Abstract
During the past decade there has been a revolution in cancer therapeutics by the emergence of antibody-based and cell-based immunotherapies that modulate immune responses against tumors. These new therapies have extended and improved the therapeutic efficacy of chemo-radiotherapy and have offered treatment options to patients who are no longer responding to these classic anti-cancer treatments. Unfortunately, tumor eradication and long-lasting responses are observed in a small fraction of patients, whereas the majority of patients respond only transiently. These outcomes indicate that the maximum potential of immunotherapy has not been reached due to incomplete knowledge of the cellular and molecular mechanisms that guide the development of successful anti-tumor immunity and its failure. In this review, we discuss recent discoveries about the immune cellular composition of the tumor microenvironment (TME) and the role of key signaling mechanisms that compromise the function of immune cells leading to cancer immune escape.
Keywords: Tumor microenvironment, Signaling pathways, Checkpoint inhibitors
1. Cancer immunosurveillance pathways and the tumor microenvironment
The exploitation of the immune system for the treatment of cancer has been investigated for many years. However, enthusiasm waxed and waned based on discoveries that supported or opposed the hypothesis that cancer can be subjected to immune-mediated control. It is currently understood that not only the immune system can control cancer growth but has an important role in shaping the immunogenicity of cancer cells via immunoediting [1]. During early stages of cancerous differentiation of normal cells, continuous immune surveillance results in the identification and elimination of these malignant populations through generation of adaptive anti-tumor immune responses. Tumor-associated macrophages (TAMs) have a central role in this process by mediating phagocytosis and clearance of cancer cells [2] and the presentation of cancer neoantigens to T cells [3]. Through continuous control, the immune system keeps cancer under check achieving equilibrium, even without complete elimination. The adaptive antitumor immune system can efficiently recognize neoantigens resulting from tumor-specific somatic mutations, antigens derived from oncogenic viruses, and antigens whose expression is shared with tissues at immune-privileged sites [4]. When cancer antigens yield peptides capable of binding to an individual’s HLA alleles (neoepitopes), they can elicit CD4+ and CD8+ T cell responses [5,6] as evidenced by the presence and prognostic significance of immune infiltrates in human tumors [7–9]. However, under the continuous immune pressure cancer cells develop alterations to overcome immune attack, resulting in escape and growth of tumors that are resistant to the physiological immune mechanisms utilized to recognize and present antigens thereby engaging adaptive immune responses.
Escape mechanisms include tumor cell-intrinsic adaptations such as downregulation of tumor neoantigens and induction of protective mechanisms rendering tumors resistant to cytotoxic cells of adaptive immunity. In that regard, it has been determined that genomic alterations and changes of neoantigen load are linked to diminished immune responses [10]. Escape may also result from the development of immunosuppressive mechanisms of the TME, with the production of soluble factors such as IDO, VEGF, and TGF-β [11,12]. Alterations in the cellular populations of the TME such as recruitment of immunosuppressive myeloid-derived suppressor cells (MDSCs) that are produced during cancer-driven emergency myelopoiesis, regulatory T cells (Tregs) and tumor associated macrophages (TAMs) have important roles in shaping the properties of the TME (Fig. 1) [13]. Changes also occur in dendritic cells (DC) which lose their ability to process and present tumor antigens to T cells [14]. Expression of co-inhibitory molecules in these immune populations shapes the signaling landscape of the TME, generating primarily pro-tumorigenic cues by suppressing critical functions of myeloid cells and TAMs, such as phagocytosis and antigen presentation, and compromising the ability of T cells to mount immune responses. These signaling pathways have a key role in cancer immune escape but also represent targets for immune-based treatments that have re-shaped modern cancer therapy. Among the coinhibitory receptors, inhibitory targeting PD-1 and its ligands with blocking antibodies has been the cornerstone of cancer immunotherapy and together with CTLA-4 blocking agents have revolutionized cancer treatment. In this review, we provide a concise summary of how key coinhibitory receptors shape the function of the immune components of the TME and discuss the role of immune populations during tumorigenesis and cancer evolution.
Fig. 1.
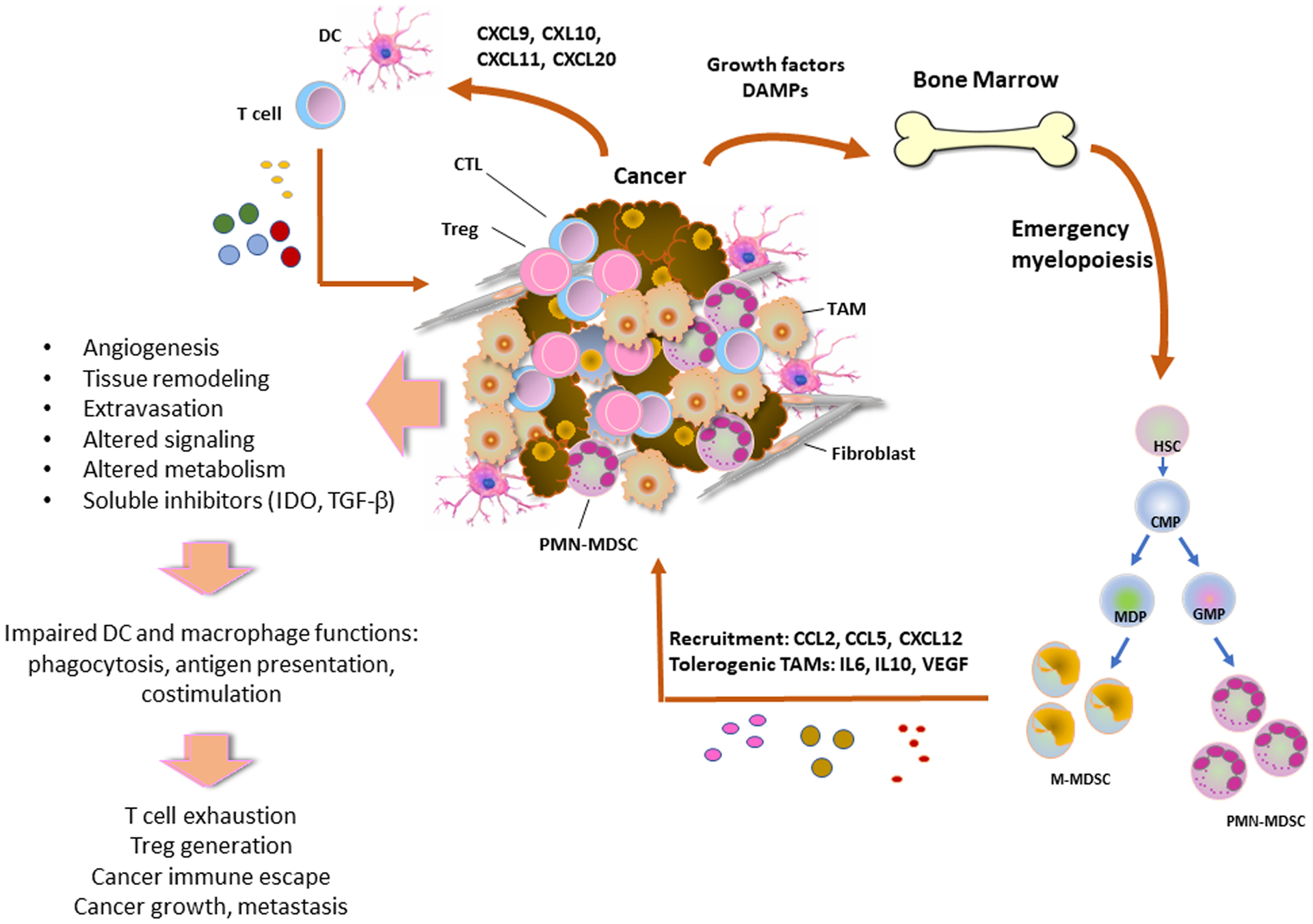
During tumorigenesis, growth factors, such as M-CSF and GM-CSF, and damage-associated molecular patterns (DAMPs) produced by cancer cells due to rapid replication and apoptosis, act on bone marrow myeloid progenitors inducing emergency myelopoiesis and output of immature myelosuppressive PMN-MDSCs and M-MDSCs, which are recruited to the tumor via chemokines such as CCL2, CCL5 and CXCL12. In the TME, PMN-MDSCs and M-MDSCs directly induce immunosuppression, whereas M-MDSCs are also converted into tolerogenic and pro-tumorigenic TAMs by the function of soluble factors such as IL-6, IL-10 and VEGF. TAMs lose the physiologic properties of macrophages such as phagocytosis and together with MDSCs and soluble factors produced in the TME suppress the antigen presenting function of DCs, leading to impaired activation of tumor-specific T cells and generation of Treg cells. These orchestrated changes in the properties of immune cells promote cancer immune escape.
2. Dendritic cells are key mediators of tumor antigen presentation
Dendritic cells (DC) are professional antigen-presenting cells (APC) that play an important role in the tumorigenic and non-tumorigenic environment by activating CD8+ T cells and enhancing antitumor responses. DCs make up a small minority of the tumor-infiltrating leucocytes [15]. In both human and mice, four different major subsets of DCs have been characterized: the classical DC1 (cDC1), the heterogeneous population cDC2, the monocyte-derived DC (mo-DC), and the plasma-cytoid DC (pDC) [16]. cDC1 expresses a distinct gene expression profile and specific markers [17]. It has been reported that cDC1 are critical for priming CD8+ T cells whereas cDC2 are responsible for priming CD4+ T cells [18] and support Th17 responses [19]. The DC2 group is a more heterogeneous population consisting of two different types of DCs, cDC2 and DC3 [20]. In agreement to that, another study revealed two distinct cDC2 populations by transcriptional analysis, cDC2A and cDC2B which express T-bet and RORγt respectively and are conserved across both human and mice [21] and might correspond to cDC2 and DC3 [22]. In humans, DC2 and DC3 derive from progenitors with differential expression of IRF8, where high expression led to the development of cDC1 and DC2, whereas low expression of IRF8 led to the development of DC3 and monocytes [23]. In contrast to cDC2 which require FLT3L for expansion, DC3 expanded and differentiated in the presence of granulocyte-macrophage stimulating factor (GM-CSF) but not FLT3L, providing further evidence that DC3s forms a distinct population within the cDC2 compartment [24]. Mo-DC can either derive directly from CD34+ precursors or from monocytes [25,26]. Studies have indicated that IRF4 might be required for the differentiation of cDC2 and mo-DC, however, mo-DC do not share the same precursors as cDC2 [27,28]. Lastly, pDC are a distinct subpopulation of DC deriving from the common DC progenitors, secrete IFN-α/β and play an important role in viral infections [28].
Chemokines are mainly responsible for cDC1 recruitment and retention in the TME (Fig. 1). Tumors secrete several chemokines including CXCL9, CXCL10, and CXCL11, which can recruit Th1 and CD8+ T cells, CCL20 which recruits Th17 and immature DC, and CCL2 or CCL5 which recruit monocytes [29]. NK cells can also recruit cDC1 to the TME by secretion of CCL5 and XCL1, an effect abrogated by prostaglandin E2 (PGE2) production [30]. Inhibition of PGE2 production by ablation of COX1 or COX2 leads to increased accumulation of cDC1, tumor eradication, and increased sensitivity of tumor towards anti-PD-1 treatment [31]. Cytokines also play a role in the positioning of the DC in the TME. FLT3L secreted by NK cells and lymphocytes in mouse and human tumors has an active role in the recruitment and localization of cDC1 [32].
Generation of anti-tumor CD8+ T cell responses requires cross-presentation of tumor peptides to naïve CD8+ T cells on MHC class I molecules by the DC (Fig. 2A). Although priming of naïve T cells occurs in the tumor-draining lymph nodes, studies have shown that cross-presentation can also occur within the TME [33]. The antigens can travel to the lymph node either on cell debris or after their engulfment by migratory CD103+ cDC1 found in the TME, which then migrate to the tumor-draining lymph node and cross-present tumor antigens to naïve T cells, either directly or through antigen exchange to resident myeloid cells [34]. However, only the migratory CD103+ cDC1, but not the lymph node resident CD8+ cDC1, can prime naïve CD8+ T cells [35]. CCR7 is required for cDC1 migration to the tumor-draining lymph node and ablation of CCR7 expression results in a significantly smaller migratory effect of cDC1 to tumor-draining lymph node and a diminished T cell activation profile [36]. In human tumors, CCR7 expression positively correlates with greater DC infiltration and increased patient survival [36].
Fig. 2.
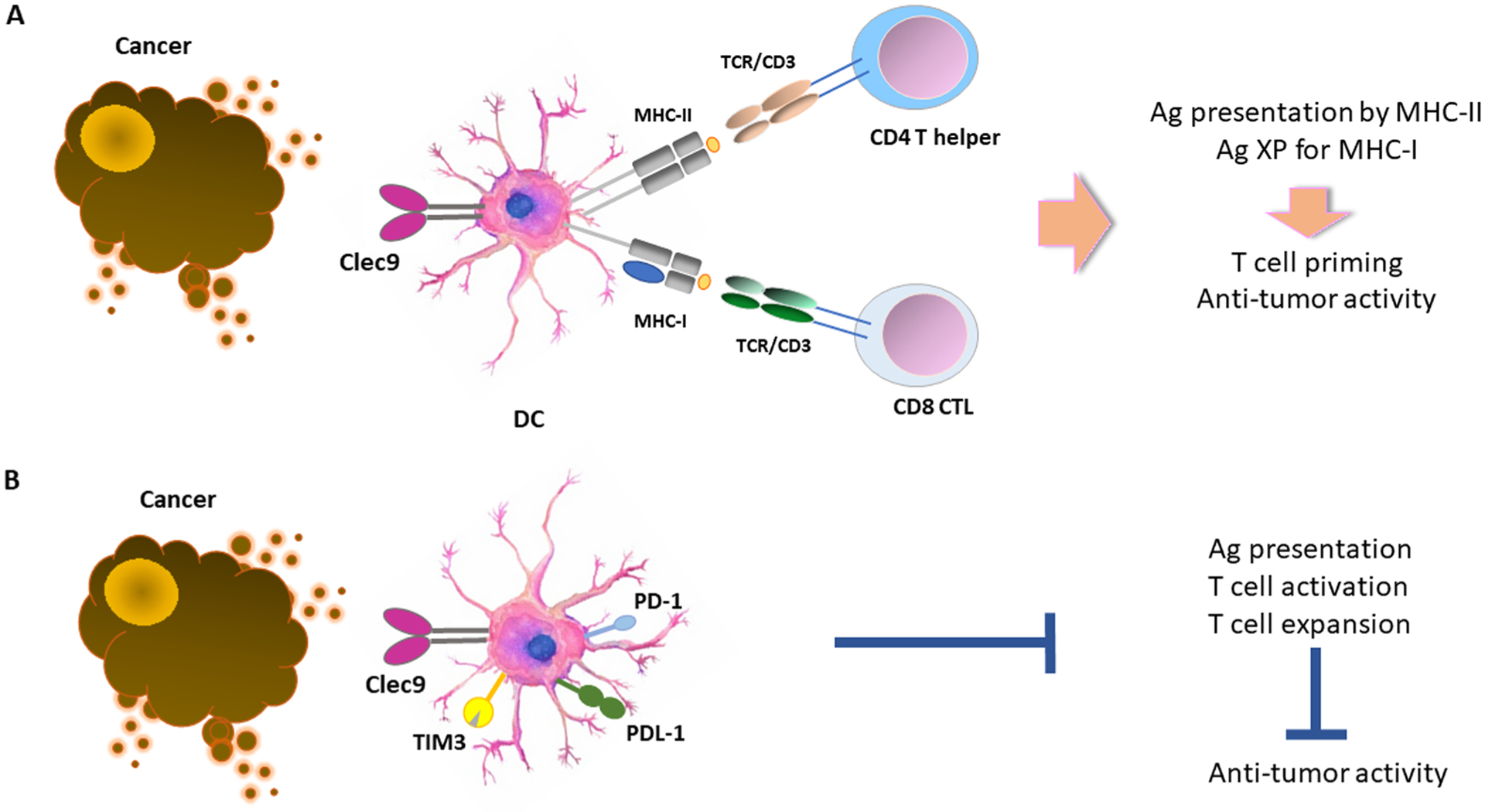
DCs, particularly the cDC1 subset, can capture and process tumor-associated antigens and present them to CD4+ T cells by MHC-II molecules. cDC1 can also cross-present (XP) tumor-associated antigens to CD8+ T cells by MHC-I molecules (A). Expression of checkpoint inhibitors such PD-1, PD-L1 and TIM-3, compromise these functions leading to impaired T cell activation, T cell expansion and anti-tumor immunity.
