Abstract
Diverse kinesin motor proteins are involved in spindle function; however, the mechanisms by which they are targeted to specific sites within spindles are not well understood. Here, we show that a fusion between yellow fluorescent protein (YFP) and a minus-end–directed Kinesin-14 (C-terminal family) from Arabidopsis, ATK5, localizes to mitotic spindle midzones and regions rich in growing plus-ends within phragmoplasts. Notably, in Arabidopsis interphase cells, YFP::ATK5 localizes to microtubules with a preferential enrichment at growing plus-ends; indicating ATK5 is a plus-end tracking protein (+TIP). This +TIP activity is conferred by regions outside of the C-terminal motor domain, which reveals the presence of independent plus-end tracking and minus-end motor activities within ATK5. Furthermore, mitotic spindles of atk5 null mutant plants are abnormally broadened. Based on these data, we propose a model in which ATK5 uses plus-end tracking to reach spindle midzones, where it then organizes microtubules via minus-end–directed motor activity.
INTRODUCTION
During cell division, the proper segregation of genetic material into daughter cells requires the action of the microtubule spindle apparatus and its associated proteins. The spindle consists of two opposing sets of microtubules oriented with the minus-ends at the poles and the plus-ends at the midzone. The midzone represents the region of overlap between the two halves of the spindle, where microtubule plus-ends terminate at chromosomal kinetochores (kinetochore microtubules) or interdigitate in an antiparallel manner with microtubules from the opposite pole (interpolar microtubules). The spindle midzone is the site of force generation during anaphase spindle elongation (Leslie and Pickett-Heaps, 1983; Khodjakov et al., 2004), and in plants it persists through telophase to form the cytokinetic microtubule apparatus, the phragmoplast (Euteneuer et al., 1982).
The assembly and functioning of spindles involve the highly orchestrated activities of diverse microtubule motor proteins. Kinesins convert the energy derived from ATP hydrolysis into translational movement along microtubules (Dagenbach and Endow, 2004; Lawrence et al., 2004). Kinesin-14 family members (previously referred to as C-terminal kinesins), such as Ncd from Drosophila and Kar3p from budding yeast, are unique in that they translocate exclusively toward microtubule minus-ends (Walker et al., 1990). Several Kinesin-14 family members contain microtubule binding sites in their tail regions, which specifies the ability to carry microtubules as cargo along other microtubules; in effect, moving microtubules in relation to one another (Walczak et al., 1997; Narasimhulu and Reddy, 1998; Karabay and Walker, 1999; Matuliene et al., 1999). This finding, in conjunction with subcellular localization and loss-of-function studies, has revealed two distinct roles for Kinesin-14s in spindle functioning. The first role is inferred from studies showing that loss or depletion of various Kinesin-14 family members rescues the spindle collapse phenotypes resulting from loss of bipolar BimC kinesins (Hoyt et al., 1993; O'Connell et al., 1993; Pidoux et al., 1996; Mountain et al., 1999; Sharp et al., 1999b). During spindle formation, Kinesin-14s cross-link antiparallel microtubules and slide them together (thereby generating inward forces) to balance the outward forces generated by plus-end–directed kinesins of the Kinesin-5 family (previously referred to as BimC kinesins). The second role of Kinesin-14 family members is to gather microtubule minus-ends and focus them into spindle poles by cross-linking parallel microtubules and motoring toward the minus-end. In support of this, mutation or inhibition of Kinesin-14 family members often results in disordered or splayed meiotic spindle poles (Hatsumi and Endow, 1992; Walczak et al., 1998; Chen et al., 2002), and many Kinesin-14 family members are localized to microtubules near the spindle poles (Kuriyama et al., 1995; Walczak et al., 1997; Smirnova et al., 1998). Hence, Kinesin-14s seem to function in two ways: 1) to generate inward forces at the spindle midzone to pull the spindle halves together, and 2) to gather and focus microtubule minus-ends at the spindle poles. However, it is unclear as to how Kinesin-14s reach these specific sites of action within spindles.
The Arabidopsis genome encodes a predicted 61 kinesins, 21 of which belong to the Kinesin-14 family (Reddy and Day, 2001). Despite this abundance of Kinesin-14 family members, little is known about their roles with respect to spindle organization and function. It has been hypothesized that because plants do not contain classical microtubule organizing centers, such as centrosomes, the activity of motor proteins may play a central role in microtubule organization (Smirnova and Bajer, 1998).
The Arabidopsis Kinesin-14 family member ATK1 is involved in mitotic and meiotic spindle function (Liu et al., 1996; Chen et al., 2002; Marcus et al., 2002, 2003). Atk1-1 null mutants exhibit splayed meiotic spindle poles, which result in defective chromosome segregation and reduced male fertility. During mitosis, atk1-1 mutants exhibit reduced spindle bipolarity in prophase and metaphase, although by anaphase, normal bipolar spindles resolve and chromosomes segregate normally. To identify additional kinesins involved in mitosis, we cloned ATK5, which encodes a protein with 83% amino acid sequence identity to ATK1. Here, we report the observation that ATK5 is a minus-end–directed motor as well as a plus-end tracking protein (+TIP). We show that ATK5 is a mitotic motor based on its localization to mitotic spindle midzones and the abnormal spindle morphology observed in atk5 null mutants. Based on these data, we present a model in which plus-end tracking facilitates midzone localization and ultimately spindle morphogenesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Columbia) were grown on Arabidopsis medium (Granger and Cyr, 2001) or on soil under 18-h/6-h day/night at 250 μE/m2/s in growth chambers (Perceivel Scientific, Perry, IA) at 20–23°C. For Agrobacterium-mediated transformation, inflorescences were sprayed with a suspension of Agrobacterium tumefaciens (strain C58C1; OD = 1.2) carrying appropriate transgenes in 5% sucrose solution supplemented with 0.02% Silwet-77 (Helena Chemical, Fresno, CA). T1 seeds were collected and plated onto medium containing appropriate antibiotics, screened at 7 d for fluorescence, and transplanted to soil. T2 and T3 plants were used for experiments.
Tobacco BY-2 cells were maintained in liquid culture and on plates as calli for long-term maintenance. Liquid cultures were subcultured at 7-d intervals (diluted 1:50 into fresh BY-2 medium, containing 4.3 g l–1 Murishige and Skoog's salts, 100 mg l–1 inositol, 1 mg l–1 thiamine, 0.2 mg l–1 2,4-D, 255 mg l–1 KH2PO4, and 3% sucrose, pH 5.0). For Agrobacterium-mediated transformation, 2 ml of 4-d BY-2 cells were mixed with 10 μl from an overnight culture of Agrobacterium (strain LBA4404, containing the appropriate transgene) and 20 μM acetosyringone and then incubated in darkness for 4 d. Transformants were selected on BY-2 medium (0.35% Phytagel) containing appropriate antibiotics, and individual calli were screened by fluorescence microscopy for suitable transgene expression.
