Abstract
Escherichia coli responds to its environment by means of a network of intracellular reactions which process signals from membrane-bound receptors and relay them to the flagellar motors. Although characterization of the reactions in the chemotaxis signaling pathway is sufficiently complete to construct computer simulations that predict the phenotypes of mutant strains with a high degree of accuracy, two previous experimental investigations of the activity remaining upon genetic deletion of multiple signaling components yielded several contradictory results (M. P. Conley, A. J. Wolfe, D. F. Blair, and H. C. Berg, J. Bacteriol. 171:5190–5193, 1989; J. D. Liu and J. S. Parkinson, Proc. Natl. Acad. Sci. USA 86:8703–8707, 1989). For example, “building up” the pathway by adding back CheA and CheY to a gutted strain lacking chemotaxis genes resulted in counterclockwise flagellar rotation whereas “breaking down” the pathway by deleting chemotaxis genes except cheA and cheY resulted in alternating episodes of clockwise and counterclockwise flagellar rotation. Our computer simulation predicts that trace amounts of CheZ expressed in the gutted strain could account for this difference. We tested this explanation experimentally by constructing a mutant containing a new deletion of the che genes that cannot express CheZ and verified that the behavior of strains built up from the new deletion does in fact conform to both the phenotypes observed for breakdown strains and computer-generated predictions. Our findings consolidate the present view of the chemotaxis signaling pathway and highlight the utility of molecularly based computer models in the analysis of complex biochemical networks.
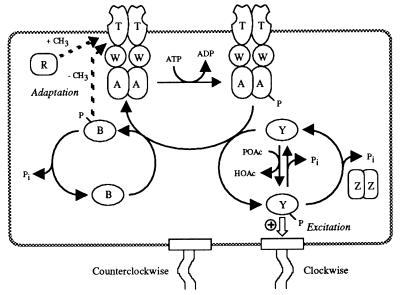
Bacteria use sophisticated information-processing systems to monitor their environment and respond appropriately to newly arising conditions. One of the best-characterized signal transduction pathways is that controlling chemotaxis in Escherichia coli. E. coli swims up gradients of favorable chemicals and down gradients of unfavorable chemicals by altering the amounts of time spent in the run (swim straight) and tumble (change direction) swimming modes (8). These behaviors correspond to counterclockwise (CCW) and clockwise (CW) flagellar rotation, respectively (24). The performance of individual flagellar motors may be quantified by observing the rotation of bacteria tethered to microscope slides by a single flagellar filament with anti-flagellar antibodies (43). The fraction of time spent in CCW rotation, called the bias, ranges between 0 and 1. A summary of the current view of the signal transduction pathway from cell surface receptors to flagellar motors, based on biochemical, genetic, and physiological data, is given in Fig. 1. For convenience, each signaling protein has been assigned a single-letter abbreviation in this report.
FIG. 1.
The signaling pathway that controls chemotaxis in E. coli. Chemotactic behavior is controlled by a family of transmembrane receptor proteins, termed transducers, in conjunction with six cytoplasmic Che proteins (reviewed in reference 44). E. coli has five known transducers (Aer, Tap, Tar, Trg, and Tsr) with differing ligand specificities. The transducers (collectively T) form stable ternary complexes with the CheW (W) coupling protein and the CheA (A) sensor kinase and regulate A autophosphorylation activity depending on the degree of ligand binding. In the excitation pathway, which generates the initial response to a stimulus, phosphoryl groups are transferred from phosphorylated A (Ap) to the CheY (Y) response regulator. Phosphorylated Y (Yp) in turn binds to the FliM flagellar switch protein to induce CW flagellar rotation. Dephosphorylation of Yp is stimulated by the CheZ (Z) protein. Ap and Z provide the primary routes of Y phosphorylation and dephosphorylation, respectively, in wild-type cells. Y also has autophosphorylation and autodephosphorylation activities. In the adaptation pathway, which returns the cell to its prestimulus behavior, phosphoryl groups are transferred from Ap to the CheB (B) response regulator. CheR (R) constitutively methylates the cytoplasmic domain of T, whereas phosphorylation enhances the methylesterase activity of B to remove methyl groups from T. This forms a feedback inhibition loop because the influence that T exerts on A autophosphorylation reflects both the ligand-binding and methylation status of T. Note that T, A, and Z are dimers whereas W, Y, B, and R are monomers. Modified from reference 14 with permission of the publisher.
Genetic analysis has played a crucial role in deciphering the chemotaxis signal transduction pathway. Two deletion analysis strategies are relevant here (Table 1). One (termed breakdown) is to remove one or several genes and see what signaling activity remains (26, 32, 33). The other (termed buildup) is to first remove all of the chemotaxis genes, creating a “gutted” (g) strain, and then to add back one or more genes to see what signaling activity is restored (17, 48). Although there should logically be no difference in the results obtained from these complementary strategies, an apparent paradox exists in the literature where the two approaches meet in the middle. A T−W−Z− strain (i.e., a strain lacking transducers, CheW, and CheZ) has an intermediate flagellar rotational bias comparable to that of wild-type cells (25, 26). In contrast, an A+Y+(g) strain (i.e., a gutted strain expressing wild-type concentrations of CheA and CheY) has a fully CCW bias (17). This is puzzling, because as designed the two strains should contain the same elements of the excitation pathway (Table 1). However, technical details of the construction of the gutted strain (see below) are known to have resulted in slight but potentially significant differences (17). With regard to the adaptation pathway, the presence of R in T−W−Z− should be irrelevant because there is no T upon which to act. The presence of B in T−W−Z− should, by competition for CheA-P (Ap), reduce CheY-P (Yp) and thus further decrease CW flagellar rotation in comparison to A+Y+(g), yet the opposite is observed. The flagellar rotational phenotypes reported for each of the other three buildup strains listed in Table 1 are also inconsistent with the observed behaviors of the analogous breakdown strains (17, 26).
TABLE 1.
Equivalent strains in two deletion analysis strategies
| Strain in buildup strategy | Presence of excitation pathway proteina:
|
Strain in breakdown strategy | ||||
|---|---|---|---|---|---|---|
| T | W | A | Y | Z | ||
| + | + | + | + | + | Wild type | |
| T+W+A+Y+(g) | + | + | + | + | − | Z− |
| T+A+Y+(g) | + | − | + | + | − | W−Z− |
| W+A+Y+(g) | − | + | + | + | − | T−Z− |
| A+Y+(g) | − | − | + | + | − | T−W−Z− |
| Gutted | − | − | − | − | − | |
Note that the R and B proteins of the adaptation pathway are present in the breakdown but not the buildup strains.