Within the TME, CD103+ cDC1 secrete CXCL9 and CXCL10 which recruit CXCR3+CD8+ effector T cells to the tumor [37]. In non-tumor models, CXCR3 has been previously shown to drive CD4+ and CD8+ T cell recruitment into the lymph nodes and in close contact to DC for T cell priming [38]. In a melanoma mouse model, CXCL9 and CXCL10 secreted by CD103+ cDC1 were responsible for the recruitment of effector T cells and for controlling tumor growth [39]. CD103+ cDC1 are located at distal regions of the tumor and despite their sparsity, they are good CTL activators [15]. Furthermore, cDC1 can uptake and cross-present tumor antigens to T cells more efficiently than other myeloid cells, which can also engulf tumor cells and uptake tumor antigens [15]. These CD103+ cDC1 also secrete increased levels of IL-12 but not IL-10 suggesting a dominant role in facilitating increased cytotoxic function of intratumoral CD8+ T cells [15].
Tumors have developed several mechanisms of immune evasion which restrict the function of cDC1 [40]. One such mechanism involves reducing the number of cDC1 in the TME. Tumors in breast and pancreatic cancers secrete the G-CSF thereby inhibiting the expression of IRF8 which is responsible for the generation of DC progenitors [41]. Similarly, reduction of FLT3L production within the tumor limits the differentiation, expansion, and survival of intratumoral cDC1 [35]. The reduced tumor infiltration by DC might also result from an impaired chemoattractant profile in the TME. PGE2 production leads to impaired NK viability and consequently impaired CCL5 and CXCL1 chemokine production, both of which act as chemoattractants for cDC1 [30].
Impairment of cDC1 function is also possible through direct inhibition of cDC1. In a breast cancer model, IL-10 which is secreted mainly by tumor-associated macrophages in the TME, resulted in low levels of IL-12 cDC1-mediated secretion [42]. Similarly, PGE2 reduced the production of IL-12 by cDC1 and downregulated the expression of co-stimulatory molecules [31]. In murine tumor models, lipid accumulation led to decreased ability of DC to translocate the peptide-MHC class I complex to the cell surface resulting in decreased antigen-presentation ability and T cell stimulation [43]. Tumor-derived TGF-β also inhibited the antigen presentation capacity of DC, impaired their ability to stimulate T cells, and decreased their migration capacity to the draining lymph nodes [44].
Suppression of cDC1 responses in the context of cancer is also driven by the inhibitory receptors present on the surface of DC (Fig. 2B). In patients with hepatocellular carcinoma, PD-1+ DC circulate in their peripheral blood, making them potentially prone to inhibition by PD-L1, the ligand of PD-1, present in the TME [45]. PD-L1 is highly expressed on both tumor cells and immune cells present in the TME [46]. PD-L1 expression in tumor infiltrating DC might be stabilized by phosphorylation mediated by the enzyme casein kinase 2 (CK2) [47], which is highly expressed in multiple cancers and is linked to increased cell growth, proliferation and inhibition of apoptosis [48]. Inhibition of CK2 decreased PD-L1 expression on DC and resulted in tumor suppression through release of CD80 from DC thereby allowing T cells to receive CD80-mediated co-stimulatory signals and become activated [47]. In PD-L1-deficient tumors, tumor-associated DC upregulatedPD-L1 expression [49]. The authors hypothesized that this upregulation could be an escape mechanism of the tumor by which immune cells receive inhibitory signals through the PD-L1-PD-1 axis. PD-L1 deletion on DC enhanced CD8+ anti-tumor T cell responses despite the higher number of tumor-associated macrophages in the TME [50]. In addition to its canonical interaction with PD-1 in trans, PD-L1 can also bind to CD80 in cis [51,52] and this interaction disrupts the PD-1: PD-L1 axis and prevents T cell inactivation [53,54]. Blocking of PD-L1 on DC allows CD80 to re-engage with CD28 on T cells and enhance T cell priming [55]. In ovarian tumor-infiltrating DC, PD-1 inhibited the secretion of TNFα and IL-6, and downregulated the expression of CD80, CD40, and MHC class I molecules [56]. PD-1 regulated multiple NF-κB targets by recruiting the SHP-2 to the cytoplasmic domain tyrosine-based switch motif (ITSM) of PD-1 [56]. Anti-PD-1 reversed the suppressive effect of PD-1 on cytokines but had no effect on the expression of the co-stimulatory and antigen-presenting molecules [56]. Together these studies provide evidence about the critical role of DC-expressed PD-L1 in the induction of PD-1-mediated T cell immunosuppression in the context of cancer.
TIM-3, a T cell checkpoint inhibitory receptor, is highly expressed on DCs in the TME compared to normal tissues and inhibits innate immune responses through recognition of nucleic acids via the pattern recognition receptors TLR3, TLR7, and TLR9 [57]. A combination of CK2 and a TIM-3 inhibitors led to greater tumor suppression and longer survival times for the mice [47]. TIM-3 was also highly expressed on intratumoral cDC1 in a mouse model for breast cancer [58]. Antibody blockade of TIM-3 led to increased granzyme B expression by CD8+ T cells and enhanced expression of CXCL9 by cDC1 [58]. Moreover, deletion of TIM-3 on DCs resulted in strong anti-tumoral CD8+ T cell responses and inflammasome activation [59]. TIM-4, a receptor responsible for engulfment of dead cells, is highly expressed in normal lung cDC1, however, inhibition of this receptor results in incomplete activation of anti-tumorigenic CD8+ T cells and increased tumor growth [60].
cDC2 cells have been historically considered to derive from monocytes and are recognized as cells responsible for Th2 and Th17 responses [61,62]. As mentioned earlier, they are recognized as a heterogeneous group which can be broken down into two different subsets, DC2 and DC3 [22]. In the context of cancer, these cells can travel from the tumor to the tumor-draining lymph nodes and present tumor antigens to CD4+ T cells [63]. Depletion of Treg leads to increased cDC2 ability to induce strong T cell responses and increased ratio of cDC2 to Treg is correlated with better clinical outcome and responsiveness to anti-PD-1 therapy [63]. DC3 are poorly characterized in the context of cancer. They rise from granulocyte-monocyte-DC progenitors and express low levels of IRF8 [23]. They are present in human papillomavirus-associated oropharyngeal squamous cell carcinoma tumors, are potent Th1 activators, and can secrete high levels of IL-12 and IL-18 which have the potential to drive anti-tumor responses [64]. It has been reported that DC3 bear signatures of both DC and monocyte-derived DC [22]. DC3 can stimulate memory T cells to produce IL-17 and differentiate naïve T cells into Th17 [65]. The differentiation program of these cells is regulated by GM-CSF [24]. Similarly, mo-DC can be generated in vitro from CD34+ precursors by culture with GM-CSF and TNF-α or monocytes by culture with GM-CSF and IL-4 [25,26]. However, only Mo-DC generated in vitro express the DC marker Zbtb46, providing evidence that these in vitro generated populations might not be the true counterparts of mo-DC differentiated in vivo [66]. The role of mo-DC in the presentation of tumor antigens is currently unclear. It was reported that although mo-DC can uptake antigen they have impaired capacity to induce potent T cell responses due to increased nitric oxide (NO) production [67]. However, in a different experimental system it was found that mo-DC could induce strong anti-tumor CD8+ T cell responses, which were impaired by blockade of monocyte entry to tumors [68]. Further studies are needed to fully characterize the cDC2, DC3 and mo-DC roles in the context of cancer.
pDCs are found in higher frequencies in human cancers and their presence is associated with poor prognosis and decreased survival [69]. Although pDCs normally produce IFN-α, in tumor settings including ovarian cancer, breast cancer, and melanoma, cancer-associated pDC have impaired IFN-α secretion [70]. This effect might be mediated by TNF-α and TGF-β present in the TME, which affect production of other cytokines such as IL-6, MIP-1b (CCL4) and RANTES [70]. In human ovarian cancer, pDCs are present in the tumor -but not ascites- can induce IL-10 secretion by naïve allogeneic T cells, and correlate with relapse [70]. Furthermore, when pDCs were cultured in medium from head and neck squamous cell carcinoma their ability to secrete IFN-α after TLR stimulation was significantly limited by IL-10 present in the medium, supporting the hypothesis that immunosuppressive cytokines in the TME contribute to the anti-inflammatory profile of tumor infiltrating immune cells [71]. Lastly, pDCs express the ICOS-L which can stimulate ICOS-expressing Foxp3+ Treg which in turn secrete IL-10 in the TME and suppress immune responses [72]. Together, these immunological, signaling and functional features of DCs underline the pivotal role of this immune cell population in the development of anti-tumor responses and explain why DC-targeting signaling alterations in the TME might have a significant impact in anti-tumor immunity.
3. Myeloid derived suppressor cells (MDSC) inhibit anti-tumor T cell responses
The constant and prolonged secretion of danger-associated molecular patterns (DAMPs) and cytokines from the tumor cells has a direct effect on the bone marrow, through a process that is called in emergency myelopoiesis. During emergency myelopoiesis, immature myeloid cells, named MDSCs, with potent immunosuppressive properties, are produced and accumulate in the tumor microenvironment and peripheral organs. High numbers of these cells are associated with poor treatment outcome and worse clinical prognosis in various cancer types [73]. Two major subsets of MDSCs have been described in mice: Polymorphonuclear MDSCs (PMN-MDSCs) monocytic-MDSCs (M-MDSCs), which resemble morphologically and phenotypically to neutrophils and monocytes, respectively. PMN-MDSCs are identified as CD11b+Ly6G+Ly6Clo in mice and CD11b+CD14−CD15+/CD66b+ in humans, while M-MDSCs are identified as CD11b+Ly6G−Ly6Chi in mice and CD14+CD15−HLA-DRlo/− in humans [74].
Among the mechanisms proposed for MDSC-mediated suppression function and subsequent cancer immune evasion, checkpoint inhibitors might have an important role. MDSCs isolated from patients with AML express high levels of VISTA and siRNA-mediated VISTA deletion attenuated MDSC-mediated inhibition of CD8+ T cells [75]. VISTA, a B7 family ligand, is predominantly expressed on the hematopoietic compartment, with highest expression observed in myeloid cells, including monocytic and granulocytic cells, and weaker expression on T cells [76]. VISTA was also identified as a homolog of PD-1, a member of the CD28 superfamily, indicating a potential function both as a ligand and a receptor [77]. VISTA transcription can be induced by HIF-1α suggesting that hypoxia is implicated in VISTA upregulation in MDSCs located in hypoxic regions of the TME [78]. VISTA deletion in MDSCs had no effects in T cell proliferation in vitro. However, VISTA deletion resulted in increased T cell proliferation under hypoxia, suggesting that regulation of MDSC function by VISTA might occur only under oxygen deprivation [78]. Furthermore, VISTA blockade in tumor-bearing mice altered the tumor infiltrating immune cell populations by decreasing MDSCs and increasing T cells with anti-tumor properties [79].
The MDSCs suppression capacity is also affected by the PD-1/PD-L1 pathway. Compared to MDSCs localized in non-cancerous tissues, tumor infiltrating MDSCs express high levels of PD-L1, which is regulated by the COX2/mPGES1/PGE2 pathway, pSTAT1-IRF1 axis and hypoxia-induced HIF-1α [80–82]. Consistent with an active role of these pathways in MDSC function, PD-L1 blockade under hypoxia diminished the ability of MDSCs to suppress T cell activation by reducing the production of IL-10 and IL-6 [82]. Notably, conditional deletion of PD-1 in the myeloid compartment in tumor-bearing mice, resulted in diminished NO production and attenuated suppressive capacity of M-MDSCs indicating that PD-1 blockade or deletion counteracts the generation of immunosuppressive immature myeloid cells [83].
Due to their potent immunosuppressive properties, MDSCs have been associated with the limited efficacy of immunotherapy and standard chemotherapy in mouse tumor models and cancer patients. Conversely, selective depletion of MDSC reversed resistance to anti-CTLA4 Ab treatment in tumor bearing mice and increased the numbers of cytotoxic CD8+ T cells in tumor-deraining lymph nodes and TME [84]. Doxycyline-induced suppression of MDSCs improved the efficacy of PD-1 inhibitors [85]. Similarly, in a mouse model of gastric cancer, 5-flurouracil and oxaliplatin combination, which decreased MDSCs, augmented CD8+ tumor infiltrating T cells and improved the therapeutic efficacy of anti-PD-1 treatment [86]. Furthermore, epigenetic targeting of MDSC in mice bearing immunotherapy-resistant tumors resulted in tumor eradication after anti-PD-1/anti-CTLA4 combined immunotherapy [87].