Cloning and Construct Design
Polymerase chain reaction (PCR) products were cloned using the Topo TA cloning system (Invitrogen, Carlsbad, CA). Fragments were then excised with the appropriate restriction enzymes for cloning into the plant transformation vector pCambia1300 (Cambia Institute, Canberra, Australia) containing a CaMV35S- or HSP18.2-driven mYFP (Haseloff, 1999) or into the bacterial expression vector pGEX-5 × -3 (Novagen, Madison, WI). ATK5 (accession no. AtC17L17.110/At4g05190) cDNA was amplified from an Arabidopsis floral cDNA library with the following primers: ATK5fwd (5′-GGATCCATGCCACTTCGCAACCAGA-3′) and ATK5rev (5′-GGATCCTTAACCGTAACTTAGGCGA-3′). BamHI fragments containing ATK5 were ligated into either pCambia1300 for plant transformation or the pGEX5 × -3 for in vitro motility experiments. ATK5 tail/stalk was amplified using the following primers: ATK5fwd and ATK5TSrev (5′-GAGCTCGCGGCCGCTTAAACATGAGTTTCATCCAC-3′). The tail/stalk fragment was excised using BamHI and SacI and cloned into pCambia 1300.
Genotyping
T-DNA insertional mutants were obtained from the Salk collection (La Jolla, CA). SALK_001544 (T-DNA insertion at 2211 in coding sequence) was designated atk5-1, and SALK_065546 (T-DNA insertion at 2213 in coding sequence) was designated atk5-2. Genotyping was performed with two PCR reactions: 1) T-DNA specific and 2) gene specific. For the T-DNA–specific reaction, the following primers were used: ATK5fwd and LBb1 (5′-AACCAGCGTGGACCGCTTGCTG-3′). For the gene-specific reaction, the following primers were used: 38 (5′-TCACTACAAGATCAATTAG-3′) and 43 (5′-TCAAAGAATACTTCCTCC-3′).
Reverse Transcriptase (RT)-PCR
RNA was extracted from wild-type, atk5-1, and atk5-2 seedlings and treated with RQ1 DNase (Promega, Madison, WI). cDNA was synthesized by using Superscript II (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. RT-PCR was performed by combining gene-specific primers oMC 772 (5′-GGCCTCTTGGCGTCCTCCATCA-3′) and oMC 795 (5′-AGCGAGCTGCAACAAGTCCGTG-3′). Expression of the constitutive adenine phosphoribosyltransferase (APT1) gene was examined as a control (Moffatt et al., 1994).
Immunofluorescence
Five-day Arabidopsis seedlings grown on vertical agar plates were fixed for 1 h (4% formaldehyde [freshly prepared from paraformaldehyde], 50 mM PIPES, pH 6.9, 5 mM EGTA, 1 mM MgSO4, and 1% glycerol), rinsed, and digested with cell wall-degrading enzymes (0.5% cellulose, 0.05% pectolyase, and 0.5% Triton X-100) for 15 min. Roots were excised and squashed on poly-l-lysine (Sigma-Aldrich, St. Louis, MO)-coated slides. The squashed cells were treated with 3% bovine serum albumin for 1 h in a humidity chamber, rinsed, and detergent extracted for 20 min in 0.5% Triton X-100 before incubation in fluorescein isothiocyanate (FITC)-conjugated DMA1 anti-tubulin antibody (1:75 dilution; Sigma-Aldrich) for 1.5 h. After rinsing in phosphate-buffered saline, 40 μl of fluorescent mounting medium (0.1 M Tris, pH 9.0, 50% glycerol, and 1 mg ml–1 phenylenediamine, supplemented with 1 μg ml–1 Hoescht 33258 for chromosomal staining) was added, coverslips were applied, and the specimens were sealed with nail polish.
Fluorescence Microscopy
Images were collected using a Plan-Neofluar 40× (1.3 numerical Aperture) oil immersion objective (Carl Zeiss, Thornwood, NY). Wide-field microscopy was conducted using a shutter-equipped Zeiss Axiovert, and images were captured with a Coolsnap HQ charge-coupled device camera (Roper Industries, Duluth, GA) controlled by ESEE software (Inovision, Durham, NC). The following filter sets were used to distinguish between fluorophores: green fluorescent protein (GFP)/FITC (460- to 500-nm excitation, 510- to 560-nm emission); yellow fluorescent protein (YFP) (490- to 510-nm excitation, 520- to 550-nm emission); DsRed (530- to 560-nm excitation, 575- to 645-nm emission); and Hoescht (340- to 380-nm excitation, 435- to 485-nm emission). The construct MBD::DsRed was used to visualize microtubules in BY-2 cells, as reported previously (Dixit and Cyr, 2003). Five-second intervals were used for time-lapse imaging of microtubule dynamics in Arabidopsis interphase cells, and 1-min intervals were used for monitoring cell division in BY-2 cells. Typical exposure times were 1–2 s.
In Vitro Motility Assays
Cleared bacterial lysates from BL21 (DE3) cells (Novagen) expressing glutathione S-transferase (GST)::ATK5 or GST::YFP::ATK5 were used. Polarity marking of microtubules and microtubule gliding assays were done as described previously (Marcus et al., 2002).