Our molecularly based computer simulation of the chemotaxis signal transduction system, developed over the past several years, both highlights this experimental enigma and points the way to a potential resolution. The BCT (bacterial chemotaxis) model is built from units representing the molecular components in the pathway, almost all of which can be assigned experimentally determined intracellular concentrations and enzymatic rate or binding constants (14, 15). The program has been used to evaluate some 65 different chemotactic mutants and currently predicts the correct swimming behavior in the vast majority of cases, including the breakdown strains (25). However, the predictive accuracy of BCT appears to be reduced for the category of strains constructed by the buildup strategy. BCT predicts that an A+Y+(g) strain will exhibit alternating episodes of CW and CCW flagellar rotation, in contrast to the observed exclusively CCW behavior (17). Furthermore, BCT predicts that an elevated concentration of Y alone in a gutted strain will lead to sufficient Yp formation by autophosphorylation with acetyl phosphate to generate a bias well below 1, yet this has not been observed (47).
Thus, both logical analysis of different mutant strains and quantitative predictions by means of a computer model indicate an inconsistency in the published literature. In the particular gutted strain used for these experimental observations, Z was removed by deleting the promoter rather than the gene, and some residual Z expression was observed (17). Could a small amount of Z plausibly have a significant impact on the behavioral phenotype of these cells? The measured biases of wild-type and T−W−Z− strains are comparable (25, 26), reflecting comparable Yp concentrations, because in a wild-type cell the enormous enhancement of the A autophosphorylation rate caused by T and W and the substantial enhancement of Yp dephosphorylation by Z balance one another. In the absence of T and W, where A autophosphorylation and hence Yp formation are relatively slow, BCT predicts that the dephosphorylation stimulating activity of Z is so potent that a small percentage of the wild-type concentration of Z would be sufficient to shift the bias of A+Y+(g) from the expected intermediate value to the observed CCW value. Similarly, simulated trace Z contamination is enough to counteract the modest rate of Yp formation by autophosphorylation in BCT simulations of Y++(g) (i.e., a gutted strain expressing a larger-than-wild-type amount of Y).
To test the BCT prediction that the unexpectedly high biases observed experimentally in A+Y+(g) and Y++(g) strains could be attributable to residual Z, we created a new gutted strain completely lacking chemotaxis proteins, reintroduced the chemotaxis proteins in various combinations, and determined the flagellar rotational biases of the resulting strains. The results of our analysis provide a resolution of both the experimental paradox involving T−W−Z− and A+Y+(g), which exists entirely independently of the computer simulation, and the discrepancy between computer simulation and experimental observation for Y++(g). Furthermore, the new gutted strain permits examination of several features of the chemotaxis signaling pathway not previously observable.
MATERIALS AND METHODS
Bacterial strains.
The primary bacterial strains used in this work are described in Table 2. A large number of secondary strains bearing plasmids are not explicitly listed in Table 2 but, rather, are identified in Tables 4 and 5 as a combination of a bacterial strain from Table 2 and a plasmid from Table 3. P1vir transductions were conducted as described by Silhavy et al. (42). Zeocin (Invitrogen) was used at 15 μg/ml in low-salt Luria broth (10 g of tryptone, 5 g of NaCl, and 5 g of yeast extract per liter [pH 7.5]). The manufacturer reports that salt concentration and pH critically affect Zeocin activity. A consideration in the strain construction strategy was that in addition to Kanr, Tn5 [and the XhoI fragment of Tn5 in Δ(cheA-cheY)::XhoI(Tn5) as well] contains a bleomycin resistance gene that confers Zeor.
TABLE 2.
Primary bacterial strains used in this study
| Strain | Relevant genotype | Source, reference, or constructiona |
|---|---|---|
| CP875 | thr(Am)-1 leuB6 his-4 rpsL136 thi-1 lacΔX74 λlacY | 34 |
| D301 | recD1903::mini-Tn10 | 40 |
| HCB56 | RP437 trg::Tn10 | = RP1131 (48) |
| HCB317 | RP437 Δ(tsr)7021 | 48 |
| HCB721 | HCB317 Δ(cheA-cheY)::XhoI(Tn5) trg::Tn10 | 47 |
| HCB722 | HCB721 (λDFB19) | 47 |
| RBB382 | RP437 Δ(cheA)m102-11 zec::Tn10-992 Δ(recA)SstII-EcoRI | 13 |
| RBB1031 | RP437 recD1903::mini-Tn10 | P1.D301→RP437; Tetr |
| RBB1041 | RP437 Δ(cheA-cheZ)::Zeor | (Step 1) PstI-BamHI fragment of pWA24→RBB1031, Zeor; (Step 2) P1.RBB1031 Zeor transformant→RP437; Zeor |
| RBB1049 | HCB317 trg::Tn10 | P1.HCB56→HCB317, Tetr |
| RBB1050 | RBB1049 Δ(cheA-cheZ)::Zeor | P1.RBB1041→RBB1049, Zeor |
| RBB1051 | RBB1049 Δ(cheA-cheY)::XhoI(Tn5) | P1.HCB721→RBB1049, Kanr |
| RBB1052 | RBB1050 (λDFB19) | This work |
| RBB1053 | RBB1051 (λDFB19) | This work |
| RBB1106 | CP875 Δ(cheA-cheZ)::Zeor | 5 |
| RBB1109 | CP875 Δ(cheA-cheZ)::Zeor Φ(Δacs::Km-1) Δ(ackA pta hisJ hisP dhu)zej-223::Tn10 | 5 |
| RBB1174 | RP437 Δ(cheA-cheY)::XhoI(Tn5) | P1.HCB721→RP437, Kanr |
| RBB1175 | RBB1041 (λDFB19) | This work |
| RBB1176 | RBB1174 (λDFB19) | This work |
| RP437 | thr(Am)-1 leuB6 his-4 metF(Am)159 eda-50 rpsL136 thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78 | 32 |
| RP437recA | RP437 Δ(recA)SstII-EcoRI srl::Tn10 | 13 |
For strains constructed by P1 transduction, the donor, recipient, and selection used are indicated.
