse studies of SI have included full transcriptome gene expression analysis of the molecular changes caused by SI in the hippocampus. Previously, transcriptomic analysis of the basolateral amygdala in socially-isolated mice identified genes associated with aggressive behaviour and found evidence of upregulated ion channel function, while genes related to limbic system development and cognition were downregulated. Furthermore, in the ventral tegmental area, genes related to neuropeptide signalling were downregulated and genes related to synaptic signalling were upregulated [33]. Other transcriptomic work using microarray in rat cortex [29] also found dysregulation of genes related to synaptic structure and function, particularly in relation to inhibitory GABAergic synapses.
Previous work by our group [10] showed that mice exposed to post-weaning SI exhibited a significant increase in anxiety related behaviours (males and females) and decreased sociability (females) compared to animals that were group-housed. We now extend our previous investigation by performing RNA sequencing (RNA-seq) to identify differentially expressed genes (DEGs) in the brains of the same animals–specifically in the hippocampus, a region that regulates stress response and emotion [34, 35]. Transcriptomic changes were found to be induced in both male and female hippocampus by SI. These differentially expressed genes (DEGs) encoded protein involved in synaptic structure and function. We further considered how these transcriptomic changes converge with the biological basis of human psychiatric disorders and behaviours by investigating if the DEGs were enriched for common heritability contributing to neurodevelopmental phenotypes and enriched for genes harbouring rare de novo variants contributing to neurodevelopmental disorders. Finally, we explored which specific cell types were enriched for DEGs induced by SI.
Materials and methods
Ethics statement
All procedures received ethical approval from the local Animal Care Research Ethics Committee (ACREC) and the Health Products Regulatory Authority in Ireland (licence number AE19125/P083).
Social isolation procedure
Tissues used in this analysis were obtained directly from the animals used in our previous study [10], consisting of post-weaning (P21-25) adult male and female C57 BL/6J mice (Charles River Laboratories, UK) assigned to either group-housed cages (3–4 animals per cage) or single housed cages (SI; n = 7–8 per group) until animals were sacrificed by brief exposure to CO2 (90s) followed by immediate decapitation after a 60 day period. Whole brain was dissected on ice and hippocampus was snap-frozen in dry-ice before being stored at −80 °C.
Sample preparation
Total RNA was extracted from frozen hippocampus using the Absolutely RNA Miniprep kit (Agilent; product code: #400800) following appropriate standard procedure for quantity of tissue. Once purity and integrity (RIN > 6) was confirmed, isolated RNA-seq was performed on the Illumina NovaSeq (paired-end; 2x150 bp reads) sequencing platform producing a minimum of > 20M reads/sample (Genewiz Germany GmbH).
Differential expression analysis
Raw data was received through SFTP in FASTQC format. Trimmomatic v0.39 [36] was used to remove low quality and adapter sequences from the paired-end reads (LEADING:3, TRAILING:3, MINLEN:36). Salmon v1.8.0 [37] was used to quasi-map and quantify reads. The DEseq2 v1.24.0 [38] R package was used to test genes for differential expression. The sva v3.32.1 svaseq [39] function was used to detect batch effects in individual groups. Significant surrogate variable (SVs) were introduced into the differential expression model to control for technical batch effects. DEGs were defined at a false discovery rate (FDR) of < 0.05. Fold changes were shrunk using apeglm v1.6.0 [40]. Genes were converted to human orthologues using biomaRt v2.40.5 [41] where necessary.
Gene ontology analysis
Gene ontology (GO) analysis was done using ConsensusPathDB (http://cpdb.molgen.mpg.de/) [42] over-representation analysis. Ontologies with GO term levels 2–5 were tested and ontologies with a FDR-corrected p-value < 0.05 were considered significantly enriched. In order to limit GO analysis to ontologies relevant to the synapse, sets of DEGs also tested for enrichment using SynGO (https://www.syngoportal.org/) [43], an expert-curated resource for synaptic GO analysis. Ontologies with an FDR corrected p-value < 0.05 were considered significantly enriched.
Testing genes for enrichment of common genetic risk variants associated with neurodevelopmental phenotypes
Data on common variants (SNPs) associated with human phenotypes were accessed in the form of genome-wide association study (GWAS) summary stats for a range of phenotypes relevant to neurodevelopment and psychiatric disorders including schizophrenia (SCZ; GWAS based on 67,390 cases and 94,015 controls) [44], intelligence (IQ; 269,867 individuals) [45], educational attainment (EA; 766,345 individuals) [46], bipolar disorder (BPD; 41,917 cases and 371,549 controls) [47], major depressive disorder (MDD; 246,636 cases and 561,190 controls) [48] and anxiety-tension (Anx-Ten; 270,059 individuals) [49]. As control phenotypes, GWAS data for three brain-related disorders: Attention deficit/hyperactivity disorder (ADHD; 20,183 cases and 35,191 controls) [50], Alzheimer’s disease (AlzD; 71,880 cases and 383,378 controls) [51], stroke (40,585 cases and 406,111 controls) [52] and two non-brain related disorders: type-2 diabetes (T2D; 74,124 cases and 824,006 controls) [53] and coronary artery disease (CAD; 22,233 cases and 64,762 controls) [54] were used. Stratified LD Score regression (sLDSC) was used to investigate if gene-sets were significantly enriched for SNP heritability contributing to the test and control phenotypes [55, 56]. Gene start and stop coordinates of each DEG on GRCh37 were found using biomaRt v2.40.5 [41]. Annotation files were generated for each chromosome in each set of DEGs using 1000 genomes European cohort SNPs and a window size of 100kb extended to coordinates [57]. LD scores were estimated within a 1cM window using 1000 Genomes Phase 3 European reference panel. Heritability was stratified in a joint analysis between 53 previous function genomic annotations [56] and each set of DEGs. Only SNPs from HapMap Project phase 3 SNPs with a MAF > 0.05 were considered in this analysis.
For phenotypes with significantly enriched heritability from the LDSC analysis, MAGMA [58] was used to test individual genes for association. First, we annotated the SNP data to genes using the build 37 gene locations (https://ctg.cncr.nl/software/MAGMA/aux_files/NCBI37.3.zip) and 1000 Genomes European Panel reference (https://ctg.cncr.nl/software/MAGMA/ref_data/g1000_eur.zip) files, the latter which MAGMA uses to account for linkage disequilibrium (LD) between SNPs. Second, MAGMA generated p-values for individual gene reflecting their level of association with the test and control phenotypes. Genes with a Bonferroni-corrected p-value of < 0.05 (correcting for the number of genes tested) were considered genome-wide significant for association with each phenotype.