4. Generation of pro-tumorigenic tumor-associate macrophages (TAMs)
After egress from the bone marrow, monocytes (or M-MDSCs) are recruited to the TME via chemokines of the CC and CXC families, such as CCL2, CCL5, and CXCL12 [88] produced by cancer cells at early stages of the cancer immunity cycle and convert to bone marrow-derived tumor associated macrophages (Fig. 1), which, together with embryonically-derived tissue resident macrophages, form the population of TAMs. TAMs represent the most abundant immune population of the TME, consisting ~50% of hematopoietic cells and have predominantly pro-tumorigenic functions [89]. Pro-tumorigenic TAMs are immunosuppressive, lose the natural ability of macrophages to mediate phagocytosis and antigen presentation [83,90,91] and are characterized by expression of inhibitory molecules such as PD-1, PD-L1, VISTA, B7-H4 and TIM-3 [83,90–96].
The TME has a causative role in promoting the differentiation of pro-tumorigenic TAMs in a stepwise manner [97]. As a consequence of the malignant transformation of normal epithelial cells into cancerous cells by oncogenes and driver mutations, soluble factors, such as hematopoietic growth factors (e.g., M-CSF, GM-CSF) cytokines (e.g., IL-6, IL-1, and IL-8) and chemokines (e.g., CCL2, CCL5, CXCL12), are produced during cancer evolution (Fig. 1). For example, in pancreatic adenocarcinoma, KRASG12D mutation induces secretion of GM-CSF, which is correlated with an increased tumor infiltration by myeloid cells and immunosuppressive function [98,99]. p53 mutation has been documented to induce cancer-related inflammation by suppressing the production of IL-1 receptor antagonist [100]. In human melanoma, BRAFV600E, a highly oncogenic mutated form of BRAF, and STAT3, a potent transcription factor often linked to oncogenic signaling, have been shown to drive expression of IL-6, IL-10 and VEGF, which promote a tolerogenic differentiation of bone marrow derived monocytes [101]. BRCA1-associated triple negative breast cancer induces pro-tumorigenic TAMs, by reprogramming glycolysis and SREBP1-mediated lipid metabolism, a metabolic signature associated with resistance to PARP inhibitors [102]. These mechanisms induce a unique differentiation program of TAMs, which lose the normal properties of healthy macrophages and obtain pro-tumorigenic features.
The immunophenotypic and metabolic properties of TAMs were previously streamlined in the simplified concept that TAMs have a differentiation program resembling M2 macrophages, characterized by expression of CD206, CD204, VEGF, CD163 and Arg-1 [103]. This is in contrast to the expression of classical markers of M1 polarized macrophage including CD80, CD86, MHC-II, iNOS and CD68, which correlated with effector function of macrophages and tumoricidal function of TAMs [103]. The M1/M2 programs rely mainly on metabolism, since proinflammatory M1 macrophages are supported by glycolysis, whereas anti-inflammatory M2 macrophages utilize mainly fatty acid oxidation (FAO)[104]. This concept is no further considered appropriate, but such differentiation profiles remain useful in the characterization of TAMs because there is extensive experience regarding the correlation between the expression of such immune marker combinations and prognosis in various cancers [97,105].
Live cell imaging allowing assessment of the spatial interaction of TAMs with other cells of the TME has significantly improved our understanding about the role of TAMs in tumor evolution. In a breast cancer mouse model, multiphoton microscopy studies showed that TAMs interact with mammary cancer cells, facilitating their intravasation and subsequent metastasis. Tumor cells closer to TAMs are more motile and are directed toward TAMs of the perivascular area where they interact with blood vessels and enter the blood stream [106, 107]. Real time imaging provided evidence about the role of perivascular Tie2hi TAMs in promoting transient vascular permeabilization and tumor intravasation, thereby facilitating metastasis [108]. In a model of GBM, intravital 2-photon microscopy showed two distinct TAM subtypes with morphological and functional differences: microglial cells which are large, highly branched and less motile, and bone marrow-derived macrophages which are smaller, less branched and less motile [109]. The differential properties of the spatially distinct subsets of TAMs might form attractive targets for therapeutic intervention.
Beyond the impact of oncogenes in regulating the production of tumor-derived soluble factors that regulate monocyte recruitment and TAM generation, the mutational landscape of tumor cells alter other immune cell types that are recruited in the TME. Colorectal cancer (CRC) serves as a paradigm for this process. Based on gene expression studies, CRC can be classified into four molecular subtypes (CMS1–4) [110]. CRC-CMS1 has DNA mismatch-repair defects, which cause microsatellite instability and hypermutation. These tumors are densely infiltrated by CD4+ and CD8+ T cells with high expression of immune checkpoint inhibitors including CTLA-4, PD-1, and PD-L1 and display favorable responses to checkpoint immunotherapy [111]. In contrast, CRC-CMS4 is characterized by tumor cells with a mesenchymal-like phenotype, an RNA sequencing profile dominated by signatures of the TGF-β pathway, Th17 pathway, monocyte/macrophage infiltration, and resistance to checkpoint immunotherapy [111]. Thus, cancer-specific properties have a significant impact on the quantitative and qualitative inflammatory infiltrate of the TME and in regulating the crosstalk between myeloid cells and the adaptive immune system.
5. Tumor-associated T cells and signaling pathways shaping their function
A critical determinant of tumor containment and progression over time is the number and function of T cells within the TME. For this reason, T cells are the key targets of antibody-based and cell-based immunotherapies in cancer. During the phase of cancer escape from immune control, T cells that can recognize tumor-associated antigens and control tumor growth, lose the ability to mediate this function due to mechanisms related with tumor-induced tolerance and immunosuppression (Fig. 1) [112]. These dysfunctional T cells are characterized by features of T cell exhaustion (TEX) observed in chronic viral infections [113] such as high surface expression of inhibitory receptors CTLA-4, PD-1, TIM-3, LAG-3, loss of expansion ability, and impaired effector function as determined by the diminished production of cytokines such as IFNγ and TNF-α (Fig. 3) [114]. The TEX state might be reversible or irreversible [115] and a central goal of novel immunotherapies is to achieve re-invigoration of tumor-specific TEX cells. The development of TEX state is mediated primarily by T cell-extrinsic factors that lead to persistent T cell activation by tumor antigens [116].
Fig. 3.
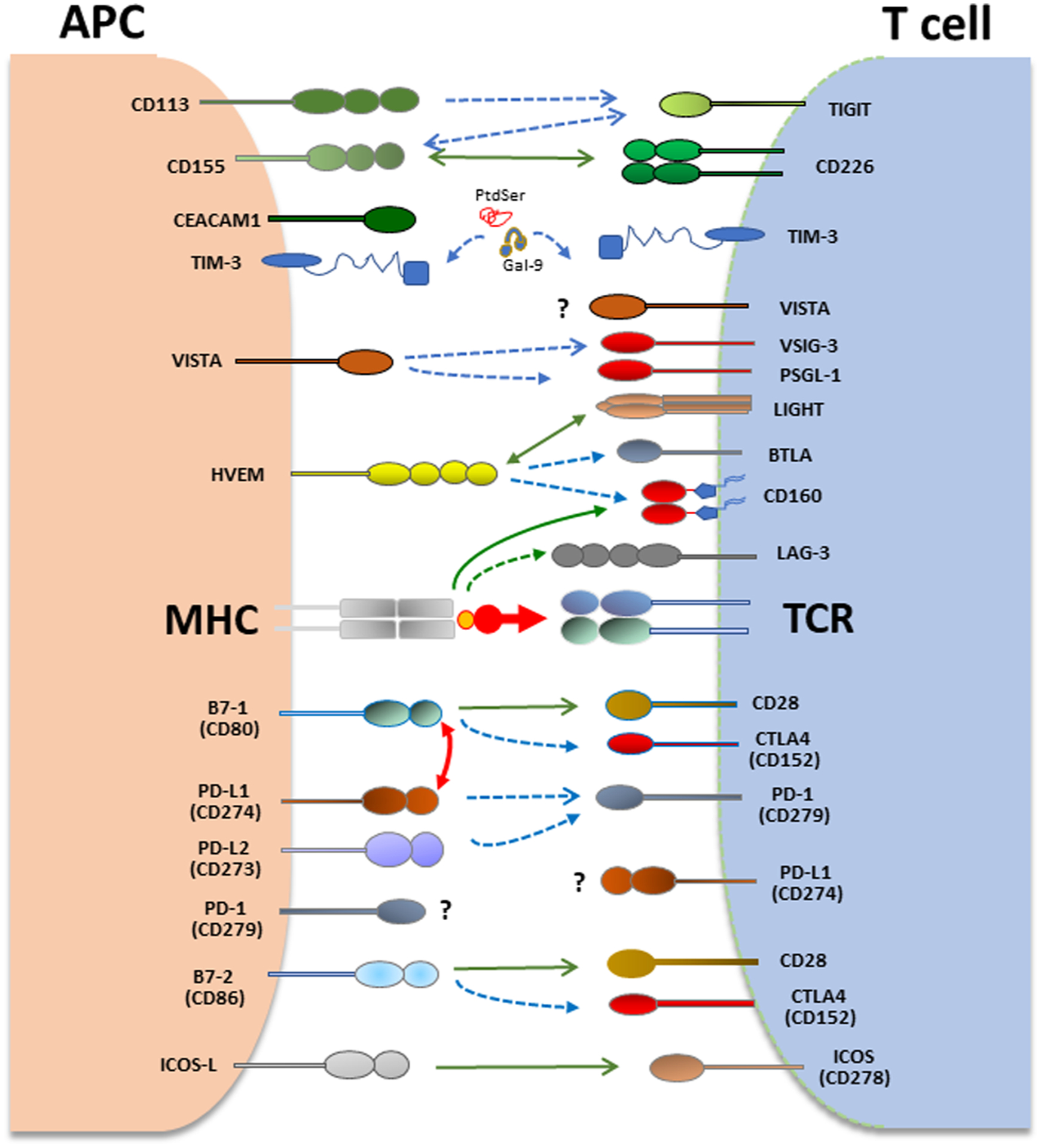
Coinhibitory pathways in T cells and APC. T cell activation is initiated by recognition of antigens presented by antigen-presenting cells (APCs) to the T cell receptor (TCR)/CD3 complex. CD28 is the prototype costimulatory receptor in T cells and interacts with CD80 and CD86. Many coinhibitory receptors are upregulated upon T cell activation and can attenuate TCR and costimulatory signals. CTLA4 and PD-1 are the prototype co-inhibitory receptors expressed in T cells. CTLA-4 interacts with B7–1 (CD80) and B7–2 (CD86) whereas PD-1 (CD279) interacts with PD-L1 (CD274) and PD-L2 (CD273) to inhibit T cell responses. In addition to canonical interaction of PD-L1 with PD-1 in trans, PD-L1 interacts with B7–1 (CD80) in cis, when co-expressed on the same APC leading to diminished availability of PD-L1 for canonical interaction with PD-1 in trans. VISTA, TIM-3 and PD-1 are expressed both in T cells and APC and can regulate immune responses by altering the properties of myeloid cells and T cells.
The co-inhibitory molecules such as PD-1 (CD279) and CTLA-4 (CD152) are induced during physiologic T cell stimulation to tame activation signals. CTLA-4 being upregulated and acting early during T cell activation, is the high affinity receptor of CD80/CD86. CTLA-4 directly competes with CD28 for binding on CD80/CD86, and its binding on CD80/CD86 ligands on APC can result in the depletion of CD80/CD86 by trans-endocytosis [117] while simultaneously releasing free PD-L1 by eliminating availability of CD80 for PD-L1 engagement in PD-L1: CD80 interaction [118]. PD-1 is also induced upon TCR-mediated activation. Engagement of PD-1 by its ligands PD-L1 (B7-H1 or CD274) and/or PD-L2 (B7-DC or B7-H3) counteracts TCR signaling and CD28-mediated co-stimulation [119,120]. PD-1 and CTLA-4 are the prototype inhibitory receptors, known as immune checkpoints, and are the most extensively utilized therapeutic targets in cancer immunotherapy [121].
After engagement with its ligands PD-L1 or PD-L2, but also via tonic signaling [122], PD-1 inhibits T cell functions by recruiting phosphatases, predominantly SHP-2 but also SHP-1, to the immunoreceptor tyrosine-based switch motif (ITSM) and immunoreceptor tyrosine-based inhibitory motif (ITIM), expressed in PD-1 cytoplasmic tail, which antagonize TCR and CD28-mediated activation signals [123–126]. This TCR proximal signaling blockade results in inhibition of key downstream pathways including PI3K/Akt and Ras leading to altered biochemical, transcriptional and metabolic T cell reprograming [127–129]. Constitutive PD-1 expression on T cells induced by persistent antigen stimulation during cancer promotes immune evasion, and blockade of the PD-1 pathway can improve T cell function and reduce tumor burden in multiple experimental tumor models and multiple types of human cancers [130,131].
The expression status of PD-L1 on tumor cells has served as a factor for patient stratification for anti-PD-1 therapy. However, in several clinical studies, there were patients with undetectable PD-L1 on tumors, who also responded to anti-PD-1 therapy [132]. This effect can be explained by the fact that, other cell types in the TME, such as macrophages, DCs, tumor-associated fibroblasts and myeloid cells also express PD-L1, creating an immunosuppressive TME for T cells and supporting tumor cell growth. Therefore, the relative contribution of PD-L1 on tumor cells and other cell types in limiting anti-tumor responses in the TME remains under investigation. PD-L1 expression on tumor cells can locally inhibit CD8+ T cell activation and protect PD-L1+, but not PD-L1− tumors from eradication by the immune system, indicating a critical role for tumor PD-L1 in suppressing antitumor immunity [133]. By genetic deletion of PD-L1 in tumor cells and host mice, a different study showed comparable contribution of PD-L1 on each of these compartments to immune suppression, suggesting that PD-L1 expression in either of these compartments can be predictive of responses to PD-1/PD-L1 blockade therapy [134]. However by using conditional deletion of PD-L1 in tumor cells and dendritic cells-, transplantation chimeras, as well as various approaches of antibody blockade, it was determined by several investigators that PD-L1 expressed in APC, rather than tumor cells, has a causative role in compromising anti-tumor T cell responses and this APC-mediated PD-1 ligation and T cell immunosuppression is the dominant interaction targeted by checkpoint blockade therapy [95,135]. Since DCs mediate CD4+ T cell priming and cross-presentation of tumor antigens to CD8 + T cells, upregulation of PD-L1 on dendritic cells can attenuate cytotoxic T cell activity and compromise antitumor responses [136,137].