RESULTS
In Vivo Localization of ATK5
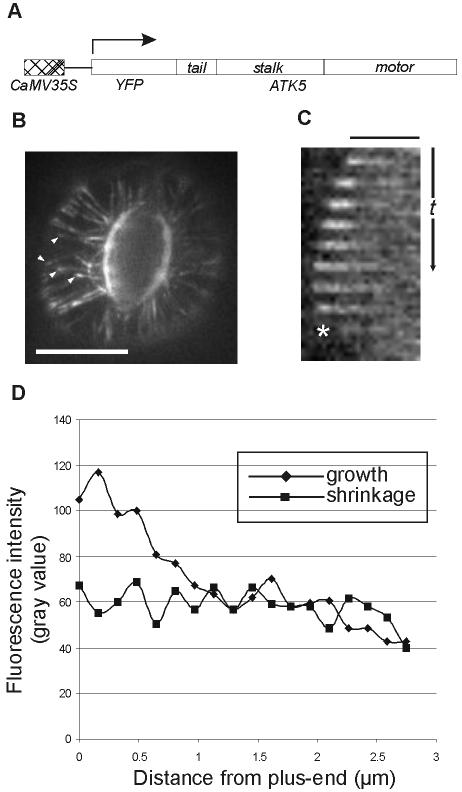
We first sought to elucidate the mechanism of ATK5 functioning by examining the dynamics of ATK5 subcellular localization in vivo. To this end, full-length ATK5 cDNA was fused in-frame to the 3′ end of mYFP (Figure 1A). The fusion gene was stably expressed in Arabidopsis plants and tobacco BY-2 cells under the control of constitutive CaMV35S or inducible HSP18.2 promoters. In Arabidopsis plants, YFP::ATK5 localized to microtubules in dividing cells of root tips, and interphase cells of hypocotyls, roots, and leaves. Notably, in interphase cells, fluorescence of YFP::ATK5 was not uniform along the length of microtubules but was enriched specifically at growing microtubule plus-ends (Figure 1, B–D; see Supplementary Movie 1). Figure 1C shows a time sequence tracking a single microtubule over time. The microtubule is initially growing and then switches to shrinkage phase (denoted by an asterisk). As the microtubule switches from growth to shrinkage, fluorescence accumulation at the plus-end decreases to a level comparable with that seen along the microtubule sidewall. The plus-end enrichment of YFP::ATK5 was dependent on the dynamic state of the microtubule; accumulation of fluorescence at the plus-end is lost upon microtubule catastrophe, and restored upon rescue (see Supplemental Movie 1). This fluorescence pattern is consistent with that of a +TIP (Carvalho et al., 2003). Figure 1D shows typical YFP::ATK5 fluorescence intensity profiles along microtubules in both growth and shrinkage phases.
Figure 1.
YFP::ATK5 localizes to microtubules in vivo and is concentrated at growing plus-ends. (A) Cartoon representation of the YFP::ATK5 construct used for transformation and stable expression in A. thaliana plants and cultured BY-2 cells. (B) YFP::ATK5 localization in Arabidopsis leaf guard cell. YFP::ATK5 localizes along the length of microtubules and is enriched at microtubule plus ends (arrowheads). (C) Slices from a time series showing plus-end enrichment of YFP::ATK5 fluorescence on a microtubule. Plus-end enrichment is lost upon microtubule catastrophe (asterisk). (D) Fluorescence profiles of YFP::ATK5 along a microtubule in growth state (diamonds) and shrinkage state (squares). Bars, 10 μm (B), 2.5 μm (C). Time scale, 35 s (C).
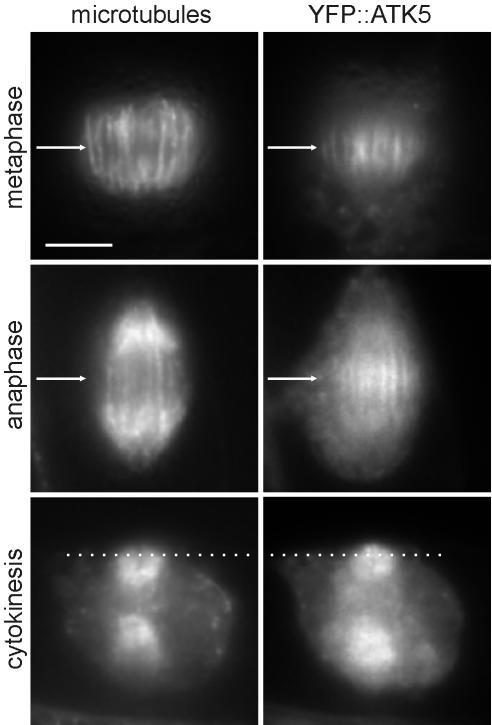
In dividing tobacco BY-2 cells, YFP::ATK5 localized to mitotic spindles and cytokinetic phragmoplasts (Figure 2). Notably, YFP::ATK5 fluorescence accumulated in spindle midzones from early prometaphase (just after nuclear envelope breakdown) to telophase (see Supplementary Movie 2). During metaphase, YFP::ATK5 was enriched on microtubules of the midzone region (Figure 2, top row). As the cells progressed from metaphase to anaphase, fluorescence remained concentrated on the overlapping region of midzone interpolar microtubules, but it was not detected on kinetochore microtubules (Figure 2, middle row). As anaphase progressed, the region of midzone fluorescence narrowed, presumably corresponding to the known reduction in overlap between interdigitating interpolar microtubules as they slide apart during spindle elongation (McDonald et al., 1977; McIntosh et al., 1979; Euteneuer et al., 1982). In centrifugally expanding phragmoplasts (i.e., during cytokinesis), YFP::ATK5 was localized predominately at the leading edge, just in front of the bulk of the microtubule population (Figure 2, bottom row). Phragmoplasts consist of two opposing sets of microtubules oriented with the plus-ends facing inward, where they often overlap in the middle (Hepler and Jackson, 1968; Euteneuer and McIntosh, 1980; Euteneuer et al., 1982). As phragmoplast expansion progresses, microtubule depolymerization in the center is accompanied by polymerization at the leading edges, indicating that the leading edge is rich in growing microtubule plus-ends (Staehelin and Hepler, 1996).
Figure 2.
YFP::ATK5 localizes to mitotic spindles and phragmoplasts in BY-2 cells, with enrichment at spindle midzones and phragmoplast leading edges. During metaphase (top row), YFP::ATK5 localizes preferentially to microtubules in the spindle midzone (arrow). During anaphase (middle row), YFP::ATK5 remains localized on overlapping sectors of interpolar microtubules in the spindle midzone (arrow), whereas the kinetochore microtubules have moved to the poles. During cytokinesis (bottom row), YFP::ATK5 localizes to the phragmoplast advancing edge (dotted line), which contains numerous polymerizing microtubule plus-ends. Bar, 10 μm.
These data show that ATK5 behaves as a +TIP and that it accumulates at spindle midzones and phragmoplast leading edges. The observed plus-end accumulation is inconsistent with the predicted minus-end motor activity of ATK5; therefore, we sought biochemical confirmation of minus-end–directed motor activity.