Testing genes for enrichment of de novo mutations reported in neurodevelopmental disorders
Rare de novo mutations (DNMs) contributing to a phenotype can be detected using exome sequencing of trios including an affected proband and their biological parents. To test if DEGs were enriched for DNMs that contribute to neurodevelopmental disorders, we analysed DNMs reported in studies of autism spectrum disorder (ASD), intellectual disability (ID), SCZ and developmental disorders (DD). The functional class and gene location of DNMs identified in patients with ASD (n = 6,430), ID (n = 192) and in unaffected siblings (n = 1,995) and controls (n = 54) based on exome sequencing of trios were sourced from [59] and [60]. Genes harbouring DNMs reported in affected SCZ trios (n = 3,394) were taken from [61] and [62]. DNMs identified in developmental disorders (DD) were sourced from [63]. Mutations used for DD were subject to additional filtering based on posterior probability of de novo mutations, as described in [63]. To account for underlying mutational burden associated with the test phenotypes, results were then subject to a competitive test against background de novo mutation rate using a two-sample Poisson rate ratio test. Results with a Bonferroni-adjusted p-value of < 0.05 (correcting for the number of mutational classes and disorders tested) were considered significantly enriched.
Cell type enrichment
Data from single cell RNA-seq (scRNA-seq) of the mouse brain (565 cell types) [64] was used to test if different cell types were enriched for DEGs. Analysis was performed using the expression-weighted cell type enrichment (EWCE) R package [65], which investigated whether the cell types were significantly enriched for a gene-set when weighted by gene expression. Cell types were considered significantly enriched at a Bonferroni-corrected p-value of < 0.05 (correcting for the number of cell types tested).
Results
Differential gene expression
Table 1 summarises the numbers of DEGs induced by SI detected in male and female hippocampal tissue at FDR < 0.05. In total, 1,215 significant DEGs were identified in females and 400 in males. In both sexes, approximately twice as many DEGs were found to be downregulated than upregulated. Among the downregulated DEGs, 98 were common between males and females. Of the upregulated DEGs, 30 were common between males and females. Expression log 2 fold change (Log2FC) was highly consistent among DEGs shared between male and female samples, producing a R2 correlation coefficient of 0.88 (p-value < 2.2e-16). Full differential expression results can be found in S1 Table.
Table 1. Summary of DEGs induced by SI in hippocampus.
| Set | DEGs at FDR < 0.05 | Downregulated | Upregulated |
|---|---|---|---|
| Female | 1215 | 810 (67.7%) | 405 (33.3%) |
| Male | 400 | 259 (64.7%) | 141 (35.3%) |
GO and SynGO analysis
GO analysis was performed in order to gain insight into the functional roles of DEGs. ConsensusPathDB was used to identify GO biological processes (BP), cellular compartments (CC) and molecular functions (MF) terms enriched for DEGs. A total of 738 GO terms were found to be enriched for the female DEGs and 306 GO terms were enriched for the male DEGs at FDR < 0.05. (S2 Table). Of these enriched terms, 72 (9.8%) of the female and 36 (11.8%) of the male contained any of "neuro", "synap", "dendr" or "axon", indicating their relation to having neuronal, axonal or synaptic structure or function. Female DEGs were highly enriched for a number of neurodevelopmental GO terms. The most enriched level 4 and level 5 terms were nervous system development (GO:0007399, FDR-adjusted p-value = q-value = 2.76E-16) neurogenesis (GO:0022008, q-value = 9.02E-16), neuron development (GO:0048666, q-value = 1.26E-15), generation of neurons (GO:0048699, q-value = 1.57E-15), neuron projection development (GO:0031175, q-value = 1.57E-15) and neuron differentiation (GO:0030182, q-value = 2.57E-15). The significantly enriched GO terms for the male DEGs included nervous system development (GO:0007399, q-value = 6.79E-6), neuron part (GO:0097458, q-value = 9.82E-5), central nervous system development (GO:0007417, q-value = 3.31e-4) and brain development (GO:0007420, q-value = 4.61e-4).
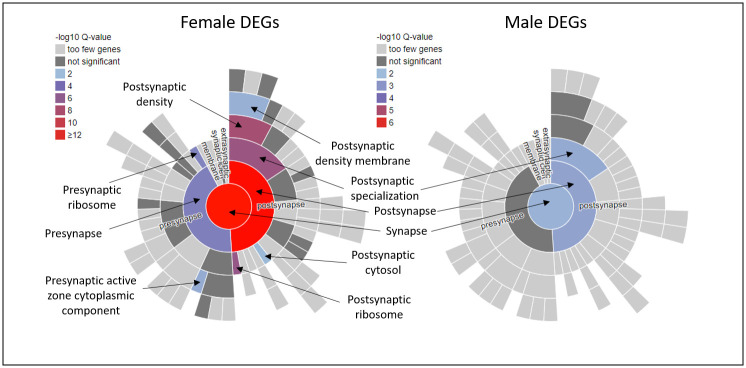
Following up these neuronal GO term enrichments, additional GO analysis was performed using SynGO [43] to gain insight into the synaptic CCs and BPs enriched for DEGs (Fig 1; S3 Table). All enriched SynGO analysis terms were related to presynaptic and postsynaptic structure (CC) and function (BP) and provide greater focus on the precise neurobiological changes brought about by SI. The female DEG set showed significant enrichments in 39 SynGO terms. Both postsynapse (GO:0098794, q-value = 5.14e-17) and presynapse (GO:0098793, q-value = 3.15e-4) CCs were significantly enriched as well as a number of specific postsynaptic ontologies including postsynaptic density (GO:0014069, q-value = 2.25e-8), postsynaptic specialisation (GO:0099572, q-value = 1.56e-7), translation at postsynapse (GO:0140242, q-value = 5.76e-4) and postsynaptic organisiation (GO:0099173, q-value = 2.19e-3). The male DEGs showed significant enrichment in the postsynapse (GO:0098794, q-value = 3.67e-3) but not presynapse CCs. Similar to that of the females, male DEGs were also significantly enriched for processes of postsynaptic specialisation (GO:0099572, q-value = 0.003667) and postsynaptic density (GO:0014069, q-value = 0.012).
Fig 1. SynGO cellular compartment (CC) enrichments of female and male DEG sets.
Annotated sunburst of enriched SynGO synaptic CC ontologies. Enrichments were detected for both pre and post-synaptic terms, however most significantly-enriched ontologies are post-synaptic. DEGs from females showed more enrichments in SynGO terms than DEGs from males.
