Abstract
The TPS1 gene from Candida albicans, which encodes trehalose-6-phosphate synthase, has been cloned by functional complementation of a tps1 mutant from Saccharomyces cerevisiae. In contrast with the wild-type strain, the double tps1/tps1 disruptant did not accumulate trehalose at stationary phase or after heat shock. Growth of the tps1/tps1 disruptant at 30°C was indistinguishable from that of the wild type. However, at 42°C it did not grow on glucose or fructose but grew normally on galactose or glycerol. At 37°C, the yeast-hypha transition in the mutant in glucose-calf serum medium did not occur. During growth at 42°C, the mutant did not form hyphae in galactose or in glycerol. Some of the growth defects observed may be traced to an unbalanced sugar metabolism that reduces the cellular content of ATP. Mice inoculated with 106 CFU of the tps1/tps1 mutant did not show visible symptoms of infection 16 days after inoculation, while those similarly inoculated with wild-type cells were dead 12 days after inoculation.
The dimorphic yeast Candida albicans is commonly found as a commensal in the human population. This organism is associated with various types of lesions, mainly in cutaneous and mucosal surfaces, and it can cause deep systemic infections in humans with diminished defenses. Different properties of C. albicans have been considered putative virulence factors (11), prominent among them the ability to switch from the yeast to the filamentous form, although both forms of the organism have been found in infected tissues (38). Several antimycotic agents are available for the treatment of candidiasis, but the search for new specific targets is an issue of current pharmacological interest. Because C. albicans is a eukaryote, the number of specific targets is reduced since the host may be sensitive to certain drugs acting on the fungus.
Trehalose is a disaccharide present in microorganisms and absent from mammals. Saccharomyces cerevisiae and Kluyveromyces lactis strains mutant in the TPS1 gene encoding trehalose-6-phosphate synthase (trehalose-6-P synthase) do not grow in glucose (1, 20, 32). However, tps1− mutants of Schizosaccharomyces pombe grow in glucose and show only a defect in spore germination (7). Also, S. cerevisiae strains mutant in the TPS2 gene encoding trehalose-6-P phosphatase show a thermosensitive phenotype and are unable to grow at 37°C (12, 41). We reasoned that if C. albicans strains mutant with respect to trehalose metabolism were affected in their growth characteristics, enzymes of the trehalose biosynthetic pathway could be considered potential targets for the design of specific therapeutic agents. We therefore decided to isolate the TPS1 gene from C. albicans and study the physiological effects of its interruption. We show here that disruption of both copies of the C. albicans TPS1 gene impairs development of hyphae and decreases the infectivity of the organism.
MATERIALS AND METHODS
Yeast strains, growth and transformation conditions.
The following yeast strains were used: S. cerevisiae WDC-3A (MATa ade2-1 his3-11,15 ura3-1 leu2-1 trp1-1 tps1::HIS3) (6) and its isogenic parental strain W303 (48), and C. albicans SC5314 (19) and its derivative CAI4 (ura3Δ::imm434/ura3Δ::imm434) (16) (provided by C. Nombela, Madrid, Spain). The yeasts were grown with shaking at 30°C in 1% yeast extract–2% peptone (YP) or in a synthetic medium (Difco yeast nitrogen base, 0.67%) with adequate auxotrophic requirements. As the carbon source, 2% glucose, fructose, or galactose or 3% glycerol was added. For formation of hyphae, C. albicans strains were grown at 30°C in YP-glucose until stationary phase and then shifted to the same medium containing 10% newborn calf serum (Gibco BRL) and the desired carbon source at 37°C. S. cerevisiae (26) and C. albicans (28) were transformed as described previously.
Bacterial strains and plasmids.
Escherichia coli TG1 and DH5α were used for transformations and preparation of plasmid DNA. E. coli JM101 was used for M13 propagation (35). Plasmids YEp352 (23) and pRS316 (46) were used for constructions in S. cerevisiae, and plasmid pRM1 (42) was used for work with C. albicans. A genomic library from C. albicans in vector YEp352 was provided by C. Nombela and J. Pla (Madrid, Spain).
DNA and RNA manipulations.
Recombinant DNA manipulations were done by standard techniques (44). DNA probes were labeled (13) and genomic DNA was obtained (24) as described elsewhere. Total RNA from C. albicans was extracted from 50-mg (wet weight) samples with the Gibco Trizol reagent (10) and fractionated on a 1.5% formaldehyde agarose gel. Nucleic acids were transferred to nylon membranes as recommended by the manufacturer. As an internal probe, the 1.5-kb ClaI-SalI fragment from the C. albicans actin gene (30) was used.
DNA sequencing.