ATK5 Is a Minus-End–directed Motor
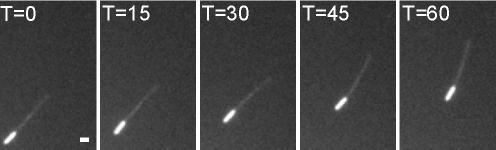
To exclude the possibility that ATK5 uses plus-end–directed motor activity to reach growing microtubule plus-ends, full-length ATK5 cDNA was cloned into a GST expression vector for bacterial expression and in vitro microtubule gliding assays. The GST control did not support microtubule motility (our unpublished data). Figure 3 shows that in the presence of ATP, GST::ATK5 supports the minus-end directed motility of microtubules. Microtubules land and glide along the ATK5-coated surface with their plus-ends leading (minus-ends are brighter), showing that ATK5 is a minus-end–directed motor. This finding is consistent with the properties of all other Kinesin-14 family members studied to date (Dagenbach and Endow, 2004). By measuring the velocity of microtubule translocation, the speed of ATK5 was determined to be 6.30 ± 1.36 μm min–1 at 20°C (n = 6), which is similar to ATK1 and other Kinesin-14 family members (McDonald et al., 1990; Endow et al., 1994b; Marcus et al., 2002). Additionally, bacterially expressed GST::YFP::ATK5 also supported microtubule motility; which confirms the viability of the fusion protein. These data show that ATK5 is a minus-end–directed motor, thereby excluding the possibility that ATK5 motors toward microtubule plus-ends. This suggests that the plus-end tracking of ATK5 activity is independent of its motor activity.
Figure 3.
ATK5 is a minus-end–directed motor. Minus-end–labeled taxol-stabilized microtubules move with their plus-ends leading over a surface coated with cleared bacterial lysates from Escherichia coli expressing recombinant GST::ATK5, indicating the bound kinesin exhibits minus-end–directed motility. Bar, 1 μm.
Plus-End Tracking of ATK5 Is Independent of the Motor Domain
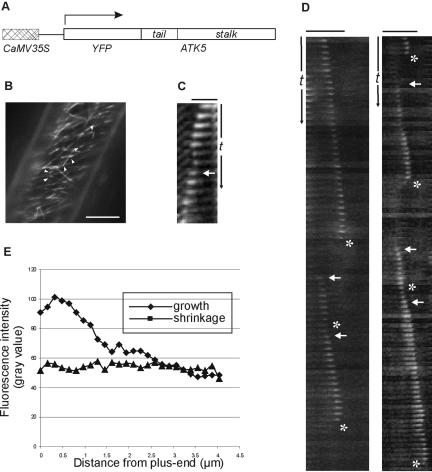
To determine whether the plus-end tracking behavior was conferred by regions outside the motor domain, a truncation was made consisting of only the tail and coiled-coil stalk domains (YFP::TS; amino acids 1–423; Figure 4A). This construct was stably expressed in Arabidopsis plants and BY-2 cells under control of the CaMV35S promoter. In Arabidopsis plants, YFP::TS localized to microtubule plus-ends in a manner similar to the full-length YFP::ATK5 (Figure 4, B–D; see Supplementary Movie 3). The time-series plots depicted in Figure 3, C and D, track single, dynamic microtubule plus-ends exhibiting loss of plus-end fluorescence enrichment concurrent with catastrophe, and restoration upon rescue. Figure 3E shows the typical YFP::TS fluorescence intensity profiles along microtubules in growth or shrinkage phases. These data indicate that the tail-stalk region of ATK5 is responsible for the accumulation at growing microtubule plus-ends. To exclude the possibility of cryptic plus-end–directed motor activity in the Tail/Stalk, YFP::TS also was expressed as a GST fusion (GST::YFP::TS). GST::YFP::TS did not support gliding of microtubules in vitro (our unpublished data).
Figure 4.
YFP::TS localizes to microtubules in vivo and is concentrated at growing plus-ends. (A) Cartoon representation of YFP::TS construct used for transformation and stable expression in Arabidopsis plants and cultured BY-2 cells. (B) YFP::TS localization in an Arabidopsis hypocotyl epidermal cell. YFP::TS localizes to microtubules and is enriched at microtubule plus-ends (arrowheads). (C) Slices from a time series showing plus-end enrichment of YFP::TS fluorescence on a microtubule. Plus-end accumulation decreases upon microtubule catastrophe (asterisk) and is restored with rescue (arrow). (D) Slices from a time series showing localization of YFP::TS to dynamic microtubules. Arrows indicate rescue, asterisks indicate catastrophe. Bar, 10 μm (B), 2.5 μm (C), and 5 μm (D). Time scale (t), 25 s (C), 75 s (D).
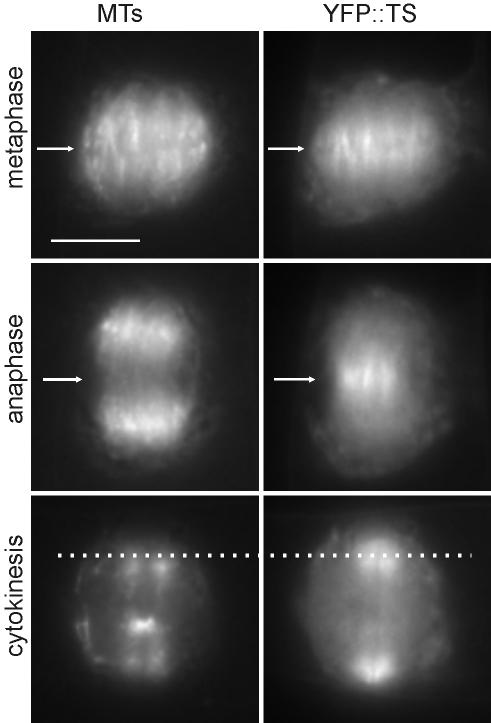
When stably expressed in BY-2 cells, YFP::TS localized to spindles and phragmoplasts in a manner similar to the full-length YFP::ATK5 (Figure 5; see Supplementary Movie 4), thus excluding the possibility that midzone targeting of ATK5 is achieved by cross-linking microtubules via motor and nonmotor microtubule binding sites. These data indicate that the Tail/Stalk region is responsible for targeting ATK5 to spindle midzones and phragmoplasts and suggest that ATK5 exerts force in these regions.
Figure 5.
YFP::TS localizes to mitotic spindles and phragmoplasts in BY-2 cells, with enrichment at spindle midzones and phragmoplast leading edges. All localization patterns are similar to that of full-length YFP::ATK5. During metaphase (top row), YFP::TS localizes preferentially to microtubules in the spindle midzone (arrow). During anaphase (middle row), YFP::TS remains localized on overlapping sectors of interpolar microtubules in the spindle midzone (arrow), whereas the kinetochore microtubules have moved to the poles. During cytokinesis (bottom row), YFP::TS localizes to the phragmoplast's advancing edge (dotted line). Bar, 10 μm.