Enrichment for genes contributing to human neurodevelopmental phenotypes
To assess whether genes dysregulated in mouse hippocampal tissue by SI harboured genetic risk for human neurodevelopmental phenotypes, enrichments for common heritability (SNPs) and genes harbouring rare DNMs contributing to neurodevelopmental disorders and related phenotypes were investigated. sLDSC [55, 56] was used to test common SNPs. The 1,215 female DEGs accounted for 9.5% of the total SNPs in the analysis, while the smaller male set of 400 DEGs accounted for 3.5% of total SNPs. Results can been seen in Table 2, showing percentage of heritability (% h2) and enrichment p-value in SNP-based heritability for SCZ, EA, IQ and BPD for the female DEG set and enrichments in SCZ, IQ and BPD for the male DEG set. Of the five control phenotypes tested, just one (coronary artery disease) showed a marginal enrichment for the female DEG set. Full results from the sLDSC analysis can be found in S4 Table.
Table 2. Enrichment of SNP-based heritability for test phenotypes.
| DEG Set | % of Total SNPs | SCZ | IQ | EA | MDD | BPD | Anx-Ten | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % h2 | P* | % h2 | P | % h2 | P | % h2 | P | % h2 | P | % h2 | P | ||
| Female | 9.5% | 13.3% | 2.27E-05 | 12.9% | 0.000109 | 13.3% | 2.60E-07 | 11.7% | 0.0472 | 14.2% | 0.000388 | 12.3% | 0.0422 |
| Male | 3.5% | 6.2% | 7.08E-05 | 5.0% | 0.00276 | 4.5% | 0.0132 | 4.6% | 0.0434 | 7.2% | 2.51E-05 | 4.1% | 0.4202 |
Results in bold survive Bonferroni correction for multiple testing (including control phenotypes)
Given there was evidence of convergence of SI-induced DEGs and genes contributing to risk of neurodevelopmental phenotypes, we investigated which genes may be driving this connection. For the test phenotypes with significant enrichments in LDSC (SCZ, EA, IQ and BPD), MAGMA [58] was used to test which genes had genome-wide significant associations. Genes were considered significantly enriched at a Bonferroni-corrected p-value of < 0.05. A total of 619 genes were genome-wide significant for SCZ, 160 for BPD, 377 for IQ and 689 for EA. Full MAGMA gene-based analysis results can be found in S5 Table. These lists were further restricted to DEGs that were common between male and female hippocampus and with consistent direction of expression change. A total of ten genes, shown in Table 3 below, matched these criteria and were significantly associated with one or more of SCZ, BPD, IQ or EA. Three of the 10 highlighted genes were genome-wide significant for two phenotypes, while 7 had a single significant gene-phenotype association from MAGMA. At least one gene was associated with SCZ, BPD, IQ or EA. All ten genes were downregulated, with male hippocampus consistently showing greater expression change.
Table 3. Genes found to be common between DEG analysis (same direction of effect in females and males) and MAGMA gene-based association analysis.
| Symbol | Gene Name | Associated Phenotypes | Direction in Females | Direction in Males |
|---|---|---|---|---|
| RIMS1 | Regulating Synaptic Membrane Exocytosis 1 | SCZ, BPD | Downregulated, Log2FC = -0.22 | Downregulated, Log2FC = -0.32 |
| ZNF365 | Zinc Finger Protein 365 | SCZ, BPD | Downregulated, Log2FC = -0.21 | Downregulated, Log2FC = -0.32 |
| AGAP1 | ArfGAP With GTPase Domain, Ankyrin Repeat And PH Domain | IQ, EA | Downregulated, Log2FC = -0.21 | Downregulated, Log2FC = -0.25 |
| TTBK1 | Tau Tubulin Kinase 1 | SCZ | Downregulated, Log2FC = -0.14 | Downregulated, Log2FC = -0.31 |
| PPP1R16B | Protein Phosphatase 1 Regulatory Subunit 16B | SCZ | Downregulated, Log2FC = -0.18 | Downregulated, Log2FC = -0.31 |
| SPTBN2 | Spectrin Beta, Non-Erythrocytic 2 | BPD | Downregulated, Log2FC = -0.21 | Downregulated, Log2FC = -0.32 |
| SHANK2 | SH3 And Multiple Ankyrin Repeat Domains 2 | BPD | Downregulated, Log2FC = -0.14 | Downregulated, Log2FC = -0.27 |
| UTRN | Utrophin | EA | Downregulated, Log2FC = -0.25 | Downregulated, Log2FC = -0.24 |
| PHF2 | PHD Finger Protein 2 | IQ | Downregulated, Log2FC = -0.12 | Downregulated, Log2FC = -0.18 |
| ZBTB4 | Zinc Finger And BTB Domain Containing 4 | EA | Downregulated, Log2FC = -0.13 | Downregulated, Log2FC = -0.33 |
Using gene-level data from these studies, the R package denovolyzeR [66] was used to test gene-sets for enrichment for genes harbouring rare DNMs contributing to SCZ, ASD, ID and DD. Categories of synonymous (syn), missense (mis) and loss-of-function (lof) mutations were considered for analysis. Following a competitive analyses, gene-sets were considered enriched at a Bonferroni-corrected p-value of <0.05. Table 4 shows enriched categories of variants in each gene-set. Female DEGs showed strong enrichment for genes containing missense variants contributing to DD. Male DEGs were also enriched for missense variants contributing to DD as well as being enriched for genes containing loss-of-function mutations contributing to DD and ASD. As a control, gene-sets were not enriched for synonymous variants in any of the disorders. No enrichments were seen in trios containing unaffected siblings or controls. Complete results from rare variant analysis can be found in S6 Table.
Table 4. Enrichments for genes harbouring rare de novo mutations in male and female gene sets.
| Set | DEGs | SCZ n = 3394 Trios |
ASD n = 6430 Trios |
ID n = 192 Trios |
DD n = 4293 Trios |
|---|---|---|---|---|---|
| Female Hippocampus | 1215 | ns | ns | ns | mis (p = 5.20e-6) |
| Male Hippocampus | 400 | ns | lof (p = 2.60e-5) | ns |
lof (p = 8.23e-8)
mis (p = 8.05e-5) |
SCZ = Schizophrenia; ASD = Autism; ID = Intellectual Disability; DD = Developmental Disorder
ns = non-significant; lof = loss of function mutations; mis = missense mutations; Results in bold survive Bonferroni correction for multiple testing
Cell-type enrichment analysis
Cell types enriched for male and female DEGs were tested using EWCE [65]. Both sets were tested in scRNA-seq gene expression data from mouse brain [64]. Although this dataset contains expression data on 565 cell types, enrichments from this analysis was restricted to cell types from the hippocampus (n = 104). Cell types were considered enriched at a Bonferroni-corrected p-value of < 0.05. The primary cell type found to be enrichment in female DEGs was of glial origin. Of the cell types in the mouse brain data [64] three hippocampal cell types, HC_7–1 (p < 0.00001), HC_7–2 (p < 0.00001) and HC_7–3 (p < 0.00001) were enriched; all three were populations of hippocampal astrocytes. No hippocampal cell types were considered significantly enriched after multiple testing correction in the male DEG set. Full cell type enrichment results found in S7 Table.
