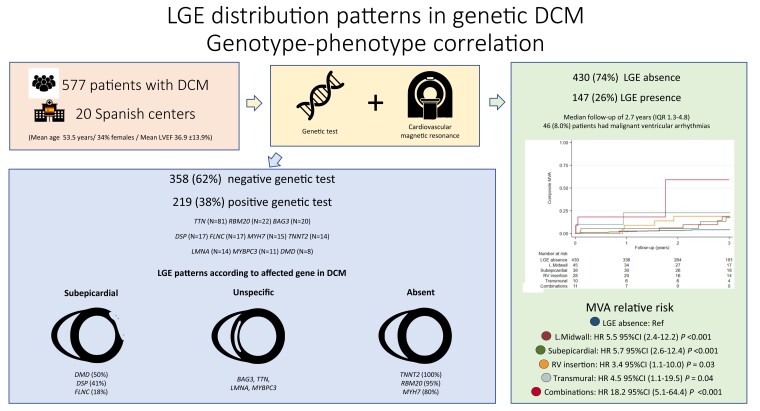
Abstract
Aims
Late gadolinium enhancement (LGE) is frequently found in patients with dilated cardiomyopathy (DCM); there is little information about its frequency and distribution pattern according to the underlying genetic substrate. We sought to describe LGE patterns according to genotypes and to analyse the risk of major ventricular arrhythmias (MVA) according to patterns.
Methods and results
Cardiac magnetic resonance findings and LGE distribution according to genetics were performed in a cohort of 600 DCM patients followed at 20 Spanish centres. After exclusion of individuals with multiple causative gene variants or with variants in infrequent DCM-causing genes, 577 patients (34% females, mean age 53.5 years, left ventricular ejection fraction 36.9 ± 13.9%) conformed to the final cohort. A causative genetic variant was identified in 219 (38%) patients, and 147 (25.5%) had LGE. Significant differences were found comparing LGE patterns between genes (P < 0.001). LGE was absent or rare in patients with variants in TNNT2, RBM20, and MYH7 (0, 5, and 20%, respectively). Patients with variants in DMD, DSP, and FLNC showed a predominance of LGE subepicardial patterns (50, 41, and 18%, respectively), whereas patients with variants in TTN, BAG3, LMNA, and MYBPC3 showed unspecific LGE patterns. The genetic yield differed according to LGE patterns. Patients with subepicardial, lineal midwall, transmural, and right ventricular insertion points or with combinations of LGE patterns showed an increased risk of MVA compared with patients without LGE.
Conclusion
LGE patterns in DCM have a specific distribution according to the affected gene. Certain LGE patterns are associated with an increased risk of MVA and with an increased yield of genetic testing.
Keywords: dilated cardiomyopathy, late gadolinium enhancement, cardiac magnetic resonance, genetics, sudden cardiac death
Graphical Abstract
Graphical Abstract.
A cohort of 577 individuals with DCM phenotyped with genetic testing and CMR was analysed. A causative genetic variant was identified in 219 (38%) patients, and 147 (25.5%) had LGE. LGE patterns in genetic DCM have a specific distribution. Patients with subepicardial, lineal midwall, transmural, and right ventricular insertion points or with combinations of LGE patterns showed an increased risk of MVA compared with patients without LGE. Abbreviations: CMR, cardiovascular magnetic resonance; DCM, dilated cardiomyopathy; HR, hazard ratio; IQR, interquartile range; LGE, late gadolinium enhancement; L.Midwall, lineal midwall; LVEF, left ventricular ejection fraction; MVA, major ventricular arrhythmias; RV, right ventricle.
Introduction
Cardiovascular magnetic resonance (CMR) has become a key tool in the assessment of patients with dilated cardiomyopathy (DCM), allowing cardiologists to obtain detailed tissue characterization of the myocardium with a special focus on areas of fibrosis based on late gadolinium enhancement (LGE) presence. Recent observational studies have shown the pivotal role of LGE in sudden cardiac death (SCD) risk assessment, placing CMR in the spotlight of evaluation of patients with DCM.1–6
During the last decade, several publications with gene-specific cohorts have provided relevant clues to describe genotype–phenotype correlations in the most prevalent genes associated with DCM including TTN,7DSP,8,9 and LMNA.10 Nevertheless, most cohorts neglected detailed information about CMR findings, particularly regarding LGE patterns, or this information was available only in a small subset of patients.11,12
The relevance of establishing genotype–phenotype associations is bidirectional. First of all, it could provide more evidence about different mechanisms of disease pathogenesis based on the genetic substrate. Conversely, defining specific features could improve variant classification in genes involved in DCM based on phenotype-enhanced tools. In addition, determining LGE patterns is important because certain LGE patterns like the ring-like pattern have been associated with a more adverse clinical course and a higher susceptibility to ventricular arrhythmias.13 Despite its relevance, so far, only one cohort of 89 individuals with detailed information regarding LGE patterns in genetic DCM has been published.14 The limited sample size of that study did not allow a detailed description of LGE patterns associated with multiple genes and to evaluate the relationship of LGE patterns with clinical outcomes.
With this background, in the present study, we sought to describe CMR genotype–LGE phenotype associations in a large cohort of genetically studied DCM patients. Additionally, we examined the frequency of malignant ventricular arrhythmias (MVA) during follow-up according to CMR LGE patterns.
Methods
Study design
This is a subanalysis of a multicentre registry of 600 patients with non-ischaemic DCM evaluated at 20 Spanish hospitals. Detailed information about cohort composition has been previously published.1 In summary, DCM patients were included in the study if they had a CMR and had undergone genetic testing with a panel of at least 50 DCM-associated genes at participating institutions between 2015 and 2021 or had a DCM-causing genetic variant previously identified in a relative with DCM using targeted next-generation sequencing (NGS) panels. DCM was defined as the presence of left ventricular ejection fraction (LVEF) < 50% on the echocardiogram at diagnosis in the absence of abnormal loading conditions, coronary artery disease (by coronary angiography or computed tomography), excessive alcohol consumption, or any other identifiable causes (inflammation, toxic exposure). Additionally, we included consecutive relatives with DCM who had CMR performed at participating institutions and who harboured a pathogenic or likely pathogenic genetic variant previously identified in a DCM proband. Only patients with age ≥ 15 years at the time of diagnosis were included. The study was approved by the Hospital Universitario Puerta de Hierro ethics committee and conformed to the principles of the Helsinki Declaration. The authors from each participating centre guarantee the integrity of the data.