TABLE 4.
Comparison of flagellar rotational bias of isogenic Δ(cheA-cheZ)::Zeor and Δ(cheA-cheY)::XhoI(Tn5) strains expressing various chemotaxis proteins
| Chemotaxis genes expresseda (plasmids and bacteriophages) | Mean flagellar rotational bias ± SEM (strain) in:
|
|
|---|---|---|
| Δ(cheA-cheZ)::Zeor background | Δ(cheA-cheY)::XhoI(Tn5) background | |
| None | 1.00 ± 0.00 (RBB1050) | 1.00 ± 0.00 (RBB1051) |
| Y+ (λDFB19) | 1.00 ± 0.00 (RBB1052) | 1.00 ± 0.00 (RBB1053) |
| T+Y+ (λDFB19) | 0.98 ± 0.02 (RBB1175) | 0.99 ± 0.00 (RBB1176) |
| A+Y+ (pWA27, λDFB19) | 0.51 ± 0.09 (RBB1122) | 0.88 ± 0.06 (RBB1123) |
| W+A+Y+ (pWA28, λDFB19) | 0.20 ± 0.06 (RBB1124) | 0.93 ± 0.04 (RBB1125) |
| T+A+Y+ (pWA27, λDFB19) | 0.72 ± 0.07 (RBB1183) | 0.96 ± 0.03 (RBB1185) |
| T+W+A+Y+ (pWA28, λDFB19) | 0.00 ± 0.00 (RBB1184) | 0.10 ± 0.05 (RBB1186) |
| Y++ (pRBB38) | 0.96 ± 0.03 (RBB1068) | 1.00 ± 0.00 (RBB1069) |
| Y+++ (pRL22ΔZ) | 0.22 ± 0.07 (RBB1167) | 1.00 ± 0.00 (RBB1168) |
| Y++++ (pDFB19) | 0.01 ± 0.00 (RBB1066) | 0.30 ± 0.06 (RBB1067) |
T+ cells express Tsr and Trg in either the RBB1041 or the RBB1174 strain background. Strains without T are built up in the RBB1050 and RBB1051 strain backgrounds. +, approximately wild-type amounts of protein; ++, +++, and ++++, increasing amounts of protein.
TABLE 5.
Effect of acetate metabolism on the bias of strains expressing elevated concentrations of CheY
| Parent strain without plasmid | Relevant phenotype
|
Mean flagellar rotational bias ± SEM (strain)
|
||
|---|---|---|---|---|
| Chemo- taxis | Acetate metab- olism | With pRL22ΔZ | With pDFB19 | |
| CP875 | + | + | 0.01 ± 0.01 (RBB1169) | 0.02 ± 0.02 (RBB1113) |
| RBB1106 | − | + | 0.50 ± 0.09 (RBB1161) | 0.07 ± 0.03 (RBB1115) |
| RBB1109 | − | − | 0.46 ± 0.08 (RBB1162) | 0.05 ± 0.02 (RBB1111) |
TABLE 3.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pBAD18 | araC oripBR322 Ampr | 19 |
| pDFB19 | plac cheY oripUC9 Ampr | D. F. Blair and H. C. Berg |
| pGD2 | ′flhC motAB cheA′ oripBR322 Tetr | 18 |
| pRL22 | ptrp cheYZ oripBR322 Ampr | 30 |
| pRL22ΔZ | ptrp cheY oripBR322 Ampr | 39 |
| pRBB28 | ptrp cheA oripBR322 Ampr | 11 |
| pRBB30 | ptrp cheAW oripBR322 Ampr | 11 |
| pRBB38 | ptrp cheY oripBR322 Ampr | 3 |
| pRBB48 | plac cheY oripBR322 Ampr | This work |
| pUC19 | oripUC19 Ampr | 50 |
| pWA24 | ′flhC motAB cheA′-Zeor-′cheZ flhB′ oripUC19 Ampr | This work |
| pWA27 | araC paraBAD cheA oripBR322 Ampr | This work |
| pWA28 | araC paraBAD cheAW oripBR322 Ampr | This work |
| pZEro-1 | oripUC Zeor | Invitrogen |
Construction of Δ(cheA-cheZ)::Zeor.
Plasmid pWA24, constructed in a series of manipulations involving standard techniques, contains the following elements: (i) the 2.7-kb BamHI-PstI fragment of pUC19 (50) bearing Ampr and ori, (ii) the 2.2-kb PstI-BsmI fragment of pGD2 (18) carrying ′flhC-motA-motB-cheA′, (iii) a custom-synthesized BsmI-XbaI linker that creates an in-frame stop codon at codon 44 of cheA, (iv) the XbaI-HincII segment of the pUC19 polylinker, (v) the 607-bp SspI-DraI Zeor cassette from vector pZEro-1 (Invitrogen) oriented opposite to cheA, (vi) the HincII-HindIII segment of the pUC19 polylinker with a 12-bp synthetic sequence inserted to destroy the PstI site, (vii) a custom-synthesized HindIII-Bsu36I linker that creates a stop codon in frame with ′cheZ, and (viii) the 1.0-kb Bsu36I-BamHI fragment of pRL22 (30) containing ′cheZ-flhB′. For nucleotide sequence details, see Fig. 2B.
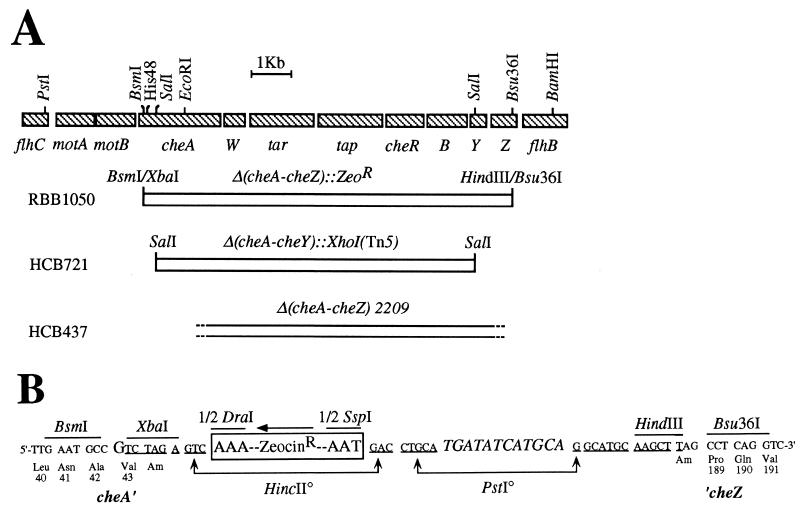
FIG. 2.