Recently, in cis interaction of PD-L1 and B7–1, on the same cell surface of APC, has been reported by several studies [51–54]. The in cis interaction of B7–1 with PD-L1 impairs the interaction of PD-L1 with PD-1 in trans (Fig. 3) resulting in reduced PD-1 signaling (reviewed in [138]). Blockade of the in cis interaction of PD-L1/B7–1 by B7–1-specific antibodies could increase PD-1-mediated T cell suppression and alleviate autoimmunity in several mouse models [139]. Among tumor-infiltrating myeloid cells, PD-L1-expressing DCs have a dominant role in regulating antitumor T cell responses via in cis interaction of PD-L1/B7–1 on DCs and PD-L1/PD-1 interactions between DCs and T cells. These findings suggest a new therapeutic mechanism of PD-L1 checkpoint blockade acting on DCs to induce enhanced antigen presentation for T cell priming and simultaneous clonal expansion [137]. A more comprehensive understanding of these mechanisms will be necessary to optimize PD-1/PD-L1 targeting immunotherapy.
While the PD1/PD-L1 axis has been extensively investigated, other inhibitory pathways are also exploited by tumor cells, contributing to the generation of an immunosuppressive TME and escape of immunosurveillance [140]. VISTA (B7-H5, PD-1 H, Gi24, Dies1, SISP1 and DD1α) has increasingly become a promising target for overcoming resistance to anti-PD-1 immunotherapy [141]. In addition to the ability of VISTA to regulate the immunosuppressive function of MDSC under hypoxia, as mentioned above [78], VISTA can function as an inhibitory regulator of naïve T cells, critical for steady-state maintenance of immune quiescence and peripheral tolerance (Fig. 3) [142]. Notably, VISTA can mitigate pathogenesis and progression of murine lupus by transmitting inhibitory signals on both T cells and myeloid cells [143, 144].
Although few studies have reported VISTA expression on tumor cells [145,146], VISTA expression on intratumoral myeloid cells and stromal cells has been recognized as a potential mediator of acquired resistance to anti-PD-1 and anti-CTLA-4 immunotherapies in patients with metastatic melanoma and pancreatic ductal adenocarcinoma [147–149]. Therapeutic blockade of the VISTA pathway has been limited by its unknown binding partners and their function. A recent study identified VSIG-3 as a putative ligand of VISTA to inhibit T cell activation and cytokine production [150]. While at physiological pH VISTA interacts mainly with VSIG-3, in acidic pH, VISTA serves as a selective ligand for PSGL-1 receptor on T cells suppressing T cell function (Fig. 3) [151]. This unique pH-dependent VISTA/PSGL-1 interaction suggests that engineering pH-sensitive antibodies might enable selective targeting of VISTA within the acidic TME to reverse cancer-mediated immune suppression without compromising self-tolerance and inducing autoimmunity [151]. Thus, targeting the VISTA pathway can be achieved in a context-dependent manner to overcome the immunosuppressive TME and reinvigorate responses of tumor-specific T cells.
5.1. T regulatory cells of the TME and implications in cancer immunotherapy
It has been previously established that inhibition of Akt and its downstream target mTOR is required to induce Treg differentiation and sustain Treg suppressor function [152,153]. PD-1 ligation inhibits Akt activation [127] and synergizes with TGF-β to induce FoxP3+ iTreg cells with potent suppressive capacity [154]. Moreover, PD-1 promotes fatty acid oxidation [128], a metabolic program that supports the differentiation, survival and function of Treg cells [155]. Based on these, one would anticipate that PD-1 promotes Treg generation and suppressor function. However, recent studies revealed an unexpected connection between PD-1 expression in Treg cells and the outcome of PD-1 pathway blockade [156]. In humans, FoxP3+CD4+ Treg cells represent a heterogenous population that can be subdivided into three functionally and phenotypically distinct subsets [157]. Thymus derived naïve Tregs (nTreg: CD45RA+ FOXP3lo) and activated effector-Tregs (eTreg: CD45RA− FOXP3hi) are highly suppressive while the CD45RA− FOXP3lo Treg cells represent a non-Treg population with a potential of inflammatory cytokine production. Among these Treg subsets, FoxP3+ Tregs infiltrating the TME in most cancers are predominantly the eTreg cells [158]. Expression of PD-1 was found in eTreg cells and the frequency of PD-1+ eTreg cells was higher in tumor samples from patients who did not respond to PD-1-blocking immunotherapy. Notably, blockade of the PD-1: PD-L1 pathway induced activation of eTreg in TILs, as determined by upregulation of CTLA-4, GITR and ICOS and these Treg obtained a more potent suppressor function. These results highlight a previously unappreciated role of PD-1 signaling in Tregs and suggest that PD-1 blockade enhances the suppressive activity of Tregs that express high levels of PD-1. While blocking PD-1 in PD-1+ CD8+ T cells converts them to CD8+ Teff cells that have potent effector function leading to tumor regression, blocking PD-1 in PD-1+ Treg converts them to activated eTreg that have potent suppressor function leading to tumor progression. Thus, PD-1 expression balance between CD8+ Teff cells and eTreg in the TME that might predict the clinical efficacy of PD-1 blocking immunotherapy.
6. T cell differentiation and cancer immunosurveillance
CD8+ T cells play an important role in the adaptive immune response to intracellular pathogens and cancer [159]. Therapeutic responses to PD-1 checkpoint immunotherapy correlate with expansion of CD8+ memory T cells in mouse tumor models and patients [160]. Immunometabolic programs have a causative role in T cell differentiation and their immune function [161,162]. This is particularly important in the metabolically stressful TME where nutrient competition, hypoxia, excess ROS and metabolic byproducts of cancer cells create uniquely hostile conditions under which T cell activation by and cytotoxic function should be operated [163–165]. Although glycolysis has been intimately linked to effector function [166,167], augmenting glycolytic flux drives CD8+ T cells toward a terminally differentiated state, while its inhibition preserves the formation of long-lived TM CD8+ T cells [168]. Thus, multiple signaling pathways and mediators of metabolic reprogramming, such as mTOR, SREBP, Myc [169–172], have important roles in immune-mediated tumor control by guiding differentiation and function of T cells in the context of cancer [172–175]. After stimulation with antigen, CD8+ naive T cells expand and differentiate into TEFF cells and distinct TM cell subsets, including TSCM, TCM, and TEM [176]. Preclinical studies using adoptive transfer of purified CD8+ T cell populations revealed that less-differentiated T cells with features of TSCM and TCM mediate enhanced antitumor and antiviral responses compared with more-differentiated TEM and TEFF cells [177–179]. Preservation of TSCM and TCM cells with a quiescent phenotype and increased proliferative and survival capacities enhance T cell ability to maintain sustained anti-tumor control [180]. For example, Wnt signaling prevents TEFF differentiation, while promoting the generation of TSCM that maintain stemness and pluripotency and display long-lasting potent anti-tumor properties [179].
7. Tissue resident memory cells (TRM) are novel regulators of anti-tumor immunity
Memory T cells have been classically divided into two subsets [181]. TEM cells create immediate effector function to inflamed tissues whereas central memory T cells (TCM) accumulate in secondary lymphoid organs and generate protective immunity [181,182]. Recently, a distinct T memory cell population, named tissue-resident memory T cells (TRM) were identified and classified by their unique phenotype and properties. These cells reside permanently in peripheral non-lymphoid tissues and provide a protective effect against infections and cancer [183,184]. TRM cells have been identified and investigated both in mice and humans in many tissues including skin, gut, brain and liver [185–187]. Although TRM cells were defined as either CD4+ or CD8+, only CD8+ TRM play a major role in cancer immunosurveillance and tumor prognosis [188–190]. Traditionally, CD8+ TRM are defined by the co-expression of CD103, CD69 and/or CD49 [191], and downregulation of receptors that promote T cell recirculation including S1pr1, CD62L (L-selectin) and CCR7 [192–194]. CD103 integrin, formed by αE (CD103) and β7 subunits, promotes TRM cells retention and homing to epithelial tissue and tumors [195–197]. TRM cells have an established role in protective immunity in bacterial, viral infections and autoimmune diseases, including Listeria monocytogenes and Herpes Simplex infections and vitiligo.
An important role of TRM in cancer is currently evolving. In a model of melanoma-associated vitiligo (MAV), using adoptive pmel cells which have a TCR that can detect melanoma gp100 antigen, it was shown that adoptively transferred pmel T cells acquired a TRM immunological profile with a CD103+CD69+CD62Llo phenotype in the skin, draining lymph nodes, lung, and liver [198]. In this context, TRM cells had a protective function against metastasis and expressed transcripts different from TCM cells, characterized by features of effector function and lipid metabolism. In a mouse model of epicutaneous melanoma, it was also determined that TRM have a role in promoting melanoma immune equilibrium [189]. Mice that did not develop melanoma (non-developer group) had higher numbers of CD69+CD103+ TRM cells than tumor-developer mice. In addition, intact skin from the non-developer group, peritumoral area, and tumoral area showed higher number of TRM cells. Notably, both CD69 and CD103 knock-out mice are more susceptible to melanoma formation than wild type counterparts, indicating the causative role of CD69 and CD103 proteins in regulating TRM development and anti-tumor function [189]. Other studies have shown that CD103 + TRM cells can protect against melanoma re-exposure providing evidence for the long-lasting ability of TRM to provide cancer immunosurveilance [199]. In many types of cancer, CD8 + TRM cells display robust production of cytokines such as granzyme B, granzyme A, perforin, and IFN-γ [198,200–203]. Importantly, recent work, using a VHL deficiency mouse model, provided evidence for a link between HIF-1α and CD103 expression and showed that HIF-1α expression supported enhanced differentiation of TRM cells with cytotoxic function [200,203]. These findings link HIF-1α, a key signature molecule of the TME, with TRM differentiation and function and underline the direct relevance of TRM in anti-tumor immunity.
TRM cells express checkpoint receptors, and might serve as markers for prognosis prediction In melanoma patients, TRM express PD-1, TIM-3, and PD-L1 at higher levels than peripheral blood T cells. Single-cell RNA sequencing of T cells isolated from human breast cancer demonstrated a high number of TRM cells with a high level of TIM-3, PD-1, CTLA-4, TIGIT, and LAG-3 expression [190]. In lung cancer, PD-1, TIM − 3, and CD137 are highly expressed [202,204]. After ex vivo pharmacologic stimulation of PD-1+CD103+ CD8 TILs from ovarian cancer, these TRM-like cells highly expressed TIM-3, CTLA-4, and LAG-3 [205, 206]. In lung cancer patients, after anti-PD-1 treatment, TRM cells upregulated PD-1, CTLA-4, TIM-3, TIGIT, and CD39 [207], whereas in melanoma patients PD-1 checkpoint immunotherapy resulted in TRM-specific upregulation of PD-1 and LAG-3 [208]. Notably, the number of TRM cells is correlated with a better response to PD-1 checkpoint immunotherapy and a favorable prognosis in multiple cancers including melanoma, breast cancer, lung cancer, urothelial cancer, ovarian cancer and cervical cancer [205,208–215]. The unravelling role of TRM cells in cancer evolution and response to immunotherapy suggest that TRM might be a promising novel target for therapeutic exploitation in cancer.
8. Innate lymphoid cells and their role in cancer immunotherapy
Innate lymphoid cells (ILCs), which lack antigen-specific receptors, are considered as the innate equivalents of T cells. The most recent nomenclature classifies ILCs into five groups, namely ILC1s, ILC2s, ILC3s which are the equivalent of Th1, Th2, and Th17 CD4+ T helper (Th) cells, respectively, natural killer (NK) cells, which are the equivalent of CD8+ cytotoxic T cells, and lymphoid tissue-inducer cells (LTis), a unique subset involved in the development of secondary lymphoid organs [216]. With the exception of the highly mobile and constantly circulating NK cells, ILCs are largely tissue-resident cells. Besides their main role of immune surveillance and tissue homeostasis, ILCs have emerged as critical players in cancer growth and therapy [217]. ILCs express a plethora of activating and inhibitory receptors [218]. Among the latter, checkpoint inhibitory receptors increasingly raise attention due to their role in tumor immunotherapy. Here, we summarize the current knowledge on the role of checkpoint inhibitory receptors in ILCs in the context of cancer.
8.1. Natural Killer (NK) cells
NK cells belong to a highly diverse subset of ILCs that circulate between peripheral organs, exerting cytotoxic activity that can directly eliminate cancer cells or cells undergoing microbial infections [219–221]. In healthy adults, NK cells comprise about 5–15% of circulating lymphocytes with the majority of healthy tissues mainly consisting of the mature highly cytolytic CD56dim NK cell subset, while the immature and poorly cytolytic CD56bright subset, although also present in healthy tissues, it becomes significantly enriched in cancer [222]. A plethora of activating and inhibitory receptors regulate NK cell development and function, with the dominant signal being inhibitory, resulting primarily from the interactions of certain NK cell inhibitory receptors with major histocompatibility complex class I (MHC-I) molecules [223,224]. These “classical” NK inhibitory receptorsinclude the group of Killer Ig-like receptors (KIRs) in humans and the LY49 group of receptors in rodents, as well as the CD94/NKG2A receptor (Fig. 4A), which also recognizes HLA-E in humans and Qa-1b in mice [218, 223–228]. According to the “missing self” hypothesis [229], these inhibitory receptors have the unique ability to prevent NK cell responses against self, and allow NK cells to exert their cytotoxic activity against cells that have reduced or absent expression of MHC-I molecules. However, in the context of cancer, these NK inhibitory receptors hamper NK cell responses and contribute to tumor cell escape from elimination. Blocking antibodies, usually termed “immune checkpoint inhibitors” against KIRs (lirilumab) [230] and NKG2A (monalizumab) [231], which aim to improve NK cell function, are in phase I/II clinical trials against a range of hematologic malignancies and solid tumors either as mono- or combination therapy [232,233]. In addition, adoptive transfer of NK cells engineered to downregulate NKG2A in immunodeficient mice, efficiently prevented HLA-E+ tumor-mediated suppression leading to increased anti-tumor activity, suggesting this as a promising approach to be tested in a clinical setting [234].
Fig. 4.
