Abstract
Proteins are selectively packaged into vesicles at specific sites and then delivered correctly to the various organelles where they function, which is critical to the proper physiology of each organelle. The precursor form of the vacuolar hydrolase aminopeptidase I is a selective cargo molecule of the cytoplasm to vacuole targeting (Cvt) pathway and autophagy. Precursor Ape1 along with its receptor Atg19 forms the Cvt complex, which is transported to the pre-autophagosomal structure (PAS), the putative site of Cvt vesicle formation, in a process dependent on Atg11. Here, we show that this interaction occurs through the Atg11 C terminus; subsequent recruitment of the Cvt complex to the PAS depends on central regions within Atg11. Atg11 was shown to physically link several proteins, although the timing of these interactions and their importance are unknown. Our mapping shows that the Atg11 coiled-coil domains are involved in self-assembly and the interaction with other proteins, including two previously unidentified partners, Atg17 and Atg20. Atg11 mutants defective in the transport of the Cvt complex to the PAS affect the localization of other Atg components, supporting the idea that the cargo facilitates the organization of the PAS in selective autophagy. These findings suggest that Atg11 plays an integral role in connecting cargo molecules with components of the vesicle-forming machinery.
INTRODUCTION
In eukaryotic cells, intracellular proteins are transported to their proper organellar destinations via specific pathways, and vesicular carriers typically mediate this transit. To ensure proper organelle function each protein must be delivered to the correct location. Accordingly, proteins are first selectively incorporated into appropriate vesicles as cargoes, and then these vesicles transport to, and fuse with, an acceptor organelle. Two resident vacuolar hydrolases, aminopeptidase I (Ape1) and α-mannosidase (Ams1), are known to be transported directly from the cytosol to the vacuole in the yeast Saccharomyces cerevisiae, via the cytoplasm to vacuole targeting (Cvt) pathway (Klionsky et al., 1992; Hutchins and Klionsky, 2001). In the Cvt pathway, these two cargo molecules are packaged inside a double-membrane–bound Cvt vesicle. The outer membrane of the vesicle fuses with the vacuole membrane, and the vesicle inner membrane is then broken down to release the hydrolases into the vacuole lumen (Baba et al., 1997; Scott et al., 1997). The process of the Cvt pathway is mechanically and topologically similar to that of autophagy, even though the functions of the two pathways are physiologically opposite; the Cvt pathway is a constitutive biosynthetic process, whereas autophagy is used for bulk degradation of cytoplasmic proteins and is induced under specific environmental conditions such as nutrient starvation (Klionsky and Ohsumi, 1999). Most of the components needed for the Cvt pathway and autophagy function in both pathways and have been termed autophagy-related (Atg) proteins (Klionsky et al., 2003).
Although >20 Atg proteins have been identified, little is known about their specific functions. One of the best-characterized Atg proteins is Atg19, which acts as a receptor for vacuolar hydrolase aminopeptidase I (prApe1) in both the Cvt and autophagy pathways (Scott et al., 2001). Recently, the mechanism for the selective sorting of cargo molecules by Atg19 in the Cvt pathway has been elucidated (Shintani et al., 2002; Suzuki et al., 2002). Precursor Ape1 is assembled into a homo-dodecamer and then a larger oligomeric Ape1 complex in the cytosol in a process that depends on its propeptide (Oda et al., 1996; Kim et al., 1997). Atg19 binds to the Ape1 complex through interaction with the prApe1 propeptide to form the Cvt complex. The prApe1–Atg19 complex is transported to the perivacuolar site termed the pre-autophagosomal structure (PAS) where most Atg components localize, and the Cvt and autophagic vesicles may be formed (Suzuki et al., 2001; Kim et al., 2002). Finally, the encapsulated Cvt complexes are targeted to the vacuole. Microscopy analyses of fluorescent protein-tagged prApe1 and Atg19 reveal that the two proteins colocalize at the PAS, whereas prApe1 is located away from the vacuole in the absence of Atg19, and Atg19 is dispersed in the cytosol in the absence of prApe1 or when the prApe1 propeptide is mutated (Shintani et al., 2002). Moreover, with regard to the relationship between cargo recruitment and vesicle formation, a unique model was recently proposed that cargo assembly (i.e., formation of the Cvt complex) facilitates the organization of the PAS and the formation of Cvt vesicles (Shintani and Klionsky, 2004). Atg proteins such as Atg1, Atg2, Atg8, and Atg20 localize at the PAS in wild-type cells, whereas the localization of these proteins at the PAS was much less efficient when the Cvt complex was depleted by deletion of ATG19 or APE1.