Details of the new che gene deletion Δ(cheA-cheZ)::Zeor. (A) Map of the chromosomal deletions in the mocha and meche operons of the gutted E. coli strains RBB1050, HCB721, and HCB437. In addition to their che deletions, RBB1050 and HCB721 carry Δ(tsr)7021 trg::Tn10 whereas HCB437 carries Δ(tsr)7021 Δ(trg)100 zbd::Tn5. W, B, Y, and Z represent cheW, cheB, cheY, and cheZ, respectively. Deletions are represented by open boxes. Δ(cheA-cheZ)::Zeor extends from codon 44 of the cheA (654 total) reading frame to codon 188 of cheZ (214 total). This deletion removes the 3′-terminal 611 codons of cheA (which includes the site of phosphorylation, His48), cheW, tar, tap, cheR, cheB, cheY, and more than 85% of cheZ. Δ(cheA-cheY)::XhoI(Tn5) leaves the cheZ gene intact and apparently promoterless. Δ(cheA-cheZ)2209 expresses a CheA-CheZ hybrid fusion protein with residual kinase activity. (B) Nucleotide sequence of the junctions around Δ(cheA-cheZ)::Zeor. The cheA and cheZ reading frames are shown with their respective codons. Custom-synthesized linkers were ligated at the BsmI and Bsu36I junctions of the Δ(cheA-cheZ) deletion to create XbaI and HindIII restriction sites, respectively. These linkers also created in-frame amber codons at codons 44 of cheA and 188 of cheZ to prevent the expression of any potential fusion proteins. The G in codon 43 of cheA is a substitution from the native A, which changes the codon from ATC (Ile) to GTC (Val). This change was made to eliminate a dam methylation site from the palindromic BsmI-XbaI linker that would have blocked XbaI digestion during construction. A 607-bp DraI-SspI fragment containing a Zeor cassette was first cloned into the HincII site of pUC19, and then a slightly larger XbaI-HindIII fragment was used to replace the deleted che genes. Expression of the Zeor cassette (boxed) proceeds in the opposite orientation to transcription from the mocha and meche operons. The pUC19 plasmid sequence is underlined. The PstI site adjacent to the HincII site of pUC19 was disrupted by an adapter (italics) to facilitate the recombination of a PstI-BamHI fragment (A) into the chromosome.
Plasmid pWA24 was digested with PstI and BamHI to generate a 3.8-kb DNA fragment containing ′flhC motAB cheA′-Zeor-′cheZ flhB′. This linear DNA fragment was gel purified and used to transform the recD strain RBB1031 by the RbCl method (29). Zeor transformants were isolated and screened on motility plates. Several Zeor Che− isolates were recovered. Tethering assays confirmed that these isolates were Fla+ Mot+. P1vir was grown on one RBB1031 Zeor isolate and used to transduce RP437 to Zeor, creating RBB1041. The Che− phenotype was found to be 100% cotransducible with the Zeor marker, consistent with insertion of the Zeor cassette in the che locus. The putative Δ(cheA-cheZ)::Zeor deletion and Δ(cheA-cheY)::XhoI(Tn5) from HCB721 were moved by P1 transduction in parallel into RBB1049, a Δ(tsr)7021 trg::Tn10 derivative of RP437, to create strains RBB1050 and RBB1051. Note that strain RBB1051 should be identical to HCB721 (47).
The expected structure of Δ(cheA-cheZ)::Zeor in strain RBB1050 (Fig. 2B) was confirmed by determining the nucleotide sequence of an 802-bp PCR-generated DNA fragment with chromosomal DNA and oligonucleotides which anneal to the remnants of the cheA and cheZ reading frames. Furthermore, Western blots of whole-cell lysates of RBB1050 failed to detect T, A, or Y.
Plasmids.
The plasmids used in this work are described in Table 3. pRBB48 was constructed by replacing the 0.7-kb EcoRI-SalI fragment of pRBB38 (3) containing ptrp ′cheB cheY′ with the 0.8-kb EcoRI-SalI fragment of pDFB19 containing plac ′cheB cheY′. The previously described (4, 12, 13) cheY alleles 57DA, 57DE, 57DQ, and 56SA 57DN were incorporated into pRBB48 by swapping appropriate restriction fragments between plasmids and confirmed by DNA sequencing.
Plasmid pWA27 expresses cheA from paraBAD of expression vector pBAD18 (19), whereas pWA28 expresses both cheA and cheW from the same promoter. The araBAD promoter is under control of the plasmid-borne araC gene product. pWA27 was constructed by cloning the 2.8-kb NheI-HindIII fragment of plasmid pRBB28 (11), which contains the entire cheA gene, into the NheI-HindIII sites of the pBAD18 multicloning site. Similarly, pWA28 was constructed by cloning the 3.9-kb NheI-HindIII fragment of pRBB30 (11), which contains both the cheA and cheW genes, into the NheI-HindIII sites of pBAD18.
The l-arabinose concentrations that yielded approximately wild-type levels of CheA and CheW were determined by both complementation studies and Western blotting. The ΔcheA strain RBB382 was transformed with pWA27 or pWA28, and the che+ strain RP437recA was transformed with pBAD18. The behavior of the three strains was examined on motility plates containing various concentrations of l-arabinose. pWA28 complements RBB382 for chemotaxis, with maximal swarming rates at l-arabinose concentrations near 10 μM. Immunoblots of whole-cell lysates of RP437 (wild type), RBB1122 [A+Y+(g)], and RBB1124 [W+A+Y+(g)] grown with various inducer concentrations revealed comparable amounts of A at 10 μM l-arabinose (data not shown), the concentration used in all experiments reported here. l-Arabinose at 10 μM also induces approximately wild-type levels of A from the closely related paraBAD motAB cheA plasmid pCS31 (46, 49).
Bacteriophages.