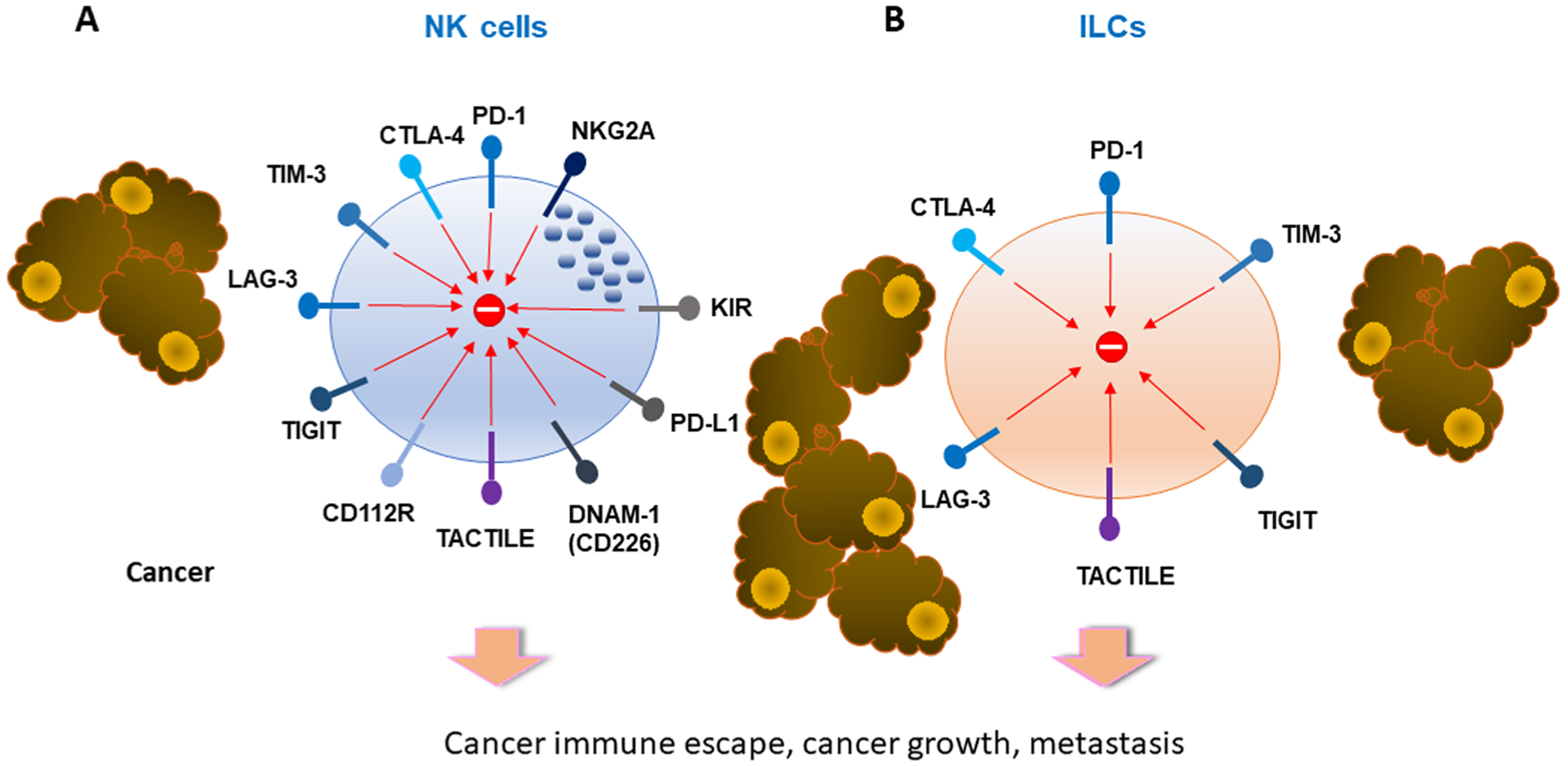
NK cells and ILCs express multiple inhibitory receptors. NK cells express the classical inhibitory receptors KIR and NKG2A. Both NK cells and ILC express multiple checkpoint inhibitors. These receptors and their ligands offer new therapeutic opportunities to improve NK cell functions for cancer immunotherapy.
Besides the classical inhibitory receptors, NK cells also express checkpoint inhibitory receptors (Fig. 4A). Expression of programmed cell death 1 (PD-1) was found on NK cells from healthy individuals correlating with prior HCMV infection [235]. Notably, PD-1 expression has been found to be restricted to the fully mature CD56dim or to the CD56neg NK compartment and not to the immature CD56bright NK cell subset [236]. PD-1 is also expressed on NK cells in the context of cancer. PD-1+ NK cells were detected in the peripheral blood of patients with multiple myeloma, Kaposi sarcoma and lung cancer patients [237–239], in pleural effusions from primary mesothelioma or metastatic adenocarcinoma patients [240], in the peritoneal fluid of high-grade peritoneal carcinomatosis patients [241], and in the peripheral blood and ascitic fluid of ovarian carcinoma patients, with both mRNA and protein PD-1 levels to be higher in the CD56dim NK cell subset [242]. In contrast, PD-1 expression was found to be higher in CD56bright than CD56dim mature NK cells from blood and tissues from patients with Hodgkin lymphoma [243]. As in T cells, blockade of the PD-1/PD-L1 axis in NK cells could improve anti-tumor activity [243,244]. Interestingly, glucocorticoids present in the TME were found to be indispensable for PD-1 induction on human NK cells, particularly when combined with the inflammatory cytokines interleukin (IL)− 12, IL-15 and IL-18, which are abundant at the tumor site [245]. In addition, expression of the PD-1 ligand, PD-L1, on NK cells has been associated with a regulatory function, limiting DC-mediated tumor antigen cross-presentation to CD8+ cells and resulting in impaired memory responses [246].
The inhibitory receptor cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) has also been detected on NK cells from mice [247], in the blood from healthy donors and in the blood and tissues from cancer patients [248,249]. Similarly to PD-1, CTLA-4 was found to be predominantly expressed on CD56dim NK cells from the blood of healthy individuals and was associated with decreased NK cell functionality [249]. These findings explain why CTLA-4 blockade, besides T cells, was also found to favor NK cell infiltration and antitumor function [250]. Of note, treatment with anti-CTLA-4 antibody could favor NK cell anti-tumor function indirectly, by depleting CTLA-4-expressing intratumoral regulatory T cells (Treg) [247].
Other checkpoint inhibitory receptors such as TIM-3, LAG-3, TIGIT), CD96 (TACTILE) have also been identified in NK cells (Fig. 4A). Although mostly considered as an inhibitory receptor [251,252], TIM-3 has also been reported to have an activation role in NK cells [253]. Higher TIM-3 expression has been observed on the CD56dim NK cell subset, while on the CD56bright cell subset it may be upregulated upon cytokine stimulation [251]. In the context of cancer, TIM-3 has been detected in NK cells from patients with advanced melanoma [254,255], gastric cancer [256], lung adenocarcinoma [257], and bladder cancer [258]. Increased TIM-3 expression was found in both CD56dim and CD56bright NK cell subsets, correlating with shorter overall survival [257]. Conversely, blockade of TIM-3 signals with TIM-3-specific antibodies improved NK cell functions in vitro [254,257].
LAG-3 acts synergistically with PD-1 and contributes to T cell and NK cell dysfunction, exhaustion [259] and tumor escape [260]. Several years of efforts testing LAG-3 as promising checkpoint blockade therapy recently led to the first approval of LAG-3 blocking antibody (relatlimab-rmbw) for combination therapy together with PD-1 antibody nivolumab (Opdualag) for the treatment of patients with unresectable or metastatic melanoma. An alternative approach used a LAG-3-Ig fusion protein (IMP321) as a soluble high affinity MHC-II agonist to enhance antigen presenting function. Treatment with IMP321 as monotherapy enhanced IFN-γ and TNF-α production in NK cells from control donors and cancer patients ex vivo [261], and was also shown to increase the number and activation of NK cells in the blood of breast cancer patients in combination with standard chemotherapy [262].
A separate group of receptors, also referred to as novel NK cell immune checkpoints, includes the inhibitory receptors TIGIT and CD112R [263] as well as the costimulatory receptor CD226 (DNAM-1) [264], and CD96 (TACTILE) for which both inhibitory and costimulatory functions have been reported [265,266]. These receptors interact with molecules of the nectin family CD155 (PVR) and CD112 (Nectin-2) (Fig. 4A) [267], two ligands with ubiquitous low level expression, often up-regulated in cancer cells [268,269] and myeloid suppressor cells within the TME [270,271]. TIGIT is expressed by activated T cells and NK cells and binds its ligands CD155 and CD112 expressed on antigen presenting cells (APC) [272]. Expression of these ligands by several cancer types results in T cell and NK cell exhaustion, impaired tumor immune infiltration and compromised antitumor function [273–275]. The TIGIT pathway and its related receptors and ligands offer new options for antibody-based blocking strategies, which have shown to improve tumor control in mice particularly when combined with PD-1/PD-L1 or CTLA-4 blockade [276].
8.2. Helper ILCs
Recent studies suggest that checkpoint receptors are involved in the immune responses of helper ILC subsets and have important implications in anti-tumor immunity (Fig. 4B). However, little information is currently available regarding the mechanisms by which these receptors regulate helper ILC functions. Although not detected in helper ILCs from healthy human tissue samples [277], PD-1 is expressed on tumor-associated helper ILC populations [278]. It was recently reported that helper ILC subsets are present in human malignant pleural effusions, with a notable PD-1 expression level on the ILC3 subset [240]. Another study, using single-cell RNA sequencing and a CRC mouse model associated with AOM/DSS-induced colitis, identified high-PD-1-expressing ILC2 infiltrates and demonstrated the importance of this specific subset in tumor progression [279]. Furthermore, PD-1+ ILC2 cells were present in most human PDAC tissues as well as in orthotopic PDAC tumors in mice, in which PD-1 blockade, resulted in ILC2 expansion and improved antitumor immunity [280]. These findings suggest the potential role of ILC2 cells as tissue-specific enhancers of cancer immunity that may amplify the efficacy of anti-PD-1 immunotherapy.
Similarly to PD-1, CTLA-4 expression was found to be very low in helper ILCs from healthy donors [281], but is increased in helper ILC subsets within tumor tissues [278]. High CTLA-4 expression has also been reported on intratumoral ILC1 subsets in mice [282]. Although TIM-3 has been detected on ILC3s from human decidua during pregnancy and inhibiting IL-22 production [283], currently very limited information is known regarding TIM-3 expression on helper ILCs in the context of cancer. Transcriptomic and immunophenotyping analyses in mouse and human cSCCs identified infiltration of functionally impaired NK and ILC1 cells characterized by reduced cytotoxicity and IFN-γ secretion, which correlated with decreased expression of activating receptors and increased expression of exhaustion markers including TIGIT on NK cells, and PD-1 and TIM-3 on ILC1s [284]. LAG-3 expression has not been reported in resting ILCs, but in the context of cancer and upon TGF-β-mediated conversion of NK to ILC1 cells, several checkpoint receptors were upregulated, among which LAG-3, TIGIT and CD96 expression has been documented [282]. The functional effects of blocking these receptors on helper ILCs remain to be determined.
9. Concluding remarks
Profiling the immune cells in the TME of patients with evolving techniques has revealed significant information regarding the immune cell composition of the TME in experimental animal models and patients with various types of cancer. It is increasingly understood that the immune cellular components of the TME, including DCs, TAMs and MDSCs have decisive roles in regulating cancer evolution and immune escape but also the outcomes of cancer therapy including chemotherapy, checkpoint immunotherapy and cancer vaccines. Although in healthy tissues these myeloid cells provide defense against insults mediated by pathogens, in the TME these cells lose their protective immune functions and convert to pro-tumorigenic mediators that support cancer growth and metastasis. Via such changes, myeloid cell populations fail to recruit T cells, present tumor antigens and mediate anti-tumor T cell responses. Instead, these myeloid cells suppress T cell activation by blunting recognition of tumor antigens, eliminating engagement of costimulatory pathways, upregulating the expression of inhibitory receptors and their ligands, and producing inhibitory soluble mediators thereby creating an immunosuppressive milieu. Engagement of inhibitory receptors on T cells of the TME impairs signaling events mediated by T cell receptor and costimulatory pathways and promotes generation of TEX tumor-specific cells that are unable to mount anti-tumor responses. The TME also promotes differentiation of iTreg by T cell receptor engagement by tumor antigens and concomitant ligation of coinhibitory receptors, and production of Treg-promoting soluble factors such as TGF-β and IL-10. Together these mechanisms abrogate cancer immunosurveillance and support cancer evolution and immune escape. Although checkpoint blockade and cell-based therapies have achieved significant progress, challenges ahead include advancing the outcome of immunotherapy by targeting changes of the TME that compromise the function of key immune populations and preclude their ability to recognize and respond to tumor antigens. Such tentative targets for intervention include metabolic alterations, vasculature abnormalities and soluble inhibitors produced in the TME. Such efforts might repurpose available drugs previously used in the clinic, which can be administered together or sequentially with immunomodulating immunotherapies and cell-based therapies. In doing so, an additional challenge will be to achieve cancer elimination while preserving self-tolerance and preventing autoimmunity.
Acknowledgments
This work was supported by NIH grants R01CA238263, R01CA229784, R01CA212605 (VAB); and grants JDRF (International 1-INO-2002–1119-A-N) and Sanofi (iAwards SRA 2020–0033) (NP).