It has been reported that another protein, Atg11, is involved in the delivery of the Cvt complex to the PAS (Shintani et al., 2002). Atg11 was originally identified as a component specifically required for the Cvt pathway and pexophagy, the latter defining the pathway for the selective degradation of peroxisomes; Atg11 is not absolutely required for nonselective bulk autophagy (Kim et al., 2001b). Like most Atg components, Atg11 also is localized at the PAS. Atg11 interacts with Atg1 (Figure 1A), a kinase essential for both the Cvt and autophagy pathways (Kim et al., 2001b). When ATG11 is deleted, the Cvt complex is not localized at the PAS, although Atg19 and prApe1 can still form a complex. Moreover, ATG11 deletion causes a defect in the localization of other Atg components to the PAS similar to the effects of eliminating the Cvt complex (Shintani et al., 2002). Because Atg11 can interact with the C terminus of Atg19 (Figure 1A), these results suggest that Atg11 has some critical role in recruiting the Cvt complex to the PAS. The function of Atg11, however, and the mechanism by which it recruits the Cvt complex to the PAS is still unclear.
Figure 1.
Atg11 interacts with multiple Atg proteins. (A) Schematic drawing of interactions involving Atg11 and its interacting proteins (except for the Ape1 complex, drawn in approximately relative scales). The proteins shown in yellow are specific to import of prApe1, whereas those shown in green are needed primarily for specific types of autophagy such as the Cvt pathway and/or pexophagy. The protein depicted in purple is required only for nonspecific autophagy, whereas those shown in blue are needed for both specific and nonspecific types of autophagic sequestration. 8, Atg8; PAS, pre-autophagosomal structure. (B) Atg11 is predicted to have four coiled-coil domains. Double-headed arrows show the interactions of Atg11 with Atg components based on previous and present studies. See the text for details and references.
In the present study, we have mapped functional domains in Atg11 (Figure 1B). We found that separate functional regions exist within Atg11 and have identified a role for the central domains in targeting the Cvt complex to the PAS, and the C terminus in interacting with Atg19. Furthermore, a yeast two-hybrid analysis has shown that Atg11 can interact with Atg17 and Atg20, previously unidentified binding partners, and particular regions of Atg11 are involved in forming complexes with multiple proteins in addition to Atg17 and Atg20, including Atg1, Atg11 itself, and Atg19 (Figure 1B). Finally, analyses of Atg11 mutants suggest that Atg11 forms a homodimer or homo-oligomer complex whose localization or assembly/disassembly at the PAS is influenced by the Atg1–Atg13 complex.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae strains used in this study are listed in Table 1. Yeast strains were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic medium (SD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as required).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3,112 his3-200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | Robinson et al., 1988 |
| PJ69-4A | MATα leu2-3,112 trp1-Δ901 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| AHY001 | SEY6210 atg11Δ::HIS3 | Kim et al., 2001b |
| CWY233 | SEY6210 atg13Δ::KAN | This study |
| D3Y109 | SEY6210 atg20Δ::HIS5 | Nice et al., 2002 |
| SSY31 | SEY6210 atg19Δ::HIS5 | Scott et al., 2001 |
| YTS107 | SEY6210 ape1Δ::LEU2 | Shintani et al., 2002 |
| YTS108 | SEY6210 ape1Δ::LEU2 atg19Δ::HIS5 | This study |
| YTS157 | SEY6210 atg1Δ::HIS5 atg11Δ::LEU2 | This study |
| YTS192 | SEY6210 atg11Δ::HIS3 GFP-ATG8::URA3 | Shintani and Klionsky, 2004 |
| TYY014 | SEY6210 ATG20-YFP::HIS5 vps38Δ::LEU2 atg11Δ::HIS3 | Shintani and Klionsky, 2004 |
| YTS121 | PJ69-4A atg11Δ::KAN | This study |
| WHY1 | SEY6210 atg1Δ::HIS5 | Shintani et al., 2002 |
Plasmids
Plasmids expressing protein A fused to Atg19 (pCuProtA-CVT19) and Atg20 (pProtA-Atg20), hemagglutinin (HA)-tagged Atg11 (pCuHA-CVT9), and myc-tagged Atg11 (pmyc-Atg11), and two-hybrid plasmids expressing Atg1 lacking a kinase domain (pGBD-Atg1Δk), Atg19 (pGAD-CVT19), Atg20 (pGBDU-CVT20), and Atg17 (pGBDU-ATG17) have been described previously (Kim et al., 2001b; Nice et al., 2002; Shintani et al., 2002; Abeliovich et al., 2003). The kinase-defective Atg1K54A mutation was described previously (Kamada et al., 2000; Abeliovich et al., 2003). For the deletion of Atg11 domains, the truncated open reading frames were amplified by polymerase chain reaction and ligated into the BamHI and SalI sites of pGBDU-C1 or pGAD-C1 for yeast two-hybrid analysis. For plasmids expressing HA-Atg11 truncations, fragments of two-hybrid plasmid-based truncations generated by restriction digestion with BamHI and SalI were introduced into the corresponding sites of pCuHA-CVT9. For tagging cyan fluorescent protein (CFP), pCuHA-CVT9 or its variants were cut by BamHI and the DNA fragment of CFP was inserted into this site.
Fluorescence Microscopy
Yeast cells expressing fluorescent protein-fused chimeras were grown to mid-log phase. To label the vacuolar membrane, we incubated cells in medium containing 20 μg/ml N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4-64; Molecular Probes, Eugene, OR) at 30°C for 15 min. After being washed with medium, the cells were incubated in medium at 30°C for 30 min. Fluorescence microscopy observation was carried out as described previously (Shintani et al., 2002).