CMR
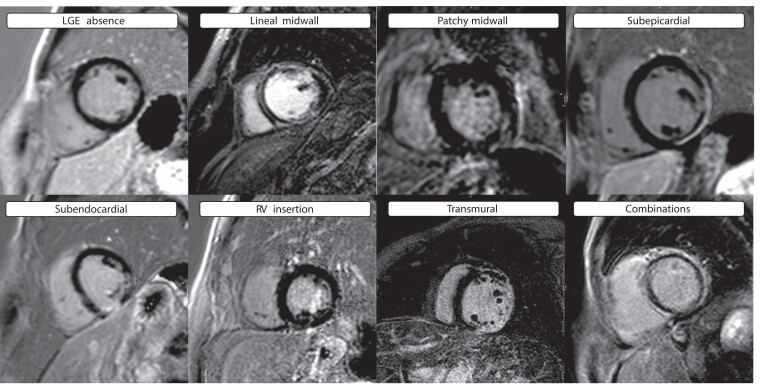
All patients underwent CMR imaging on a 1/1.5/3.0 T CMR scanner for assessment of LVEF and myocardial scars. Steady-state free precession cine images were acquired in multiple short-axis and three long-axis views. LGE was obtained using a segmented inversion recovery gradient echo technique in identical views as cine CMR 10–15 min after 0.15 mmol/kg of gadolinium contrast administration. Cine and LGE images from all centres were centrally evaluated in a core laboratory blinded to genotypes and outcomes. Patients who received an implantable cardioverter defibrillator (ICD) or cardiac resynchronization therapy (CRT) had images acquired before device implantation. The presence and location of hyperenhanced tissue on LGE, which was interpreted as representing the scarred myocardium, were determined by visual inspection. LGE patterns were categorized in eight categories: absence, lineal midwall, patchy midwall, subepicardial, subendocardial, right ventricular (RV) insertion, transmural, and combinations. This classification has been used by other groups, and it is commonly used in the clinical setting.6 Examples of these patterns extracted from the cohort are displayed in Figure 1. A simplified categorization with the four main groups (absence, midwall, subepicardial, and others) was also applied to summarize results and reduce granularity. Scar size (extent) as a percentage of the left ventricular (LV) myocardium was quantified with semi-automated planimetry (manually corrected) using the full-width half-max thresholding method.15 Furthermore, the association between LGE presence according to LVEF (LVEF ≤ 35% vs. >35%) and according to LV end-diastolic volume (LVEDV) was assessed for each gene. Lastly, we analysed the yield of genetic testing according to LGE patterns in probands based on simplified LGE categories.
Figure 1.
Examples of LGE patterns extracted from the cohort. Abbreviations: LGE, late gadolinium enhancement; RV, right ventricle.
Genotype
Genetic variant interpretation was centrally curated following the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) recommendations as previously described.1,16 Genetic variants were centrally classified as pathogenic (P), likely pathogenic (LP), unknown significance (VUS), or likely benign/benign (LB/B) after a systematic review by a cardiologist expert in cardiovascular genetics (J.P.O.). A variant was considered disease-causing if it affected a DCM-related gene and was classified P/LP. The frequencies of variants in the general population were extracted from the gnomAD database v2.1.1.22.
Patients harbouring variants of unknown significance and likely benign/benign were clustered in a genotype-negative group that served as a reference. To establish clear genotype–phenotype correlations, six patients were excluded from analysis because they had ≥2 disease-causing variants in more than one gene. In addition, 17 patients with DCM-causing variants in genes that were present in <8 individuals in the cohort were excluded, as 8 was considered the minimum number of patients required to draw conclusions regarding the LGE pattern associated with a certain gene.
Outcomes
MVA during follow-up were registered. MVA included SCD or aborted SCD, sustained ventricular tachycardia, and appropriate ICD interventions. Only appropriate ICD shocks to terminate ventricular tachycardia or ventricular fibrillation episodes were considered for the purpose of this study (anti-tachycardia pacing therapy was not considered). Cardiovascular mortality and overall mortality were also retrieved. Follow-up started on the date of CMR and finished at the last follow-up. Follow-up was censored in case of heart transplantation, non-MVA-related death, or last follow-up.
Statistical analyses
Continuous variables are expressed as mean ± standard deviation (SD) or median [interquartile range (IQR)], as appropriate. Groups were compared using Student’s t-test, the Mann–Whitney test, the Analysis of Variance (ANOVA) test, or the Kruskal-Wallis test when comparing more than two groups. Non-continuous categorical variables were expressed as counts (percentages) and compared using the χ2 test or Fisher’s exact test, as appropriate. The cumulative probability of an event during follow-up was estimated using the Kaplan–Meier method and Cox regression, and the log-rank test was used to compare survival between groups. Analyses were performed using Stata Statistics version 15 (StataCorp LLC, TX, USA). A two-sided P value < 0.05 defined statistical significance.
Results
Study participants and baseline characteristics
A total of 577 patients with DCM conformed to the final study cohort including 516 (89.4%) DCM probands and 61 (10.6%) relatives with DCM diagnosed through cascade screening. A causative genetic variant in DCM-related genes was identified in 219 (38%), whereas 358 (62.0%) were genotype negative. Table 1 shows the distribution of patients according to genes and baseline characteristics. Patients with truncating variants in TTN (n = 81) represented one-third of the genotype-positive cohort, with the remaining genes ranging from 8 to 22 patients. The mean age was 53.5 ± 14.1 years, with significant differences between genes (P < 0.001). Patients with variants in TNNT2 (43.9 ± 17.4) and LMNA (44.5 ± 11.9) were younger, and those with variants in MYBPC3 (57.0 ± 11.5) and MYH7 (56.4 ± 13.5) were older. Left bundle branch block (LBBB) was unequally distributed among groups (P < 0.001), with patients in the genotype-negative group having a higher prevalence (n = 157, 43.9%), followed by LMNA (n = 4, 28.6%). Significant differences were also found in the prevalence of low QRS voltage in limb leads, with DSP and FLNC patients showing a higher prevalence (n = 7, 41.2% and n = 6, 35.3%, respectively).
Table 1.