Abbreviations:
- HLA
human leukocyte antigens
- IDO
indoleamine 2,3-dioxygenase
- VEGF
vascular endothelial growth factor
- TGF-β
transforming growth factor-β
- PD-1
programmed cell death protein 1
- CTLA-4
cytotoxic T-lymphocyte-associated protein 4
- RORγt
retinoic acid-related orphan receptor γ
- FLT3L
FMS-like tyrosine kinase 3 ligand
- IRF4
interferon regulatory factor 4
- CXCL9
CXC chemokine ligand 9
- CXCL10
CXC chemokine ligand 10
- CXCL11
CXC chemokine ligand 11
- CCL2
C-C motif chemokine ligand 2
- CCL5
C–C motif chemokine ligand 5
- XCL1
X-C motif chemokine ligand 1
- COX1
cyclooxygenase 1
- COX2
cyclooxygenase 2
- CXCR3
CXC chemokine receptor 3
- G-CSF
granulocyte colony-stimulating factor
- PGE2
prostaglandin E2
- CXCL1
CXC motif chemokine ligand 1
- TNFα
tumor necrosis factor alpha
- NF-κB
nuclear factor-kappaB
- SHP-2
Src homology domain-containing phosphatase-2
- TIM-3
T-cell immunoglobulin mucin-3
- TLR3
Toll-like receptor 3
- TLR7
Toll-like receptor 7
- TLR9
Toll-like receptor 9
- TIM-4
T-cell immunoglobulin mucin-4
- IRF8
interferon regulatory factor 8
- Th17
T helper 17 cell
- MIP-1b
macrophage inflammatory protein-1β
- CCL4
C–C motif chemokine ligand 4
- RANTES
regulated on activation, normal T cell expressed and secreted
- IFN-α
interferon alpha
- ICOS-L
inducible costimulator ligand
- AML
acute myeloid leukemia
- VISTA
V-domain Ig suppressor of T-cell activation
- mPGES1
microsomal prostaglandin E synthase-1
- pSTAT1-IRF1
phosphorylated signal transducer and activator of transcription 1- interferon regulatory factor 1
- HIF-1a
hypoxia-inducible factor 1
- CXCL12
CXC chemokine ligand 12
- B7-H4
B7 homolog 4
- SREBP1
sterol regulatory element-binding protein-1
- PARP
poly adenosine diphosphate-ribose polymerase
- Arg-1
arginase 1
- iNOS
inducible nitric oxide synthase
- GBM
glioblastoma multiform
- LAG-3
lymphocyte activation gene-3
- VSIG-3
V-set and immunoglobulin domain containing 3
- PSGL-1
P-selectin glycoprotein ligand-1
- GITR
glucocorticoid-induced tumor necrosis factor receptor family-related receptor
- mTOR
mammalian target of rapamycin
- CCR7
C-C chemokine receptor type 7
- VHL
von Hippel Lindau
- TIGIT
T cell immunoreceptor with immunoglobulin and ITIM domain
- HCMV
human cytomegalovirus
- AOM/DSS
azoxymethane/dextran sodium sulfate
- PDAC
pancreatic ductal adenocarcinoma
- cSCCs
cutaneous squamous cell carcinomas
Footnotes
CRediT authorship contribution statement
SY, KA, QW, AC, RS wrote the main sections of the manuscript. NP generated sections of the manuscript and prepared figures. VAB generated sections of the manuscript, prepared figures, guided the co-authors and was responsible for the organization of the document.
Conflict of interest
VAB has patents on the PD-1 pathway licensed by Bristol-Myers Squibb, Roche, Merck, EMD-Serono, Boehringer Ingelheim, AstraZeneca, Novartis, and Dako. The authors declare no other competing interests.
References
- [1].Schreiber RD, Old LJ, Smyth MJ, Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion, Science 331 (6024) (2011) 1565–1570. [DOI] [PubMed] [Google Scholar]
- [2].Lecoultre M, Dutoit V, Walker PR, Phagocytic function of tumor-associated macrophages as a key determinant of tumor progression control: a review, J. Immunother. Cancer 8 (2020) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hirayama D, Iida T, Nakase H, The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis, Int J. Mol. Sci 19 (1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schumacher TN, Scheper W, Kvistborg P, Cancer neoantigens, Annu Rev. Immunol 37 (2019) 173–200. [DOI] [PubMed] [Google Scholar]
- [5].Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer, Science 348 (6230) (2015) 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Snyder A, et al. , Genetic basis for clinical response to CTLA-4 blockade in melanoma, New Engl. J. Med 371 (23) (2014) 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bindea G, et al. , Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer, Immunity 39 (4) (2013) 782–795. [DOI] [PubMed] [Google Scholar]
- [8].Herbst RS, et al. , Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients, Nature 515 (7528) (2014) 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Galon J, et al. , Type, density, and location of immune cells within human colorectal tumors predict clinical outcome, Science 313 (5795) (2006) 1960–1964. [DOI] [PubMed] [Google Scholar]
- [10].Hahn WC, et al. , An expanded universe of cancer targets, Cell 184 (5) (2021) 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Spranger S, et al. , Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells, Sci. Transl. Med 5 (200) (2013) 200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rooney MS, et al. , Molecular and genetic properties of tumors associated with local immune cytolytic activity, Cell 160 (1–2) (2015) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Binnewies M, et al. , Understanding the tumor immune microenvironment (TIME) for effective therapy, Nat. Med 24 (5) (2018) 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Murphy TL, Murphy KM, Dendritic cells in cancer immunology, Cell Mol. Immunol 19 (1) (2022) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Broz M, et al. , Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity, Cancer Cell 26 (5) (2014) 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Amon L, et al. , The ontogenetic path of human dendritic cells, Mol. Immunol 120 (2020) 122–129. [DOI] [PubMed] [Google Scholar]
- [17].Collin M, Bigley V, Human dendritic cell subsets: an update, Immunology 154 (1) (2018) 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hildner K, et al. , Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity, Science 322 (5904) (2008) 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Leal Rojas I, et al. , Human blood CD1c+ dendritic cells promote Th1 and Th17 effector function in memory CD4+ T Cells, Front Immunol. 8 (2017) 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Villani A, et al. , Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors, Science 356 (6335) (2017) eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brown C, et al. , Transcriptional basis of mouse and human dendritic cell heterogeneity, Cell 179 (4) (2019) 846–863.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kvedaraite E, Ginhoux F, Human dendritic cells in cancer, Sci. Immunol 7 (70) (2022) eabm9409. [DOI] [PubMed] [Google Scholar]
- [23].Cytlak U, et al. , Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans, Immunity 53 (2) (2020) 353–370.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bourdely P, et al. , Transcriptional and functional analysis of CD1c+ human dendritic cells identifies a CD163+ subset priming CD8+CD103+ T Cells, Immunity 53 (2) (2020) 335–352.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Caux C, et al. , GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells, Nature 360 (6401) (1992) 258–261. [DOI] [PubMed] [Google Scholar]
- [26].Sallusto F, Lanzavecchia A, Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha, J. Exp. Med 179 (4) (1994) 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lehtonen A, et al. , Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages, J. Immunol 175 (10) (2005) 6570–6579. [DOI] [PubMed] [Google Scholar]
- [28].Geissmann F, et al. , Development of monocytes, macrophages, and dendritic cells, Science 327 (5966) (2010) 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nagarsheth N, Wicha M, Zou W, Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy, Nat. Rev. Immunol 17 (9) (2017) 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Böttcher J, et al. , NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control, Cell 172 (5) (2018) 1022–1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zelenay S, et al. , Cyclooxygenase-dependent tumor growth through evasion of immunity, Cell 162 (6) (2015) 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Barry K, et al. , A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments, Nat. Med 24 (8) (2018) 1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shrikant P, Mescher M, Control of syngeneic tumor growth by activation of CD8 + T cells: efficacy is limited by migration away from the site and induction of nonresponsiveness, J. Immunol 162 (5) (1999) 2858–2866. [PubMed] [Google Scholar]
- [34].Asano K, et al. , CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens, Immunity 34 (1) (2011) 85–95. [DOI] [PubMed] [Google Scholar]
- [35].Salmon H, et al. , Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition, Immunity 44 (4) (2016) 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Roberts E, et al. , Critical role for CD103(+)/CD141(+) dendritic cells bearing ccr7 for tumor antigen trafficking and priming of T cell immunity in melanoma, Cancer Cell 30 (2) (2016) 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mikucki M, et al. , Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints, Nat. Commun 6 (2015) 7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kurachi M, et al. , Chemokine receptor CXCR3 facilitates CD8(+) T cell differentiation into short-lived effector cells leading to memory degeneration, J. Exp. Med 208 (8) (2011) 1605–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Spranger S, et al. , Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy, Cancer Cell 31 (5) (2017) 711–723. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hanahan D, Hallmarks of cancer: new dimensions, Cancer Discov. 12 (1) (2022) 31–46. [DOI] [PubMed] [Google Scholar]
- [41].Meyer M, et al. , Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance, Nat. Commun 9 (1) (2018) 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ruffell B, et al. , Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells, Cancer Cell 26 (5) (2014) 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Herber D, et al. , Lipid accumulation and dendritic cell dysfunction in cancer, Nat. Med 16 (8) (2010) 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kobie J, et al. , Transforming growth factor beta inhibits the antigen-presenting functions and antitumor activity of dendritic cell vaccines, Cancer Res 63 (8) (2003) 1860–1864. [PubMed] [Google Scholar]
- [45].Lim T, et al. , PD-1 expression on dendritic cells suppresses CD8+ T cell function and antitumor immunity, Oncoimmunology 5 (3) (2016), e1085146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Boussiotis V, Molecular and biochemical aspects of the PD-1 checkpoint pathway, New Engl. J. Med 375 (18) (2016) 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhao X, et al. , Phosphorylation and stabilization of PD-L1 by CK2 suppresses dendritic cell function, Cancer Res (2022) p. canres.2300.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gapany M, et al. , Association of elevated protein kinase CK2 activity with aggressive behavior of squamous cell carcinoma of the head and neck, Mol. Med 1 (6) (1995) 659–666. [PMC free article] [PubMed] [Google Scholar]
- [49].Lau J, et al. , Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice, Nat. Commun 8 (2017) 14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Oh S, et al. , PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer, Nat. Cancer 1 (7) (2020) 681–691. [DOI] [PubMed] [Google Scholar]
- [51].Chaudhri A, et al. , PD-L1 Binds to B7–1 Only In Cis on the Same Cell Surface, Cancer Immunol. Res 6 (8) (2018) 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Haile S, et al. , Tumor cell programmed death ligand 1-mediated T cell suppression is overcome by coexpression of CD80, J. Immunol 186 (12) (2011) 6822–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhao Y, et al. , Antigen-presenting cell-intrinsic PD-1 neutralizes PD-L1 in cis to attenuate PD-1 signaling in T cells, Cell Rep. 24 (2) (2018) 379–390.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sugiura D, et al. , Restriction of PD-1 function by cis-PD-L1/CD80 interactions is required for optimal T cell responses, Science 364 (6440) (2019) 558–566. [DOI] [PubMed] [Google Scholar]
- [55].Mayoux M, et al. , Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy, Sci. Transl. Med 12 (534) (2020) eaav7431. [DOI] [PubMed] [Google Scholar]
- [56].Karyampudi L, et al. , PD-1 blunts the function of ovarian tumor-infiltrating dendritic cells by inactivating NF-κB, Cancer Res 76 (2) (2016) 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chiba S, et al. , Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1, Nat. Immunol 13 (9) (2012) 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].de Mingo Pulido Á, et al. , TIM-3 regulates CD103+ dendritic cell function and response to chemotherapy in breast cancer, Cancer Cell 33 (1) (2018) 60–74.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dixon K, et al. , TIM-3 restrains anti-tumour immunity by regulating inflammasome activation, Nature 595 (7865) (2021) 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Caronni N, et al. , TIM4 expression by dendritic cells mediates uptake of tumor-associated antigens and anti-tumor responses, Nat. Commun 12 (1) (2021) 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Anderson D, et al. , Genetic models of human and mouse dendritic cell development and function, Nat. Rev. Immunol 21 (2) (2021) 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Amon L, et al. , Transcriptional control of dendritic cell development and functions, Int. Rev. Cell Mol. Biol 349 (2019) 55–151. [DOI] [PubMed] [Google Scholar]
- [63].Binnewies M, et al. , Unleashing type-2 dendritic cells to drive protective antitumor CD4+ T cell immunity, Cell 177 (3) (2019) 556–571.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mittag D, et al. , Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status, J. Immunol 186 (11) (2011) 6207–6217. [DOI] [PubMed] [Google Scholar]
- [65].Segura E, et al. , Human inflammatory dendritic cells induce Th17 cell differentiation, Immunity 38 (2) (2013) 336–348. [DOI] [PubMed] [Google Scholar]
- [66].Wu X, et al. , Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells, J. Exp. Med 213 (12) (2016) 2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Laoui D, et al. , The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity, Nat. Commun 7 (2016) 13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kuhn S, Yang J, Ronchese F, Monocyte-derived dendritic cells are essential for CD8(+) T cell activation and antitumor responses after local immunotherapy, Front Immunol. 6 (2015) 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Treilleux I, et al. , Dendritic cell infiltration and prognosis of early stage breast cancer, Clin. Cancer Res 10 (22) (2004) 7466–7474. [DOI] [PubMed] [Google Scholar]
- [70].Labidi-Galy S, et al. , Quantitative and functional alterations of plasmacytoid dendritic cells contribute to immune tolerance in ovarian cancer, Cancer Res 71 (16) (2011) 5423–5434. [DOI] [PubMed] [Google Scholar]
- [71].Bruchhage K, et al. , IL-10 in the microenvironment of HNSCC inhibits the CpG ODN induced IFN-α secretion of pDCs, Oncol. Lett 15 (3) (2018) 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Conrad C, et al. , Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells, Cancer Res 72 (20) (2012) 5240–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Veglia F, Sanseviero E, Gabrilovich DI, Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity, Nat. Rev. Immunol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bronte V, et al. , Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards, Nat. Commun 7 (2016) 12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang L, et al. , VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML, Oncoimmunology 7 (9) (2018), e1469594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang L, et al. , VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses, J. Exp. Med 208 (3) (2011) 577–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Flies DB, et al. , Cutting edge: a monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models, J. Immunol 187 (4) (2011) 1537–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Deng J, et al. , Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment, Cancer Immunol. Res 7 (7) (2019) 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Le Mercier I, et al. , VISTA regulates the development of protective antitumor immunity, Cancer Res 74 (7) (2014) 1933–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Prima V, et al. , COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells, Proc. Natl. Acad. Sci. USA 114 (5) (2017) 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lu C, et al. , The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells, Oncoimmunology 5 (12) (2016), e1247135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Noman MZ, et al. , PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation, J. Exp. Med 211 (5) (2014) 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Strauss L, et al. , Targeted deletion of PD-1 in myeloid cells induces antitumor immunity, Sci. Immunol 5 (43) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Clavijo PE, et al. , Resistance to CTLA-4 checkpoint inhibition reversed through selective elimination of granulocytic myeloid cells, Oncotarget 8 (34) (2017) 55804–55820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Li Y, et al. , Targeting myeloid-derived suppressor cells to attenuate vasculogenic mimicry and synergistically enhance the anti-tumor effect of PD-1 inhibitor, iScience 24 (12) (2021), 103392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kim W, et al. , PD-1 signaling promotes tumor-infiltrating myeloid-derived suppressor cells and gastric tumorigenesis in mice, Gastroenterology 160 (3) (2021) 781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kim K, et al. , Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells, Proc. Natl. Acad. Sci. USA 111 (32) (2014) 11774–11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mantovani A, et al. , Chemokines in the recruitment and shaping of the leukocyte infiltrate of tumors, Semin Cancer Biol. 14 (3) (2004) 155–160. [DOI] [PubMed] [Google Scholar]