ATK5 Functions in Spindle Morphogenesis
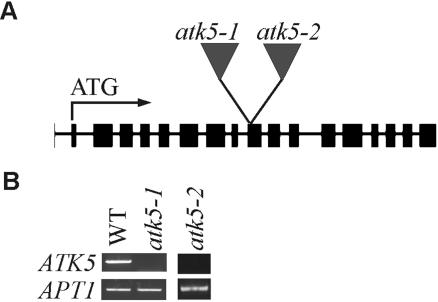
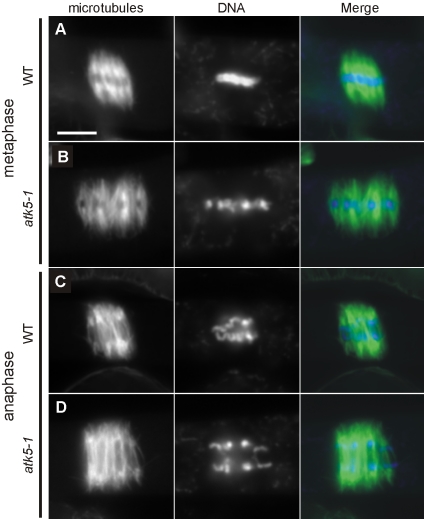
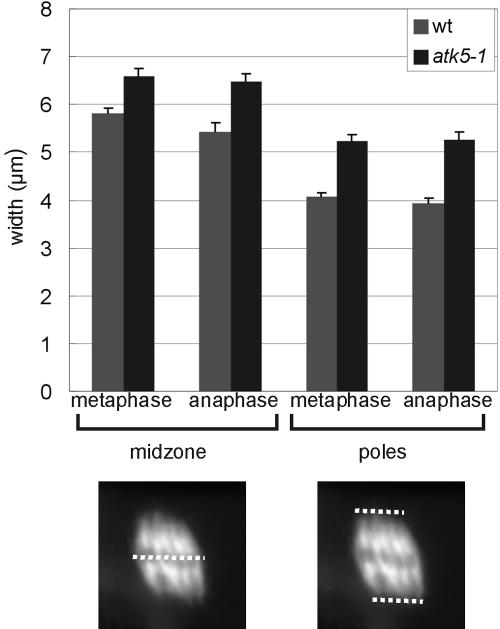
To explore the role of ATK5 in mitotic spindles, mutant atk5 plants containing T-DNA insertions in coding regions (exon 9; Figure 6A) of ATK5 were analyzed for defects in mitosis. RT-PCR showed loss of transcript in mutant plants (Figure 6B). Two alleles, designated atk5-1 and atk5-2, containing independent insertions in exon 9 of ATK5 were studied. Plants, homozygous for either allele, seem normal and exhibit developmental and reproductive characteristics indistinguishable from wild-type. All data presented herein were obtained from atk5-1, but comparable results were seen for atk5-2 (our unpublished data). Using anti-tubulin antibodies, immunofluorescence was performed in wild-type and atk5-1 root mitotic cells (Figure 7). Although interphase cells contained normal microtubule arrays, dividing cells in the root tips of atk5 mutant plants exhibited abnormally broadened mitotic spindles in metaphase (compare rows A and B) and anaphase (compare rows C and D). Spindle microtubules as well as chromosomes were laterally broadened throughout mitosis in atk5 plants, resulting in a greater distance between chromosomes at the metaphase plate; however, this did not affect the ability of the chromosomes to align during metaphase and segregate normally during anaphase. During metaphase, mean spindle width was significantly larger in atk5-1 compared with wild type (Figure 8), as measured at the spindle midzone (6.58 ± 0.12 μm for atk5-1, n = 45 cells vs. 5.80 ± 0.18 μm for wild type, n = 45 cells; p < 0.0002, Student's t test), and at spindle poles (5.21 ± 1.58 μm for atk5-1, n = 90 cells vs. 4.08 ± 1.12 μm for wild type, n = 75 cells, p < 7.23 e–13, Student's t test). During anaphase, mean spindle width was also significantly greater compared with wild-type as measured both at the spindle midzone (6.48 ± 0.17 μm for atk5-1, n = 47 cells vs. 5.42 ± 0.19 μm for wild type, n = 26 cells; p < 0.0002, Student's t test), and at the poles (5.33 ± 1.21 μm for atk5–1, n = 45 cells; vs. 3.70 ± 1.32 μm for wild-type, n = 30 cells; p < 3.89 e–13, Student's t test). Because ATK5 is localized predominately to the spindle midzone, the spindle pole broadening is interpreted as largely a result of the loss of forces generated in the spindle midzone, which links together rigid microtubules emanating from the poles.
Figure 6.
atk5-1 and atk5-2 are insertional null alleles of ATK5. (A) Diagram showing locations of each T-DNA insertion. Exons are indicated by boxes, introns by lines. (B) RT-PCR showing loss of ATK5 transcript in atk5-1 and atk5-2 null mutants, relative to control (APT1).
Figure 7.
Plants lacking ATK5 contain abnormally broadened mitotic spindles. (A) Metaphase spindle from a wild-type plant. (B) Metaphase spindle from an atk5-1 mutant. (C) Anaphase spindle from a wild-type plant. (D) Anaphase spindle from an atk5-1 mutant. Bar, 5 μm. Microtubules, green; chromatin, blue.
Figure 8.
ATK5 mutant spindles are wider at both the midzone and the poles. (A) Mean spindle widths during metaphase and anaphase in wild-type and atk5-1 mutant plants. Dotted lines indicate measured parameters. Error bars indicate SE.
DISCUSSION
ATK5 Is a +TIP That Functions in Spindle Morphogenesis
With the advent of GFP technology and continuing advances in the ability to observe dynamic subcellular events, a large number of proteins have been found to be associated with microtubule plus-ends (Carvalho et al., 2003; Galjart and Perez, 2003; Mimori-Kiyosue and Tsukita, 2003; Vaughan, 2004). The +TIPs include members from a diverse array of microtubule-associated protein classes and have so far been shown to function in the regulation of microtubule dynamics (Brunner and Nurse, 2000), delivery of proteins to specific sites (Browning et al., 2003), and attachment to other structures in the cell, such as kinetochores and the cell cortex (Tirnauer et al., 2002; Mimori-Kiyosue and Tsukita, 2003). Here, we show that the Kinesin-14 family member ATK5 is a +TIP that functions in spindle morphogenesis. The C-terminal motor domain of ATK5 confers motility toward microtubule minus-ends, whereas the N-terminal Tail/Stalk domain targets it to growing plus-ends. Similarly, the Tail/Stalk domain is responsible and sufficient for localization of ATK5 to mitotic spindle midzones and to phragmoplast leading edges. Null mutations in ATK5 result in abnormally broadened spindles throughout mitosis, consistent with its hypothesized role as a mitotic motor.
