The chemical composition of many fungal cell walls is known, but we have not understood the interactions of the various macromolecules nor the assembly processes. The biochemistry and molecular genetics of biosynthesis have been comprehensively reviewed (9, 42), but recent results obtained with the yeast Saccharomyces cerevisiae have confirmed and extended a structural model that explains many results and points out new directions for research.
Cell walls of fungi share with plant and bacterial cell walls, and indeed with extracellular matrix material of mammalian cells, an anionic surface and a reliance on β1,4- and β1,3-linked polysaccharides as fibrous components. These glycans have all of the non-hydrogen ring constituents in an equatorial position and form ribbon-like (cellulose and chitin) or helical (β1,3 glucan) structures. Other characteristics of fungal, plant, and bacterial cell walls differ markedly. Where the glycans of eubacterial walls are cross-linked by peptides, those in plants have cross-linking phenolics and polysaccharides that promote cross-associations by hydrogen bonding (hemicelluloses) or gel properties (pectins) (10, 15).
Composition of cell walls.
In S. cerevisiae, the cell wall makes up 15 to 30% of the dry weight of the cell (42) and 25 to 50% of the volume based on calculations from electron micrographs. The walls are composed mostly of mannoprotein and fibrous β1,3 glucan (Table 1). There is also branched β1,6 glucan that links the other components of the wall (25, 28, 42). An important minor component is chitin, which contributes to the insolubility of the fibers. The β1,3 glucan-chitin complex is the major constituent of the inner wall. β1,6 glucan links the components of the inner and outer walls. On the outer surface of the wall are mannoproteins, which are extensively O and N glycosylated. They are densely packed and limit wall permeability to solutes (12, 57). Covalent linkages between each of these components have now been identified (28).
TABLE 1.
Major components of S. cerevisiae cell walls
| Component (degree of polymerization) | Mean molecular mass (kDa) | % of wall mass | Relative molar ratio |
|---|---|---|---|
| β1,3 glucan (1,500) | 240 | 50 | 1.0 |
| β1,6 glucan (150) | 24 | 10 | 2 |
| Mannoprotein | 100–200 | 40 | 1.2–2.4 |
| Chitin (120) | 25 | 1–3 | 0.1–0.3 |
Modular construction.
Many of the wall components are present in low molar ratios (Table 1). β1,3 glucan is the major component and forms the fibrous scaffold of the wall. Dividing the polymer size into the cellular glucan content yields a figure of about 1 × 106 to 3 × 106 glucan chains per cell. There is a similar number of β1,6 glucan molecules attached to the β1,3 glucan. If we estimate the average size of mannoproteins as 100 to 200 kDa, the number of mannoproteins is also similar (14, 52). The small amount of chitin (1% of the dry weight exclusive of the bud scar) is in linear chains of about 120 units, present in a molar ratio of 0.1 to 0.3 (27).
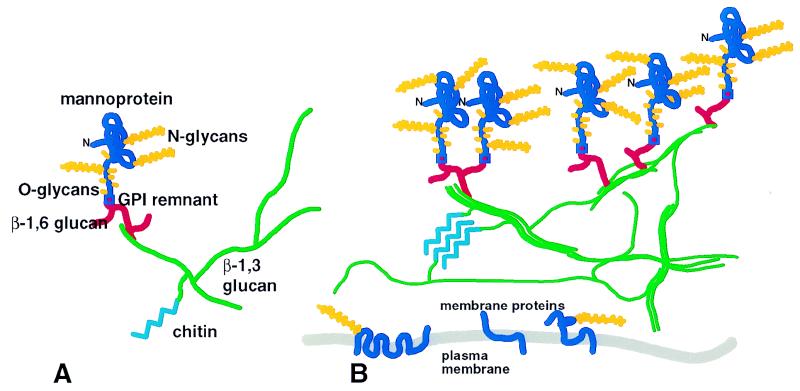
These components are covalently linked to form macromolecular complexes, which are assembled to form the intact wall. A team (including the Cabib, Klis, and Ashwell groups) has now identified linkages between all of these components (28). These authors have called the covalent complex a “flexible building block.” However, because the cell wall components occupy only 10 to 20% of the wall volume, a better analogy is that the wall is a latticework, rather than a solid structure. The lattice is an assembly of unit modules, each built around a molecule of β1,3 glucan (Fig. 1A). A prototypical module would have a β1,3 glucan chain with 40 to 50 branch points and would also include one or two β1,6 glucan and mannoprotein moieties as well. A minority of modules have chitin chains attached to the β1,3 or β1,6 glucan (27, 28). Note that these molar ratios are only averages; there is no evidence for a fixed stoichiometry of the various components. The modules are associated by noncovalent interactions in the glucan-chitin layer and by covalent cross-links in the mannoprotein layer (Fig. 1B), including disulfide bonds between mannoproteins (12, 42, 57) and perhaps novel mannoprotein-glucan links that are as yet uncharacterized (28).