Baseline characteristics according to genotypes
| Overall (n = 577) | Gene negative (n = 358) | TTN (n = 81) | RBM20 (n = 22) | BAG3 (n = 20) | DSP (n = 17) | FLNC (n = 17) | MYH7 (n = 15) | TNNT2 (n = 14) | LMNA (n = 14) | MYBPC3 (n = 11) | DMD (n = 8) | P value* | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 53.5 (±14.1) | 55.2 (±13.5) | 53.6 (±15.3) | 50.2 (±14.4) | 49.7 (±11.8) | 47.5 (±12.5) | 46.8 (±13.4) | 56.4 (±13.5) | 43.9 (±17.4) | 44.5 (±11.9) | 57.0 (±11.5) | 45.7 (±15.1) | <0.001 |
| Female sex | 196 (34.0%) | 113 (31.6%) | 26 (32.1%) | 13 (59.1%) | 5 (25%) | 12 (70.6%) | 8 (47.1%) | 8 (53.3%) | 5 (35.7%) | 5 (35.7%) | 0 (0%) | 2 (25.0%) | 0.66 |
| Clinical status | |||||||||||||
| NYHA | 0.03 | ||||||||||||
| NYHA I | 219 (38.0%) | 114 (31.8%) | 36 (44.4%) | 11 (50%) | 7 (35%) | 10 (58.8%) | 13 (76.5%) | 7 (46.7%) | 7 (50%) | 9 (64.3%) | 2 (18.2%) | 3 (37.5%) | |
| NYHA II | 178 (30.8%) | 129 (36.0%) | 14 (17.3%) | 5 (22.7%) | 7 (35%) | 4 (23.5%) | 3 (17.7%) | 5 (33.3%) | 2 (14.3%) | 2 (14.3%) | 5 (45.5) | 2 (25.0%) | |
| NYHA III | 154 (26.7%) | 97 (27.1%) | 27 (33.3%) | 6 (27.3%) | 5 (25%) | 3 (17.7%) | 1 (5.9%) | 3 (20%) | 2 (21.4%) | 3 (21.4%) | 4 (36.4%) | 2 (25.0%) | |
| NYHA IV | 26 (4.5%) | 18 (5.0%) | 4 (4.9%) | 0 (0%) | 1 (5%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (14.3%) | 0 (0%) | 0 (0%) | 1 (12.5%) | |
| NTProBNP (pg/mL) | 2637.6 (±4715.2) | 2962.9 (±5044.8) | 3282.7 (±5794.3) | 267.7 (±155.3) | 2315.1 (±3114.7) | 1166.0 (±1379.7) | 235.7 (±346.5) | 716.7 (±771.3) | 2540.6 (±3063.9) | 1326.9 (±1758.7) | 1042.1 (±684.0) | 2116.0 (±1474.4) | 0.04 |
| Skeletal muscle disease | 16 (2.8%) | 8 (2.2%) | 0 (0%) | 0 (0%) | 1 (5%) | 0 (0%) | 0 (0%) | 3 (20%) | 0 (0%) | 2 (14.3%) | 2 (18.2%) | 0 (0%) | 0.04 |
| Devices | |||||||||||||
| ICD | 161 (27.9%) | 89 (24.9%) | 19 (23.5%) | 6 (27.3%) | 8 (40%) | 9 (52.9%) | 8 (47.1%) | 4 (26.7%) | 2 (14.3%) | 9 (64.3%) | 4 (36.4%) | 3 (37.5%) | 0.01 |
| CRT | 63 (10.9%) | 51 (14.2%) | 4 (4.94%) | 2 (9.1%) | 2 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (21.4%) | 1 (9.1%) | 0 (0%) | 0.05 |
| ECG | |||||||||||||
| AF | 46 (8.0%) | 34 (9.5%) | 6 (7.41%) | 1 (4.5%) | 0 (0%) | 2 (11.8%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (7.1%) | 2 (18.2%) | 0 (0%) | 0.06 |
| LBBB | 179 (31.0%) | 157 (43.9%) | 6 (7.41%) | 4 (18.2%) | 1 (5%) | 1 (5.9%) | 0 (0%) | 2 (13.3%) | 2 (14.3%) | 4 (28.6%) | 2 (18.2%) | 0 (0%) | <0.001 |
| Complete AVB | 55 (9.5%) | 38 (10.6%) | 8 (9.9%) | 1 (4.5%) | 1 (5%) | 0 (0%) | 0 (0%) | 1 (6.7%) | 1 (7.1%) | 5 (35.7%) | 0 (0%) | 0 (0%) | 0.56 |
| Abnormal TWI | 221 (38.3%) | 141 (39.4%) | 33 (40.8%) | 3 (13.6%) | 7 (35%) | 8 (47.1%) | 6 (35.3%) | 7 (46.7%) | 6 (42.9%) | 3 (21.4%) | 5 (45.5%) | 2 (25.0%) | 0.21 |
| Low QRS V limb leads | 60 (10.4%) | 18 (5.0%) | 14 (17.3%) | 0 (0%) | 4 (20%) | 7 (41.2%) | 6 (35.3%) | 1 (6.7%) | 3 (21.4%) | 3 (21.4%) | 3 (27.3%) | 1 (12.5%) | <0.001 |
| Low QRS V precordial leads | 15 (2.6%) | 6 (1.7%) | 3 (3.7%) | 0 (0%) | 0 (0%) | 1 (5.9%) | 3 (17.6%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (9.1%) | 1 (12.5%) | 0.13 |
Abbreviations: AF, atrial fibrillation; AVB, atrioventricular block; CRT, cardiac resynchronization therapy; ECG, electrocardiogram; ICD, implantable cardioverter defibrillator; LBBB, left bundle branch block; NYHA, New York Heart Association; TWI, T wave inversion; V, voltage.
* P value refers to overall analysis between groups.
CMR measurements
Table 2 shows the main CMR findings according to genotypes. The overall mean LVEF assessed by CMR was 36.9 ± 13.5% without significant differences between groups (P = 0.13). In contrast, significant differences were found comparing indexed RV end-diastolic volume (RVEDV) (P = 0.001) and indexed RV mass (P = 0.001) with variants in LMNA and TNNT2 showing higher values for the former parameter and TNNT2 and DMD for the latter. No differences were found regarding other parameters including indexed LVEDV, indexed LV mass, left and right atrial volumes, and RV ejection fraction (RVEF).
Table 2.
CMR findings according to genotypes
| Overall (n = 577) | Gene negative (n = 358) | TTN (n = 81) | RBM20 (n = 22) | BAG3 (n = 20) | DSP (n = 17) | FLNC (n = 17) | MYH7 (n = 15) | TNNT2 (n = 14) | LMNA (n = 14) | MYBPC3 (n = 11) | DMD (n = 8) | P value* | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVEDV, mL | 241 (±81.1) | 249 (±86.4) | 225 (±66.5) | 221 (±72.8) | 248 (±74.9) | 217 (±84.1) | 195 (±42.3) | 220 (±53.9) | 255 (±75.6) | 243 (±72.3) | 238 (±83.4) | 276 (±75.0) | 0.50 |
| LVEDV index, mL/m2 | 128 (±39.9) | 130 (±42.3) | 75.7 (±20.6) | 124 (±38.0) | 132 (±48.0) | 121 (±36.2) | 110 (±21.6) | 120 (±24.1) | 137 (±39.8) | 130 (±34.1) | 121 (±40.7) | 148 (±42.9) | 0.97 |
| LV mass, g | 145 (±44.1) | 153 (±46.0) | 130 (±34.3) | 130 (±40.5) | 143 (±44.2) | 106 (±39.3) | 123 (±26.6) | 123 (±27.9) | 148 (±30.6) | 134 (±33.8) | 149 (±38.4) | 159 (±52.1) | 0.01 |