Protein A-Affinity Isolation
Protein A-affinity isolation was carried out essentially as described previously (Shintani et al., 2002). Cells expressing HA-Atg11 and harboring a plasmid encoding protein A or PrtA-Atg19, or those expressing myc-Atg11 and harboring a plasmid encoding protein A or PrtA-Atg20, were converted to spheroplasts and lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], and Complete EDTA-free protease inhibitor [Roche Diagnostics, Indianapolis, IN]). After centrifugation to remove the insoluble material, the detergent extract was incubated with IgG-Sepharose for 1 h at 4°C. The beads were washed with lysis buffer five times and then eluted in SDS-PAGE sample buffer by boiling. The resulting eluate was resolved by SDS-PAGE followed by immunoblotting with anti-HA antibody or anti-Ape1 serum.
Coimmunoprecipitation
Coimmunoprecipitation was carried out essentially as described above. Cells expressing myc-Atg11 and transformed with a plasmid encoding HA-Atg11 or the empty vector were converted to spheroplasts and then resuspended in lysis buffer (20 mM HEPES-KOH, pH 6.8, 150 mM KOAc, 5 mM MgOAc, 250 mM sorbitol, 0.5% Triton X-100, 1 mM PMSF, and Complete EDTA-free protease inhibitor [Roche diagnostics]). The detergent extract was incubated with anti-HA antibody and protein A-Sepharose overnight at 4°C. The beads were washed with lysis buffer three times and then eluted in SDS-PAGE sample buffer by boiling. The resulting eluate was resolved by SDS-PAGE followed by immunoblotting with anti-HA or anti-myc antibody.
RESULTS
Atg11 Localization to the PAS Requires the prApe1-Atg19 Complex
The cargo–receptor complex in the Cvt pathway consisting of prApe1 and Atg19 (the Cvt complex) is delivered into the vacuolar lumen through a transient localization at the perivacuolar, pre-autophagosomal structure, where the complex becomes enwrapped within a vesicle. It has been shown that the proper localization and delivery of prApe1 and Atg19 are dependent on each other; without prApe1, Atg19 is dispersed in the cytosol and without Atg19, prApe1 is located away from the PAS (Shintani et al., 2002). Atg11 also localizes at the PAS and has some critical role in transporting the Cvt complex to the PAS through its interaction with Atg19; in atg11Δ cells, prApe1 and Atg19 colocalize and assemble together in the Cvt complex, but this complex does not localize at the PAS (Shintani et al., 2002). To examine whether the localization of Atg11 at the PAS was dependent on the Cvt complex, we observed CFP-Atg11 with YFP-Atg19 or YFP-Ape1 in ape1Δ or atg19Δ cells, respectively.
In wild-type cells, CFP-Atg11 colocalized to the PAS with YFP-Atg19 and YFP-Ape1 in every cell observed, and in the absence of Ape1, YFP-Atg19 was not able to assemble into a higher order structure as described previously (Shintani et al., 2002; Figure 2, A and B). In addition, in ape1Δ cells CFP-Atg11 was not localized at the PAS but instead was dispersed in the cytosol in 100% of the cells examined (Figure 2A). In contrast, without Atg19, YFP-Ape1 could form a large complex as reported previously (Shintani et al., 2002); however, there was again no CFP-Atg11 punctate structure in any of the cells (Figure 2B). The absence of a CFP-Atg11 punctate signal was not due to instability of the chimera because deletion of either ATG19 or APE1 did not affect the expression level of CFP-Atg11 (our unpublished data). These findings indicate that Atg11 localization to the PAS requires both prApe1 and Atg19, suggesting that Atg11 is assembled with the prApe1–Atg19 complex to be recruited together to the PAS; however, Atg11 is not localized to the PAS before the delivery of the Cvt complex to this site.
Figure 2.
Atg11 interacts with the Cvt complex via Atg19. Atg11 does not localize to a punctate structure in the absence of prApe1 (A) or Atg19 (B). (A) Wild-type (SEY6210) or ape1Δ (YTS107) cells expressing CFP-Atg11 and YFP-Atg19 were grown to mid-log phase and observed by fluorescence microscopy. DIC, differential interference contrast. (B) Wild-type or atg19Δ (SSY31) cells expressing CFP-Atg11 and YFP-Ape1 were grown to mid-log phase and observed by fluorescence microscopy. (C) Atg11 interacts with the Cvt complex via Atg19. Detergent extracts of atg19Δ or ape1Δ atg19Δ (YTS108) cells expressing HA-Atg11 and protein A-fused Atg19 (PrtA-Atg19) were incubated with IgG-Sepharose. Precipitated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-HA antibody or anti-Ape1 antiserum. T and IP lanes show total lysates and IgG precipitates, respectively.