Discussion
This study builds on our previous behavioural work [10] using post-weaning SI mice and here investigates the molecular consequences of the environmental stressor on the mouse brain. The use of RNA-seq to investigate the transcriptomic changes in mouse hippocampus caused by SI presents a novel insight into the underlying molecular underpinnings of altered behaviour. Furthermore, we used data from studies of rare and common risk variants for neurodevelopmental phenotypes to investigate the relevance of the SI model to human illness and behaviour.
Differential expression analysis detected hundreds of dysregulated genes with approximately three times the number of DEGs in female (n = 1,215) compared to male (n = 400) hippocampal tissue. This is consistent with our behavioural data in these animals [10], which showed females to be more susceptible to SI-induced behavioural measures of anxiety and sociability. GO analysis identified that DEGs in both males and females were enriched in structural and functional synaptic ontologies with DEGs from females implicated in both overall presynapse and postsynapse ontologies, while DEGs from males were only implicated postsynapse ontologies. In the context of our behavioural data [10], this suggests that presynaptic processes may play a role in facilitating SI-induced behaviours of anxiety and decreased sociability observed in the females. Synaptic biology is implicated in many neurodevelopmental disorders. In SCZ, the latest GWAS implicates genes involved in the organisation, differentiation and function of synapses [44]. For ASD, rare variants in genes involved in synapse structure including from the SHANK [67], NRXN [68] and NLGN [69] gene families are linked with the disorder.
In our rare variant analysis, enrichment for genes containing loss-of-function mutations contributing to ASD were found in the DEGs from males but not females. DEGs from both males and females were enriched for genes harbouring DNMs contributing to DD, however the DEGs from males showed a greater level of enrichment. Our sLDSC analysis detected enrichments in SNP heritability associated with SCZ, BPD and IQ in both male and female DEG sets with again males displaying stronger enrichments in all of these phenotypes. Together, the rare variant and heritability analyses indicate that the set of DEGs from male SI mice, despite only totalling a third of the number of DEGs in the set from female SI mice, converge more strongly with the genes that underpin risk for neurodevelopmental disorders and associated phenotypes in humans.
Anxiety was the most prominent phenotype induced by SI in our behavioural work [10]. Although a nominal enrichment in heritability was seen in the Anx-Ten phenotype for the DEGs from females (p = 0.042), this result did not survive multiple-testing correction. However, it is important to note that anxiety disorders are under less genetic influence (heritability of 20–60%) [70] than for example SCZ and BPD (both with heritability of 60–80% heritable) [44, 47]. As a result and despite a very large sample size, the GWAS of Anx-Ten identified just fourteen independent loci [49] and therefore we were unlikely to detect that our sets of DEGs were strongly enriched for common variants associated with this phenotype. Based on the pretext that no SI-induced cognitive changes were detected in these mice [10], the enrichment in IQ and EA heritability in the set of DEGs from female mice suggests that a subclinical effect of SI on cognition may have been present in this circumstance.
Ten genes were prioritised based on being consistently differentially expressed in males and females, as well as having genome-wide significant associations with any of the phenotypes that were enriched in the sLDSC analysis. Three genes, RIMS1, ZNF365 and AGAP1, were associated with two of the four phenotypes tested. RIMS1 plays an important role regulating and localising calcium channels and neurotransmission [71–75], regulating synaptic plasticity [76–79] as well as being crucial for learning and memory in mouse [80]. In humans, RIMS1 is also an ASD candidate risk gene implicated in rare variant studies [81–83]. ZNF365 (also called DBZ) is involved in neurogenesis, especially regarding basket cells in the somatosensory cortex [84]. It is also involved in oligodendrocyte differentiation [85] and regulating dendritic spine density in pyramidal neurons [86]. ZNF365 interacts with the SCZ candidate gene, DISC1, which through its role on oligodendrocyte differentiation could be a contributor to SCZ and MDD [87]. AGAP1 (also called CENTG2) regulates dendritic spine morphology [88] and is involved in neurotransmitter release in dopaminergic neurons [89]. In humans, variation in AGAP1 has been associated with to ASD [90], SCZ [91]and ASD/ID [92]. Further human data from PsychEncode is consistent with these prioritised mouse DEGs, with RIMS1 downregulated in post-mortem brain samples SCZ (Log2FC = -0.05, FDR = 0.004) and ZNF365 downregulated in post-mortem brain samples of ASD (Log2FC = -0.24, FDR = 0.002) [93].
Although GO analysis primarily implicated changes in synaptic biology caused by SI, the primary findings from cell type enrichment analysis were glial cells. Given the number of cell types analysed here, there is a strict Bonferroni threshold for cell types to be enriched (104 x 2 independent tests). Only cell types with a number highly unique genes overrepresented in our analysis will show up an enriched, which may explain the lack of neuronal cell types showing to be significantly enriched after correction. However, it is now well established that there is a non-neuronal component of neurodevelopment [94]. Astrocytes, enriched in this analysis, play a number of crucial functions to maintain a normal early developmental trajectory. These include maintaining a balanced extracellular environment, protecting neurons during neuroinflammation, and promoting synaptogenesis [95]. In the context of these results, altered expression of genes crucial to astrocyte function may impair the brain’s ability to deal with other biological effects of isolation, including neuroinflammation [96] and increased oxidative stress [97]. Human data also highlights the importance of astrocyte function. As discussed in Kruyer, Kalivas [98], human post-mortem data widely implicate morphological and molecular changes in multiple psychiatric disorders including SCZ, MDD and BPD discussed here.