λDFB19 is λgt-4 cI857 plac cheY (47). Lysates of λDFB19 were prepared by heat induction of HCB722 (47). To isolate lysogens, λDFB19 was plated on the recipient strain at 30°C. Candidate lysogens were streak purified from the center of turbid plaques and identified by screening for the ability to grow at 30°C but not at 42°C. As described by Conley et al. (17), 25 μM isopropyl-β-d-thiogalactopyranoside IPTG was used to induce the expression of approximately wild-type concentrations of Y from λDFB19 lysogens. Immunoblotting of whole-cell lysates with affinity-purified anti-Y antibody (a kind gift from Phil Matsumura, University of Illinois at Chicago) confirmed that the λDFB19 lysogens RBB1052 and RBB1053 expressed approximately the same amount of Y as the wild-type strain RP437 when induced with 25 μM IPTG (data not shown).
Quantitative measurement of flagellar rotational bias.
Bacteria were prepared and tethered as previously described (15). Their behavior was observed by dark-field microscopy, recorded on videotape, and quantified in a semi-automated manner with The Observer 3.1 Video Tape Analysis System software (Noldus Information Technology, Wageningen, The Netherlands). The videotape was played back at low speed, and a key was pressed on the computer keyboard to simultaneously mark the start of each CCW, CW, or Pause episode and the end of the previous event. A vertical-interval time code recorded on the videotape concurrently with the cell images was recovered from the video cassette recorder by the computer, allowing accurate assignment of the duration of various behavioral episodes. Note that bias is defined to be the fraction of time spent in CCW rotation and thus is the ratio of the CCW time to the sum of the CCW, CW, and Pause times. It is therefore possible for a cell to have a bias slightly below 1, due to pausing, without ever exhibiting CW rotation. Cells that paused for more than 5% of the quantitation period were excluded from bias calculations.
A series of tests were performed to determine the optimum quantitation parameters and validate the tethering assay.
(i) Playback speed.
The choice of videotape playback speed reflected a balance between assay accuracy and rapidity. The same 100 s of videotaped behavior of a RP437 cell was scored at playback speeds of about 1/20×, 1/10×, 1/5×, 1/2×, and 1× real time. Bias measurements made at the four lowest speeds fell within a range of 0.02, whereas playback at real time yielded a significantly different value. The videotape was routinely scored at 1/5× playback speed, with appropriate adjustment for exceptional individual cells. It is worth noting that even though obvious coding errors were made during trials at 1/2× playback speed, they had little effect on the overall bias measurement because the missed reversal events were of such short duration. Thus, the method is suited for accurate measurement of the overall fraction of time spent in various behavioral modes but not for determining the lengths of individual CCW, CW, or Pause episodes.
(ii) Reproducibility of scoring.
The bias values obtained when the same segment of tape was scored either by different observers or multiple times by a single observer typically fell within a range of 0.02, providing a measure of assay reproducibility.
(iii) Number of cells/length of time.
To determine the dependence of the mean bias value on the number of cells and the observation time included in the calculation, bias measurements were made of 50 RP437 cells for 100 s each and the data were analyzed in increments of 5 cells or 10 s. Mean biases converged most of the way to their final values by the point at which 25 cells or 40 s of data had been included in the calculations. However, the effects were relatively small, since calculated biases differed by a maximum of 10% over all permutations of cell number and observation time examined. Bias determinations for each strain described in this paper were computed by scoring 30 s of behavior for each of 30 cells, unless otherwise noted.
(iv) Culture density.
The physiological state of the cell affects flagellar expression, motility, and bias (2, 21, 35). The dependence of bias upon culture density was therefore examined. Four independent cultures of RP437 harvested at optical densities at 600 nm (OD600) of 0.26, 0.44, 0.68, and 0.76 gave mean bias measurements between 0.78 and 0.84 (a range of <10%) with no obvious correlation between OD600 and bias. Therefore, cultures were harvested for tethering at OD600s of 0.4 to 0.8 (∼1 × 108 to 5 × 108 CFU/ml).
Computer simulation.
The current version of BCT and extensive supporting documentation are available at http://www.zoo.cam.ac.uk/zoostaff/levin/chemotaxis.html.
RESULTS
Construction of a new gutted strain.
The genes encoding the six Che proteins and two of the transducers are contiguous in the E. coli chromosome. Two deletions spanning this region have previously been used to construct gutted strains HCB437 and HCB721 (Fig. 2A). Although studies performed with these strains have been valuable (17, 48), both deletions have features recognized by previous investigators to potentially complicate the interpretation of experimental results. Δ(cheA-cheZ)2209 leads to expression of a CheA-CheZ fusion protein that retains partial kinase activity (22). Δ(cheA-cheY)::XhoI(Tn5) leaves the cheZ gene intact, although without the usual promoter. We made a new deletion, Δ(cheA-cheZ)::Zeor, and recombined it into a strain lacking the Tsr and Trg transducers to make the new gutted strain RBB1050 (see Materials and Methods). Δ(cheA-cheZ)::Zeor removes all the genes encoding W, Tar, Tap, R, B, and Y and most of the genes encoding A and Z. The Zeor marker is transcribed in the opposite direction to the che genes and is flanked by in-frame stop codons to eliminate expression of potential fusion peptides containing the remnants of A or Z (Fig. 2B). The N-terminal 7% of A presumably is expressed from the mocha promoter in RBB1050, but it should not participate in phosphorylation reactions because the site of A autophosphorylation and the kinase catalytic domain are both absent. Coding sequence for the C-terminal 12% of Z, which contains the Yp binding site (10), is present in RBB1050, but there is no obvious promoter to transcribe it, no ATG codon to initiate translation, and no way to make a fusion protein containing these sequences. Note that RBB1050 does retain the recently discovered Aer transducer (9, 38).
Reconstitution of the chemotaxis excitation signaling pathway.
The chemotaxis excitation pathway was systematically rebuilt in parallel in isogenic strains containing Δ(cheA-cheZ)::Zeor (referred to as the new che deletion) or Δ(cheA-cheY)::XhoI(Tn5) (referred to as the old che deletion). Various combinations of T, W, A, and Y were expressed at approximately wild-type concentrations via bacteriophages, plasmids, and/or absence of chromosomal mutations. The flagellar rotational biases of the resulting buildup strains were quantified (Table 4) and are compared below both to one another and to previously reported observations of analogous breakdown strains.