FIG. 1.
Relationships among components of S. cerevisiae cell walls. (A) Prototypical module with components individually labeled and colored. The mannoprotein polypeptide is blue, and oligosaccharides are shown in yellow, labelled as N or O linked. Only a few of the branch points of the glucans are shown. Chitin can also be linked to the β1,6 glucan. (B) Association of modules to form a wall lattice. Colors are as in panel A. The β1,3 glucan chains are intertwined to designate triple helices, and chitin is shown as a crystalline microdomain. Cross-linking of mannoproteins through disulfide and other bonds is not depicted.
STRUCTURE OF CELL WALL COMPONENTS
Glucans.
β1,3 glucan forms a fibrous network visible by scanning electron microscopy of the inner surface of walls and forms amorphous components as well (30). Its average degree of polymerization of 1,500 corresponds to a molecular mass of 240,000 and a maximum fiber length of about 600 nm. This length is roughly three to six times the average wall thickness, or 1/10 of a cell circumference. Larger complexes have been occasionally reported (40, 55). Branching of the polymer (about 3% branch points) might substantially reduce this length, depending on the branch length (37, 40). Much of the β1,3 glucan has a helical conformation, based on in vitro studies, now confirmed by solid state nuclear magnetic resonance of intact yeast cells (31). Such helices are composed of a single polysaccharide chain or of three hydrogen-bonded chains (a triple helix) (50, 55). In electron micrographs fibers are 10 to 30 nm in diameter, consistent with lateral associations of multiple chains, each with a diameter of 0.5 to 1 nm (29, 30).
There is no direct data about the length of the branches (37). The branch points are the 6-hydroxy groups, and substituents at this position do not interfere with formation of either single or triple helices (50, 55). Long branch lengths would result in a “bushy” polysaccharide with the reducing end at the base of the stalk, consistent with the Stokes radius of yeast glucan, which is 20 to 30 nm per 106 Da (40). This value is about 10% of the observed length of model glucans that form linear fibers. It is also much shorter than the predicted length for an unbranched helical structure and therefore implies that the branches are of significant length (50). If there are long branches, the association of neighboring chains might form an anastomosing network of fibers (Fig. 1B). On the other hand, short branches would promote formation of the triple helices (46, 55). The fibrous network would then consist of alternating regions of single helices and triple helices formed from glucan chains of three different modules (46, 55). Such a structure could serve a role similar to that of the hemicellulose-cellulose interactions in plant cell walls (15).
β1,3 glucan synthase is located in the plasma membrane (42). Electron microscopy of regenerating spheroplasts shows that the polysaccharide product is extracellular (29). Thus, the complex acts as a glycosyl transferase and transporter. Branches may be formed extracellularly by a putative branching enzyme, Bgl2p, which has activity analogous to that of the starch branching enzymes (18).
β1,6 glucan.
β1,6 glucan is a highly branched polysaccharide that links the components of each module together (28). Despite extensive genetic and biochemical analyses, the site and mode of synthesis of β1,6 glucan are unclear (42). Because the glucan is the primary receptor for yeast K1 killer factor, mutations in genes necessary for glucan synthesis lead to toxin resistance (KRE genes) (42). KRE genes and their extragenic suppressors and synthetic-lethal partners encode a variety of intracellular and extracellular proteins. Many of these proteins participate in N and O glycosylation of mannoproteins (see below). Of the other KRE gene products, no in vitro assays for function are known, so that biochemistry and localization of β1,6 glucan synthesis and cross-linking to β1,3 glucan remain obscure.