Plasmid pSecCA1 was constructed by insertion of the 3-kb SmaI-XbaI fragment from plasmid pOZ31 (Fig. 1A) into pUC18 digested with the same enzymes. Nested deletions were generated from this plasmid with ExoIII-mung bean nuclease (21), using a Stratagene deletion kit. Sequence upstream of the DNA inserted in pSecCA1 was obtained from a plasmid that carried the 1.2-kb EcoRV-BalI fragment from pOZ11 (Fig. 1A) into pUC18. Sequencing was performed by the dideoxy-chain termination method (45), using either double- or single-stranded DNA as the template. Sequences were derived from both strands. Computer analyses were carried out by using the University of Wisconsin Genetics Computer Group software on a Digital 5000/200 workstation.
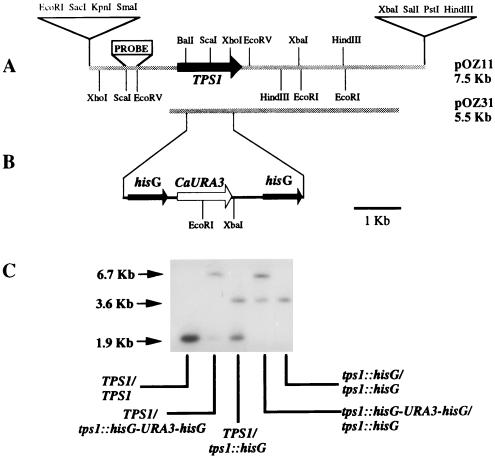
FIG. 1.
Structure of the TPS1 region from C. albicans and Southern blot analysis of tps1 disruptants. (A) Restriction maps of the C. albicans DNA inserts in pOZ11 and pOZ31. Dotted lines indicate yeast DNA. The region corresponding to the CaTPS1 gene and its direction of transcription are indicated. (B) Disruption of CaTPS1 with the hisG-URA3-hisG cassette (see Materials and Methods for details of the construction). (C) Southern analysis of the chromosomal disruption of CaTPS1. The probe used was the 0.3-kb ScaI-EcoRV fragment and is indicated in panel A as PROBE. Genomic DNA was digested with ScaI and HindIII. The relevant genotypes of the strains used for DNA analysis are indicated under the lanes. Sizes of the fragments are indicated at the left.
Chromosomal deletion of TPS1.
To disrupt the chromosomal copy of the C. albicans TPS1 gene, the following constructs were made. The 1.2-kb XhoI-HindIII fragment from plasmid pSecCA1 was inserted into pUC18 digested with SalI and HindIII to give plasmid pCA1. The KpnI site in the polylinker from pSecCA1 was eliminated by digestion with KpnI and made blunt with the Klenow fragment to give pSecCA2. A 0.3-kb fragment comprising the most external 5′ region of the cloned yeast DNA was obtained by PCR using pSecCA2 as the template, the reverse primer, and the oligonucleotide 5′GTAATGCGGTACCGAGTCCACC3′, which introduces a KpnI site at the 3′ end of the fragment. The 0.3-kb EcoRI-KpnI fragment was cloned into pCA1 digested with the same enzymes to produce pCA3. The cassette hisG-URA3-hisG carrying the C. albicans URA3 gene flanked by Salmonella typhimurium hisG direct repeats was excised by digestion with BamHI and BglII from pCUB-6K1 (a derivative of pCUB6 [16] constructed by J. Pla) and inserted into the BamHI site of pCA3 to produce plasmid pTDU. The 5.5-kb SacI-HindIII fragment of plasmid pTDU was used to disrupt the chromosomal copy of the C. albicans TPS1 gene (Fig. 1B and C).
Excision of the disruption cassette from the chromosome was performed by growing the cells in YP-glucose supplemented with uridine (15 μg/ml) for several generations and then plating the cells on minimal medium with proline (100 μg/ml) as the nitrogen source (34), supplemented with uridine (15 μg/ml) and 5-fluoro-orotic acid (200 μg/ml). Correct insertion and deletion of the disruption cassette was checked by PCR using adequate primers. The colonies producing a correct PCR pattern were also checked by Southern analysis (Fig. 1C). After excision of the disruption cassette from one chromosomal copy of TPS1, the other one was disrupted and treated similarly (Fig. 1C).
Heat shock.
Cells growing exponentially at 30°C were shifted to 37 or 42°C, and samples were taken at different times after the transfer.
Other methods.
Rapid sampling of yeast and extraction of metabolites was done as described elsewhere (43). Determinations of glucose, glycolytic intermediates, and ATP were done spectrophotometrically as described by Bergmeyer (2). Trehalose was determined by measuring the released glucose after treatment with trehalase (7).
Infectivity test.
Male Swiss CD-1 mice (specific pathogen free; Charles River), 6 weeks old and weighing approximately 25 to 30 g, were used. They were inoculated in the lateral caudal vein with 200 μl of suspensions with different cell concentrations of each strain tested, and survival was scored over 16 days. A group of six mice per condition was used.