Taking the new che deletion series first, the RBB1050 gutted strain exhibits the default (exclusively CCW) behavior as expected. Introduction of Y alone did not change this behavior, nor did the joint presence of T and Y, since there is no known direct connection between these two signaling species (Fig. 1). Joint expression of A and Y led to significant CW flagellar rotation, because A is a good source of phosphoryl groups for Y. Addition of W to A+Y+(g) lowered the bias further, consistent with the observation of Liu and Parkinson (26) that T−Z− has a lower bias than T−W−Z−. The biochemical basis for the CW effect of W may involve a higher affinity for ATP when A is complexed with W than when A is free (31). Alternatively, W might promote CW flagellar rotation by coupling Aer (a transducer retained in the T− and gutted strains) to A. In contrast to the effect of W, adding T to A+Y+(g) raised the bias. This W-independent effect of T was not observed in the analogous breakdown experiments unless T was overexpressed; i.e., W−Z− and T−W−Z− had similar biases, which were lower than that of T++W−Z− (26). The reduction in CW flagellar rotation upon introduction of T may be accounted for by T-mediated inhibition of A autophosphorylation in the absence of W (1). Finally, completion of the excitation pathway resulted in exclusively CW flagellar rotation, as is seen for a Z− strain (32).
In contrast to strains containing the new che deletion, only T+W+A+Y+(g) had a bias significantly below 1 among buildup strains constructed in the old che deletion background (Table 4). These results are entirely consistent with previous observations made in the same strain background but with different expression vectors. Specifically, Conley et al. (17) reported biases of 1.0, 0.80, 1.0, and 0.43 for A+Y+, W+A+Y+, T+A+Y+, and T+W+A+Y+ strains, respectively, whereas we measured biases of 0.88, 0.93, 0.96, and 0.10, respectively. A closely similar result was also obtained by computer modeling when a low level of Z (equivalent to 3% of wild-type activity) was added to a series of gutted strains (25).
The difference in observed biases for pairs of isogenic strains that differ only by whether they carry the old or new che gene deletion is striking. In each case where the bias of strains with Δ(cheA-cheZ)::Zeor was significantly below 1, the bias of the matched strain with Δ(cheA-cheY)::XhoI(Tn5) was higher (Table 4). The consistent disparity between the two strain backgrounds is strong evidence in support of the prediction that strains containing the old che deletion must express some Z. Genetic and biochemical evidence for low-level expression of Z in a strain carrying the old che deletion has been reported previously (17). We also directly confirmed the presence of Z in RBB1051 by immunoblotting (data not shown). Use of the new che deletion, which also removes the gene for Z, brings the results with buildup and breakdown strategies largely into agreement, thus resolving the apparent paradox posed in the introduction and in agreement with the results of computer-simulated predictions.
High levels of Y cause CW flagellar rotation in the absence of other chemotaxis proteins.
Overexpression of Y changes the behavior of wild-type bacteria from reversing to exclusively CW (16). There have been multiple reports of a similar CW effect of excess Y in strains bearing Δ(cheA-cheZ)2209 (16, 23, 48). Because Δ(cheA-cheZ)2209 codes for a CheA-CheZ fusion protein with kinase activity (22), the CW behavior observed in these two cases may plausibly be attributed to an increased concentration of Yp formed from Ap or A-Zp. However, the phenotype of a strain simultaneously lacking A and expressing high concentrations of Y has never been reported. The new gutted strain provides the opportunity to determine whether Y can cause CW rotation in an intact cell lacking all other Che proteins. Expression of increasing amounts of Y in the RBB1050 strain background does in fact result in decreasing bias (Table 4). Expression of Y from the para cheY plasmid pJH120 (48) in the HCB721 gutted strain also results in CW behavior (46). These observations, made with intact cells, are consistent with the work of Ravid et al. (37), who found that incorporation of purified Y protein into tethered cell envelopes washed free of cytoplasmic constituents resulted in CW flagellar rotation. The biases of strains expressing Y in the Δ(cheA-cheY)::XhoI(Tn5) background are again consistently higher than those of the analogous Δ(cheA-cheZ)::Zeor strains (Table 4).
There are three plausible mechanisms by which high concentrations of Y might cause CW behavior in the absence of A. The most familiar is to increase the concentration of Yp. In the absence of A, the source of phosphoryl groups would be either small-molecule phosphodonors such as acetyl phosphate (28) or cross talk from other sensor kinases in the cell (45). The difference in bias noted above for the new and old che deletion backgrounds implies that Z affects the behavior of strains overexpressing Y, which in turn suggests a role for Yp. Acetylation by acetyl coenzyme A synthetase is a second covalent modification of Y that results in CW signal generation (7, 36). Finally, nonphosphorylated Y has been reported to possess ∼1% of the CW rotation-generating activity of Yp (6). The current version of BCT, which includes both phosphorylation of Y by acetyl phosphate and an effect of nonphosphorylated Y on flagellar rotation, gives bias values in excellent quantitative agreement with the data presented in Table 4 (25). A series of experiments described below were performed in an attempt to more directly determine which mechanism(s) might account for the CW activity of Y in the gutted strain.
Acetate and acetyl coenzyme A can be interconverted by either of two metabolic pathways that pass through acetyl-phosphate (a phosphodonor for Y) or acetyl-AMP (an acetyl donor for Y) intermediates. To determine whether activated acetate compounds contribute to the CW-generating ability of Y in the absence of A, plasmids expressing high concentrations of Y were introduced into a series of three isogenic strains—wild type for both chemotaxis and acetate metabolism, mutated by deletion of the che genes with intact acetate metabolism, or mutated by deletion of the genes encoding both chemotaxis functions as well as the two acetate pathways. The wild-type strains exhibited strongly CW behavior (Table 5). Removing the che genes raised the bias to an intermediate value for cells containing pRL22ΔZ (Table 5), implying that at least part of the CW activity in che+ cells comes from Yp synthesized through Ap. The bias of cells containing pDFB19 was unaffected by the presence or absence of che genes, suggesting that there is enough Y in these cells to generate saturating CW signals. Removing the genes encoding both acetate metabolic pathways had no further effect on biases (Table 5), indicating that neither acetyl-phosphate nor acetylation of Y is responsible for the CW behavior of gutted cells overexpressing Y.