We next decided to examine whether the cargo protein affected the interaction between Atg11 and Atg19. Accordingly, we expressed HA-Atg11 and Atg19 fused to protein A, PrtA-Atg19, in cells with or without prApe1 and carried out protein A-affinity isolation with IgG-Sepharose (Figure 2C). Without PrtA-Atg19, there was neither HA-Atg11 nor prApe1 in the IgG precipitates (IPs). In contrast, PrtA-Atg19 precipitated HA-Atg11 with precursor Ape1 as shown previously (Shintani et al., 2002). In the absence of prApe1, HA-Atg11 was still coprecipitated with PrtA-Atg19 at a level comparable with that seen in the presence of prApe1. Ams1 was characterized as another cargo molecule in the Cvt pathway (Hutchins and Klionsky, 2001; Shintani et al., 2002). We also examined the effect of Ams1 on the Atg19–Atg11 interaction. As with prApe1 alone, the absence of both Ams1 and prApe1 did not affect the coprecipitation of PrtA-Atg19 with HA-Atg11 (our unpublished data). These results indicate that Atg11 interacts with Atg19 independent of the cargo proteins.
Atg11 Recruits the Cvt Complex to the PAS
It has been reported that Atg11 interacts with the C terminus of Atg19. The Atg19Δ388–395 mutant lacking the amino acids from 388 to 395, which loses the interaction with Atg11, retains the ability to assemble with prApe1 but cannot target it to the PAS (Shintani et al., 2002), suggesting that the Atg19–Atg11 interaction is important for delivery of the Cvt complex to the PAS. To further analyze the Atg19–Atg11 interaction, we generated truncated versions of Atg11 and investigated its Atg19 binding site. A yeast two-hybrid assay was performed between Atg11 fused with the Gal4 DNA binding domain, BD-Atg11, and activation domainfused Atg19, AD-Atg19 (Figure 3A). Cells containing two-hybrid mutants BD-Atg11Δ1–817 (lacking coiled-coils 1–3) and Δ1–850 (also lacking coiled-coils 1–3, and hereafter referred to as ΔCC1–3), which lack the N-terminal and central Atg11 regions, allowed the cells to grow in the presence of AD-Atg19 similar to the full-length BD-Atg11. In contrast, Atg11Δ627–1178 (lacking coiled-coils 3–4) and Δ859–1178 (lacking coiled-coil 4, and hereafter referred to as ΔCC4), which have in common the absence of the C-terminal region, could not grow on selective adenine-minus plates. To further examine the interaction between truncated Atg11 and Atg19, we expressed wild-type or mutant HA-Atg11 with PrtA-Atg19 and performed protein A affinity isolation as described above. HA-Atg11ΔCC1–3 was coprecipitated with PrtA-Atg19 similar to the wild-type protein, whereas HA-Atg11ΔCC4 was not able to bind the PrtA-Atg19 protein (Figure 3B). These results suggest that Atg19 binds to the C-terminal region of Atg11.
Figure 3.
Atg19 interacts with the C terminus of Atg11. (A) Mapping of the Atg19 binding site within Atg11 by a yeast two-hybrid assay. A schematic of Atg11 is shown indicating the location of the coiled-coil domains. The two-hybrid atg11Δ (YTS121) strain was transformed with plasmids containing the activation domain (AD)-fused Atg19 and the binding domain (BD)-fused wild-type or mutant Atg11, and transformants were grown on plates lacking uracil, leucine, and adenine at 30°C for 2 d. (B) Atg19 binds the C terminus of Atg11. Detergent extracts of atg11Δ (AHY001) cells expressing wild-type or mutant HA-Atg11 and PrtA-Atg19 were incubated with IgG-Sepharose. Precipitated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-HA antibody. T and IP lanes show total lysates and IgG precipitates, respectively. (C) The ability to bind Atg19 is not sufficient for Atg11 to facilitate prApe1 maturation. Total cell extracts of atg11Δ cells expressing wild-type or mutant Atg11 were separated by SDS-PAGE followed by immunoblotting with anti-Ape1 antiserum. (D) Multiple Atg11 domains are required to recruit prApe1 to the PAS. atg11Δ cells expressing GFP-Ape1 and wild-type Atg11 or Atg11 mutant proteins were grown to mid-log phase and labeled with FM 4-64. (E) Colocalization of Atg11 with Atg19 requires CC4. atg11Δ cells expressing wild-type or mutant CFP-Atg11 and YFP-Atg19 were grown to mid-log phase and examined by fluorescence microscopy. DIC, differential interference contrast.
Previous results show that Atg19Δ388–395, defective for the interaction with Atg11, does not allow prApe1 to be matured (Shintani et al., 2002). To examine the importance of the Atg19–Atg11 interaction in the Cvt pathway, we checked prApe1 maturation in cells that expressed Atg11 mutants. Total extracts were prepared from cells expressing wild-type Atg11, Atg11ΔCC4, or Atg11ΔCC1–3, and immunoblotting was carried out with anti-Ape1 antiserum. Cells expressing wild-type Atg11 showed the mature form of Ape1, whereas those expressing either Atg11ΔCC4 or Atg11ΔCC1–3 accumulated only prApe1 (Figure 3C). This result indicates that the Atg11ΔCC1–3 mutant retains the ability to interact with Atg19 but is nonetheless unable to facilitate maturation of prApe1.