Nucleotide sequence accession number.
The sequence obtained for the C. albicans TPS1 gene has been submitted to the EMBL databank and assigned accession no. Y07918.
RESULTS
Isolation of the C. albicans TPS1 gene.
S. cerevisiae tps1 mutants do not grow in glucose (1, 20). To isolate the TPS1 gene from C. albicans, we screened for growth in glucose about 15,000 transformants obtained from a transformation on galactose of an S. cerevisiae tps1 mutant with a C. albicans genomic library. Two plasmids, pOZ11 and pOZ31, were recovered from the transformed S. cerevisiae tps1 cells growing in glucose. They had a piece of inserted DNA with an overlapping region (Fig. 1A), and their expression in a S. cerevisiae tps1 mutant increased the trehalose level at the stationary phase of growth from <1 to 22 nmol/mg of yeast. This result indicated that plasmids pOZ11 and pOZ31 likely contain the C. albicans TPS1 gene. The C. albicans DNA inserted into plasmid pOZ11 was subcloned, and a smaller DNA insert complementing the tps1 glucose-negative phenotype was sequenced. The DNA sequence obtained, to which we will refer as CaTPS1, reveals a single open reading frame encoding a putative protein of 478 amino acids with a calculated molecular mass of 54 kDa. The predicted amino acid sequence exhibited high similarity with the sequences of trehalose-6-P synthases from other organisms (Fig. 2A). The highest identity was observed with the proteins of S. cerevisiae and K. lactis (about 70%), although there were also high homologies in certain regions with trehalose-6-P synthases from other origins.
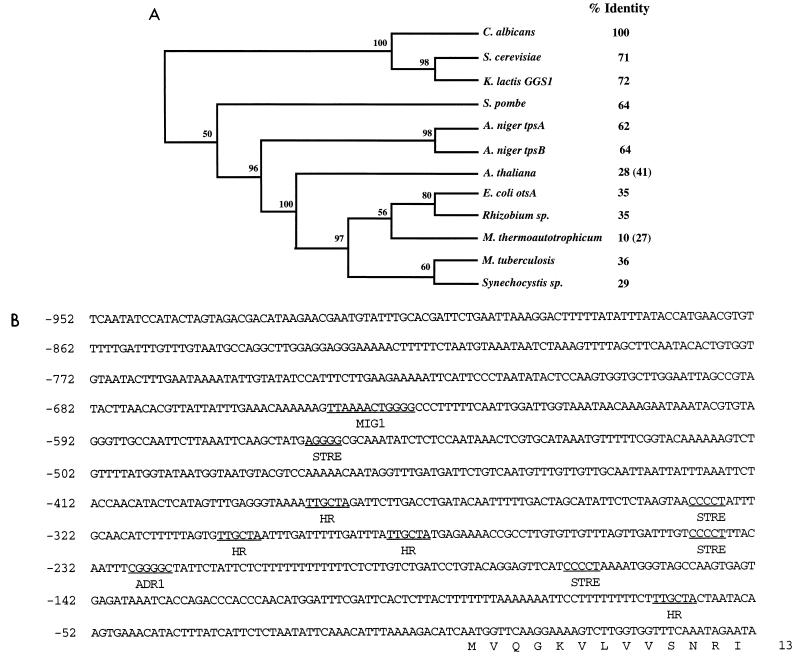
FIG. 2.
Phylogenetic tree of trehalose-6-P synthase from different organisms and regions of putative regulatory importance in the CaTPS1 promoter. (A) Amino acid sequences of the various proteins were aligned with the CLUSTAL V program (22). A maximum parsimony consensus tree (100 bootstrap resamplings) was obtained with the PHYLIP 3.5 package (14). The sequences were retrieved from the SWISSPROT database. Numbers at the intersections indicate the bootstrap value as a percentage. The percentage of identity with respect to the C. albicans amino acid sequence is shown at the right. Figures in brackets were obtained by comparing only the 500 initial amino acids in the case of Arabidopsis thaliana or using only amino acids 290 to 415 from C. albicans in the case of Methanobacterium thermoautotrophicum. (B) Part of the 5′ noncoding region of the CaTPS1 gene. Putative regulatory sequences are underlined.
The 5′ noncoding region of CaTPS1 showed several interesting features (Fig. 2B). One was the existence of several C4T stretches (STRE sequences [27]) usually implicated in stress-controlled transcription (33, 36). A possible binding site for Mig1 (31), a protein involved in catabolite repression of certain genes (37), and four copies of the hexameric HR repeat (47) were also found.
Effects of the disruption of the CaTPS1 gene.