With acetylation ruled out, either Yp or nonmodified Y could conceivably be responsible for generating CW flagellar rotation. An ideal way to distinguish between these possibilities would be to measure the concentration of Yp in vivo as a function of increasing Y expression (i.e., to determine whether elevated levels of Y lead to elevated levels of Yp). However, the phosphoryl group on Yp is highly labile, with a half-life of less than 20 s (27); therefore, such an experiment may not be technically feasible. Instead, the ability of high concentrations of mutant Y proteins bearing amino acid substitutions at the phosphorylation site to support CW flagellar rotation was assessed. Support of CW flagellar rotation by a nonphosphorylatable Y mutant protein would provide direct evidence of phosphorylation-independent signaling. There is precedence for such activity in the related response regulator protein OmpR, where high-level expression of the OmpR55DQ mutant resulted in a pattern of omp gene transcription characteristic of OmpR-P (phosphorylated OmpR) (20). The primary site of Y phosphorylation is Asp57 (41). When Asp57 is replaced specifically by asparagine, phosphorylation can occur on a secondary site, Ser56 (4, 13). Introduction of plasmid pRBB48 expressing wild-type CheY shifted the bias of the RBB1050 gutted strain from 1 to 0. In contrast, pRBB48 derivatives encoding mutant Y proteins with alanine, glutamate, or glutamine substitutions at residue 57, or a double substitution of alanine at residue 56 and asparagine at position 57, did not alter the CCW bias of RBB1050 (data not shown). The failure of the mutant Y proteins to generate CW behavior was not due to a lack of Y protein in vivo, as measured by immunoblotting (data not shown). Abolition of CW activity by removal of the Y phosphorylation site is entirely consistent with the hypothesis that the CW behavior observed in the presence of wild-type Y is due to Yp.
DISCUSSION
Observed behavior is affected by the particular che gene deletion used.
The primary motivation for this work was to resolve the apparent paradox in the literature between the bias values reported for T−W−Z− and A+Y+(g) strains. Liu and Parkinson (26) proposed two possible explanations for the more CW bias of the T−W−Z− strain. One possibility is that R and B, present only in the T−W−Z− strain, might somehow assist A in activating Y in the absence of T. If true, this hypothesis would imply the existence of as yet uncharacterized biochemical activities for R or B. The other conjecture was that the presence of some Z only in the A+Y+(g) strain might override Y activation. Our computer simulations with the BCT program supported the latter hypothesis and demonstrated that trace amounts of Z in the gutted strain HCB721 carrying Δ(cheA-cheY)::XhoI(Tn5) could give rise to the observed bias values. This prediction was directly tested by constructing a new che gene deletion, Δ(cheA-cheZ)::Zeor, which could not express Z, and then reconstituting the chemotaxis excitation signaling pathway. In each of the five matched pairs of strains described in Table 4 where both did not exhibit the same bias of ∼1 or ∼0, the bias of the Δ(cheA-cheZ)::Zeor strain was less than that of the corresponding Δ(cheA-cheY)::XhoI(Tn5) strain. These results provide compelling experimental support for the validity of the BCT prediction and demonstrate the utility of this computer simulation as a research tool.
The data reported in Table 4 support BCT not only in terms of the comparison between the two che gene deletion backgrounds examined (i.e., between the two data columns) but also in terms of the different biases generated by the various incomplete signaling pathways tested (i.e., within the leftmost data column). There is complete qualitative agreement between BCT and experiment with regard to the relative biases reported in this paper for various strains, but some quantitative discrepancies remain. It is not yet clear whether these second-order problems represent technical difficulties in the experiment (e.g., expression of Che proteins at other than precisely wild-type concentrations) or flaws in the simulation. The overall success rate of BCT in predicting mutant phenotypes is high and is described in detail elsewhere (25).
Some results obtained with the new che deletion (Table 4) reveal features of the chemotaxis signaling pathway that were not observable in gutted strains based on previously available che gene deletions. The increase or decrease in phosphorylation due to the expression of the CheA-CheZ fusion protein from Δ(cheA-cheZ)2209 or small amounts of Z from Δ(cheA-cheY)::XhoI(Tn5), respectively, masks such subtleties as the differences between A+Y+(g), W+A+Y+(g), and T+A+Y+(g). Reasonable agreement has now been achieved between the results obtained from the buildup and breakdown deletion analysis strategies.
Overexpression of Y in the absence of A nevertheless results in CW flagellar rotation.
High-level expression of Y in a strain lacking Ap, the primary source of phosphoryl groups for Y, nevertheless results in a strongly CW flagellar rotational bias, the phenotype associated with Yp. The observed CW rotation could be due either to Yp formation from sources other than Ap (28, 45) or to CW-signaling activity of nonphosphorylated Y (6). Higher (more-CCW) biases were observed when Y was overexpressed in the old rather than the new che deletion background (Table 4). This comparison suggests that the observed behavior is sensitive to catalytic amounts of Z and therefore must at least partly be due to Yp. The failure of mutant Y proteins lacking the phosphorylation site to cause CW behavior upon overexpression is also consistent with the notion that CW signals originate from Yp rather than Y. However, it remains possible that the mutant Y proteins do not support CW flagellar rotation because Asp57 is required for Y to achieve the active signaling conformation in the absence of phosphorylation.
The BCT simulation provides further insight into the basis of the CW consequences of Y overexpression. Overexpression of Y in wild-type cells, which do contain A, is predicted to result in a surprisingly modest increase in the level of Yp. This is because the Ap-to-Y phosphotransfer reaction is so fast that cells are predicted to contain very low concentrations of Ap at steady state, providing few additional phosphoryl groups to donate to an increased supply of Y. In other words, the phenotype of a Y overexpression strain apparently cannot be accounted for by the known phosphorylation reactions of the chemotaxis signaling pathway, even when A is present. Another source of activated Y, either Yp formed by cross talk with other signaling systems or phosphorylation-independent signaling, is required. Thus, the primary mechanism by which Y overexpression causes CW behavior may well be the same in both cheA+ and cheA cells. The current version of BCT achieves excellent agreement with experimental observations of behavioral phenotypes stemming from Y overexpression (25), in strains either lacking or containing A, by incorporating the suggestion of Barak and Eisenbach (6) that nonphosphorylated Y has ∼1% of the signaling ability of Yp. The same effect could equally well be achieved by including a simulated reaction in BCT which instead generated Yp by cross talk from unspecified sources other than Ap. Determination of the relative contributions of Yp and Y to CW signaling in strains overexpressing Y will have to await additional experimental investigation.