Recent work in the SI field further summarises the effects of SI on the brain [4]. The authors reported overall changes in neuron biology caused by SI in rodents, including changes in neurogenesis, synaptogenesis, neurotransmission and cell morphology. Between GO results highlighting synaptic ontologies and prioritised genes being strongly implicated in neurotransmitter release, neurogenesis and cell neuron morphology, our findings generally align with these conclusions. Furthermore, there was an emphasis placed on the importance of glial cell types mediating the effects of SI [4]. This is also generally supported by our findings in two ways. First, the prioritised ZNF365 gene (downregulated in both males and females) plays a role in oligodendrocyte differentiation, which as discussed [4] is a process clearly playing an important role in dysfunction in an SI context. Second, we found that genes are dysregulated as a consequence of SI in hippocampal astrocytes, which as noted [4] are significantly activated as a result of early life SI in females [99].
There are a number of limitations to this present study. First, focusing on a single region limits the ability to generalise these results. Second, these data are generated from a stressor at a single timepoint. Investigating a number of developmental timepoints could help detect transient gene expression changes and could facilitate investigation into the impacts of earlier or later social stressors on gene expression in the mouse brain. Finally, we use a single technique to measure the effect of SI. Other techniques, especially epigenetic profiling using ATAC-seq, or the use of single-cell RNA-seq methods could add valuable context to gene expression profiles found in this data. Addressing these limitations in future studies could help shed light on the relationship between gene expression changes, altered behavioural phenotypes and known human risk variation for neurodevelopmental disorders.
In summary, we provide novel insight into how SI affects the mouse brain. The use of RNA-seq highlights gene expression changes related to synaptic biology that may underpin the changes in behaviour previously found in these animals. We show convergence of DEGs in socially isolated mice with genes containing genetic variation contributing to neurodevelopmental phenotypes in humans. We highlight genes such as RIMS1, ZNF365 and AGAP1 as candidates for studying disrupted developmental trajectories in disorders such as SCZ, BPD and ASD. Further, these data provide support for the molecular validity of the SI mouse model to study neurodevelopmental phenotypes with further behavioural, molecular and interventional investigations.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting information files. Raw RNA-seq data is available under GEO accession number GSE246551.
Funding Statement
This work was funded by grants from the European Research Council (ERC-2015- STG-677467 to GD; https://erc.europa.eu/), Science Foundation Ireland (SFI- 16/ERCS/3787 to GD; https://www.sfi.ie/) and the Irish Research Council (GOIPG/2019/1932 to AL; https://research.ie/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Esposito G, Azhari A, Borelli JL. Gene x Environment Interaction in Developmental Disorders: Where Do We Stand and What’s Next? Front Psychol. 2018;9:2036. Epub 2018/11/13. doi: 10.3389/fpsyg.2018.02036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister RF, Leary MR. The need to belong: desire for interpersonal attachments as a fundamental human motivation. Psychol Bull. 1995;117(3):497–529. Epub 1995/05/01. . [PubMed] [Google Scholar]
- 3.Li DC, Hinton EA, Gourley SL. Persistent behavioral and neurobiological consequences of social isolation during adolescence. Semin Cell Dev Biol. 2021;118:73–82. Epub 2021/06/12. doi: 10.1016/j.semcdb.2021.05.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y, Hong H, Liu C, Zhang YQ. Social isolation and the brain: effects and mechanisms. Mol Psychiatry. 2023;28(1):191–201. Epub 2022/11/27. doi: 10.1038/s41380-022-01835-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santini ZI, Jose PE, York Cornwell E, Koyanagi A, Nielsen L, Hinrichsen C, et al. Social disconnectedness, perceived isolation, and symptoms of depression and anxiety among older Americans (NSHAP): a longitudinal mediation analysis. Lancet Public Health. 2020;5(1):e62–e70. Epub 2020/01/09. doi: 10.1016/S2468-2667(19)30230-0 . [DOI] [PubMed] [Google Scholar]
- 6.Beutel ME, Klein EM, Brahler E, Reiner I, Junger C, Michal M, et al. Loneliness in the general population: prevalence, determinants and relations to mental health. BMC Psychiatry. 2017;17(1):97. Epub 2017/03/23. doi: 10.1186/s12888-017-1262-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreu-Bernabeu A, Diaz-Caneja CM, Costas J, De Hoyos L, Stella C, Gurriaran X, et al. Polygenic contribution to the relationship of loneliness and social isolation with schizophrenia. Nat Commun. 2022;13(1):51. Epub 2022/01/12. doi: 10.1038/s41467-021-27598-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cacioppo JT, Hawkley LC. Perceived social isolation and cognition. Trends Cogn Sci. 2009;13(10):447–54. Epub 2009/09/04. doi: 10.1016/j.tics.2009.06.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Ding Z, Fan T, Wang K, Li S, Zhao J, et al. Childhood social isolation causes anxiety-like behaviors via the damage of blood-brain barrier in amygdala in female mice. Front Cell Dev Biol. 2022;10:943067. Epub 2022/09/03. doi: 10.3389/fcell.2022.943067 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desbonnet L, Konkoth A, Laighneach A, McKernan D, Holleran L, McDonald C, et al. Dual hit mouse model to examine the long-term effects of maternal immune activation and post-weaning social isolation on schizophrenia endophenotypes. Behav Brain Res. 2022;430:113930. Epub 2022/05/25. doi: 10.1016/j.bbr.2022.113930 . [DOI] [PubMed] [Google Scholar]
- 11.Yu L, Wang Y, Zhang H, Li M, Chen G, Hao J, et al. Involvement of purinergic P2Y1R in antidepressant-like effects of electroacupuncture treatment on social isolation stress mice. Purinergic Signal. 2022. Epub 2022/01/31. doi: 10.1007/s11302-021-09827-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim GS, Lee H, Jeong Y. Altered dorsal functional connectivity after post-weaning social isolation and resocialization in mice. Neuroimage. 2021;245:118740. Epub 2021/11/23. doi: 10.1016/j.neuroimage.2021.118740 . [DOI] [PubMed] [Google Scholar]