| LV mass index, g/m2 | 76.7 (±20.7) | 80.2 (±21.7) | 69.3 (±14.9) | 72.9 (±21.9) | 75.5 (±22.7) | 58.8 (±15.8) | 68.9 (±12.1) | 67.3 (±10.5) | 78.9 (±16.5) | 71.8 (±13.3) | 76.2 (±19.9) | 84.9 (±27.2) | 0.06 |
| LVEF, % | 36.9 (±13.5) | 36.1 (±13.5) | 36.3 (±13.0) | 39.7 (±16.5) | 39.3 (±15.5) | 40.7 (±10.2) | 44.1 (±9.2) | 40.2 (±11.1) | 36.9 (±14.6) | 42.9 (±13.6) | 32.0 (±12.6) | 31.2 (±13.4) | 0.13 |
| RVEDV, mL | 150 (±49.9) | 151 (±51.5) | 142 (±44.1) | 147 (±44.2) | 162 (±55.7) | 150 (±36.0) | 141 (±34.5) | 138 (±53.2) | 175 (±53.3) | 180 (±61.7) | 143 (±39.6) | 164 (±49.2) | 0.05 |
| RVEDV index, mL/m2 | 79.1 (±22.9) | 78.2 (±23.2) | 75.7 (±20.6) | 81.2 (±17.8) | 85 (±28.3) | 84.6 (±18.7) | 79.6 (±18.5) | 74.8 (±23.4) | 92.9 (±25.9) | 94.9 (±23.3) | 73.6 (±21.1) | 87.4 (±26.2) | 0.001 |
| RV mass, g | 40.0 (±13.1) | 40.1 (±13.0) | 37.6 (±16.2) | 41.0 (±15.3) | 42.4 (±14.7) | 36.2 (±12.0) | 35.7 (±6.8) | 40.5 (±14.8) | 50.1 (±13.9) | 44.4 (±18.8) | 38.5 (±16.4) | 43.9 (±14.3) | 0.04 |
| RV mass index, g/m2 | 21.1 (±5.9) | 20.9 (±5.74) | 20.0 (±4.91) | 22.5 (±6.35) | 22.2 (±7.0) | 20.3 (±5.8) | 20.1 (±3.2) | 22.1 (±7.7) | 26.7 (±7.3) | 23.3 (±7.2) | 19.6 (±7.7) | 23.6 (±8.8) | 0.001 |
| RVEF, % | 45.8 (±15.3) | 46.0 (±15.1) | 45.4 (±14.1) | 46.5 (±14.7) | 40.5 (±18.5) | 47.4 (±16.2) | 50.2 (±10.3) | 52.1 (±16.4) | 42.6 (±20.6) | 46.8 (±18.4) | 41.5 (±16.6) | 41.4 (±13.0) | 0.34 |
| LA max vol, mL | 95.0 (±49.7) | 97.4 (±53.8) | 99.0 (±43.1) | 70.7 (±32.8) | 84.8 (±40.7) | 76.6 (±38.9) | 68.1 (±28.0) | 85.8 (±35.1) | 87.4 (±42.1) | 108 (±43.2) | 116 (±50.2) | 111 (±41.3) | 0.34 |
| LAEF, % | 39.8 (±17.2) | 39.2 (±17.3) | 37.3 (±16.2) | 42.1 (±19.0) | 42.2 (±17.4) | 50.2 (±9.3) | 54.6 (±7.7) | 46.0 (±10.5) | 45.1 (±20.7) | 33.7 (±15.3) | 26.7 (±17.1) | 36.2 (±18.4) | 0.42 |
| RA max vol, mL | 69.1 (±32.7) | 69.1 (±33.7) | 69.4 (±27.9) | 63.3 (±23.0) | 64.2 (±24.8) | 64.4 (±26.6) | 63.8 (±19.6) | 52.5 (±21.1) | 71.8 (±20.9) | 92.6 (±54.1) | 90.4 (±56.9) | 71.4 (±31.5) | 0.25 |
| RAEF, % | 35.6 (±14.8) | 34.1 (±15.0) | 35.4 (±13.1) | 43.2 (±11.8) | 37.4 (±19.4) | 43.4 (±10.4) | 44.6 (±13.4) | 45.4 (±10.5) | 38.1 (±10.1) | 30.7 (±15.2) | 26.4 (±17.1) | 41.9 (±14.9) | 0.04 |
Abbreviations: LA, left atrium; LAEF, left atrial ejection fraction; LV, left ventricle; LVEDV, left ventricular end-diastolic volume; LVEF, left ventricular ejection fraction; RA, right atrium; RV, right ventricle; RAEF, right atrial ejection fraction; RVEDV, right ventricular end-diastolic volume; RVEF, right ventricular ejection fraction.
* P value refers to overall analysis between groups.
LGE distribution
LGE distribution according to genes is presented in Table 3 and summarized in Figure 2. Overall, 147 (25.5%) patients had LGE on CMR. We did not find significant differences (P = 0.19) between groups in the proportion of patients with LGE, although some gene groups had LGE in a majority of individuals (DSP 64.7%/DMD 62.5%) and LGE was not present in any individual in a gene category (TNNT2 0%). Similarly, LGE extension was not statistically different among the different genes (Table 3).
Table 3.
LGE distribution according to genotypes
| Overall (n = 577) | Gene negative (n = 358) | TTN (n = 81) | RBM20 (n = 22) | BAG3 (n = 20) | DSP (n = 17) | FLNC (n = 17) | MYH7 (n = 15) | TNNT2 (n = 14) | LMNA (n = 14) | MYBPC3 (n = 11) | DMD (n = 8) | P value* | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LGE | 147 (25.5%) | 84 (23.5%) | 21 (25.9%) | 1 (4.5%) | 5 (25%) | 11 (64.7%) | 5 (29.2%) | 3 (20%) | 0 (0%) | 7 (50%) | 5 (45.5%) | 5 (62.5%) | 0.19 |
| Percentage of LV, % (IQR) | 4.7 (1.3–12.6) | 4.9 (1.4–11.1) | 2.6 (0.8–7.8) | 1.5 (1.5–1.5) | 1.8 (1.4–2.8) | 16.7 (2.5–23.8) | 14.0 (7.2–15.3) | 36.7 (0.3–54.0) | — | 2.2 (1.3–6.2) | 4.0 (0.6–7.9) | 6.7 (5.0–8.4) | 0.76 |
| LGE pattern | <0.001 | ||||||||||||
| − Absence | 430 (74.5%) | 274 (76.5%) | 60 (74.1%) | 21 (95.5%) | 15 (75%) | 6 (35.3%) | 12 (70.6%) | 12 (80%) | 14 (100%) | 7 (50%) | 6 (54.6%) | 3 (37.5%) | |
| − Lineal midwall | 45 (7.8%) | 30 (8.4%) | 5 (6.2%) | 0 (0%) | 1 (5%) | 2 (11.7%) | 1 (5.9%) | 0 (0%) | 0 (0%) | 4 (28.6%) | 2 (18.2%) | 0 (0%) | |
| − Patchy midwall | 11 (1.9%) | 7 (2.0%) | 2 (2.5%) | 0 (0%) | 0 (0%) | 1 (5.9%) | 0 (0%) | 1 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| − Subepicardial | 38 (6.6%) | 16 (4.5%) | 5 (6.2%) | 0 (0%) | 0 (0%) | 7 (41.2%) | 3 (17.7%) | 1 (6.7%) | 0 (0%) | 1 (7.1%) | 1 (9.1%) | 4 (50.0%) | |
| − Subendocardial | 4 (0.7%) | 3 (0.8%) | 0 (0%) | 0 (0%) | 1 (5%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| − RV insertion | 28 (4.9%) | 14 (3.9%) | 8 (9.9%) | 1 (4.5%) | 1 (5%) | 0 (0%) | 0 (0%) | 1 (6.7%) | 0 (0%) | 1 (7.1%) | 2 (18.2%) | 0 (0%) | |
| − Transmural | 10 (1.7%) | 8 (2.2%) | 1 (1.2%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (12.5%) | |
| − Combinations | 11 (1.9%) | 6 (1.7%) | 0 (0%) | 0 (0%) | 2 (10%) | 1 (5.9%) | 1 (5.9%) | 0 (0%) | 0 (0%) | 1 (7.1%) | 0 (0%) | 0 (0%) |
Abbreviations: IQR, interquartile range; LGE, late gadolinium enhancement; LV, left ventricle; RV, right ventricle.