Chitin.
The signal structural work by Cabib’s group and collaborators showed that chitin is glycosidically linked to nonreducing branches of the β1,3 glucan and β1,6 glucan (Fig. 1A) (27, 28). Presumably, the chitin chains from several modules anneal to form microdomains of crystalline α-chitin, the most common form in aqueous environments and the form in the walls of other fungi. The structure of α-chitin is similar to that of α-cellulose, with hydrogen-bonded antiparallel chains of N-acetylglucosamine units. Hydrogen bonds involving the amide groups (absent in cellulose) further stabilize the crystals. These extra bonds together with the hydrophobic core formed by the acetamido methyl groups prevent invasion by water and dissolution of the crystals (2). Although crystalline domains of chitin have not been seen in yeast, no serious X-ray work on digested walls has been attempted for about 30 years, and such domains might now be found with the improved diffraction methods and uncontaminated glucanases available (25).
Chitin synthesis is vectorial, with the substrates and regulatory sites intracellular and the product extracellular, based on enzymology, microscopy, and studies of sites of action of membrane-impermeant inhibitors (4, 42). Addition of chitin to modules is essential for insolubility of the wall, and chitin incorporation results in transfer of the wall material from the alkali-soluble to the alkali-insoluble fraction (20).
Mannoproteins.
Yeast wall mannoproteins are highly glycosylated polypeptides, often 50 to 95% carbohydrate by weight, and thus may be thought of as yeast proteoglycans (42, 52). Many of them carry N-linked glycans with a core structure of Man10–14GlcNAc2-Asn, structures very similar to mammalian high-mannose N-glycan chains. “Outer chains” present on many yeast N-glycans consist of 50 to 200 additional α-linked mannose units, with a long α1,6-linked backbone decorated with short α1,2- and α1,3-linked side chains (42). There are often several large N-glycans per glycopeptide, so that N-linked sugar can add 50,000 to 100,000 Da to the size of the mannoproteins. Phosphorylation of the mannosyl side chains gives yeast its anionic surface charge (42). N-chain elongation is not essential for wall biogenesis per se, but the lack of outer chains in mnn9 mutants increases wall permeability and decreases integrity (12).
Ser and Thr residues are often clustered within the sequences of wall mannoproteins (42, 52). Where O-linked saccharides have been mapped, most (8) or all (56) of the clustered residues are O glycosylated. The clustered O-glycans are oligosaccharides of 1 to 5 mannosyl units, creating rigid stalks that elevate protein domains from membranes or wall surfaces (6, 8, 16, 22) (Fig. 1).
O mannosylation is important for proper wall biogenesis. Disruption of O glycosylation causes not only aberrant processing of wall mannoproteins (35, 42) but also leads to significant reduction in wall content of the branched β1,6 linker glucan (42). Two explanations for this phenotype have been offered: (i) the β1,6 glucan is partly assembled intracellularly, and secretion is dependent on association with O-glycosylated mannoproteins (42); (ii) glucan synthesis or assembly is dependent on mannoproteins whose proper localization or function is dependent on O glycosylation. Kre9p and Gas1p/Ggp1p are examples of O-glycosylated proteins required for proper wall biogenesis (13, 16, 17, 43).
Cell wall anchorage of module mannoproteins.
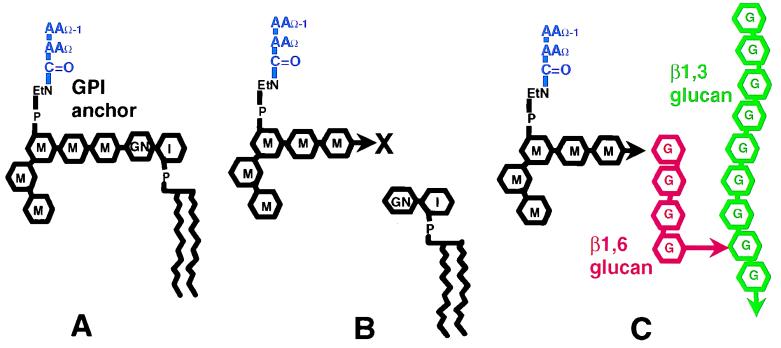
The mannoproteins of the modules are resistant to extraction in hot sodium dodecyl sulfate but can be liberated from the wall by β1,3 glucanases or β1,6 glucanases (42, 52, 53). Studies with the cell adhesion protein α-agglutinin led to the proposal of the anchorage hypothesis, which states that each outer-layer mannoprotein is posttranslationally modified by addition of a glycosyl phosphatidylinositol (GPI) anchor (11) (Fig. 2). After secretion of the GPI-anchored mannoprotein to the outer leaflet of the plasma membrane, the anchor is cleaved within the C-terminal glycan and the remnant is transferred to form a glycosidic linkage with the branched β1,6 glucan (11, 24, 25, 28, 34, 53). The β1,6 glucan is, in turn, glycosidically linked to the β1,3 glucan-chitin complex that makes up the fibers of the inner wall (25, 28).