- 13.Salihu SA, Ghafari H, Ahmadimanesh M, Gortany NK, Shafaroodi H, Ghazi-Khansari M. Glatiramer acetate attenuates depressive/anxiety-like behaviors and cognitive deficits induced by post-weaning social isolation in male mice. Psychopharmacology (Berl). 2021;238(8):2121–32. Epub 2021/04/03. doi: 10.1007/s00213-021-05836-5 . [DOI] [PubMed] [Google Scholar]
- 14.Lander SS, Linder-Shacham D, Gaisler-Salomon I. Differential effects of social isolation in adolescent and adult mice on behavior and cortical gene expression. Behav Brain Res. 2017;316:245–54. Epub 2016/09/14. doi: 10.1016/j.bbr.2016.09.005 . [DOI] [PubMed] [Google Scholar]
- 15.Ueno M, Okamura T, Mishina M, Ishizaka Y. Modulation of long interspersed nuclear element-1 in the mouse hippocampus during maturation. Mob Genet Elements. 2016;6(4):e1211980. Epub 2016/09/02. doi: 10.1080/2159256X.2016.1211980 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumari A, Singh P, Baghel MS, Thakur MK. Social isolation mediated anxiety like behavior is associated with enhanced expression and regulation of BDNF in the female mouse brain. Physiol Behav. 2016;158:34–42. Epub 2016/02/28. doi: 10.1016/j.physbeh.2016.02.032 . [DOI] [PubMed] [Google Scholar]
- 17.Benfato ID, Quintanilha ACS, Henrique JS, Souza MA, Rosario BDA, Beserra Filho JIA, et al. Effects of long-term social isolation on central, behavioural and metabolic parameters in middle-aged mice. Behav Brain Res. 2022;417:113630. Epub 2021/10/18. doi: 10.1016/j.bbr.2021.113630 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu XR, Zhang Y, Liu XD, Han WB, Xu NJ, Sun S. EphB2 mediates social isolation-induced memory forgetting. Transl Psychiatry. 2020;10(1):389. Epub 2020/11/11. doi: 10.1038/s41398-020-01051-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Wu Q, Lei L, Sun H, Michael N, Zhang X, et al. Long-term social isolation inhibits autophagy activation, induces postsynaptic dysfunctions and impairs spatial memory. Exp Neurol. 2019;311:213–24. Epub 2018/09/17. doi: 10.1016/j.expneurol.2018.09.009 . [DOI] [PubMed] [Google Scholar]
- 20.Chang CH, Kuek EJW, Su CL, Gean PW. MicroRNA-206 Regulates Stress-Provoked Aggressive Behaviors in Post-weaning Social Isolation Mice. Mol Ther Nucleic Acids. 2020;20:812–22. Epub 2020/05/29. doi: 10.1016/j.omtn.2020.05.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caruso A, Ricceri L, Caruso A, Nicoletti F, Gaetano A, Scaccianoce S. Postweaning social isolation and autism-like phenotype: A biochemical and behavioral comparative analysis. Behav Brain Res. 2022;428:113891. Epub 2022/04/15. doi: 10.1016/j.bbr.2022.113891 . [DOI] [PubMed] [Google Scholar]
- 22.Mumtaz F, Khan MI, Zubair M, Dehpour AR. Neurobiology and consequences of social isolation stress in animal model-A comprehensive review. Biomed Pharmacother. 2018;105:1205–22. Epub 2018/07/20. doi: 10.1016/j.biopha.2018.05.086 . [DOI] [PubMed] [Google Scholar]
- 23.DeVito LM, Konigsberg R, Lykken C, Sauvage M, Young WS, Eichenbaum H. Vasopressin 1b receptor knock-out impairs memory for temporal order. J Neurosci. 2009;29(9):2676–83. Epub 2009/03/06. doi: 10.1523/JNEUROSCI.5488-08.2009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrientos RM, Sprunger DB, Campeau S, Higgins EA, Watkins LR, Rudy JW, et al. Brain-derived neurotrophic factor mRNA downregulation produced by social isolation is blocked by intrahippocampal interleukin-1 receptor antagonist. Neuroscience. 2003;121(4):847–53. Epub 2003/10/29. doi: 10.1016/s0306-4522(03)00564-5 . [DOI] [PubMed] [Google Scholar]
- 25.Pugh CR, Nguyen KT, Gonyea JL, Fleshner M, Wakins LR, Maier SF, et al. Role of interleukin-1 beta in impairment of contextual fear conditioning caused by social isolation. Behav Brain Res. 1999;106(1–2):109–18. Epub 1999/12/14. doi: 10.1016/s0166-4328(99)00098-4 . [DOI] [PubMed] [Google Scholar]
- 26.Moller M, Du Preez JL, Viljoen FP, Berk M, Emsley R, Harvey BH. Social isolation rearing induces mitochondrial, immunological, neurochemical and behavioural deficits in rats, and is reversed by clozapine or N-acetyl cysteine. Brain Behav Immun. 2013;30:156–67. Epub 2012/12/29. doi: 10.1016/j.bbi.2012.12.011 . [DOI] [PubMed] [Google Scholar]
- 27.Levine JB, Youngs RM, MacDonald ML, Chu M, Leeder AD, Berthiaume F, et al. Isolation rearing and hyperlocomotion are associated with reduced immediate early gene expression levels in the medial prefrontal cortex. Neuroscience. 2007;145(1):42–55. Epub 2007/01/24. doi: 10.1016/j.neuroscience.2006.11.063 . [DOI] [PubMed] [Google Scholar]
- 28.Iwata H, Yamamuro Y. Subregional Expression of Hippocampal Glutamatergic and GABAergic Genes in F344 Rats with Social Isolation after Weaning. Comp Med. 2016;66(1):4–9. Epub 2016/02/18. . [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy KJ, Ter Horst JP, Cassidy AW, DeSouza IE, Morgunova M, Li C, et al. Temporal dysregulation of cortical gene expression in the isolation reared Wistar rat. J Neurochem. 2010;113(3):601–14. Epub 2010/01/26. doi: 10.1111/j.1471-4159.2010.06617.x . [DOI] [PubMed] [Google Scholar]
- 30.Pinna G, Agis-Balboa RC, Zhubi A, Matsumoto K, Grayson DR, Costa E, et al. Imidazenil and diazepam increase locomotor activity in mice exposed to protracted social isolation. Proc Natl Acad Sci U S A. 2006;103(11):4275–80. Epub 2006/03/16. doi: 10.1073/pnas.0600329103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karailiev P, Hlavacova N, Chomanic P, Riecansky I, Jezova D. Dopamine concentrations and dopamine receptor gene expression in emotion-related brain structures of female adult rats exposed to stress of chronic isolation from weaning. Gen Physiol Biophys. 2020;39(4):393–8. Epub 2020/09/10. doi: 10.4149/gpb_2020015 . [DOI] [PubMed] [Google Scholar]