* P value refers to overall analysis between groups.
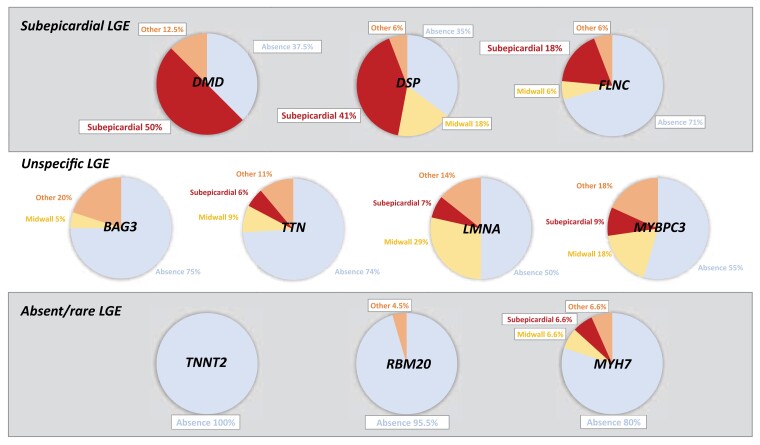
Figure 2.
LGE patterns according to the affected gene in DCM. Simplified LGE pattern distribution based on the causative underlying gene shows a cluster in three categories: subepicardial, unspecific, and absent/rare. Abbreviations: DCM, dilated cardiomyopathy; LGE, late gadolinium enhancement.
Regarding LGE pattern distribution, lineal midwall (n = 45, 7.8%) was the most frequent pattern found, followed by subepicardial (n = 38, 6.6%) and RV insertion (n = 28, 4.9%). A combination of patterns was found in 11 (1.9%) patients that included midwall + subepicardial in nine cases, midwall + RV insertion in one case, and subendocardial + RV insertion in another case. Significant differences were found comparing LGE patterns between groups (P < 0.001). Patients with variants in DMD, DSP, and FLNC showed a predominance of LGE subepicardial patterns (50, 41, and 18%, respectively), whereas the lineal midwall pattern was the most frequent pattern in patients with variants in LMNA (28.6%). On the other hand, LGE was absent or found in a low proportion of patients with variants in TNNT2, RBM20, and MYH7 (0, 5, and 20%, respectively). A simplified categorization of patterns according to the predominant LGE pattern (absent, midwall, subepicardial, and others) resulted in clustering of genes in three categories as described in Figure 2: subepicardial (DMD, DSP, and FLNC), unspecific (TTN, BAG3, LMNA, and MYBPC3), and absent/rare (TNNT2, RBM20, and MYH7).
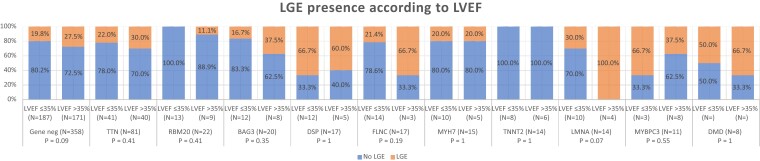
LGE presence according to LVEF
Figure 3 displays the presence of LGE according to LVEF for each gene category. No statistically significant differences in the presence of LGE were observed between patients with severe systolic dysfunction (LVEF ≤ 35%) and those with higher LVEF. In fact, a trend to the inverse association was found in LMNA and gene-negative subgroups. No significant differences were observed between patients according to LVEDV (see Supplementary data online, Appendix S2)
Figure 3.
LGE presence according to LVEF. The presence of LGE was not significantly associated with lower LVEF. Abbreviations: LGE, late gadolinium enhancement; LVEF, left ventricular ejection fraction.
Yield of genetic testing based on LGE patterns
The yield of genetic testing in probands (n = 516) was significantly different according to the LGE pattern (P = 0.007) (Table 4). More than half of probands with subepicardial patterns (18/34, 52.9%) had pathogenic or likely pathogenic variants, mostly in DSP and FLNC genes. The diagnostic yield diminished to 30.2% (16/53) and 40.4% (21/52) in case of midwall patterns and other patterns, respectively. The yield of genetic testing was only 27.3% (103/377) in patients who did not show LGE.
Table 4.
Yield of genetic testing in probands (n = 516) based on LGE patterns
| Gene negative (n = 358) | Gene positive (n = 158) | P value | |
|---|---|---|---|
| LGE pattern | 0.007 | ||
| − Absence | 274 (72.7%) | 103 (27.3%) | |
| − Midwall | 37 (69.8%) | 16 (30.2%) | |
| − Subepicardial | 16 (47.1%) | 18 (52.9%) | |
| − Other | 31 (59.6%) | 21 (40.4%) |
Abbreviation: LGE, late gadolinium enhancement.
Events by LGE patterns
During a median follow-up of 2.7 years (IQR 1.3–4.8), 46 (8.0%) patients had MVA due to appropriate ICD shocks (n = 29, 5%), aborted SCD (n = 13, 2.3%), or SCD (n = 5, 0.9%). Table 5 displays the incidence of MVA according to LGE patterns. Significant differences were found between groups comparing the incidence of composite MVA (P < 0.001) and all events separately (ICD therapy P = 0.005/aborted SCD P < 0.001/SCD P = 0.004).
Table 5.
MVA events by LGE patterns
| Overall (n = 577) | Absence (n = 430) | L.Midwall (n = 45) | Subepicardial (n = 38) | RV insertion (n = 28) | Transmural (n = 10) | Combinations (n = 11) | P value | |
|---|---|---|---|---|---|---|---|---|
| Appropriate ICD therapy | 29 (5.0%) | 11 (2.6%) | 8 (17.8%) | 6 (15.8%) | 2 (7.1%) | 1 (10.0%) | 1 (9.1%) | 0.005 |
| Aborted SCD | 13 (2.3%) | 6 (1.4%) | 0 (0%) | 2 (5.3%) | 2 (7.1%) | 0 (0%) | 3 (27.3%) | <0.001 |
| SCD | 5 (0.9%) | 2 (0.5%) | 0 (0%) | 1 (2.6%) | 1 (3.6%) | 0 (0%) | 1 (9.1%) | 0.004 |
| Composite MVA | 46 (8.0%) | 18 (4.2%) | 9 (20%) | 10 (26.3%) | 4 (14.3%) | 2 (20.0%) | 3 (27.3%) | <0.001 |
Abbreviations: ICD, implantable cardioverter defibrillator; L.Midwall, lineal midwall; MVA, malignant ventricular arrhythmias; P.Midwall, patchy midwall; RV, right ventricle; SCD, sudden cardiac death; VT, ventricular tachycardia.