FIG. 2.
Structure of a yeast GPI anchor and model for assembly of a mannoprotein into a module. (A) GPI anchor: AA, amino acid residue; EtN, ethanolamine; P, phosphate; M, mannose; GN, glucosamine; I, myoinositol. At the bottom right is a phospholipid, which may be glycerol or sphingosine based. (B) Proposed cleavage of the glycan of the GPI anchor. The arrow denotes the glycan reducing end, with an “X” denoting a hypothetical complex to a protein or other “activator.” (C) Formation of a glycosidic linkage between the GPI remnant and glucans. G, glucose.
GPI addition to mannoproteins is essential because mutations in GPI synthesis are lethal (32, 42) and restriction of GPI synthesis causes aberrant wall biogenesis and growth limitation (54). Also, cell wall synthesis ceases immediately upon inositol starvation (19). About 40 open reading frames in the yeast genome have the sequence characteristics of GPI-anchored wall proteins (7). Many of the findings for S. cerevisiae are echoed for Candida albicans and other fungi (18, 24, 38, 42).
Yeast GPIs are attached in the endoplasmic reticulum by transpeptidation to the C terminus of proteins possessing GPI signal sequences (42). Successful transport from the endoplasmic reticulum to the cell surface is dependent on the presence of sphingolipids (21, 49), and most secretion is targeted to the site of bud emergence or to the growing bud (23). Thus, cell wall mannoproteins and the enzymes mediating wall assembly are probably secreted in the same place.
Other mannoproteins are wall associated by other mechanisms. Invertase and other enzymes are physically entrapped in the wall (9). Flo1p, a component of the yeast flocculation apparatus, is initially GPI anchored but may remain unlinked to glucan (3). The cyclic AMP binding protein Gce1p is GPI anchored when synthesized but is later processed by lipolytic and proteolytic cleavage near the C terminus before cell wall association (41). In addition, mild base treatment (30 mM NaOH, 16 h, 4°C) liberates four mannoproteins that have no GPI anchor signal in their corresponding genes (39). Some mannoproteins are disulfide bonded to GPI-anchored lattice proteins (6, 9, 42).
UTILITY OF THE MODULE CONCEPT
Inferences from the model.
The modular structure hypothesis is a basis for explanations of cell wall phenotypes and predictions of functions for specific genes. Popolo et al. have shown that GAS1/GGP1/CWH52 (alternate names for the same gene) mutants have a disorganized wall structure and are resistant to digestion with the lytic enzyme mixture Zymolyase (43). They have argued that the formation of β1,3 glucan fibers is abnormal in these mutants, suggesting that the GAS1 gene product, an extracellular membrane-bound GPI-anchored protein, is necessary for proper fiber assembly. Ram et al. report that gas1Δ cells secrete wall modules into the growth medium, consistent with a lattice assembly defect (44).
We believe that these results could be due to decreased cross-linking between β1,3 and β1,6 glucans or to decreased intertwining of the β1,3 glucan chains into fibers. The yeast cell could compensate for this defect by altering the composition of the modules: there would be an increased reliance on cross-linking between β1,6 glucan and chitin and/or increased cellular content of β1,6 glucan at the expense of β1,3 glucan. Therefore, a kre6Δ mutation (affecting synthesis of β1,6 glucan) would be synthetically lethal with gas1Δ, as observed (43). gas1Δ chs3Δ (chitin synthetase III) double mutants have a severe growth defect, as expected for cells dependent on chitin synthesis for cell wall integrity (43).