Atg11 has some role in transporting the Cvt complex to the PAS. Accordingly, to check the Atg11 mutants for the ability to recruit prApe1 to the PAS, we expressed GFP-Ape1 along with wild-type or mutant Atg11 and observed the localization by fluorescence microscopy. We also labeled the cells with the dye FM 4-64 to stain the vacuoles. The Cvt complex was localized to the PAS in the presence of wild-type Atg11 and was located away from the PAS without Atg11, as shown previously (Figure 3D; Shintani et al., 2002). Both the Atg11ΔCC4 and Atg11ΔCC1–3 mutants exhibited a Cvt complex localization apart from the vacuole similar to the ATG11 deletion in 100% of the cells examined (Figure 3D). From this result, we concluded that the inability of the Atg11ΔCC1–3 mutant to facilitate maturation of prApe1 was due to the loss of transport to the PAS.
Finally, we investigated the subcellular localization of Atg11ΔCC4 and Atg11ΔCC1–3. CFP fusions to these proteins were generated, and cells expressing these chimeras were observed along with YFP-Atg19 by fluorescence microscopy (Figure 3E). CFP-Atg11ΔCC1–3 showed up as a single dot and colocalized with YFP-Atg19 similar to the wild-type CFP-Atg11, whereas CFP-Atg11ΔCC4 was dispersed similar to the result with wild-type CFP-Atg11 in the absence of Atg19 or Ape1 in 100% of the observed cells (Figure 2, A and B). Together, these results suggest that Atg11ΔCC1–3, retaining the Atg11 C-terminal domain, can be assembled with the Cvt complex but is not competent to transport the complex to the PAS. This finding supports the idea that Atg11 is assembled with the prApe1–Atg19 complex before the complex is recruited to the PAS.
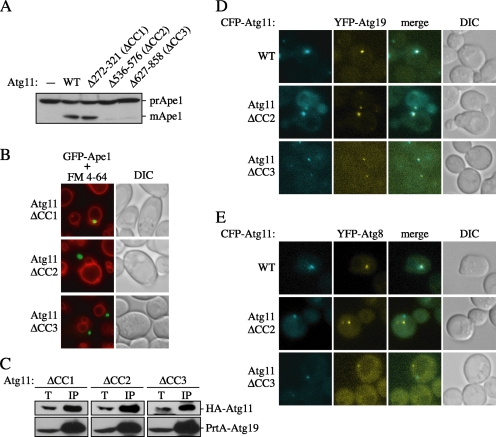
These results also led us to predict that the N-terminal and/or central regions of Atg11 contain information necessary for the proper transport of the Cvt complex to the PAS. To test this possibility, we carried out further analyses of these regions. Atg11 is predicted to contain four coiled-coil motifs (CC1–4), and three of them are located within the region that is absent in the Atg11Δ1–850 (ΔCC1–3) mutant. To examine the functions of these coiled-coil motifs, we constructed three deletion mutants lacking each motif, Δ272-Δ321 (ΔCC1), Δ536–576 (ΔCC2), and Δ627–858 (ΔCC3). First, we examined prApe1 maturation in cells expressing each of these mutants (Figure 4A). Atg11ΔCC1 allowed maturation at the same level as wild-type Atg11. In contrast, Atg11ΔCC2 and ΔCC3 mutants did not mature prApe1.
Figure 4.
Coiled-coil domains 2 and 3 of Atg11 are required for the transport of prApe1 to the PAS. (A) Total cell extracts of atg11Δ (AHY001) cells expressing wild-type or mutant Atg11 proteins were separated by SDS-PAGE followed by immunoblotting with anti-Ape1 antiserum. (B) Deletion of Atg11 CC2 or CC3 prevents localization of prApe1 to the PAS. atg11Δ cells expressing GFP-Ape1 and wild-type or mutant Atg11 proteins were grown to mid-log phase, labeled with FM 4-64, and examined by fluorescence microscopy. (C) Atg11ΔCC2 and ΔCC3 mutants can interact with Atg19. Detergent extracts of atg11Δ cells expressing mutant HA-Atg11 and PrtA-Atg19 were incubated with IgG-Sepharose. Precipitated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-HA antibody. T and IP lanes show total lysates and IgG precipitates, respectively. (D) Colocalization of Atg11ΔCC2 and ΔCC3 with Atg19. atg11Δ cells expressing wild-type or mutant CFP-Atg11 and YFP-Atg19 were grown to mid-log phase and examined by fluorescence microscopy. (E) Atg11ΔCC2 and ΔCC3 do not localize to the PAS. atg11Δ cells expressing wild-type or mutant CFP-Atg11 and YFP-Atg8 were grown to mid-log phase and examined by fluorescence microscopy. DIC, differential interference contrast.
To investigate the ability of these mutants to recruit prApe1 to the PAS, we observed the localization of GFP-Ape1 in cells expressing the mutant Atg11 proteins while simultaneously staining the vacuole with FM 4-64 (Figure 4B). GFP-Ape1 localized at the PAS with Atg11ΔCC1, corresponding to the result seen with prApe1 maturation. In contrast, Atg11ΔCC2 and ΔCC3 exhibited the GFP-Ape1 localization away from the vacuole. These phenotypes were constant in essentially every cell that we observed. This result suggests that the loss of prApe1 maturation is due to the inability of the ΔCC2 and ΔCC3 mutants to promote delivery of the Cvt complex to the PAS.