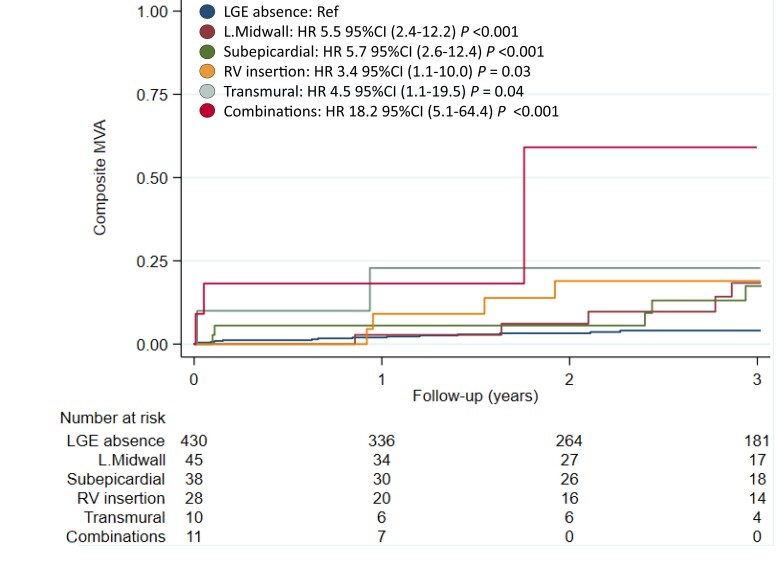
Figure 4 shows Kaplan–Meier curves for MVA according to LGE patterns using the absence of LGE as the reference group. Patients with LGE patterns of combinations [hazard ratio (HR) 18.2, 95% confidence interval (CI) (5.1–64.4), P < 0.001], subepicardial [HR 5.7, 95% CI (2.6–12.4), P < 0.001], lineal midwall [HR 5.5, 95% CI (2.4–12.2), P < 0.001], transmural [HR 4.5, 95% CI (1.1–19.5), P = 0.04], and RV insertion [HR 3.4, 95% CI (1.1–10.0), P = 0.03] showed increased risk for MVA compared with patients without LGE. In the other end of the spectrum, patients with subendocardial or patchy midwall patterns did not exhibited MVA during follow-up. Patients with lineal midwall, RV insertion, and combinations of LGE also had a higher risk of cardiovascular and overall mortality (see Supplementary data online, Appendices S3 and S4).
Figure 4.
Kaplan–Meier curve of MVA according to LGE patterns. Certain LGE patterns are associated with an increased risk of MVA. Abbreviations: CI, confidence interval; HR, hazard ratio; LGE, late gadolinium enhancement; L.Midwall, lineal midwall; MVA, major ventricular arrhythmias; RV, right ventricle.
Among the 18 patients who presented a MVA and who did not have LGE in CMR, 7 patients (38.9%) were gene negative, while 11 patients (61.1%) had pathogenic variants. Of note, 8 out of the 11 individuals with MVA and without LGE had genetic variants in genes considered associated with increased susceptibility to ventricular arrhythmias (LMNA n = 3, RBM20 n = 3, FLNC n = 1, and DSP n = 1).
Discussion
To our knowledge, this study represents the largest cohort of DCM patients phenotyped using genetic testing and detailed CMR. Our study with central CMR analysis found significant differences in LGE pattern distribution according to the underlying affected gene, despite relatively small numbers in each group. Based on our findings, DCM genes could be classified into three categories based on predominant LGE patterns: subepicardial (DMD, DSP, and FLNC), unspecific (TTN, BAG3, LMNA, and MYBPC3), and absent/rare (TNNT2, RBM20, and MYH7). Moreover, our study provides the first cohort description of CMR patterns in DCM caused by genetic variants in RBM20, BAG3, MYBPC3, and TNNT2. Lastly, it provides additional data supporting that the risk of MVA varies according to LGE patterns.
Patterns of LGE according to gene characteristics have been seldom described so far, with the largest cohort previously reported containing 89 individuals.14 In that study, authors found a specific subepicardial pattern in a combined group of patients with variants in DSP and FLNC compared with the rest of the cohort. Due to the reduced sample size for each gene, authors were unable to describe specific patterns in specific genes. Our study confirms that the subepicardial LGE pattern is the most common LGE pattern in patients with DSP and FLNC when analysed separately, although the percentage of patients with LGE was highly different between both genes (64.7% in DSP vs. 29.2% in FLNC). Interestingly, in our study, 35.3% of DCM patients with DSP variants did not show LGE, which is a slightly higher proportion than previous reports from Smith et al.8 (n = 10; 26%), Wang et al.9 (n = 63; 17%), and Augusto et al.14 (n = 25; 8%).
Although the subepicardial LGE pattern has also been described as a hallmark of FLNC cardiomyopathy and we found this pattern in 18% of individuals with variants in this gene, our value was much lower than the one found by Augusto et al.14 that reported 85.7% and is in line with a recently published cohort of 23 patients with FLNC variants that showed 25% prevalence.12 In our study, a third gene (DMD) also exhibited an increased prevalence of subepicardial patterns. Interestingly, DMD-associated DCM is not considered an entity with high arrhythmic risk in contrast with FLNC- and DSP-associated DCM, and current guidelines maintain standard recommendations for primary prevention ICD insertion based on LVEF ≤ 35% for DMD-associated DCM.17 Nevertheless, it is worth noting that in the largest cohort of DMD-associated DCM (n = 112) published,18 a significant proportion of patients (9%) suffered a MVA during follow-up and that the presence of LGE (transmural pattern) has been associated with worse outcomes even among patients with LVEF > 45% in other studies.19 Whether patients with LVEF 35–50% and subepicardial/transmural LGE patterns might benefit from ICD insertion in primary prevention remains to be elucidated. Along this line, two additional genes with high susceptibility to arrhythmias like LMNA and RBM20 showed different LGE patterns in our cohort, with the midwall pattern being the most frequently found among LMNA patients and mostly the absence of LGE (only 1 patient showed LGE) in the 22 patients with RBM20 variants included in the study. Interestingly, patients with the most frequent cause of genetic DCM, TTN truncating variants, exhibited a variety of LGE patterns, with a similar number of patients showing midwall, subepicardial, and other patterns among the 21 individuals who had fibrosis. Remarkably, BAG3 and MYBPC3 showed no specific trend in terms of the presence, extension, and distribution of LGE. Regarding other CMR parameters analysed, no differences were found between gene groups except for RVEDV and indexed RV mass. LGE presence was not associated with severe systolic dysfunction in our cohort or in any gene group. This observation is in line with the previous cohort published by Augusto et al.,14 although they found an association of LGE presence with severe LVEF dysfunction in patients with DSP/FLNC variants. These results suggest that systolic dysfunction and fibrosis appearance follow different pathophysiological pathways that could be approached from a therapeutic perspective independently.
Lastly, our study revealed that distinct LGE patterns confer different risks of MVA, with combinations and subepicardial patterns showing the highest odds compared with the absence of LGE. These results are consistent with a previously published study of 874 DCM patients evaluated with CMR and published by Halliday et al.6 Interestingly, LGE presence at RV insertion points, traditionally considered a benign LGE pattern, was associated with an increased risk of MVA compared with the absence of LGE, although half of these patients were gene negative and the other half were mostly composed of TTN. These results contrast with a recent study reporting 72 DCM patients with this LGE pattern, where none of them experienced MVA and had similar outcomes to LGE-negative patients.20 In our opinion, this discrepancy should trigger prospective studies before the RV insertion pattern is considered a benign finding.