- 32.Yang AC, Tsai SJ. New Targets for Schizophrenia Treatment beyond the Dopamine Hypothesis. Int J Mol Sci. 2017;18(8). Epub 2017/08/05. doi: 10.3390/ijms18081689 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZJ, Shwani T, Liu J, Zhong P, Yang F, Schatz K, et al. Molecular and cellular mechanisms for differential effects of chronic social isolation stress in males and females. Mol Psychiatry. 2022;27(7):3056–68. Epub 2022/04/23. doi: 10.1038/s41380-022-01574-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron HA, Glover LR. Adult neurogenesis: beyond learning and memory. Annu Rev Psychol. 2015;66:53–81. Epub 2014/09/25. doi: 10.1146/annurev-psych-010814-015006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476(7361):458–61. Epub 2011/08/05. doi: 10.1038/nature10287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. Epub 2014/04/04. doi: 10.1093/bioinformatics/btu170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. Epub 2017/03/07. doi: 10.1038/nmeth.4197 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. Epub 2014/12/18. doi: 10.1186/s13059-014-0550-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42(21). Epub 2014/10/09. doi: 10.1093/nar/gku864 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35(12):2084–92. Epub 2018/11/06. doi: 10.1093/bioinformatics/bty895 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–40. Epub 2005/08/06. doi: 10.1093/bioinformatics/bti525 . [DOI] [PubMed] [Google Scholar]
- 42.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11(10):1889–907. Epub 2016/09/09. doi: 10.1038/nprot.2016.117 . [DOI] [PubMed] [Google Scholar]
- 43.Koopmans F, van Nierop P, Andres-Alonso M, Byrnes A, Cijsouw T, Coba MP, et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron. 2019;103(2):217–34.e4. Epub 2019/06/07. doi: 10.1016/j.neuron.2019.05.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trubetskoy V, Pardinas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604(7906):502–8. Epub 2022/04/10. doi: 10.1038/s41586-022-04434-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50(7):912–9. Epub 2018/06/27. doi: 10.1038/s41588-018-0152-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50(8):1112–21. Epub 2018/07/25. doi: 10.1038/s41588-018-0147-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53(6):817–29. Epub 2021/05/19. doi: 10.1038/s41588-021-00857-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–52. Epub 2019/02/06. doi: 10.1038/s41593-018-0326-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill WD, Weiss A, Liewald DC, Davies G, Porteous DJ, Hayward C, et al. Genetic contributions to two special factors of neuroticism are associated with affluence, higher intelligence, better health, and longer life. Mol Psychiatry. 2020;25(11):3034–52. Epub 2019/03/15. doi: 10.1038/s41380-019-0387-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. Epub 2018/11/28. doi: 10.1038/s41588-018-0269-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–13. Epub 2019/01/09. doi: 10.1038/s41588-018-0311-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malik R, Rannikmae K, Traylor M, Georgakis MK, Sargurupremraj M, Markus HS, et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann Neurol. 2018;84(6):934–9. Epub 2018/11/02. doi: 10.1002/ana.25369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13. Epub 2018/10/10. doi: 10.1038/s41588-018-0241-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–8. Epub 2011/03/08. doi: 10.1038/ng.784 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. Epub 2015/02/03. doi: 10.1038/ng.3211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–35. Epub 2015/09/29. doi: 10.1038/ng.3404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–9. Epub 2018/04/11. doi: 10.1038/s41588-018-0081-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. Epub 2015/04/18. doi: 10.1371/journal.pcbi.1004219 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19(11):1433–41. Epub 2016/10/28. doi: 10.1038/nn.4402 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180(3):568–84.e23. Epub 2020/01/26. doi: 10.1016/j.cell.2019.12.036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howrigan DP, Rose SA, Samocha KE, Fromer M, Cerrato F, Chen WJ, et al. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat Neurosci. 2020;23(2):185–93. Epub 2020/01/15. doi: 10.1038/s41593-019-0564-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rees E, Han J, Morgan J, Carrera N, Escott-Price V, Pocklington AJ, et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat Neurosci. 2020;23(2):179–84. Epub 2020/01/15. doi: 10.1038/s41593-019-0565-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–8. Epub 2017/01/31. doi: 10.1038/nature21062 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell. 2018;174(4):1015–30.e16. Epub 2018/08/11. doi: 10.1016/j.cell.2018.07.028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skene NG, Grant SG. Identification of Vulnerable Cell Types in Major Brain Disorders Using Single Cell Transcriptomes and Expression Weighted Cell Type Enrichment. Front Neurosci. 2016;10:16. Epub 2016/02/10. doi: 10.3389/fnins.2016.00016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7 25 1–7 15. Epub 2015/10/07. doi: 10.1002/0471142905.hg0725s87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18(3):147–57. Epub 2017/02/10. doi: 10.1038/nrn.2016.183 . [DOI] [PubMed] [Google Scholar]
- 68.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82(1):199–207. Epub 2008/01/09. doi: 10.1016/j.ajhg.2007.09.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trobiani L, Meringolo M, Diamanti T, Bourne Y, Marchot P, Martella G, et al. The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci Biobehav Rev. 2020;119:37–51. Epub 2020/09/30. doi: 10.1016/j.neubiorev.2020.09.017 . [DOI] [PubMed] [Google Scholar]
- 70.Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2020;25(12):3292–303. Epub 2019/11/22. doi: 10.1038/s41380-019-0559-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X, Cai Q, Shen Z, Chen X, Zeng M, Du S, et al. RIM and RIM-BP Form Presynaptic Active-Zone-like Condensates via Phase Separation. Mol Cell. 2019;73(5):971–84.e5. Epub 2019/01/22. doi: 10.1016/j.molcel.2018.12.007 . [DOI] [PubMed] [Google Scholar]
- 72.Kaeser PS, Deng L, Fan M, Sudhof TC. RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci U S A. 2012;109(29):11830–5. Epub 2012/07/04. doi: 10.1073/pnas.1209318109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han Y, Kaeser PS, Sudhof TC, Schneggenburger R. RIM determines Ca(2)+ channel density and vesicle docking at the presynaptic active zone. Neuron. 2011;69(2):304–16. Epub 2011/01/26. doi: 10.1016/j.neuron.2010.12.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiyonaka S, Wakamori M, Miki T, Uriu Y, Nonaka M, Bito H, et al. RIM1 confers sustained activity and neurotransmitter vesicle anchoring to presynaptic Ca2+ channels. Nat Neurosci. 2007;10(6):691–701. Epub 2007/05/15. doi: 10.1038/nn1904 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss N, Sandoval A, Kyonaka S, Felix R, Mori Y, De Waard M. Rim1 modulates direct G-protein regulation of Ca(v)2.2 channels. Pflugers Arch. 2011;461(4):447–59. Epub 2011/02/19. doi: 10.1007/s00424-011-0926-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muller JA, Betzin J, Santos-Tejedor J, Mayer A, Oprisoreanu AM, Engholm-Keller K, et al. A presynaptic phosphosignaling hub for lasting homeostatic plasticity. Cell Rep. 2022;39(3):110696. Epub 2022/04/21. doi: 10.1016/j.celrep.2022.110696 . [DOI] [PubMed] [Google Scholar]