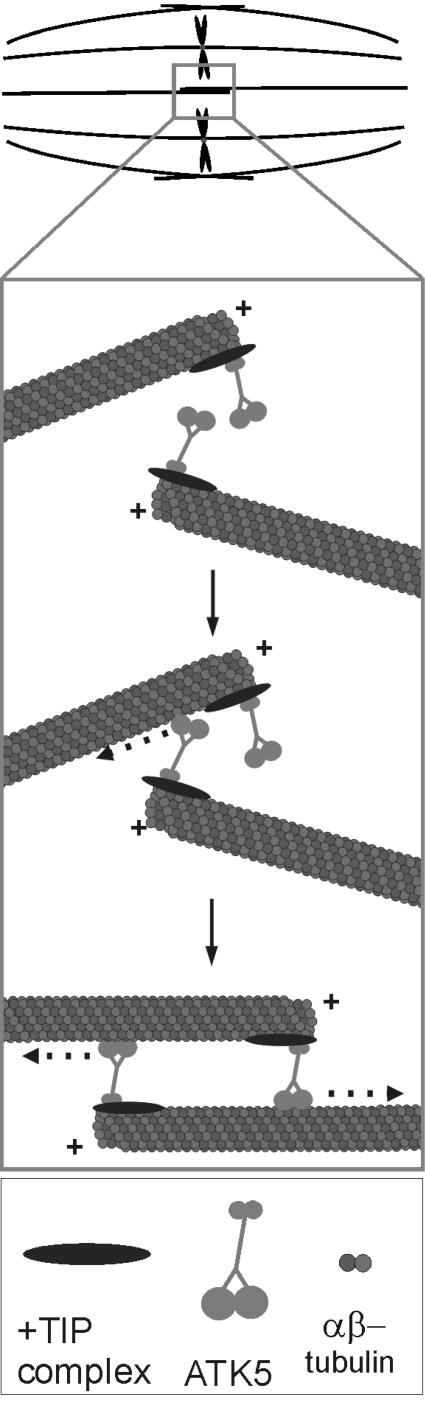
Based on these data, we present the following hypothetical model for ATK5 function (Figure 9): ATK5 binds to microtubule plus-ends via the Tail/Stalk domain, either 1) directly, through nonmotor microtubule binding sites, and/or 2) indirectly, via cooperation in a +TIP complex. This preferential association with growing plus-ends favors the accumulation of ATK5 at plus-end-rich sites of action, such as spindle midzones and phragmoplast leading edges. At these locations, ATK5 functions in at least two ways: 1) to cross-link microtubules, thereby decreasing the lateral distance between neighboring microtubules; and 2) to align obliquely oriented microtubules via minus-end motor activity. This model is consistent with the observed spindle broadening phenotype because loss of cross-linking activity would predictably result in fewer lateral microtubule-microtubule interactions, and therefore greater intermicrotubule distances. Similarly, interpolar microtubules growing from the poles toward the midzone may frequently encounter antiparallel interpolar microtubules at an angle, and ATK5 could serve to lessen this angle of encounter by zippering these microtubules together via motor-dependent microtubule sliding. This model implicates the spindle midzone as the primary site of action; however, atk5 mutants also exhibited broadened spindle poles, suggesting that ATK5 contributes to focusing of spindle poles. This may occur either 1) indirectly, by ATK5 gathering microtubules at the midzone, which leads to greater focusing of the poles due to the rigidity of the connecting microtubules; or 2) directly, by additional ATK5 molecules present near the poles, acting to gather minus-ends together, as in the case of several other Kinesin-14 family members (Hatsumi and Endow, 1992; Endow et al., 1994a; Walczak et al., 1998; Mountain et al., 1999; Goshima and Vale, 2003).
Figure 9.
Hypothetical model for ATK5 function. The midzone region of the mitotic spindle (top) is projected into the underlying box. ATK5 is targeted to spindle midzones via plus-end tracking (either in a +TIP complex and/or by direct microtubule binding). At these sites of action, ATK5 functions by decreasing the lateral distance between neighboring microtubules via cross-linking and by promoting alignment of antiparallel microtubules via motor activity. ATK5 is shown here in association with antiparallel interpolar microtubules, although these functions may apply also to parallel microtubule interactions.
Consistent with previous models of Kinesin-14 function, the above-mentioned model also predicts that ATK5 cross-links interdigitating antiparallel microtubules in the midzone and generates inward forces by sliding them past one another via minus-end motor activity. A prediction of this is that loss of an inward force-producing motor in the spindle midzone will result in longer spindles (increase in length along long axis of the spindle); however, no change in spindle length was observed in atk5 null mutants. In ncd null Drosophila embryos, prophase/prometaphase spindle pole separation occurs more rapidly and to a greater extent than in wild type (Sharp et al., 1999a), and in budding yeast loss of Klp2p results in increased metaphase spindle length (Troxell et al., 2001). Perhaps in Arabidopsis, which lacks centrosomes and astral microtubules, several redundant factors are involved in mediating spindle length.
Because this study used fixed cells to investigate atk5 defects, we may not have detected more subtle abnormalities in the kinetics of spindle formation and mitotic progress, as seen with ATK1 and other Kinesin-14 family members (Matthies et al., 1996; Prigozhina et al., 2001; Marcus et al., 2003). In support of a role in spindle formation, ATK5 localizes to midzone microtubules early in prometaphase, before the spindle is organized into two halves with a well defined midzone (see Supplementary Movies 2 and 4). This localization to prometaphase midzone microtubules poises ATK5 in the ideal location to organize microtubules into bipolar spindles. It is possible that ATK5 also functions in spindle assembly and that live-cell imaging of microtubules in atk5 mutants is required to detect these defects.
Mechanisms of ATK5 Targeting to Midzones
Based on the fact that no other known +TIPs have been demonstrated to accumulate at mitotic spindle midzones, it is unlikely that +TIP activity alone is responsible for ATK5 midzone enrichment. One possibility is that ATK5 binds to another protein or complex of proteins in the midzone, and the +TIP activity of ATK5 serves as a mechanism to limit the diffusion of ATK5 away from this static complex. Indeed, several proteins have been shown to localize to spindle midzones, and the presence of a proteinaceous “spindle matrix” is a long-standing idea in the field (Wells, 2001). Although we were unable to detect individual plus-ends within spindles expressing the fusion proteins due to a low signal-to-noise ratio (all stable cell lines we recovered only weakly expressed this construct, presumably due to lethality at higher expression levels), evidence that +TIP activity is involved in ATK5 midzone localization comes from the observation that during anaphase, accumulation of ATK5 fluorescence was not detectable on kinetochore microtubules, which in mammalian cells have been shown to depolymerize as chromosomes move poleward (Gorbsky and Borisy, 1989), but it was retained on overlapping regions of interpolar microtubules, which are in a state of polymerization to facilitate spindle elongation (Schuyler et al., 2003). Therefore, although not directly demonstrating the necessity of plus-end tracking for midzone localization, this does show that differences in the dynamic state of microtubule subpopulations correlate with the localization of ATK5.