To determine the role of CaTPS1 in the physiology of C. albicans, we disrupted both chromosomal copies of the gene (Fig. 1B and C; Materials and Methods). Growth rates of the wild type and the double disruptant in glucose were similar at 30°C (160-min generation time). On plates, no striking differences in growth on fructose, galactose, or glycerol were observed. At 37°C, the disruptant grew more slowly than the wild type in glucose (180-min versus 90-min generation time), but both had generation times of 90 min in galactose. At 42°C, the double disruptant did not grow on glucose but grew on galactose or glycerol. Generation times in these latter carbon sources could not be compared at this temperature because the wild type flocculated (Fig. 3B). However, the wild type and disruptant appeared to grow at similar rates on glycerol and galactose.
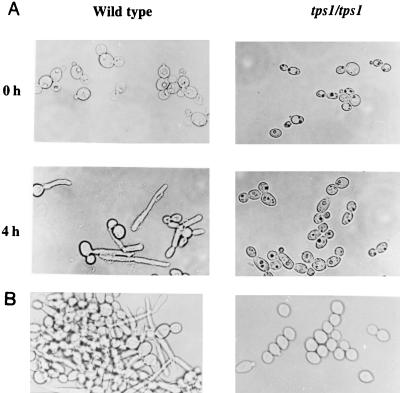
FIG. 3.
Influence of disruption of the CaTPS1 gene on formation of hyphae. (A) Yeasts were grown in YP-glucose until stationary phase and suspended in the same medium with 10% newborn calf serum. The pictures were taken at the time of serum addition (0 h) and 4 h later. (B) Morphologies of cultures of wild-type and tps1/tps1 disruptant on YP-glycerol during the exponential phase of growth at 42°C.
Trehalose accumulation upon entrance into the stationary phase varied with the temperature of the culture, being higher at 37°C than at 30°C (Table 1). However, heat shock at 37°C per se did not elicit a noticeable increase in trehalose. A heat shock at 42°C increased the levels of trehalose (Table 1) and expression of the TPS1 gene (results not shown). Trehalose accumulation in the double mutant was never observed (Table 1), indicating that no other significant activities contribute to trehalose synthesis in C. albicans. Trehalose levels in the strain carrying only one copy of CaTPS1 were between those of the wild type and the double mutant (results not shown). We observed that in S. cerevisiae tps1 mutants expressing the CaTPS1 gene, trehalose accumulated to a level higher than that observed in the C. albicans strain in the same conditions. Curiously, the trehalose values measured were not significantly different when the CaTPS1 gene was expressed from a centromeric or a multicopy plasmid (Table 1). A noteworthy result was that after heat shock, C. albicans CAI4 (TPS1 ura−) accumulated about 75% less trehalose than its parental counterpart SC5314 (TPS1 ura+) (Table 1).
TABLE 1.
Trehalose accumulation in stationary phase and after heat shock in C. albicans and in S. cerevisiae strains expressing the CaTPS1 gene
| Strain (relevant genotype) | Growth temp (°C) | Trehalose (nmol/mg [wet wt] of yeast)a
|
|
|---|---|---|---|
| Stationary phase | Heat shock | ||
| C. albicans SC5314 (wild type) | 30 | 5 | 50 (17b) |
| 37 | 20 | NT | |
| C. albicans (tps1/tps1) | 30 | 2 | 1 |
| 37 | 2 | NT | |
| S. cerevisiae W303 (wild type) | 30 | 20 | 29 |
| S. cerevisiae WDC-3A (tps1) | 30 | <1 | <1 |
| S. cerevisiae (tps1/CaTPS1) | 30 | 18c/22d | 14 |
C. albicans and S. cerevisiae strains were grown in YPD and minimal medium with 2% galactose as the carbon source (S. cerevisiae tps1 strains do not grow in glucose), respectively. Heat shock was performed in exponentially growing cells by a shift of the culture to 42°C during 1 h. In exponential-phase cells of all strains tested, trehalose production was <1 nmol/mg. All C. albicans strains were prototrophic for uracil, but see footnote b. NT, not tested.
Refers to strain CAI4, which is auxotrophic for uracil.
CaTPS1 expressed in the centromeric plasmid pRS316.
CaTPS1 expressed from the multicopy plasmid YEp352.
Metabolite profile.
In S. cerevisiae, the failure of the tps1 mutant to grow on glucose is associated with a severe imbalance of glycolytic metabolites and a decrease in ATP (20). In the case of C. albicans, the profile of internal metabolites in the tps1/tps1 mutant in the presence of glucose was altered in all circumstances tested. A lower level of ATP and an accumulation of hexose phosphates with respect to the wild type were measured even during growth at 30°C, a condition in which the growth rate was identical to that of the wild type (Table 2). A similar pattern was observed in cells suspended in buffer. At 42°C the mutant accumulated a high amount of glucose-6-P, but the level of fructose-1,6-bisphosphate was not higher than that at 30°C. However, the level of ATP dropped drastically at 42°C, being barely detectable 1 min after glucose addition (Table 2). In spite of the lower level of ATP, glucose consumption was higher in the mutant than in the wild type (about 30% higher at 30°C and about 10% higher at 42°C).