- 77.Powell CM. Gene targeting of presynaptic proteins in synaptic plasticity and memory: across the great divide. Neurobiol Learn Mem. 2006;85(1):2–15. Epub 2005/10/19. doi: 10.1016/j.nlm.2005.08.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blundell J, Kaeser PS, Sudhof TC, Powell CM. RIM1alpha and interacting proteins involved in presynaptic plasticity mediate prepulse inhibition and additional behaviors linked to schizophrenia. J Neurosci. 2010;30(15):5326–33. Epub 2010/04/16. doi: 10.1523/JNEUROSCI.0328-10.2010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castillo PE, Schoch S, Schmitz F, Sudhof TC, Malenka RC. RIM1alpha is required for presynaptic long-term potentiation. Nature. 2002;415(6869):327–30. Epub 2002/01/18. doi: 10.1038/415327a . [DOI] [PubMed] [Google Scholar]
- 80.Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, et al. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42(1):143–53. Epub 2004/04/07. doi: 10.1016/s0896-6273(04)00146-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–8. Epub 2015/05/12. doi: 10.1038/ng.3303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9(1):16–23. Epub 2014/10/07. doi: 10.1016/j.celrep.2014.08.068 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peter B, Dinu V, Liu L, Huentelman M, Naymik M, Lancaster H, et al. Exome Sequencing of Two Siblings with Sporadic Autism Spectrum Disorder and Severe Speech Sound Disorder Suggests Pleiotropic and Complex Effects. Behav Genet. 2019;49(4):399–414. Epub 2019/04/06. doi: 10.1007/s10519-019-09957-8 . [DOI] [PubMed] [Google Scholar]
- 84.Koyama Y, Hattori T, Shimizu S, Taniguchi M, Yamada K, Takamura H, et al. DBZ (DISC1-binding zinc finger protein)-deficient mice display abnormalities in basket cells in the somatosensory cortices. J Chem Neuroanat. 2013;53:1–10. Epub 2013/08/06. doi: 10.1016/j.jchemneu.2013.07.002 . [DOI] [PubMed] [Google Scholar]
- 85.Shimizu S, Koyama Y, Hattori T, Tachibana T, Yoshimi T, Emoto H, et al. DBZ, a CNS-specific DISC1 binding protein, positively regulates oligodendrocyte differentiation. Glia. 2014;62(5):709–24. Epub 2014/02/01. doi: 10.1002/glia.22636 . [DOI] [PubMed] [Google Scholar]
- 86.Koyama Y, Hattori T, Nishida T, Hori O, Tohyama M. Alterations in dendrite and spine morphology of cortical pyramidal neurons in DISC1-binding zinc finger protein (DBZ) knockout mice. Front Neuroanat. 2015;9:52. Epub 2015/05/20. doi: 10.3389/fnana.2015.00052 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miyata S, Hattori T, Shimizu S, Ito A, Tohyama M. Disturbance of oligodendrocyte function plays a key role in the pathogenesis of schizophrenia and major depressive disorder. Biomed Res Int. 2015;2015:492367. Epub 2015/02/24. doi: 10.1155/2015/492367 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arnold M, Cross R, Singleton KS, Zlatic S, Chapleau C, Mullin AP, et al. The Endosome Localized Arf-GAP AGAP1 Modulates Dendritic Spine Morphology Downstream of the Neurodevelopmental Disorder Factor Dysbindin. Front Cell Neurosci. 2016;10:218. Epub 2016/10/08. doi: 10.3389/fncel.2016.00218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC, Sulzer D, et al. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO J. 2010;29(16):2813–26. Epub 2010/07/29. doi: 10.1038/emboj.2010.154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wassink TH, Piven J, Vieland VJ, Jenkins L, Frantz R, Bartlett CW, et al. Evaluation of the chromosome 2q37.3 gene CENTG2 as an autism susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2005;136B(1):36–44. Epub 2005/05/14. doi: 10.1002/ajmg.b.30180 . [DOI] [PubMed] [Google Scholar]
- 91.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753–7. Epub 2009/07/03. doi: 10.1038/nature08192 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pacault M, Nizon M, Pichon O, Vincent M, Le Caignec C, Isidor B. A de novo 2q37.2 deletion encompassing AGAP1 and SH3BP4 in a patient with autism and intellectual disability. Eur J Med Genet. 2019;62(12):103586. Epub 2018/11/26. doi: 10.1016/j.ejmg.2018.11.020 . [DOI] [PubMed] [Google Scholar]
- 93.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(v). Epub 2018/12/14. doi: 10.1126/science.aat8127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lago-Baldaia I, Fernandes VM, Ackerman SD. More Than Mortar: Glia as Architects of Nervous System Development and Disease. Front Cell Dev Biol. 2020;8:611269. Epub 2021/01/01. doi: 10.3389/fcell.2020.611269 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26(9):891–907. Epub 2012/05/03. doi: 10.1101/gad.188326.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Al Omran AJ, Shao AS, Watanabe S, Zhang Z, Zhang J, Xue C, et al. Social isolation induces neuroinflammation and microglia overactivation, while dihydromyricetin prevents and improves them. J Neuroinflammation. 2022;19(1):2. Epub doi: 10.1186/s12974-021-02368-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huong NT, Murakami Y, Tohda M, Watanabe H, Matsumoto K. Social isolation stress-induced oxidative damage in mouse brain and its modulation by majonoside-R2, a Vietnamese ginseng saponin. Biol Pharm Bull. 2005;28(8):1389–93. Epub 2005/08/05. doi: 10.1248/bpb.28.1389 . [DOI] [PubMed] [Google Scholar]
- 98.Kruyer A, Kalivas PW, Scofield MD. Astrocyte regulation of synaptic signaling in psychiatric disorders. Neuropsychopharmacology. 2023;48(1):21–36. Epub 2022/05/17. doi: 10.1038/s41386-022-01338-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakamoto K, Aizawa F, Kinoshita M, Koyama Y, Tokuyama S. Astrocyte Activation in Locus Coeruleus Is Involved in Neuropathic Pain Exacerbation Mediated by Maternal Separation and Social Isolation Stress. Front Pharmacol. 2017;8:401. Epub 2017/07/14. doi: 10.3389/fphar.2017.00401 . [DOI] [PMC free article] [PubMed] [Google Scholar]