This interpretation has been validated by Kapteyn et al. (26), who investigated gas1Δ and fks1Δ cells. The latter have a reduced content of β1,3 glucan due to mutation in the β1,3 glucan synthase. In both mutants there is a 15- to 30-fold increase in chitin content and in cross-linking of chitin to the β1,6 glucan. This alteration maintains the insolubility and integrity of the wall in the face of loss or faulty assembly of the β1,3 glucan. Module structure implies that gas1Δ fks1Δ double mutants should have a phenotype similar to the gas1Δ cells, because the latter mutation itself reduces the role of the β1,3 glucan in wall structure. Indeed the single and double mutants are similar (26). The results illustrate the flexibility of modular structure and suggest a structure for modules in fungi with chitin instead of β1,3 glucan as the major fibrous wall component: direct linking of β1,6 glucan-glycoprotein complexes to chitin fibers (1, 42).
Challenges.
There is little understanding of the processes that result in extracellular assembly of the components into a wall. A start on the problem might be based on the timing of the cross-linking of the wall components to form modules. The kinetics of anchorage of α-agglutinin offer some initial clues, with the caution that this case represents pheromone-induced incorporation and may not be typical (34). Within 5 min of appearance of the GPI-anchored protein at the cell surface, membrane anchorage is lost, with concomitant loss of label in fatty acids and inositol. A transient soluble form appears and is rapidly chased into the wall-bound form associated with β1,6 glucan. In the next hour, the α-agglutinin becomes less soluble and more difficult to extract.
A model consistent with this result and the structure of modules is that the GPI-anchored protein is released from the membrane by action of a transglycosidase that cuts between the first mannose and the glucosamine residue (28) (Fig. 2). By analogy to other transglycosidases, there must be an “activated” intermediate form of the glycosyl donor, which would preserve bond energy to allow formation of a new glycosidic bond. The glycoprotein moves to the outer layer of the wall, where it is linked to β1,6 glucan already associated with insoluble β1,3 glucan (25, 28). The amount of glycoprotein extractable by treatment with β1,3 glucanase then decreases as cross-links and chitin are added to the modules later in the cell cycle and the complex becomes more insoluble (20). This scenario predicts that association of the β1,3 and β1,6 glucans precedes bonding to mannoproteins, that chitin addition is a late event, and that there are modules without associated mannoproteins or chitin, as already demonstrated (20, 28). Validation of this or other models must await development of suitable cell-free assays for cell wall anchorage, so that substrates and products can be defined and individual steps can be dissected.
How are module components localized in walls before cross-linking? Two intriguing results hint at a role for mannoproteins. Flo1p, a component of yeast flocculins is seen in linear transwall fibers or channels when overexpressed (3). Thus, there may be transport routes through the wall to facilitate assembly. A chaperone-like protein in the walls of C. albicans has been reported (33). Such a protein might be involved in transwall transport or in delaying transglycosylation until the mannoprotein reaches an appropriate venue.
Another challenge will be the description of the processes and reactions leading to assembly and alteration of wall structures. The wall is plastic in many ways. It is “softened” for bud emergence, expands during bud growth, is modified by addition of bud scars, and becomes more refractory as it ages. The wall is remodeled during mating, cell fusion, pseudohypha formation, and formation of spore walls with phenolic cross-links (5, 42). This problem is analogous to that of wall softening in plant cells for growth and maturation (10, 15, 47, 48, 51). Genetic approaches now suggest that the number of genes involved in wall synthesis, assembly, and remodeling will be in the hundreds (36, 42). This degree of complexity is expected for synthesis and assembly of this complex, plastic organelle, which involves a major commitment of cellular resources (9, 36, 42, 45).
CONCLUSION
The discovery of a defined covalent complex composed of yeast mannoprotein, β1,6 glucan, β1,3 glucan, and chitin has changed our thinking about cell wall structure and assembly. The resulting modular model (28), along with the database of gene sequences and genetic studies of the biogenesis of the glycoconjugates (7, 35, 36), allows us to make testable predictions for cross-linking reactions and assembly pathways (26). Specifically, the structure of the modules shows us that there must be enzymes that link each pair of components and others that interlink the modules. The extracellular locations of the products of β1,3 glucan and chitin synthesis and of cross-linking of α-agglutinin to modules suggest that these processes occur exterior to the plasma membrane.