- [89].Robinson A, et al. , Monocyte regulation in homeostasis and malignancy, Trends Immunol. 42 (2) (2021) 104–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gordon SR, et al. , PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumor immunity, Nature 545 (7655) (2017) 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Qorraj M, et al. , The PD-1/PD-L1 axis contributes to immune metabolic dysfunctions of monocytes in chronic lymphocytic leukemia, Leukemia 31 (2017) 470–478. [DOI] [PubMed] [Google Scholar]
- [92].Chiba S, et al. , Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1, Nat. Immunol 13 (9) (2012) 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Dixon KO, et al. , TIM-3 restrains anti-tumour immunity by regulating inflammasome activation, Nature 595 (7865) (2021) 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Seo WI, et al. , Expression of VISTA on tumor-infiltrating immune cells correlated with short intravesical recurrence in non-muscle-invasive bladder cancer, Cancer Immunol. Immunother 70 (11) (2021) 3113–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lin H, et al. , Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression, J. Clin. Invest 128 (2) (2018) 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dangaj D, et al. , Novel recombinant human b7-h4 antibodies overcome tumoral immune escape to potentiate T-cell antitumor responses, Cancer Res 73 (15) (2013) 4820–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sica A, et al. , Macrophage polarization in tumour progression, Semin Cancer Biol. 18 (5) (2008) 349–355. [DOI] [PubMed] [Google Scholar]
- [98].Pylayeva-Gupta Y, et al. , Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia, Cancer Cell 21 (6) (2012) 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Bayne LJ, et al. , Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer, Cancer Cell 21 (6) (2012) 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ubertini V, et al. , Mutant p53 gains new function in promoting inflammatory signals by repression of the secreted interleukin-1 receptor antagonist, Oncogene 34 (19) (2015) 2493–2504. [DOI] [PubMed] [Google Scholar]
- [101].Sumimoto H, et al. , The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells, J. Exp. Med 203 (7) (2006) 1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mehta AK, et al. , Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer, Nat. Cancer 2 (1) (2021) 66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Martinez FO, et al. , Macrophage activation and polarization, Front Biosci. 13 (2008) 453–461. [DOI] [PubMed] [Google Scholar]
- [104].O’Neill LA, Pearce EJ, Immunometabolism governs dendritic cell and macrophage function, J. Exp. Med 213 (1) (2016) 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jayasingam SD, et al. , Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice, Front Oncol. 9 (2019) 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wyckoff JB, et al. , Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors, Cancer Res 67 (6) (2007) 2649–2656. [DOI] [PubMed] [Google Scholar]
- [107].Pittet MJ, Behavior of immune players in the tumor microenvironment, Curr. Opin. Oncol 21 (1) (2009) 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Harney AS, et al. , Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA, Cancer Disco 5 (9) (2015) 932–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chen Z, Ross JL, Hambardzumyan D, Intravital 2-photon imaging reveals distinct morphology and infiltrative properties of glioblastoma-associated macrophages, Proc. Natl. Acad. Sci. USA 116 (28) (2019) 14254–14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Guinney J, et al. , The consensus molecular subtypes of colorectal cancer, Nat. Med 21 (11) (2015) 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Picard E, et al. , Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer, Front Immunol. 11 (2020) 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Willimsky G, Blankenstein T, Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance, Nature 437 (7055) (2005) 141–146. [DOI] [PubMed] [Google Scholar]
- [113].Blank CU, et al. , Defining ‘T cell exhaustion’, Nat. Rev. Immunol 19 (11) (2019) 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pauken KE, et al. , Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade, Science 354 (6316) (2016) 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Im SJ, et al. , Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy, Nature 537 (7620) (2016) 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Schietinger A, et al. , Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis, Immunity 45 (2) (2016) 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Qureshi OS, et al. , Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4, Science 332 (6029) (2011) 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tekguc M, et al. , Treg-expressed CTLA-4 depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on antigen-presenting cells, Proc. Natl. Acad. Sci. USA 118 (30) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hui E, et al. , T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition, Science 355 (6332) (2017) 1428–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Mizuno R, et al. , PD-1 primarily targets TCR signal in the inhibition of functional T cell activation, Front Immunol. 10 (2019) 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Baumeister SH, et al. , Coinhibitory pathways in immunotherapy for cancer, Annu Rev. Immunol 34 (2016) 539–573. [DOI] [PubMed] [Google Scholar]
- [122].Fernandes RA, et al. , Immune receptor inhibition through enforced phosphatase recruitment, Nature 586 (7831) (2020) 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chemnitz JM, et al. , SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation, J. Immunol 173 (2) (2004) 945–954. [DOI] [PubMed] [Google Scholar]
- [124].Yokosuka T, et al. , Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2, J. Exp. Med 209 (6) (2012) 1201–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Patsoukis N, et al. , Interaction of SHP-2 SH2 domains with PD-1 ITSM induces PD-1 dimerization and SHP-2 activation, Commun. Biol 3 (1) (2020) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Celis-Gutierrez J, et al. , Quantitative interactomics in primary T cells provides a rationale for concomitant PD-1 and BTLA coinhibitor blockade in cancer immunotherapy, Cell Rep. 27 (11) (2019) 3315–3330.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Patsoukis N, et al. , Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation, Sci. Signal 5 (230) (2012) ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Patsoukis N, et al. , PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation, Nat. Commun 6 (2015) 6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Boussiotis VA, Patsoukis N, Effects of PD-1 signaling on immunometabolic reprogramming, Immunometabolism 4 (2) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Bardhan K, Anagnostou T, Boussiotis VA, The PD1:PD-L1/2 pathway from discovery to clinical implementation, Front Immunol. 7 (2016) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Topalian SL, et al. , Immunotherapy: the path to win the war on cancer? Cell 161 (2) (2015) 185–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zou W, Wolchok JD, Chen L, PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations, Sci. Transl. Med 8 (328) (2016) 328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Juneja VR, et al. , PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity, J. Exp. Med 214 (4) (2017) 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lau J, et al. , Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice, Nat. Commun 8 (2017) 14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Tang H, et al. , PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression, J. Clin. Invest 128 (2) (2018) 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Peng Q, et al. , PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade, Nat. Commun 11 (1) (2020) 4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Oh SA, et al. , PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer, Nat. Cancer 1 (7) (2020) 681–691. [DOI] [PubMed] [Google Scholar]
- [138].Patsoukis N, et al. , Revisiting the PD-1 pathway, Sci. Adv 6 (38) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sugiura D, et al. , PD-1 agonism by anti-CD80 inhibits T cell activation and alleviates autoimmunity, Nat. Immunol 23 (3) (2022) 399–410. [DOI] [PubMed] [Google Scholar]
- [140].Andrews LP, Yano H, Vignali DAA, Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups, Nat. Immunol 20 (11) (2019) 1425–1434. [DOI] [PubMed] [Google Scholar]
- [141].Yuan L, et al. , VISTA: a mediator of quiescence and a promising target in cancer immunotherapy, Trends Immunol. 42 (3) (2021) 209–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].ElTanbouly MA, et al. , VISTA is a checkpoint regulator for naive T cell quiescence and peripheral tolerance, Science 367 (6475) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Han X, et al. , PD-1H (VISTA)-mediated suppression of autoimmunity in systemic and cutaneous lupus erythematosus, Sci. Transl. Med 11 (522) (2019). [DOI] [PubMed] [Google Scholar]
- [144].ElTanbouly MA, et al. , VISTA Re-programs Macrophage Biology Through the Combined Regulation of Tolerance and Anti-inflammatory Pathways, Front Immunol. 11 (2020), 580187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Mulati K, et al. , VISTA expressed in tumour cells regulates T cell function, Br. J. Cancer 120 (1) (2019) 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Rosenbaum SR, et al. , FOXD3 Regulates VISTA Expression in Melanoma, Cell Rep. 30 (2) (2020) 510–524, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Gao J, et al. , VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer, Nat. Med 23 (5) (2017) 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Blando J, et al. , Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer, Proc. Natl. Acad. Sci. USA 116 (5) (2019) 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Kakavand H, et al. , Negative immune checkpoint regulation by VISTA: a mechanism of acquired resistance to anti-PD-1 therapy in metastatic melanoma patients, Mod. Pathol 30 (12) (2017) 1666–1676. [DOI] [PubMed] [Google Scholar]
- [150].Wang J, et al. , VSIG-3 as a ligand of VISTA inhibits human T-cell function, Immunology 156 (1) (2019) 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Johnston RJ, et al. , VISTA is an acidic pH-selective ligand for PSGL-1, Nature 574 (7779) (2019) 565–570. [DOI] [PubMed] [Google Scholar]
- [152].Haxhinasto S, Mathis D, Benoist C, The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells, J. Exp. Med 205 (3) (2008) 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Delgoffe GM, et al. , The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment, Immunity 30 (6) (2009) 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Francisco LM, et al. , PD-L1 regulates the development, maintenance, and function of induced regulatory T cells, J. Exp. Med 206 (13) (2009) 3015–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Shi LZ, et al. , HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells, J. Exp. Med 208 (7) (2011) 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kumagai S, et al. , The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies, Nat. Immunol (2020). [DOI] [PubMed] [Google Scholar]
- [157].Miyara M, et al. , Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor, Immunity 30 (6) (2009) 899–911. [DOI] [PubMed] [Google Scholar]
- [158].Wing JB, Tanaka A, Sakaguchi S, Human FOXP3(+) Regulatory T Cell Heterogeneity and Function in Autoimmunity and Cancer, Immunity 50 (2) (2019) 302–316. [DOI] [PubMed] [Google Scholar]
- [159].Williams MA, Bevan MJ, Effector and memory CTL differentiation, Annu Rev. Immunol 25 (2007) 171–192. [DOI] [PubMed] [Google Scholar]
- [160].Ribas A, et al. , PD-1 blockade expands intratumoral memory T cells, Cancer Immunol. Res 4 (3) (2016) 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].MacIver NJ, Rathmell JC, Editorial overview: metabolism of T cells: integrating nutrients, signals, and cell fate, Curr. Opin. Immunol 46 (2017) viii–xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Gerriets VA, et al. , Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression, Nat. Immunol 17 (12) (2016) 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Herbel C, et al. , Clinical significance of T cell metabolic reprogramming in cancer, Clin. Transl. Med 5 (1) (2016) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Chang CH, et al. , Metabolic competition in the tumor microenvironment is a driver of cancer progression, Cell 162 (6) (2015) 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Daneshmandi S, et al. , Blockade of 6-phosphogluconate dehydrogenase generates CD8(+) effector T cells with enhanced anti-tumor function, Cell Rep. 34 (10) (2021), 108831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Frauwirth KA, Thompson CB, Regulation of T lymphocyte metabolism, J. Immunol 172 (8) (2004) 4661–4665. [DOI] [PubMed] [Google Scholar]
- [167].Chang CH, et al. , Posttranscriptional control of T cell effector function by aerobic glycolysis, Cell 153 (6) (2013) 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Sukumar M, et al. , Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function, J. Clin. Invest 123 (10) (2013) 4479–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Porstmann T, et al. , PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP, Oncogene 24 (43) (2005) 6465–6481. [DOI] [PubMed] [Google Scholar]
- [170].Duvel K, et al. , Activation of a metabolic gene regulatory network downstream of mTOR complex 1, Mol. Cell 39 (2) (2010) 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Porstmann T, et al. , SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth, Cell Metab. 8 (3) (2008) 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Wang R, et al. , The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation, Immunity 35 (6) (2011) 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Cheng SC, et al. , mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity, Science 345 (6204) (2014) 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Zeng H, Chi H, mTOR signaling and transcriptional regulation in T lymphocytes, Transcription 5 (2) (2014), e28263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Le Bourgeois T, et al. , Targeting T cell metabolism for improvement of cancer immunotherapy, Front Oncol. 8 (2018) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kaech SM, Cui W, Transcriptional control of effector and memory CD8+ T cell differentiation, Nat. Rev. Immunol 12 (11) (2012) 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Klebanoff CA, et al. , Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells, Proc. Natl. Acad. Sci. USA 102 (27) (2005) 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Gattinoni L, et al. , Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells, J. Clin. Invest 115 (6) (2005) 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Gattinoni L, et al. , Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells, Nat. Med 15 (7) (2009) 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Klebanoff CA, Gattinoni L, Restifo NP, CD8+ T-cell memory in tumor immunology and immunotherapy, Immunol. Rev 211 (2006) 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Sallusto F, et al. , Two subsets of memory T lymphocytes with distinct homing potentials and effector functions, Nature 401 (6754) (1999) 708–712. [DOI] [PubMed] [Google Scholar]
- [182].Wherry EJ, et al. , Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment, J. Virol 77 (8) (2003) 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Masopust D, Soerens AG, Tissue-resident T cells and other resident leukocytes, Annu Rev. Immunol 37 (2019) 521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Omilusik KD, Goldrath AW, Remembering to remember: T cell memory maintenance and plasticity, Curr. Opin. Immunol 58 (2019) 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Kok L, et al. , A committed tissue-resident memory T cell precursor within the circulating CD8+ effector T cell pool, J. Exp. Med 217 (10) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Masopust D, et al. , Preferential localization of effector memory cells in nonlymphoid tissue, Science 291 (5512) (2001) 2413–2417. [DOI] [PubMed] [Google Scholar]
- [187].Steinert EM, et al. , Quantifying memory CD8 T cells reveals regionalization of immunosurveillance, Cell 161 (4) (2015) 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Menares E, et al. , Tissue-resident memory CD8(+) T cells amplify anti-tumor immunity by triggering antigen spreading through dendritic cells, Nat. Commun 10 (1) (2019) 4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Park SL, et al. , Author correction: tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin, Nature 566 (7745) (2019), E10. [DOI] [PubMed] [Google Scholar]