To check whether these Atg11 mutants can interact with Atg19, we performed a coprecipitation experiment of HA-Atg11 with PrtA-Atg19. All three of the coiled-coil HA-Atg11 mutants were precipitated with PrtA-Atg19, indicating that these mutant proteins can interact with Atg19 (Figure 4C). To further check the interaction of Atg11ΔCC2 and ΔCC3 with the Cvt complex, we examined the localization of these CFP-Atg11 mutants with YFP-Atg19 (Figure 4D). As expected from the coprecipitation results, both CFP-Atg11ΔCC2 and ΔCC3 colocalized with YFP-Atg19 in essentially 100% of the cells that we imaged. Finally, to confirm that the Cvt complex was not localized to the PAS with these mutants, we compared their localization with YFP-Atg8, a marker for the PAS (Figure 4E; Kim et al., 2001a; Suzuki et al., 2002). Wild-type CFP-Atg11 colocalized to the YFP-Atg8 dot, whereas both CFP-Atg11ΔCC2 and ΔCC3 localized away from YFP-Atg8 in all cells examined. These results indicate that the Atg11ΔCC2 and ΔCC3 mutants can assemble with the Cvt complex but are not able to transport the complex to the PAS, further supporting the model that Atg11 forms part of the Cvt complex with Atg19 and prApe1 before the localization of the complex to the PAS.
Organization of the PAS Follows the Localization of the Cvt Complex
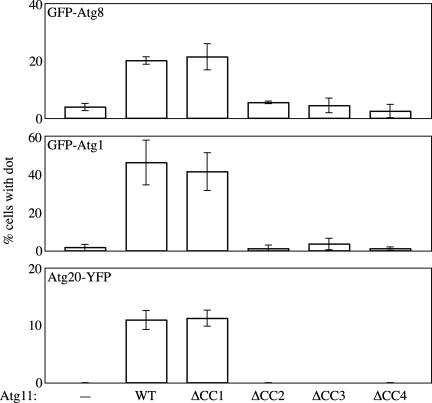
When YFP-Atg8 was observed in cells expressing Atg11ΔCC2 or ΔCC3 as the only source of Atg11, there were fewer cells containing YFP-Atg8 dots marking the PAS compared with wild-type cells, even though these mutant Atg11 proteins could assemble as part of the Cvt complex (our unpublished data; see below). Recently, it was reported that the depletion of the cargo molecule prApe1, the Atg19 receptor, or Atg11 decreases the efficient localization of some Atg components to the PAS in nutrient-rich conditions but not during starvation (Shintani and Klionsky, 2004), suggesting that the localization of the cargo complex to the PAS is required for recruitment of the Atg components and the organization of the PAS under growing conditions. Our observations indicating reduced levels of punctate YFP-Atg8 are consistent with this model. To further analyze the relationship between the locations of the Cvt complex and Atg components, we observed and quantified the localization of PAS components GFP-Atg8, GFP-Atg1, and Atg20-YFP at the PAS together with Atg11 mutants under growing conditions by fluorescence microscopy (Figure 5). With the wild-type Atg11, ∼20% of the cells contained a GFP-Atg8 punctate signal at the PAS. Similarly, GFP-Atg8 localization at the PAS was observed with 21% of the cells expressing Atg11ΔCC1. As seen above, cells expressing Atg11ΔCC2, ΔCC3, and ΔCC4, which all were defective for the transport of the Cvt complex to the PAS, showed a severe defect in the localization of GFP-Atg8 at the PAS; <6% of the cells displayed a punctate dot of GFP-Atg8, a level comparable with that seen in the complete absence of Atg11 (4%). These results suggest that efficient localization of Atg8 at the PAS requires not only assembly of Atg11 with the Cvt complex but also its recruitment to the PAS.
Figure 5.
Localization of Atg components at the PAS with Atg11 mutants. atg11Δ (AHY001) cells transformed with a plasmid expressing GFP-Atg1, atg11Δ (YTS192) cells expressing GFP-Atg8, or atg11Δ vps38Δ (TYY014) cells with Atg20-YFP containing the empty vector or a plasmid carrying the wild-type or indicated mutant Atg11 were grown in rich medium. The percentage of cells containing a punctate dot corresponding to the fluorescent protein are indicated. More than 200 cells were quantified in three individual experiments by fluorescence microscopy, and the error bars represent the SD.
A similar result on localization at the PAS was obtained with GFP-Atg1 and Atg20-YFP (Figure 5). Atg1 is a serine/threonine kinase, which is an essential component for both the Cvt pathway and autophagy and which localizes in a dot at the PAS in wild-type cells (Kamada et al., 2000; Suzuki et al., 2001; Kim et al., 2002). In the absence of Atg11, the PAS localization of GFP-Atg1 was significantly decreased compared with that seen in the presence of Atg11 (∼2 and 46%, respectively), in agreement with previous data (Shintani and Klionsky, 2004). Forty-one percent of cells with Atg11ΔCC1, which targeted prApe1 to the PAS, possessed a punctate GFP-Atg1 dot, which was comparable with the level seen with wild-type Atg11. In contrast, <4% of the cells with Atg11ΔCC2, ΔCC3, or ΔCC4 contained an Atg1-GFP dot.