ACKNOWLEDGMENTS
We thank S. Marvin Friedman and Chong K. Jue for their thoughts and comments.
This work was supported National Institute of General Medical Sciences grant GM 47176 to Janet Kurjan and by the Research Centers in Minority Institutions program of NIH, grant RR03037.
Footnotes
We dedicate this paper to Erwin Fleissner who, as Dean of Sciences and Mathematics, fostered the research environment at Hunter and founded the Institute for Biomolecular Structure and Function.
REFERENCES
- 1.Bartnicki-Garcia S, Persson J, Chanzy H. An electron microscope and electron diffraction study of the effect of calcofluor white on the biosynthesis of chitin in vitro. Arch Biochem Biophys. 1994;310:6–15. doi: 10.1006/abbi.1994.1133. [DOI] [PubMed] [Google Scholar]
- 2.Blackwell J. The macromolecular organization of cellulose and chitin. In: Brown R M Jr, editor. Cellulose and other natural polymer systems. New York, N.Y: Plenum Press; 1982. pp. 403–428. [Google Scholar]
- 3.Bony M, Thines-Semproux D, Barre P, Blondin B. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J Bacteriol. 1997;179:4929–4936. doi: 10.1128/jb.179.15.4929-4936.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabib E, Bowers B, Roberts R L. Vectorial synthesis of a polysaccharide by isolated plasma membranes. Proc Natl Acad Sci USA. 1983;80:3318–3321. doi: 10.1073/pnas.80.11.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabib E, Drgon T, Drgonova J, Ford R A, Kollar R. The yeast cell wall, a dynamic structure engaged in growth and morphogenesis. Biochem Soc Trans. 1997;25:200–204. doi: 10.1042/bst0250200. [DOI] [PubMed] [Google Scholar]
- 6.Cappellaro C, Baldermann C, Rachel R, Tanner W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and α-agglutinin. EMBO J. 1994;13:4737–4744. doi: 10.1002/j.1460-2075.1994.tb06799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caro L H P, Tettelin H, Vossen J H, Ram A F J, van den Ende H, Klis F M. In silicio identification of glycosyl-phosphoatidylinositol-anchored plasma membrane and wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1447–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Chen M-H, Shen Z-M, Bobin S, Kahn P C, Lipke P N. Structure of Saccharomyces cerevisiae α-agglutinin: evidence for a yeast cell wall protein with multiple immunoglobulin-like domains with atypical disulfides. J Biol Chem. 1995;270:26168–26177. doi: 10.1074/jbc.270.44.26168. [DOI] [PubMed] [Google Scholar]
- 9.Cid V J, Duran A, del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosgrove D J. Creeping, walls, softening fruit, and penetrating pollen tubes: the growing role of expansins. Proc Natl Acad Sci USA. 1997;94:5504–5505. doi: 10.1073/pnas.94.11.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Nobel H, Lipke P N. Is there a role for GPIs in cell wall assembly in yeast? Trends Cell Biol. 1994;4:42–45. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 12.De Nobel J G, Klis F M, Priem J, Munnik T, van den Ende H. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast. 1990;6:491–499. doi: 10.1002/yea.320060606. [DOI] [PubMed] [Google Scholar]
- 13.Dijkgraaf G J P, Brown J L, Bussey H. The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast. 1996;12:683–692. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C683::AID-YEA959%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Frevert J, Ballou C E. Saccharomyces cerevisiae structural cell wall mannoprotein. Biochemistry. 1985;24:753–759. doi: 10.1021/bi00324a033. [DOI] [PubMed] [Google Scholar]
- 15.Fry S C. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- 16.Gatti E, Popolo L, Vai M, Rota N, Alberghina L. O-linked oligosaccharides in yeast glycosyl phosphatidylinositol-anchored protein gp115 are clustered in a serine-rich region not essential for its function. J Biol Chem. 1994;269:19695–19700. [PubMed] [Google Scholar]
- 17.Gentzsch M, Tanner W. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology. 1997;7:481–486. doi: 10.1093/glycob/7.4.481. [DOI] [PubMed] [Google Scholar]