TABLE 2.
Intracellular levels of some glycolytic intermediates in C. albicans wild-type and tps1/tps1 mutant strains after addition of glucosea
| Strain (relevant genotype) | Temp (°C) | Conditions | Intracellular level (nmol/mg [dry wt] of yeast)
|
Glucose consumption (nmol/mg [dry wt] of yeast/h) | Growth yield (g [dry wt] of yeast/g of glucose) | ||
|---|---|---|---|---|---|---|---|
| Glucose-6-P | Fructose-1,6-P2 | ATP | |||||
| C. albicans CAI4 (wild type) | 30 | Buffer | 0.9, 6.4 | <0.1, 1.2 | 3.6, 1.7 | 0.56 | |
| C. albicans (tps1/tps1) | 30 | Buffer | 1.3, 8.4 | <0.1, 12 | 2.8, 0.6 | 0.9 | |
| C. albicans CAI4 (wild type) | 42 | Buffer | 0.4, 5.2 | <0.1, 1.4 | 2, 1.4 | 0.9 | |
| C. albicans (tps1/tps1) | 42 | Buffer | 0.2, 14 | <0.1, 11.6 | 1.3, <0.1 | 1 | |
| C. albicans CAI4 (wild type) | 30 | Growth | 1.12 | 2.2 | 1.8 | 1 | 0.24 |
| C. albicans (tps1/tps1) | 30 | Growth | 4.4 | 6.8 | 0.42 | 1.4 | 0.17 |
Yeasts were grown in rich medium-glycerol at 30°C and resuspended at 20 mg (dry weight)/ml in buffer as described in Materials and Methods. After equilibration at the indicated temperatures, glucose was added at a final concentration of 55 mM and samples were taken at different times. For each metabolite, the first number is the value at time zero (immediately after glucose addition), and the second is the value after 30 min, except for ATP, where the second number is the value 1 min after glucose addition. The values obtained during growth were obtained in a culture during the exponential phase of growth.
Morphological and infectivity changes.
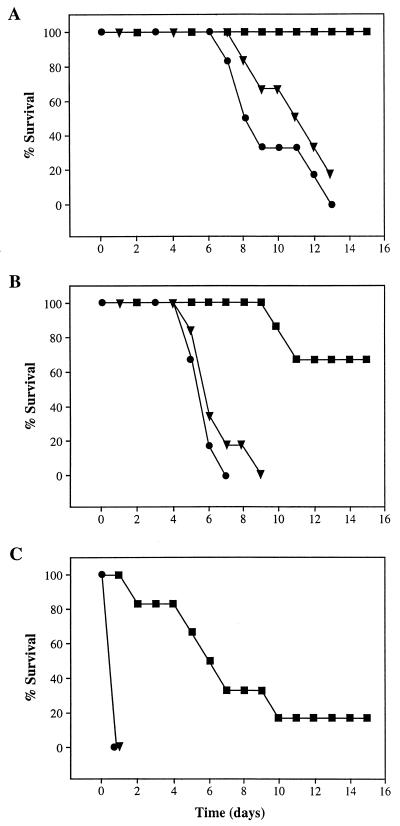
C. albicans can shift from yeast to hyphal form when the organism is cultured at 37°C in the presence of newborn calf serum. This transition was severely impaired in the tps1/tps1 mutant, where no hypha formation was observed in shake flasks with glucose as the carbon source (Fig. 3A). We observed no difference between ura− and ura+ cells with respect to this behavior. However, at this temperature, when galactose or glycerol was used instead, hyphae formed as in the wild type. At 42°C, with galactose or glycerol as the carbon source in the absence of serum, the wild type formed hyphae that aggregated and made the culture flocculent, while the mutant grew as yeast (Fig. 3B). The infectivity of the tps1/tps1 mutant was markedly less than that of the wild type (Fig. 4). Mice inoculated with the lower dose of 106 cells did not show external symptoms of infection during the 16 days of observation, while the controls died in 12 days. Even those inoculated with 108 cells had a delayed onset of symptoms. As shown in Fig. 4, strains with only one interrupted copy behaved similarly to the wild type.
FIG. 4.
Infectivity of C. albicans tps1/tps1 mutants. Mice were inoculated with different cell suspensions of the tps1/tps1 mutant (squares), the TPS1/tps1 single disruptant (triangles), and the wild type (circles), and survival was scored over time. Panels A, B, and C correspond to inocula of 106, 107, and 108 CFU. All strains were prototrophic for uracil.