Clinical implications
CMR is nowadays recognized as an essential test performed at the first evaluation of patients with DCM to elucidate possible aetiologies. Describing LGE patterns associated with specific genotypes could guide both clinicians and cardiologists specialized in CMR to prioritize genetic testing based on this information. This approach would be particularly important among patients with subepicardial patterns, as more than half of probands had disease-causing genetic variants, mostly in DSP/FLNC. These results would be actionable due to specific recommendations for ICD implantation in primary prevention in these genetic DCM subtypes.5,21 Our results also show that the subepicardial LGE pattern can be found among different subtypes of genetic DCM, many of them (DMD, MYH7, or MYBPC3) not included in arrhythmogenic cardiomyopathy gene panels and supporting a wider gene panel approach for these patients.
In addition, a fine phenotypic description of different genes responsible for DCM might help in the future for gene variant interpretation, providing supporting evidence for pathogenicity. On the other hand, our results also pose questions regarding pathophysiological pathways leading to the same LGE patterns in patients with very different genetic backgrounds that are not currently well understood.
Limitations
Some issues should be considered when interpreting our results. This is an observational retrospective study that was conducted at 20 heart failure and inherited cardiac disease units; as such, it is affected by an unavoidable degree of referral bias. This registry includes data from consecutive patients with DCM genotyped during a 5-year period who had a clinical CMR performed. Therefore, the criteria to proceed with CMR evaluation and genotyping were not uniform at the participating centres, and we cannot discard that CMR or genotype results have influenced the performance of the other test, limiting the representativity of our cohort in comparison with other cohorts of DCM patients. In line with this, it should be noted that 10% of the patients included in our study were relatives who could have had earlier stages of the disease compared with probands. Lastly, despite being the largest cohort of DCM patients phenotyped using genetic testing and CMR, the sample size of most gene groups was relatively small.
Conclusion
LGE patterns in DCM show a specific distribution according to the affected gene. Genes can be classified into three categories according to the predominant LGE pattern distribution: subepicardial (DMD, DSP, and FLNC), unspecific (TTN, BAG3, LMNA, and MYBPC3), and absent/rare (TNNT2, RBM20, and MYH7). Certain LGE patterns are associated with an increased yield of genetic testing and are associated with an increased risk of MVA.
Supplementary data
Supplementary data are available at European Heart Journal - Cardiovascular Imaging online.
Supplementary Material
Contributor Information
Fernando de Frutos, Heart Failure and Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Puerta de Hierro, Manuel de Falla, 2, Majadahonda, Madrid, 28222, Spain; CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands.
Juan Pablo Ochoa, Heart Failure and Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Puerta de Hierro, Manuel de Falla, 2, Majadahonda, Madrid, 28222, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro 3, Madrid, 28029, Spain.
Ana Isabel Fernández, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Department of Cardiology, Hospital General Universitario Gregorio Marañón, Madrid, Spain; Instituto de Investigación Sanitaria Gregorio Marañón, Madrid, Spain; Facultad de Medicina, Universidad Complutense, Madrid, Spain.
María Gallego-Delgado, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Department of Cardiology, CSUR Cardiopatías Familiares, Complejo Asistencial Universitario de Salamanca (CAUSA), Salamanca, Spain; Instituto de Investigación Biomédica de Salamanca (IBSAL), Salamanca, Spain.
Marina Navarro-Peñalver, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Virgen de la Arrixaca, El Palmar (Murcia), Spain.
Guillem Casas, European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Servicio de Cardiología, Hospital Universitario Vall Hebrón, Institut de Recerca Vall Hebrón (VHIR), Departamento de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
María Teresa Basurte, Department of Cardiology, Área del Corazón, Hospital Universitario de Navarra, Pamplona, Spain; IdiSNA—Instituto de Investigación Sanitaria de Navarra, Pamplona, Navarra, Spain.
José María Larrañaga-Moreira, Unidad de Cardiopatías Familiares, Instituto de Investigación Biomédica de A Coruña (INIBIC), Complexo Hospitalario Universitario de A Coruña, Servizo Galego de Saúde (SERGAS), Universidade da Coruña, A Coruña, Spain.
María Victoria Mogollón, Complejo Hospitalario Universitario de Cáceres, Cáceres, Spain.
Ainhoa Robles-Mezcua, Heart Failure and Familial Heart Diseases Unit, Cardiology Department, Hospital Universitario Virgen de la Victoria, IBIMA, Malaga, Spain.
Pablo Elpidio García-Granja, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Cardiology Department, Instituto de Ciencias del Corazón (ICICOR), Hospital Clínico Universitario de Valladolid, Valladolid, Spain.
Vicente Climent, Inherited Cardiovascular Diseases Unit, Department of Cardiology, Hospital General Universitario de Alicante, Institute of Health and Biomedical Research, Alicante, Spain.
Julián Palomino-Doza, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Cardiology Department, Hospital Universitario 12 de Octubre, Instituto de Investigación i+12, Madrid, Spain.
Ana García-Álvarez, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro 3, Madrid, 28029, Spain; Cardiology Department, Hospital Clínic Barcelona, IDIBAPS, Universitat de Barcelona, Barcelona, Spain.
María Brion, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Xenética Cardiovascular, Instituto de Investigación Sanitaria de Santiago, Unidad de Cardiopatías Familiares, Servicio de Cardiología, Complexo Hospitalario Universitario de Santiago de Compostela, Spain.
Ramón Brugada, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Medical Science Department, School of Medicine, University of Girona, 17003 Girona, Spain; Cardiology Service, Hospital Josep Trueta, University of Girona, 17007 Girona, Spain; Cardiovascular Genetics Center, University of Girona-IDIBGI, 17190 Girona, Spain.
Juan Jiménez-Jáimez, Department of Cardiology, Virgen de las Nieves University Hospital, 18014 Granada, Spain; Instituto de Investigacion Biosanitaria de Granada IBS, 18014 Granada, Spain.
Antoni Bayes-Genis, Heart Institute, Hospital Universitari Germans Trias i Pujol, Badalona, Barcelona, Spain.
Tomas Ripoll-Vera, Hospital Universitario Son Llatzer, IdISBa, Palma de Mallorca, Spain.
María Luisa Peña-Peña, Unidad de Imagen y Cardiopatías Familiares, Servicio de Cardiología, Hospital Universitario Virgen del Rocío, Sevilla, Spain.
José F Rodríguez-Palomares, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Servicio de Cardiología, Hospital Universitario Vall Hebrón, Institut de Recerca Vall Hebrón (VHIR), Departamento de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
Josefa Gonzalez-Carrillo, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Virgen de la Arrixaca, El Palmar (Murcia), Spain.
Eduardo Villacorta, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; Department of Cardiology, CSUR Cardiopatías Familiares, Complejo Asistencial Universitario de Salamanca (CAUSA), Salamanca, Spain; Instituto de Investigación Biomédica de Salamanca (IBSAL), Salamanca, Spain.
Maria Angeles Espinosa, CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Department of Cardiology, Hospital General Universitario Gregorio Marañón, Madrid, Spain; Instituto de Investigación Sanitaria Gregorio Marañón, Madrid, Spain; Facultad de Medicina, Universidad Complutense, Madrid, Spain.