- 18.Goldman R C, Sullivan P A, Zakula D, Copobianco J O. Kinetics of β1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur J Biochem. 1995;227:372–378. doi: 10.1111/j.1432-1033.1995.tb20399.x. [DOI] [PubMed] [Google Scholar]
- 19.Hanson B A, Lester R L. Effect of inositol starvation on the in vitro syntheses of mannan and N-acetylglucosaminylpyrophosphoryldolichol in Saccharomyces cerevisiae. J Bacteriol. 1982;151:334–342. doi: 10.1128/jb.151.1.334-342.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartland R P, Vermeulen C A, Klis F M, Sietsma J H, Wessels J G. The linkage of (1-3)-β-glucan to chitin during cell wall assembly in Saccharomyces cerevisiae. Yeast. 1994;10:1591–1599. doi: 10.1002/yea.320101208. [DOI] [PubMed] [Google Scholar]
- 21.Horvath A, Sutterlin C, Manning-Krieg U, Movva N R, Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15:291–295. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser C, Gimeno R, Shaywitz D. Protein secretion, membrane biogenesis, and endocytosis. In: Pringle J, Broach J, Jones E, editors. Molecular and cellular biology of the yeast Saccharomyces. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 91–227. [Google Scholar]
- 24.Kapteyn J C, Montijn R C, Dijkgraaf G J, Van den Ende H, Klis F M. Covalent association of β-1,3-glucan with β-1,6-glucosylated mannoproteins in cell walls of Candida albicans. J Bacteriol. 1995;177:3788–3792. doi: 10.1128/jb.177.13.3788-3792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapteyn J C, Montijn R C, Vink E, de la Cruz J, Llobell A, Douwes J E, Shimoi H, Lipke P N, Klis F M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 26.Kapteyn J C, Ram A F, Groos E M, Kollar R, Montijn R C, Van Den Ende H, Llobell A, Cabib E, Klis F M. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollar R, Petrakova E, Ashwell G, Robbins P W, Cabib E. Architecture of the yeast cell wall. The linkage between chitin and β(1→3)-glucan. J Biol Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 28.Kollar R, Reinhold B B, Petrakova E, Yeh H J, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall. β(1→6)-glucan interconnects mannoprotein, β(1→)3-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 29.Kopecka M, Kreger D R. Assembly of microfibrils in vivo and in vitro from 1-3β-D-glucan synthesized by protoplasts of Saccharomyces cerevisiae. Arch Microbiol. 1986;143:387–395. doi: 10.1007/BF00412807. [DOI] [PubMed] [Google Scholar]
- 30.Kopecka M, Phaff H J, Fleet G H. Demonstration of a fibrillar component in the cell wall in the yeast Saccharomyces cerevisiae and its chemical nature. J Cell Biol. 1974;62:66–76. doi: 10.1083/jcb.62.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krainer E, Stark R E, Naider F, Alagramam K, Becker J M. Direct observation of cell wall glucans in whole cells of Saccharomyces cerevisiae by magic-angle spinning 13C-NMR. Biopolymers. 1994;34:1627–1635. doi: 10.1002/bip.360341207. [DOI] [PubMed] [Google Scholar]
- 32.Leidich S D, Kostova Z, Latek R R, Costello L C, Drapp D A, Gray W, Fassler J S, Orlean P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J Biol Chem. 1995;270:13029–13035. doi: 10.1074/jbc.270.22.13029. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Ribot J L, Alloush H M, Masten B J, Chaffin W L. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect Immun. 1996;64:3333–3340. doi: 10.1128/iai.64.8.3333-3340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C-F, Montijn R C, Brown J L, Klis F, Kurjan J, Bussey H, Lipke P N. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β1,6-glucan in the S. cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lussier M, Sdicu A M, Winnett E, Vo D H, Sheraton J, Storms R K, Bussey H. Completion of the Saccharomyces cerevisiae genome allows identification of KTR5, KTR6, and KTR7 and definition of the nine-membered KRE2/MNT1 mannosyltransferase gene family in this organism. Yeast. 1997;13:267–274. doi: 10.1002/(SICI)1097-0061(19970315)13:3<267::AID-YEA72>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 36.Lussier M, White A M, Sheraton J, di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manners D J, Masson A J, Patterson J C. The structure of a β1,3-D-glucan from yeast cell walls. Biochem J. 1973;135:19–30. doi: 10.1042/bj1350019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montijn R C, Van Wolven P, De Hoog S, Klis F M. β-Glucosylated proteins in the cell wall of the black yeast Exophiala (Wangiella) dermatitidis. Microbiology. 1997;143:1673–1680. doi: 10.1099/00221287-143-5-1673. [DOI] [PubMed] [Google Scholar]