DISCUSSION
We have isolated and characterized the gene encoding trehalose-6-P synthase from the dimorphic yeast C. albicans. That the gene isolated encodes trehalose-6-P synthase is evidenced by the following findings: (i) it increases the trehalose content in a S. cerevisiae tps1 mutant, (ii) the sequence of the protein putatively encoded by this gene has extensive regions of identity with trehalose-6-P synthases from other organisms, and (iii) trehalose synthesis in C. albicans was absent in cells that carried a disruption of the gene in both chromosomal copies. Our results also show that in C. albicans there is only one, or at least a major, trehalose-6-P synthase implicated in the synthesis of trehalose. This is also the situation in S. cerevisiae (40), although the existence of a minor trehalose-6-P synthase activity that uses ADP-glucose as a cosubstrate has been reported (15, 39). The situation contrasts with that found in Aspergillus niger, where two trehalose-6-P synthases encoded by genes tpsA and tpsB have been detected. In this fungus, the products of these genes appear to play a different role in the physiology of the organism (49).
In the promoter of yeast genes regulated by heat shock, STRE sequences (CCCCT) (27) are usually found. In the case of CaTPS1, four copies of the STRE element are present in the 5′ nontranslated region of the gene. Although one copy may suffice for the heat shock response, the presence of multiple elements increases this response (27). The finding of a Mig1 binding site may indicate a regulatory role for this protein that is involved in catabolite repression of certain genes (37). In fact, González et al. (20) found that transcription of TPS1 (then called CIF1) in S. cerevisiae was lower in glucose-grown than in galactose-grown cultures. The existence of several copies of the hexameric repeat HR suggests some role for these DNA stretches. They have been found in the 5′ nontranslated region of the WHI1 gene from C. albicans, but their function is not known (47).
The finding that expression of CaTPS1 in S. cerevisiae produced at 30°C a level of trehalose higher than that accumulated by C. albicans in the same conditions may be due to the fermentative metabolism of S. cerevisiae. The ethanol accumulated in S. cerevisiae cultures during glucose fermentation adds a stress factor to the depletion of sugar, as shown by the finding that trehalose levels in S. cerevisiae growing in ethanol are elevated even in the exponential phase of growth (28a). We observed that C. albicans strains auxotrophic for uracil accumulated less trehalose during heat shock than congenic strains without the nutritional requirement. A possible explanation for this could be that in the uracil-requiring strains, the pool of uridine derivatives necessary for the formation of trehalose-6-P is decreased. We have not studied if other auxotrophic requirements produce the same effect, but our results indicate that it is important when comparing levels of trehalose in congenic strains with interrupted genes to ensure that the strains do not differ in nutritional requirements.
Disruption of both chromosomal CaTPS1 copies caused several defects: lack of growth on glucose at certain temperatures, impairment of hypha formation, and decrease of infectivity. The effect on growth was specific for glucose or fructose, pointing to a defect in the metabolism of these sugars. This idea was strengthened by the altered metabolite profile found either during apparently normal growth at 30°C or in resting cells at different temperatures. The severe depletion of ATP and the accumulation of hexose phosphates are similar to those found in S. cerevisiae (20) and K. lactis (32) when glucose is added to a tps1 mutant. A loss of the inhibition of hexokinase by trehalose-6-P that results in an increased glycolytic flux has been proposed as an explanation of this effect (6). In fact, the defect is alleviated in S. cerevisiae by a decrease in hexokinase activity (4, 25) or in glucose transport (5, 17). In S. pombe, where the major hexokinase is not inhibited by trehalose-6-P (7), disruption of tps1+ does not affect growth in glucose. The glucose-phosphorylating activity of C. albicans is inhibited by trehalose-6-P (3), and the phenotype observed in the CaTPS1 disruptants would be consistent with the explanation advanced for S. cerevisiae. The growth of C. albicans at 30 and 37°C (although at the latter temperature more slowly than the wild type) may be explained by the respiratory metabolism of this yeast that will make the effects of CaTPS1 disruption less severe than in a yeast with a higher demand of glycolytic flux, such as S. cerevisiae (18). It is interesting that the effect of the disruption in growth was different from that found in S. cerevisiae or K. lactis. In the latter yeasts, tps1 disruptants stopped growth at 30°C, whereas in C. albicans the effect was dependent on temperature and was total only at 42°C. Whether this is due to adaptation of C. albicans to higher temperatures or to a different mode of action of trehalose or trehalose-6-P remains to be studied.