Atg20 is a Cvt pathway-specific component and has a conserved phox domain (PX domain), which is involved in binding to phosphatidylinositol 3-phosphate [PtdIns(3)P]; Nice et al., 2002). We have found that Atg20 can bind to Atg11 (Figure 6). It also was reported that Atg20 is involved in the retrieval of proteins from the early endosome to the late Golgi apparatus (Hettema et al., 2003). Accordingly, there are two populations of Atg20 in the cell; one localizes at the PAS, dependent on components of the Cvt/autophagy-specific PtdIns 3-kinase complex I such as Atg14; and the other localizes at the endosome, dependent on subunits of PtdIns 3-kinase complex II such as Vps38 (Nice et al., 2002; Hettema et al., 2003). Atg20-YFP localization at the PAS is specifically affected by the deletion of ATG11, ATG19, and APE1 (Shintani and Klionsky, 2004). The localization of Atg20-YFP at the PAS was examined in cells deleted for VPS38 to eliminate the endosomal population (Figure 5). Eleven percent of cells expressing wild-type Atg11 contained a punctate Atg20-YFP dot at the PAS, whereas none of the cells with the empty vector displayed this phenotype. Similar to other PAS proteins, the Atg20-YFP dot at the PAS was still observed with cells expressing Atg11ΔCC1 (11%), whereas the localization was significantly decreased with Atg11ΔCC2, ΔCC3, and ΔCC4. Together, these observations suggest that in the Cvt pathway the cargo–receptor complex properly localized with Atg11 has some role in organizing the formation of the PAS.
Figure 6.
Mapping of the binding sites of Atg components within Atg11. Two-hybrid analysis of Atg1 lacking the kinase domain (Δ1–325), Atg11, Atg17, Atg19, and Atg20 with wild-type Atg11 or Atg11 mutants. The two-hybrid atg11Δ (YTS121) strain was transformed with plasmids containing BD-Atg1, BD-Atg11, BD-Atg17, or BD-Atg20 and AD-Atg11 (or AD-Atg11 with deletions), or AD-Atg19 and BD-Atg11 (or BD-Atg11 with deletions), and transformants were grown at 30°C for 7 d for all combinations with the exception of AD-Atg19 and BD-Atg11, which were grown for 2 d. + and – show an ability and inability, respectively, for cells with plasmids to grow on plates lacking uracil, leucine, and adenine.
Interaction of Atg11 with Atg Components
Atg11 interacts with at least Atg1, itself, and Atg19 (Kim et al., 2001b; Shintani et al., 2002). As a first step toward understanding the importance of these multiple interactions, we performed a yeast two-hybrid analysis by using Atg11 deletion mutants (Figure 6). A two-hybrid construct of the full-length Atg1 has autocatalytic activation; in a two-hybrid test, cells containing the full-length Atg1 on a two-hybrid plasmid could grow well on the adenine-minus–selective plates in combination with an empty vector (our unpublished data). To eliminate this problem, Atg1 lacking the kinase domain between amino acid residues 1 to 325 was used for this assay; this construct did not show autocatalytic activation. When expressed with a two-hybrid plasmid carrying Atg1, two-hybrid constructs of Atg11ΔCC1 and ΔCC4 worked as well as wild-type Atg11 to allow two-hybrid cells to grow on adenine-minus plates, whereas Atg11ΔCC2 and ΔCC3 did not. Similar results were seen when examining Atg11 self-interactions.
In contrast to the results seen for Atg11-Atg1 and Atg11-Atg11 two-hybrid interactions, cells expressing Atg19 along with Atg11 mutants that still contained C-terminal amino acids 859-1178 (CC4) showed growth on the selective plates, whereas cells expressing Atg11ΔCC4, which lacked this C-terminal domain, did not grow (Figure 6). These data are consistent with the two-hybrid analysis using the complete coiled-coil deletions (Figure 3B) that showed binding of Atg19 to Atg11ΔCC1–3, and with the affinity isolation results (Figure 4C). Thus, the C-terminal region including the fourth coiled-coil domain of Atg11 is required for an interaction with Atg19, but not with Atg1 or Atg11 itself.
Atg17 was characterized as being a component of the Atg1 kinase complex that is specific for autophagy (Kamada et al., 2000). We identified Atg17 as a binding partner of Atg11 (Figure 6). In the case of cells expressing Atg17, Atg11ΔCC1, ΔCC3, and ΔCC4 grew on the selective plates, but cells with Atg11ΔCC2 did not. These results suggest that the region including the CC2 and CC3 domains was involved in the interaction between Atg11 and Atg1 and in Atg11 self-interactions, whereas only the region encompassing the CC2 domain was required for the interaction with Atg17. It is known that Atg17 can interact with Atg1 and Atg20 (Kamada et al., 2000; Nice et al., 2002). To check whether Atg11 mediates the formation of these complexes, we examined these interactions in the atg11Δ strain by the yeast two-hybrid assay. There was no effect of ATG11 deletion on either interaction, indicating that these complexes form independent of the interaction with Atg11 (our unpublished data).