- 39.Mrsa V, Seidl T, Gentzsch M, Tanner W. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1145–1154. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Muller A, Ensley H, McNamee R, Jones E, McLaughlin E, Chandley W, Browder W, Lowman D, Williams D. The application of various protic acids to the extraction of 1-3β-D-glucan from Saccharomyces cerevisiae. Carbohydr Res. 1997;299:203–208. doi: 10.1016/s0008-6215(97)00004-9. [DOI] [PubMed] [Google Scholar]
- 41.Muller G, Gross E, Wied S, Bandlow W. Glucose-induced sequential processing of a glycosyl-phosphatidylinositol-anchored ectoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:442–456. doi: 10.1128/mcb.16.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlean P. Biogenesis of yeast wall and surface components. In: Pringle J, Broach J, Jones E, editors. Molecular and cellular biology of the yeast Saccharomyces. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- 43.Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ram A F J, Kapteyn J C, Montijn R C, Caro L H P, Douwes J E, Baginsky W, Mazur P, van den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in release of β1,3 glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ram A F, Wolters A, Ten Hoopen R, Klis F M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- 46.Saito H, Yoshioka Y, Yoloi M, Yamada J. Distinct gelation mechanisms between linear and branched 1-3β-D-glucans as revealed by high resolution solid state 13C NMR. Biopolymers. 1990;29:1689–1698. doi: 10.1002/bip.360291402. [DOI] [PubMed] [Google Scholar]
- 47.Schopfer P. Hydrogen peroxide mediated cell wall stiffening in vitro in maize coleoptiles. Planta. 1996;199:43–49. [Google Scholar]
- 48.Showalter A M. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skrzypek M, Lester R L, Dickson R C. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. J Bacteriol. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stokke B T, Elgsaeter A, Hara C, Kitamura S, Takeo K. Physicochemical properties of 1-6-branched 1-3 β-D-glucans. 1. Physical dimensions estimated from hydrodynamic and electron microscopic data. Biopolymers. 1993;33:561–573. doi: 10.1002/bip.360330406. [DOI] [PubMed] [Google Scholar]
- 51.Tabuchi A, Kamisaka S, Hoson T. Purification of xyloglucan hydrolase/endotransferase from cell walls of azuki bean epicotyls. Plant Cell Physiol. 1997;38:653–658. [Google Scholar]
- 52.Van der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Der Vaart J M, te Biesebeke R, Chapman J W, Klis F M, Verrips C T. The β-1, 6-glucan containing side-chain of cell wall proteins of Saccharomyces cerevisiae is bound to the glycan core of the GPI moiety. FEMS Microbiol Lett. 1996;145:401–407. doi: 10.1111/j.1574-6968.1996.tb08607.x. [DOI] [PubMed] [Google Scholar]
- 54.Vossen J H, Muller W H, Lipke P N, Klis F M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J Bacteriol. 1997;179:2202–2209. doi: 10.1128/jb.179.7.2202-2209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams D L, McNamee R B, Jones E L, Pretus H A, Ensley H E, Browder I W, Di Luzio N R. A method for the solubilization of 1-3β-D-glucan isolated from Saccharomyces cerevisiae. Carbohydr Res. 1991;219:203–213. doi: 10.1016/0008-6215(91)89052-h. [DOI] [PubMed] [Google Scholar]
- 56.Yen P H, Ballou C E. Partial characterization of the sexual agglutination factor from Hansenula wingei Y-2340 type 5 cells. Biochemistry. 1974;13:2428–2437. doi: 10.1021/bi00708a030. [DOI] [PubMed] [Google Scholar]
- 57.Zlotnik H, Fernandez P, Bowers B, Cabib E. Saccharomyces cerevisiae mannoproteins form an external wall layer that determines porosity. J Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]