The influence of CaTPS1 on formation of hyphae is likely to be indirect and not due to involvement of the gene product in the transition process. The yeast-hypha transition is a complex process in which different external factors produce signals that converge on specific targets to trigger the morphological changes. Although mutants that exist only as hyphae (9) or predominantly as yeast (29) have been isolated, the mechanism of the transition is not well understood. We hypothesize that the lack of hypha formation at 37°C on glucose is due to the disturbed glucose metabolism that drastically reduces the ATP content of the cells. The fact that hyphae are formed at this temperature on galactose or glycerol is compatible with this idea. The manifest lack of hypha formation at 42°C in all carbon sources may be related to the need for trehalose for maintenance of hyphae at this temperature. In fact, spores of tps1− mutants of S. pombe do not germinate because trehalose is needed for their maintenance or for initiation of germination (7).
The C. albicans tps1/tps1 mutants showed a drastic decrease in pathogenicity for mice. Injection of as much as 107 cells did not provoke visible symptoms during 8 days. In contrast, the TPS1/tps1 strains were as virulent as the wild type. We cannot determine at present whether the lower infectivity is due to the impairment of hypha formation or to poorer proliferation of the mutant in mice. In fact, both yeast and hyphal forms were detected in mice infected with the mutant (42a).
Mutations in the pathway of trehalose synthesis have pleiotropic effects in different microorganisms, affecting reactions in carbohydrate metabolism not directly related to trehalose synthesis (7, 8, 20). The effects described here on growth and hypha formation in C. albicans add another target to those observed in other organisms and indicate the important regulatory role of trehalose-6-P and trehalose in the life of fungi.
ACKNOWLEDGMENTS
We thank Juana M. Gancedo for critical reading of the manuscript and discussions during the work, W. A. Fonzi (Georgetown University, Washington, D.C.), C. Nombela, J. Pla, and F. Navarro (Fac. Farmacia, Universidad Complutense de Madrid, Madrid, Spain) for the gift of plasmids, strains, the C. albicans library, and technical hints, F. J. Gamo, E. Jiménez, and A. Martínez (Glaxo Wellcome S.A., Tres Cantos, Madrid, Spain) for help with the infectivity assays, and J. Pontón (University of Bilbao, Bilbao, Spain) and R. Lagunas (Instituto de Investigaciones Biomédicas, Madrid, Spain) for information on unpublished results.
This work was supported by grant PB94-0091-CO2-01 from the Spanish DGICYT. During part of this work, O.Z. was supported by a fellowship from the Spanish PFPI, and M.A.B. had a fellowship from the Comunidad de Madrid.
Footnotes
Dedicated to the memory of Helmut Holzer, teacher and friend, who made important contributions to the biochemistry of yeast.
REFERENCES
- 1.Bell W, Klaasen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, van der Zee P, Wiemken A. Characterization of the 56 kDA subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem. 1992;209:951–959. doi: 10.1111/j.1432-1033.1992.tb17368.x. [DOI] [PubMed] [Google Scholar]
- 2.Bergmeyer H U. Methods of enzymatic analysis. Weinheim, Germany: Verlag Chemie; 1983. [Google Scholar]
- 3.Blázquez M A. Un nuevo mecanismo de control de la glicolisis en Saccharomyces cerevisiae: la inhibición de las hexokinasas por trehalosa-6-fosfato. PhD. thesis. Madrid, Spain: Faculty of Sciences, Universidad Autónoma de Madrid; 1995. [Google Scholar]
- 4.Blázquez M A, Gancedo C. Identification of extragenic suppressors of the cif1 mutation in Saccharomyces cerevisiae. Curr Genet. 1994;25:89–94. doi: 10.1007/BF00309531. [DOI] [PubMed] [Google Scholar]
- 5.Blázquez M A, Gancedo C. Mode of action of the qcr9 and cat3 mutations in restoring the ability of Saccharomyces cerevisiae tps1 mutants to grow on glucose. Mol Gen Genet. 1995;249:665–664. doi: 10.1007/BF00418035. [DOI] [PubMed] [Google Scholar]
- 6.Blázquez M A, Lagunas R, Gancedo C, Gancedo J M. Trehalose-6-phosphate, a new regulator of yeast glycolysis. FEBS Lett. 1993;329:51–54. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- 7.Blázquez M A, Stucka R, Feldmann H, Gancedo C. Trehalose-6-P synthase is dispensable for growth on glucose but not for spore germination in Schizosaccharomyces pombe. J Bacteriol. 1994;176:3895–3902. doi: 10.1128/jb.176.13.3895-3902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgia P T, Miao Y M, Lodge C L. The orlA gene from Aspergillus nidulans encodes a trehalose-6-phosphate phosphatase necessary for normal growth and chitin synthesis at elevated temperatures. Mol Microbiol. 1996;20:1287–1296. doi: 10.1111/j.1365-2958.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 9.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 12.De Virgilio C, Bürckert N, Bell W, Jenö P, Boller T, Wiemken A. Disruption of TPS2, the gene encoding the 100 kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphatase activity. Eur J Biochem. 1993;212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 15.Ferreira J C, Thevelein J M, Hohmann S, Paschoalin V M, Trugo L C, Panek A D. Trehalose accumulation in mutants of Saccharomyces cerevisiae deleted in the UDPG-dependent trehalose synthase-phosphatase complex. Biochim Biophys Acta. 1997;1335:40–50. doi: 10.1016/s0304-4165(96)00127-4. [DOI] [PubMed] [Google Scholar]