Because the Tail/Stalk domain localizes to midzones in the absence of the motor domain, we have ruled out the possibility that this targeting is achieved by specifically recognizing and cross-linking antiparallel microtubules via motor and Tail/Stalk domain binding sites. Nevertheless we cannot rule out the possibility that ATK5 Tail/Stalk is by itself able to bind stereospecifically to overlapping antiparallel microtubules, thereby directly facilitating midzone enrichment. Both YFP fusion proteins in this study also exhibit weak labeling along the length of the microtubule during interphase, but the significance of this during mitosis is not clear.
Minus-End Motors and Plus-End Tracking
An interesting correlation to our finding that ATK5 is a +TIP is that in animals and fungi, another minus-end–directed microtubule motor, cytoplasmic dynein, also localizes to microtubule plus-ends (albeit independently of the dynamic state of the microtubule), and this serves to target dynein to the cell cortex (Carvalho et al., 2003). To date, convincing evidence for the presence of dyneins in higher plants is lacking. An intriguing possibility is that the abundant Kinesin-14 family members in Arabidopsis may take on similar roles as dynein. Indeed, both groups of motors have been detected at spindle poles (Pfarr et al., 1990; Smirnova et al., 1998), and loss of dynein in Xenopus egg extracts leads to splayed spindle poles; a defect that is enhanced in the absence of the Kinesin-14, XCTK2, suggesting functional overlap between the two groups of motors in maintaining spindle poles (Walczak et al., 1998).
Could plus-end tracking of minus-end motors be a general phenomenon used in spindle assembly and morphogenesis? The majority of Kinesin-14 family members studied to date do not exhibit enrichment at spindle midzones, but rather, have a greater accumulation at the spindle poles, consistent with their minus-end motor activity. However, the Kinesin-14 family members NCD, DSK1, Klp2p, and ATK1 are enriched at mitotic spindle midzones (Endow and Komma, 1996; Liu et al., 1996; Wein et al., 1998; Troxell et al., 2001), although plus-end tracking has not been yet demonstrated in these cases. Based on motor domain homology, the two nonplant kinesins that are most closely related to ATK5 are Xenopus XCTK2 and Schizosaccharamyces pombe Klp2p, which exhibit 49.6 and 48.8% amino acid identity, respectively, to ATK5. Although neither has been so far demonstrated to track microtubule plus-ends, a fusion between Klp2p and GFP was shown to localize to mobile cytoplasmic dots during interphase and to kinetochores during mitosis, properties consistent with that of certain +TIPs (Troxell et al., 2001). Additionally, NCD, which shares 42.2% amino acid sequence identity to ATK5 inside the motor domain was identified as an interacting partner with the +TIP EB1 in Drosophila S2 cells (Rogers et al., 2004). A fusion between Kar3p and GFP has been shown to localize to plus-ends of astral microtubules in the budding yeast schmoo tip, although in this case the enrichment was greater during microtubule depolymerization (Maddox et al., 2003). Further advances in microscopic imaging and GFP technology may reveal additional Kinesin-14 family members with +TIP activity.
What are the implications of minus-end–directed motors at microtubule plus-ends? Computer modeling of microtubule–motor interactions predicts that minus-end motor activity combined with a plus-end bias (e.g., achieved by binding plus-end kinesins or a plus-end complex) favors the establishment of antiparallel microtubule orientation (Nedelec, 2002). Perhaps the minus-end–directed kinesins involved in mediating antiparallel microtubule interactions employ plus-end tracking to remain in the vicinity of regions dense in antiparallel microtubules. It is interesting to note that mitotic spindles and phragmoplasts are similar in that they both consist of two opposing arrays of microtubules with antiparallel plus-ends overlapping in the middle. Future studies are needed to elucidate the precise mechanisms of ATK5 plus-end tracking and functions during mitosis and cytokinesis.
Supplementary Material
Acknowledgments
We thank D. Fisher for critical reading of the manuscript, Anthony Omeis for plant care, and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. This work was funded by grants from the United States Department of Agriculture and Department of Energy. J.C.A. was supported by a National Science Foundation training grant.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0935) on January 19, 2005.
Abbreviations used: ATK5, Arabidopsis thaliana kinesin 5; ATK1, Arabidopsis thaliana kinesin 1; YFP, yellow fluorescent protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Browning, H., Hackney, D. D., and Nurse, P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812–818. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., Tirnauer, J. S., and Pellman, D. (2003). Surfing on microtubule ends. Trends Cell Biol. 13, 229–237. [DOI] [PubMed] [Google Scholar]
- Chen, C., Marcus, A., Li, W., Hu, Y., Calzada, J. P., Grossniklaus, U., Cyr, R. J., and Ma, H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409. [DOI] [PubMed] [Google Scholar]
- Dagenbach, E. M., and Endow, S. A. (2004). A new kinesin tree. J. Cell Sci. 117, 3–7. [DOI] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R. (2003). Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 36, 280–290. [DOI] [PubMed] [Google Scholar]
- Endow, S. A., Chandra, R., Komma, D. J., Yamamoto, A. H., and Salmon, E. D. (1994a). Mutants of the Drosophila NCD microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107, 859–867. [DOI] [PubMed] [Google Scholar]
- Endow, S. A., Kang, S. J., Satterwhite, L. L., Rose, M. D., Skeen, V. P., and Salmon, E. D. (1994b). Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 13, 2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., and Komma, D. J. (1996). Centrosome and spindle function of the Drosophila NCD microtubule motor visualized in live embryos using NCD-GFP fusion proteins. J. Cell Sci. 109, 2429–2442. [DOI] [PubMed] [Google Scholar]
- Euteneuer, U., Jackson, W. T., and McIntosh, J. R. (1982). Polarity of spindle microtubules in Haemanthus endosperm. J. Cell Biol. 94, 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer, U., and McIntosh, J. R. (1980). Polarity of midbody and phragmoplast microtubules. J. Cell Biol. 87, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart, N., and Perez, F. (2003). A plus-end raft to control microtubule dynamics and function. Curr. Opin. Cell Biol. 15, 48–53. [DOI] [PubMed] [Google Scholar]
- Gorbsky, G. J., and Borisy, G. G. (1989). Microtubules of the kinetochore fiber turn over in metaphase but not in anaphase. J. Cell Biol. 109, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Vale, R. D. (2003). The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, C. L., and Cyr, R. J. (2001) Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma 216, 201–214. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58, 139–151. [DOI] [PubMed] [Google Scholar]