An integrated approach to the analysis of signal transduction systems.
This report describes the coordinated application of two tools, one theoretical (the BCT computer simulation) and one experimental [the new che deletion Δ(cheA-cheZ)::Zeor], to resolve a paradox that has existed in the scientific literature for almost a decade. In the process, several new aspects of the chemotaxis information-processing system were revealed that both corroborate a wide variety of previous observations and extend our knowledge of signal protein capabilities. The ability to iteratively cycle between computer simulation and experimental test is powerfully synergistic, since discrepancies between the two approaches emphasize areas of incomplete understanding. In earlier versions of BCT (14, 15), many inaccurate predictions were due to incompleteness or flaws in the simulation strategy, and so experimental results led directly to improved software. In the case reported here, benefits flowed in the reverse direction as the computer simulation drove an improved experimental design. Given the need to coherently synthesize the flood of information generated by complete genome sequencing projects, the type of integrative biology exemplified by BCT will probably find eventual application in other signaling systems, once sufficiently complete databases are available to permit the construction of other molecularly based simulations.
The next application for BCT will be to investigate the basis of the remaining discrepancies between prediction and experiment, namely, T++Z−, W++Z−, and B++Z− strains (25). The new che deletion should find use in a variety of applications in which it is important that no Che proteins are present other than those added back by the experimenter. The deletion has already been used to sort out various aspects of CheY acetylation (5, 36) and should also be valuable for in vivo investigations of the interaction between Y and the flagellar switch.
ACKNOWLEDGMENTS
We thank Alan Wolfe for providing useful advice and encouragement; Angela Seefried and Jamie Latiolais for scoring videotapes; Peter Ames, Howard Berg, David Blair, Nadim Majdalani, Phil Matsumura, Sandy Parkinson, Birgit Scharf, and Alan Wolfe for providing strains, plasmids, bacteriophages, and/or antisera; Michael Schell for giving statistical advice; and Sandy Parkinson and Alan Wolfe for commenting on the manuscript.
This work was supported by Public Health Service grant GM-50860 from the National Institute of General Medical Sciences and grant MCB-9616006 from the National Science Foundation (both to R.B.B.).
REFERENCES
- 1.Ames P, Parkinson J S. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsler C D, Cho M, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleby J L, Bourret R B. A proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleby, J. L., and R. B. Bourret. Activation of CheY mutant D57N by phosphorylation at an alternate site, Ser56. Submitted for publication. [DOI] [PubMed]
- 5.Barak R, Abouhamad W N, Eisenbach M. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in direction of flagellar rotation in Escherichia coli. J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 7.Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- 8.Berg H C, Brown D A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 9.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blat Y, Eisenbach M. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- 11.Bourret R B, Davagnino J, Simon M I. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol. 1993;175:2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 13.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bray D, Bourret R B. Computer analysis of the binding reactions leading to a transmembrane receptor-linked multiprotein complex involved in bacterial chemotaxis. Mol Biol Cell. 1995;6:1367–1380. doi: 10.1091/mbc.6.10.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray D, Bourret R B, Simon M I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clegg D O, Koshland D E., Jr The role of a signal protein in bacterial sensing: behavioral effects of increased gene expression. Proc Natl Acad Sci USA. 1984;81:5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley M P, Wolfe A J, Blair D F, Berg H C. Both CheA and CheW are required for reconstitution of chemotactic signaling in Escherichia coli. J Bacteriol. 1989;171:5190–5193. doi: 10.1128/jb.171.9.5190-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean G E, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanamaru K, Mizuno T. Signal transduction and osmoregulation in Escherichia coli: a novel mutant of the positive regulator, OmpR, that functions in a phosphorylation-independent manner. J Biochem. 1992;111:425–430. doi: 10.1093/oxfordjournals.jbchem.a123773. [DOI] [PubMed] [Google Scholar]
- 21.Khan S, Macnab R M. The steady-state counterclockwise/clockwise ratio of bacterial flagellar motors is regulated by protonmotive force. J Mol Biol. 1980;138:563–597. doi: 10.1016/s0022-2836(80)80018-0. [DOI] [PubMed] [Google Scholar]
- 22.Kuo S C, Koshland D E., Jr Multiple kinetic states for the flagellar motor switch. J Bacteriol. 1989;171:6279–6287. doi: 10.1128/jb.171.11.6279-6287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo S C, Koshland D E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987;169:1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen S H, Reader R W, Kort E N, Tso W-W, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 25.Levin, M. D., C. J. Morton-Firth, R. B. Bourret, and D. Bray. Predicting mutant phenotypes by computer: a gene-based model of bacterial chemotaxis. Submitted for publication.
- 26.Liu J D, Parkinson J S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukat G S, Lee B H, Mottonen J M, Stock A M, Stock J B. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 28.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 30.Matsumura P, Rydel J J, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNally D F, Matsumura P. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci USA. 1991;88:6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkinson J S, Houts S E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prüss B M, Nelms J M, Park C, Wolfe A J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prüss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar gene expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan R, Schuster M, Bourret R B. Acetylation at Lys92 enhances signaling by the chemotaxis response regulator protein. Proc Natl Acad Sci USA. 1998;95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravid S, Matsumura P, Eisenbach M. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1986;83:7157–7161. doi: 10.1073/pnas.83.19.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roman S J, Meyers M, Volz K, Matsumura P. A chemotactic signaling surface on CheY defined by suppressors of flagellar switch mutations. J Bacteriol. 1992;174:6247–6255. doi: 10.1128/jb.174.19.6247-6255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders D A, Gillece-Castro B L, Stock A M, Burlingame A L, Koshland D E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 42.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 43.Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 44.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 45.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe, A. J. Personal communication.
- 47.Wolfe A J, Conley M P, Berg H C. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci USA. 1988;85:6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe A J, McNamara B P, Stewart R C. The short form of CheA couples chemoreception to CheA phosphorylation. J Bacteriol. 1994;176:4483–4491. doi: 10.1128/jb.176.15.4483-4491.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]