- 16.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamo F J, Lafuente M J, Gancedo C. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–7429. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gancedo C, Serrano R. Energy yielding metabolism. In: Rose A H, Harrison J S, editors. The yeasts. 2nd ed. Vol. 3. London, United Kingdom: Academic Press; 1989. pp. 206–259. [Google Scholar]
- 19.Gillum A M, Tsay E Y, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 20.González M I, Stucka R, Blázquez M A, Feldmann H, Gancedo C. Molecular cloning of CIF1, a yeast gene necessary for growth on glucose. Yeast. 1992;8:183–192. doi: 10.1002/yea.320080304. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 22.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. Gene. 1987;57:266–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 25.Hohmann S, Neves M J, de Koning W, Alijo R, Ramos J, Thevelein J M. The growth and signalling defects of the ggs1(fdp1/byp1) deletion mutants on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet. 1993;23:281–289. doi: 10.1007/BF00310888. [DOI] [PubMed] [Google Scholar]
- 26.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat-shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz M B, Cortelyou M W, Kirsch D R. Integrative transformation of Candida albicans using a cloned Candida ADE2 gene. Mol Cell Biol. 1986;6:142–149. doi: 10.1128/mcb.6.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Lagunas, R. Personal communication.
- 29.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1725. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 30.Losberger C, Ernst J F. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 1989;22:9488. doi: 10.1093/nar/17.22.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luyten K, de Koning W, Tesseur I, Ruiz M C, Ramos J, Cobbaert P, Thevelein J M, Hohmann S. Disruption of the Kluyveromyces lactis GGS1 gene causes inability to grow in glucose and fructose and is suppressed by mutations that reduce sugar uptake. Eur J Biochem. 1993;217:701–713. doi: 10.1111/j.1432-1033.1993.tb18296.x. [DOI] [PubMed] [Google Scholar]
- 33.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCusker J H, Davis R W. The use of proline as a nitrogen source causes hypersensitivity to, and allows more economical use of 5FOA in Saccharomyces cerevisiae. Yeast. 1991;7:607–608. doi: 10.1002/yea.320070608. [DOI] [PubMed] [Google Scholar]
- 35.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 36.Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 37.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odds F C. Candida and candidosis. 2nd ed. London, England: Baillière-Tindall; 1988. [Google Scholar]
- 39.Paschoalin V M, Silva J T, Panek A D. Identification of an ADPG-dependent trehalose synthase in Saccharomyces. Curr Genet. 1989;16:81–87. doi: 10.1007/BF00393399. [DOI] [PubMed] [Google Scholar]
- 40.Petit T, François J. Accumulation of trehalose in Saccharomyces cerevisiae growing in maltose is dependent on TPS1 gene encoding the UDP-glucose linked trehalose synthase. FEBS Lett. 1994;355:309–313. doi: 10.1016/0014-5793(94)01215-6. [DOI] [PubMed] [Google Scholar]
- 41.Piper P W, Lockheart A. A temperature-sensitive mutant of Saccharomyces cerevisiae defective in the specific phosphatase of trehalose biosynthesis. FEMS Microbiol Lett. 1988;49:245–250. [Google Scholar]
- 42.Pla J, Pérez-Díaz R M, Navarro-García F, Sánchez M, Nombela C. Cloning of Candida albicans HIS1 gene by direct homologous complementation of a histidine auxotroph using an improved double-ARS shuttle vector. Gene. 1995;165:115–120. doi: 10.1016/0378-1119(95)00492-o. [DOI] [PubMed] [Google Scholar]
- 42a.Pontón, J. Personal communication.
- 43.Sáez M J, Lagunas R. Determination of intermediary metabolites in yeast. Critical examination of the effect of sampling conditions and recommendations for obtaining true values. Mol Cell Biochem. 1976;13:73–78. doi: 10.1007/BF01837056. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski R S, Hieter P. A system of shuttle vectors and yeast hosts designed for the efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srikantha T, Chandrasekhar A, Soll D R. Functional analysis of the promoter of the phase-specific WHI1 gene of Candida albicans. Mol Cell Biol. 1995;15:1797–1805. doi: 10.1128/mcb.15.3.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 49.Wolschek M F, Kubicek C P. The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase encoding genes tpsA and tpsB. J Biol Chem. 1997;272:2729–2735. doi: 10.1074/jbc.272.5.2729. [DOI] [PubMed] [Google Scholar]