- [190].Savas P, et al. , Publisher correction: single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis, Nat. Med 24 (12) (2018) 1941. [DOI] [PubMed] [Google Scholar]
- [191].Corgnac S, et al. , The emerging role of CD8(+) tissue resident memory T (T(RM)) cells in antitumor immunity: a unique functional contribution of the CD103 integrin, Front Immunol. 9 (2018) 1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Skon CN, et al. , Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells, Nat. Immunol 14 (12) (2013) 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Bai A, et al. , Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription, J. Immunol 178 (12) (2007) 7632–7639. [DOI] [PubMed] [Google Scholar]
- [194].Debes GF, et al. , Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues, Nat. Immunol 6 (9) (2005) 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Gorfu G, Rivera-Nieves J, Ley K, Role of beta7 integrins in intestinal lymphocyte homing and retention, Curr. Mol. Med 9 (7) (2009) 836–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kilshaw PJ, Murant SJ, A new surface antigen on intraepithelial lymphocytes in the intestine, Eur. J. Immunol 20 (10) (1990) 2201–2207. [DOI] [PubMed] [Google Scholar]
- [197].Schön MP, et al. , Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice, J. Immunol 162 (11) (1999) 6641–6649. [PubMed] [Google Scholar]
- [198].Molodtsov AK, et al. , Resident memory CD8(+) T cells in regional lymph nodes mediate immunity to metastatic melanoma, Immunity 54 (9) (2021) 2117–2132. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Malik BT, et al. , Resident memory T cells in the skin mediate durable immunity to melanoma, Sci. Immunol 2 (10) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Liikanen I, et al. , Hypoxia-inducible factor activity promotes antitumor effector function and tissue residency by CD8+ T cells, J. Clin. Invest 131 (7) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Han J, et al. , Resident and circulating memory T cells persist for years in melanoma patients with durable responses to immunotherapy, Nat. Cancer 2 (3) (2021) 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Ganesan AP, et al. , Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer, Nat. Immunol 18 (8) (2017) 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Mei X, et al. , The emerging role of tissue-resident memory CD8(+) T lymphocytes in human digestive tract cancers, Front Oncol. 11 (2021), 819505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Djenidi F, et al. , CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients, J. Immunol 194 (7) (2015) 3475–3486. [DOI] [PubMed] [Google Scholar]
- [205].Webb JR, Milne K, Nelson BH, PD-1 and CD103 are widely coexpressed on prognostically favorable intraepithelial CD8 T cells in human ovarian cancer, Cancer Immunol. Res 3 (8) (2015) 926–935. [DOI] [PubMed] [Google Scholar]
- [206].Komdeur FL, et al. , CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer, Oncoimmunology 6 (9) (2017), e1338230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Caushi JX, et al. , Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers, Nature 596 (7870) (2021) 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Edwards J, et al. , CD103(+) tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naïve melanoma patients and expand significantly during anti-PD-1 treatment, Clin. Cancer Res 24 (13) (2018) 3036–3045. [DOI] [PubMed] [Google Scholar]
- [209].Murray T, et al. , Very late antigen-1 marks functional tumor-resident CD8 T cells and correlates with survival of melanoma patients, Front Immunol. 7 (2016) 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Savas P, et al. , Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis, Nat. Med 24 (7) (2018) 986–993. [DOI] [PubMed] [Google Scholar]
- [211].Wang ZQ, et al. , CD103 and intratumoral immune response in breast cancer, Clin. Cancer Res 22 (24) (2016) 6290–6297. [DOI] [PubMed] [Google Scholar]
- [212].Duhen T, et al. , Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors, Nat. Commun 9 (1) (2018) 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Webb JR, et al. , Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer, Clin. Cancer Res 20 (2) (2014) 434–444. [DOI] [PubMed] [Google Scholar]
- [214].Koh J, et al. , Prognostic implications of intratumoral CD103+ tumor-infiltrating lymphocytes in pulmonary squamous cell carcinoma, Oncotarget 8 (8) (2017) 13762–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Wang B, et al. , CD103+ tumor infiltrating lymphocytes predict a favorable prognosis in urothelial cell carcinoma of the bladder, J. Urol 194 (2) (2015) 556–562. [DOI] [PubMed] [Google Scholar]
- [216].Vivier E, et al. , Innate lymphoid cells: 10 years on, Cell 174 (5) (2018) 1054–1066. [DOI] [PubMed] [Google Scholar]
- [217].Ducimetiere L, Vermeer M, Tugues S, The interplay between innate lymphoid cells and the tumor microenvironment, Front. Immunol 10 (2019) 2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Guia S, et al. , Activating and inhibitory receptors expressed on innate lymphoid cells, Semin. Immunopathol 40 (4) (2018) 331–341. [DOI] [PubMed] [Google Scholar]
- [219].Horowitz A, et al. , Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry, Sci. Transl. Med 5 (208) (2013) 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Freud AG, et al. , The broad spectrum of human natural killer cell diversity, Immunity 47 (5) (2017) 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Malmberg KJ, et al. , Natural killer cell-mediated immunosurveillance of human cancer, Semin. Immunol 31 (2017) 20–29. [DOI] [PubMed] [Google Scholar]
- [222].Carrega P, et al. , CD56(bright)perforin(low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph, J. Immunol 192 (8) (2014) 3805–3815. [DOI] [PubMed] [Google Scholar]
- [223].Moretta A, et al. , Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis, Annu. Rev. Immunol 19 (2001) 197–223. [DOI] [PubMed] [Google Scholar]
- [224].Pegram HJ, et al. , Activating and inhibitory receptors of natural killer cells, Immunol. Cell Biol 89 (2) (2011) 216–224. [DOI] [PubMed] [Google Scholar]
- [225].Sivori S, et al. , CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies, Eur. J. Immunol 26 (10) (1996) 2487–2492. [DOI] [PubMed] [Google Scholar]
- [226].Perez-Villar JJ, et al. , The CD94/NKG2-A inhibitory receptor complex is involved in natural killer cell-mediated recognition of cells expressing HLA-G1, J. Immunol 158 (12) (1997) 5736–5743. [PubMed] [Google Scholar]
- [227].Braud VM, et al. , HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C, Nature 391 (6669) (1998) 795–799. [DOI] [PubMed] [Google Scholar]
- [228].Chiossone L, et al. , Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more, Semin. Immunol 31 (2017) 55–63. [DOI] [PubMed] [Google Scholar]
- [229].Karre K, et al. , Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy, Nature 319 (6055) (1986) 675–678. [DOI] [PubMed] [Google Scholar]
- [230].Romagne F, et al. , Preclinical characterization of 1–7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells, Blood 114 (13) (2009) 2667–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].van Hall T, et al. , Monalizumab: inhibiting the novel immune checkpoint NKG2A, J. Immunother. Cancer 7 (1) (2019) 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Kohrt HE, et al. , Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies, Blood 123 (5) (2014) 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Andre P, et al. , Anti-NKG2A mAb is a checkpoint inhibitor that promotes antitumor immunity by unleashing both T and NK cells, Cell 175 (7) (2018) 1731–1743, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Kamiya T, et al. , Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells, J. Clin. Invest 129 (5) (2019) 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Della Chiesa M, et al. , Features of memory-like and PD-1(+) human NK cell subsets, Front. Immunol 7 (2016) 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Pesce S, et al. , Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization, J. Allergy Clin. Immunol 139 (1) (2017) 335–346, e3. [DOI] [PubMed] [Google Scholar]
- [237].Benson DM Jr., et al. , The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody, Blood 116 (13) (2010) 2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Beldi-Ferchiou A, et al. , PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma, Oncotarget 7 (45) (2016) 72961–72977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Niu C, et al. , PD-1-positive natural killer cells have a weaker antitumor function than that of PD-1-negative natural killer cells in lung cancer, Int. J. Med. Sci 17 (13) (2020) 1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Tumino N, et al. , Presence of innate lymphoid cells in pleural effusions of primary and metastatic tumors: Functional analysis and expression of PD-1 receptor, Int. J. Cancer 145 (6) (2019) 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Pesce S, et al. , Different features of tumor-associated NK cells in patients with low-grade or high-grade peritoneal carcinomatosis, Front. Immunol 10 (2019) 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Mariotti FR, et al. , PD-1 in human NK cells: evidence of cytoplasmic mRNA and protein expression, Oncoimmunology 8 (3) (2019) 1557030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Vari F, et al. , Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL, Blood 131 (16) (2018) 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Trefny MP, et al. , PD-1(+) natural killer cells in human non-small cell lung cancer can be activated by PD-1/PD-L1 blockade, Cancer Immunol. Immunother 69 (8) (2020) 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Quatrini L, et al. , Glucocorticoids and the cytokines IL-12, IL-15, and IL-18 present in the tumor microenvironment induce PD-1 expression on human natural killer cells, J. Allergy Clin. Immunol 147 (1) (2021) 349–360. [DOI] [PubMed] [Google Scholar]
- [246].Iraolagoitia XL, et al. , NK cells restrain spontaneous antitumor CD8+ T cell priming through PD-1/PD-L1 interactions with dendritic cells, J. Immunol 197 (3) (2016) 953–961. [DOI] [PubMed] [Google Scholar]
- [247].Stojanovic A, et al. , CTLA-4 is expressed by activated mouse NK cells and inhibits NK Cell IFN-gamma production in response to mature dendritic cells, J. Immunol 192 (9) (2014) 4184–4191. [DOI] [PubMed] [Google Scholar]
- [248].Russick J, et al. , Natural killer cells in the human lung tumor microenvironment display immune inhibitory functions, J. Immunother. Cancer 8 (2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Esen F, Deniz G, Aktas EC, PD-1, CTLA-4, LAG-3, and TIGIT: The roles of immune checkpoint receptors on the regulation of human NK cell phenotype and functions, Immunol. Lett 240 (2021) 15–23. [DOI] [PubMed] [Google Scholar]
- [250].Kohlhapp FJ, et al. , NK cells and CD8+ T cells cooperate to improve therapeutic responses in melanoma treated with interleukin-2 (IL-2) and CTLA-4 blockade, J. Immunother. Cancer 3 (2015) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Ndhlovu LC, et al. , Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity, Blood 119 (16) (2012) 3734–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Komita H, et al. , Expression of immune checkpoint molecules of T cell immunoglobulin and mucin protein 3/galectin-9 for NK cell suppression in human gastrointestinal stromal tumors, Oncol. Rep 34 (4) (2015) 2099–2105. [DOI] [PubMed] [Google Scholar]
- [253].Gleason MK, et al. , Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9, Blood 119 (13) (2012) 3064–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].da Silva IP, et al. , Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade, Cancer Immunol. Res 2 (5) (2014) 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Gallois A, et al. , Reversal of natural killer cell exhaustion by TIM-3 blockade, Oncoimmunology 3 (12) (2014), e946365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Wang Z, et al. , The clinical significance of abnormal Tim-3 expression on NK cells from patients with gastric cancer, Immunol. Invest 44 (6) (2015) 578–589. [DOI] [PubMed] [Google Scholar]
- [257].Xu L, et al. , Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma, Int. Immunopharmacol 29 (2) (2015) 635–641. [DOI] [PubMed] [Google Scholar]
- [258].Zhang W, et al. , The functional potency of natural killer cells in response to IL-2/IL-15/IL-21 stimulation is limited by a concurrent upregulation of Tim-3 in bladder cancer, Exp. Cell Res 372 (2) (2018) 92–98. [DOI] [PubMed] [Google Scholar]
- [259].Merino A, et al. , Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming, J. Clin. Invest 129 (9) (2019) 3770–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Woo SR, et al. , Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape, Cancer Res. 72 (4) (2012) 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Brignone C, et al. , A soluble form of lymphocyte activation gene-3 (IMP321) induces activation of a large range of human effector cytotoxic cells, J. Immunol 179 (6) (2007) 4202–4211. [DOI] [PubMed] [Google Scholar]
- [262].Brignone C, et al. , First-line chemoimmunotherapy in metastatic breast carcinoma: combination of paclitaxel and IMP321 (LAG-3Ig) enhances immune responses and antitumor activity, J. Transl. Med 8 (2010) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Xu F, et al. , Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions, Cancer Immunol. Immunother 66 (10) (2017) 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Bottino C, et al. , Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule, J. Exp. Med 198 (4) (2003) 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Chan CJ, et al. , The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions, Nat. Immunol 15 (5) (2014) 431–438. [DOI] [PubMed] [Google Scholar]
- [266].Chiang EY, et al. , CD96 functions as a co-stimulatory receptor to enhance CD8(+) T cell activation and effector responses, Eur. J. Immunol 50 (6) (2020) 891–902. [DOI] [PubMed] [Google Scholar]
- [267].Sanchez-Correa B, et al. , DNAM-1 and the TIGIT/PVRIG/TACTILE axis: Novel immune checkpoints for natural killer cell-based cancer immunotherapy, Cancers (Basel) 11 (6) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Fuchs A, Colonna M, The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance, Semin. Cancer Biol 16 (5) (2006) 359–366. [DOI] [PubMed] [Google Scholar]
- [269].Takai Y, et al. , Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation, Nat. Rev. Mol. Cell Biol 9 (8) (2008) 603–615. [DOI] [PubMed] [Google Scholar]
- [270].Sarhan D, et al. , Adaptive NK cells with low TIGIT expression are inherently resistant to myeloid-derived suppressor cells, Cancer Res. 76 (19) (2016) 5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Whelan S, et al. , PVRIG and PVRL2 are induced in cancer and inhibit CD8(+) T-cell function, Cancer Immunol. Res 7 (2) (2019) 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Anderson AC, Joller N, Kuchroo VK, Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation, Immunity 44 (5) (2016) 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Johnston RJ, et al. , The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function, Cancer Cell 26 (6) (2014) 923–937. [DOI] [PubMed] [Google Scholar]
- [274].Nishiwada S, et al. , Clinical significance of CD155 expression in human pancreatic cancer, Anticancer Res. 35 (4) (2015) 2287–2297. [PubMed] [Google Scholar]
- [275].Zhang Q, et al. , Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity, Nat. Immunol 19 (7) (2018) 723–732. [DOI] [PubMed] [Google Scholar]
- [276].Blake SJ, et al. , Molecular pathways: Targeting CD96 and TIGIT for cancer immunotherapy, Clin. Cancer Res 22 (21) (2016) 5183–5188. [DOI] [PubMed] [Google Scholar]
- [277].Simoni Y, et al. , Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency, Immunity 46 (1) (2017) 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Salimi M, et al. , Activated innate lymphoid cell populations accumulate in human tumour tissues, BMC Cancer 18 (1) (2018) 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Wang S, et al. , Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer, Cell Res. 30 (7) (2020) 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Moral JA, et al. , ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity, Nature 579 (7797) (2020) 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Ercolano G, et al. , Distinct and shared gene expression for human innate versus adaptive helper lymphoid cells, J. Leukoc. Biol 108 (2) (2020) 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Gao Y, et al. , Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells, Nat. Immunol 18 (9) (2017) 1004–1015. [DOI] [PubMed] [Google Scholar]
- [283].Vacca P, et al. , PD-1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua, Mucosal Immunol. 12 (3) (2019) 624–631. [DOI] [PubMed] [Google Scholar]
- [284].Luci C, et al. , Cutaneous squamous cell carcinoma development is associated with a temporal infiltration of ILC1 and NK cells with immune dysfunctions, J. Invest. Dermatol 141 (10) (2021) 2369–2379. [DOI] [PubMed] [Google Scholar]