- Hatsumi, M., and Endow, S. A. (1992). Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 101, 547–559. [DOI] [PubMed] [Google Scholar]
- Hepler, P. K., and Jackson, W. T. (1968). Microtubules and early stages of cell-plate formation in the endosperm of Haemanthus katherinae Baker. J. Cell Biol. 38, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M. A., He, L., Totis, L., and Saunders, W. S. (1993). Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 135, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabay, A., and Walker, R. A. (1999). Identification of microtubule binding sites in the NCD tail domain. Biochemistry 38, 1838–1849. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., La Terra, S., and Chang, F. (2004). Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 14, 1330–1340. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., Kofron, M., Essner, R., Kato, T., Dragas-Granoic, S., Omoto, C. K., and Khodjakov, A. (1995). Characterization of a minus end-directed kinesin-like motor protein from cultured mammalian cells. J. Cell Biol. 129, 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, R. J., and Pickett-Heaps, J. D. (1983). Ultraviolet microbeam irradiations of mitotic diatoms: investigation of spindle elongation. J. Cell Biol. 96, 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Cyr, R. J., and Palevitz, B. A. (1996). A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell 8, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P. S., Stemple, J. K., Satterwhite, L., Salmon, E. D., and Bloom, K. (2003). The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr. Biol. 13, 1423–1428. [DOI] [PubMed] [Google Scholar]
- Marcus, A. I., Ambrose, J. C., Blickley, L., Hancock, W. O., and Cyr, R. J. (2002). Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil. Cytoskeleton 52, 144–150. [DOI] [PubMed] [Google Scholar]
- Marcus, A. I., Li, W., Ma, H., and Cyr, R. J. (2003). A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 14, 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, H. J., McDonald, H. B., Goldstein, L. S., and Theurkauf, W. E. (1996). Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene, J., Essner, R., Ryu, J., Hamaguchi, Y., Baas, P. W., Haraguchi, T., Hiraoka, Y., and Kuriyama, R. (1999). Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J. Cell Sci. 112, 4041–4050. [DOI] [PubMed] [Google Scholar]
- McDonald, H. B., Stewart, R. J., and Goldstein, L. S. (1990). The kinesin-like NCD protein of Drosophila is a minus end-directed microtubule motor. Cell 63, 1159–1165. [DOI] [PubMed] [Google Scholar]
- McDonald, K., Pickett-Heaps, J. D., McIntosh, J. R., and Tippit, D. H. (1977). On the mechanism of anaphase spindle elongation in Diatoma vulgare. J. Cell Biol. 74, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, J. R., McDonald, K. L., Edwards, M. K., and Ross, B. M. (1979). Three-dimensional structure of the central mitotic spindle of Diatoma vulgare. J. Cell Biol. 83, 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., and Tsukita, S. (2003). “Search-and-capture”of microtubules through plus-end-binding proteins (+TIPs). J. Biochem. 134, 321–326. [DOI] [PubMed] [Google Scholar]
- Moffatt, B. A., McWhinnie, E. A., Agarwal, S. K., and Schaff, D. A. (1994). The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143, 211–216. [DOI] [PubMed] [Google Scholar]
- Mountain, V., Simerly, C., Howard, L., Ando, A., Schatten, G., and Compton, D. A. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhulu, S. B., and Reddy, A. S. (1998). Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell 10, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec, F. (2002). Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, M. J., Meluh, P. B., Rose, M. D., and Morris, N. R. (1993). Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J. Cell Biol. 120, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr, C. M., Coue, M., Grissom, P. M., Hays, T. S., Porter, M. E., and McIntosh, J. R. (1990). Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 345, 263–265. [DOI] [PubMed] [Google Scholar]
- Pidoux, A. L., LeDizet, M., and Cande, W. Z. (1996). Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol. Biol. Cell 7, 1639–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina, N. L., Walker, R. A., Oakley, C. E., and Oakley, B. R. (2001). Gamma-tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 12, 3161–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A. S., and Day, I. S. (2001). Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S. L., Wiedemann, U., Hacker, U., Turck, C., and Vale, R. D. (2004). Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14, 1827–1833. [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., Liu, J. Y., and Pellman, D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., McDonald, K. L., Brown, H. M., Matthies, H. J., Walczak, C., Vale, R. D., Mitchison, T. J., and Scholey, J. M. (1999a). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Yu, K. R., Sisson, J. C., Sullivan, W., and Scholey, J. M. (1999b). Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1, 51–54. [DOI] [PubMed] [Google Scholar]
- Smirnova, E. A., and Bajer, A. S. (1998). Early stages of spindle formation and independence of chromosome and microtubule cycles in Haemanthus endosperm. Cell Motil. Cytoskeleton 40, 22–37. [DOI] [PubMed] [Google Scholar]
- Smirnova, E. A., Reddy, A. S., Bowser, J., and Bajer, A. S. (1998). Minus end-directed kinesin-like motor protein, Kcbp, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil. Cytoskeleton 41, 271–280. [DOI] [PubMed] [Google Scholar]
- Staehelin, L. A., and Hepler, P. K. (1996). Cytokinesis in higher plants. Cell 84, 821–824. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J. S., Canman, J. C., Salmon, E. D., and Mitchison, T. J. (2002). EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell 13, 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell, C. L., Sweezy, M. A., West, R. R., Reed, K. D., Carson, B. D., Pidoux, A. L., Cande, W. Z., and McIntosh, J. R. (2001). pkl1(+)and klp2(+): two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol. Biol. Cell 12, 3476–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, K. T. (2004). Surfing, regulating and capturing: are all microtubule-tip-tracking proteins created equal? Trends Cell Biol. 14, 491–496. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Verma, S., and Mitchison, T. J. (1997). XCTK 2, a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 136, 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C. E., Vernos, I., Mitchison, T. J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903–913. [DOI] [PubMed] [Google Scholar]
- Walker, R. A., Salmon, E. D., and Endow, S. A. (1990). The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature 347, 780–782. [DOI] [PubMed] [Google Scholar]
- Wein, H., Bass, H. W., and Cande, W. Z. (1998). DSK1, a kinesin-related protein involved in anaphase spindle elongation, is a component of a mitotic spindle matrix. Cell Motil. Cytoskeleton 41, 214–224. [DOI] [PubMed] [Google Scholar]
- Wells, W. A. (2001). Searching for a spindle matrix. J. Cell Biol. 154, 1102–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.