Pablo Garcia-Pavia, Heart Failure and Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Puerta de Hierro, Manuel de Falla, 2, Majadahonda, Madrid, 28222, Spain; CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands; Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro 3, Madrid, 28029, Spain; Universidad Francisco de Vitoria, Carretera Pozuelo KM1800, Majadajonda 28223, Spain.
Jesus G Mirelis, Heart Failure and Inherited Cardiac Diseases Unit, Department of Cardiology, Hospital Universitario Puerta de Hierro, Manuel de Falla, 2, Majadahonda, Madrid, 28222, Spain; CIBER Cardiovascular, Instituto de Salud Carlos III, Avenida Monforte de Lemos 3-5, Madrid, 28029, Spain; European Reference Network for Rare and Low Prevalence Complex Diseases of the Heart, Amsterdam, The Netherlands.
Funding
This study has been funded by Instituto Salud Carlos III (ISCIII) through the projects ‘PI18/0004, PI19/01283, and PI20/0320’ (co-funded by the European Regional Development Fund/European Social Fund ‘A way to make Europe’/‘Investing in your future’). The Hospital Universitario Puerta de Hierro, the Hospital Universitario Vall Hebrón, the Hospital General Universitario Gregorio Marañón, and the Hospital Universitario Virgen de la Arrixaca are members of the European Reference Network for Rare, Low Prevalence, and Complex Diseases of the Heart (ERN GUARD-Heart). F.d.F. receives grant support from ISCIII (CM20/00101). R.B. receives funding from the Obra Social la Caixa Foundation. M.B. receives funding from ISCIII (PI19/01283). The CNIC is supported by the ISCIII, Ministerio de Ciencia e Innovación of the Spanish Government (MCIN), and Pro CNIC Foundation.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Mirelis JG, Escobar-Lopez L, Ochoa JP, Espinosa MÁ, Villacorta E, Navarro Met al. Combination of late gadolinium enhancement and genotype improves prediction of prognosis in non-ischaemic dilated cardiomyopathy. Eur J Heart Fail 2022;24:1183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klem I, Klein M, Khan M, Yang EY, Nabi F, Ivanov Aet al. Relationship of LVEF and myocardial scar to long-term mortality risk and mode of death in patients with nonischemic cardiomyopathy. Circulation 2021;143:1343–58. [DOI] [PubMed] [Google Scholar]
- 3. Di Marco A, Brown PF, Bradley J, Nucifora G, Claver E, de Frutos Fet al. Improved risk stratification for ventricular arrhythmias and sudden death in patients with nonischemic dilated cardiomyopathy. J Am Coll Cardiol 2021;77:2890–905. [DOI] [PubMed] [Google Scholar]
- 4. Ferrández-Escarabajal M, Islas F, Zulet Fraile P, Travieso A, Olmos C. External validation of an algorithm for risk stratification of ventricular arrhythmia in nonischemic dilated cardiomyopathy. Rev Española Cardiol (English Ed) 2022;75:684–5. [DOI] [PubMed] [Google Scholar]
- 5. Zeppenfeld K, Tfelt-Hansen J, de Riva M, Winkel BG, Behr ER, Blom NAet al. 2022 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J 2022;43:3997–4126. [DOI] [PubMed] [Google Scholar]
- 6. Halliday BP, Baksi AJ, Gulati A, Ali A, Newsome S, Izgi Cet al. Outcome in dilated cardiomyopathy related to the extent, location, and pattern of late gadolinium enhancement. JACC Cardiovasc Imaging 2019;12:1645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akhtar MM, Lorenzini M, Cicerchia M, Ochoa JP, Hey TM, Sabater Molina Met al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Hear Fail 2020:496–508. [DOI] [PubMed] [Google Scholar]
- 8. Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta ACet al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation 2020;141:1872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W, Murray B, Tichnell C, Gilotra NA, Zimmerman SL, Gasperetti Aet al. Clinical characteristics and risk stratification of desmoplakin cardiomyopathy. Europace 2022;24:268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmström M, Kivistö S, Heliö T, Jurkko R, Kaartinen M, Antila Met al. Description of A/C gene mutation related dilated cardiomyopathy with gadolinium-enhanced magnetic resonance imaging. J Cardiovasc Magn Reson 2011;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Domínguez F, Cuenca S, Bilińska Z, Toro R, Villard E, Barriales-Villa Ret al. Dilated cardiomyopathy due to BLC2-associated athanogene 3 (BAG3) mutations. J Am Coll Cardiol 2018;72:2471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gigli M, Stolfo D, Graw SL, Merlo M, Gregorio C, Nee Chen Set al. Phenotypic expression, natural history, and risk stratification of cardiomyopathy caused by filamin C truncating variants. Circulation 2021;144:1600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muser D, Nucifora G, Muser D, Nucifora G, Pieroni M, Castro SAet al. Prognostic value of nonischemic ringlike left ventricular scar in patients with apparently idiopathic nonsustained ventricular arrhythmias. Circulation 2021;143:1359–73. [DOI] [PubMed] [Google Scholar]
- 14. Augusto JB, Eiros R, Nakou E, Moura-Ferreira S, Treibel TA, Captur Get al. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: a comprehensive genotype-imaging phenotype study. Eur Heart J Cardiovasc Imaging 2019;21:326–36. [DOI] [PubMed] [Google Scholar]
- 15. Gao P, Yee R, Gula L, Krahn AD, Skanes A, Leong-Sit Pet al. Prediction of arrhythmic events in ischemic and dilated cardiomyopathy patients referred for implantable cardiac defibrillator. Circ Cardiovasc Imaging 2012;5:448–56. [DOI] [PubMed] [Google Scholar]
- 16. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster Jet al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groh WJ, Bhakta D, Tomaselli GF, Aleong RG, Teixeira RA, Amato Aet al. 2022 HRS expert consensus statement on evaluation and management of arrhythmic risk in neuromuscular disorders. Hear Rhythm 2022;19:e61–e120. [DOI] [PubMed] [Google Scholar]
- 18. Restrepo-Cordoba MA, Wahbi K, Florian AR, Jiménez-Jáimez J, Politano L, Arad Met al. Prevalence and clinical outcomes of dystrophin-associated dilated cardiomyopathy without severe skeletal myopathy. Eur J Heart Fail 2021;23:1276–86. [DOI] [PubMed] [Google Scholar]
- 19. Florian A, Ludwig A, Engelen M, Waltenberger J, Rösch S, Sechtem Uet al. Left ventricular systolic function and the pattern of late-gadolinium-enhancement independently and additively predict adverse cardiac events in muscular dystrophy patients. J Cardiovasc Magn Reson 2014;16:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claver E, Di Marco A, Brown PF, Bradley J, Nucifora G, Ruiz-Majoral Aet al. Prognostic impact of late gadolinium enhancement at the right ventricular insertion points in non-ischaemic dilated cardiomyopathy. Eur Heart J Cardiovasc Imaging 2023;24:346–53. [DOI] [PubMed] [Google Scholar]
- 21. Towbin JA, McKenna WJ, Abrams DJ, Ackerman MJ, Calkins H, Darrieux FCCet al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Hear Rhythm 2019;16:e301–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.