Atg20 has been characterized to be involved specifically in the Cvt pathway and has a conserved PtdIns(3)P-binding domain (PX domain; Nice et al., 2002). The two-hybrid analyses revealed that Atg11 could bind to Atg20 (Figure 6). We also examined the binding site of Atg20 in Atg11 by a yeast two-hybrid analysis as described above (Figure 6). Atg11ΔCC1, ΔCC3, and ΔCC4 allowed the two-hybrid strains to grow on the adenine-minus–selective plate, but Atg11ΔCC2 did not. This result suggests that the interaction between Atg11 and Atg20 also requires the region of amino acids from 536 to 576, coiled-coil 2, in Atg11. Furthermore, we found that Atg20-YFP did not localize to the PAS in the Atg11ΔCC3 and ΔCC4 mutants (Figure 5), even though these domains are not needed for the interaction between these two proteins. The Atg11ΔCC4 mutant, however, is defective in interaction with Atg19 and is unable to maintain the PAS. It is possible that the inability of the Atg11ΔCC3 mutant to recruit the Cvt complex to the PAS prevents Atg20 localization to this site, although the timing of the Atg11–Atg20 interaction is still not known.
To further confirm this previously undetected interaction, we carried out an affinity isolation between PrtA-Atg20 and wild-type myc-Atg11. We found that myc-Atg11 was precipitated together with PrtA-Atg20 (Figure 7A). In contrast, myc-Atg11ΔCC2 did not copurify with PrtA-Atg20 (Figure 7B), consistent with the two-hybrid result. It was reported that Atg20 localizes to both the PAS and the endosomal membrane (Nice et al., 2002; Hettema et al., 2003). Accordingly, we determined whether the loss of interaction with Atg11 affected the localization of Atg20 at the PAS and/or endosome. Atg20-YFP still exhibited a punctate dot without Atg11, which corresponded to localization at the endosome; this dot was no longer seen in a vps38Δ strain lacking PtdIns 3-kinase complex II that is required for localization at this organelle (Shintani and Klionsky, 2004; our unpublished data). Next, Atg20-YFP was expressed with wild-type Atg11 or Atg11ΔCC2 in atg11Δ cells. In contrast to the previous analysis and that shown in Figure 5, we used a strain deleted only for endogenous ATG11 (i.e., expressing Vps38) for this experiment. The localization of CFP-Atg11ΔCC2 was separated from that of Atg20-YFP (Figure 7C), indicating that CFP-Atg11ΔCC2, which was defective in localization at the PAS, was not colocalized with Atg20-YFP located at the endosome.
Figure 7.
Atg20 interacts with Atg11 via coiled-coil domain 2. Detergent extracts of atg11Δ (AHY001) cells expressing myc-Atg11 with or without PrtA-Atg20 (A) or myc-Atg11ΔCC2 with PrtA-Atg20 (B) were incubated with IgG-Sepharose. Precipitated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-myc antibody. T and IP lanes show total lysates and IgG precipitates, respectively. (C) atg11Δ cells expressing CFP-Atg11 or CFP-Atg11ΔCC2 with Atg20-YFP were grown to mid-log phase and observed by fluorescence microscopy. DIC, differential interference contrast.
Finally, it was shown previously that Atg20 interacts with a second PX domain-containing protein, Atg24 (Nice et al., 2002); however, we did not detect an interaction between Atg11 and Atg24 (our unpublished data). The PX domain is functionally important for Atg20 and Atg24, because substitution of a conserved tyrosine residue in this domain (Atg20Y193A or Atg24Y79A) causes a defect in prApe1 maturation, and proper localization of these proteins (Nice et al., 2002). According to our protein A isolation experiment, however, the Atg20Y193A mutant is still able to interact with Atg11 (our unpublished data). Depletion of Atg19 or prApe1 causes a defect in Atg20 localization at the PAS, and a deletion of ATG11 shows a more severe defect (Shintani and Klionsky, 2004). These results suggest that proper localization of Atg20 requires both the PAS localization of the Cvt complex and the ability to bind to PtdIns(3)P. The PAS localization of Atg20 might be mediated by the interaction with Atg11 at the PAS.
Homo-oligomerization of Atg11 at the PAS
Immunoprecipitation data suggest that Atg11 interacts with itself to form a homodimer or homo-oligomer (Kim et al., 2001b). Our yeast two-hybrid assay also showed the self-interaction of Atg11 proteins (Figure 6). We further investigated the role of the Atg11 binding domains in Atg11 self-interaction through fluorescence microscopy experiments. As shown above, when expressed in atg11Δ cells, Atg11ΔCC4, which is a C-terminal truncation, was not localized at the PAS because of the loss of interaction with Atg19. According to our yeast-two hybrid result, however, this mutant protein could bind to wild-type Atg11 (Figure 6). We hypothesized that if an Atg11 homo-oligomer formed between these two proteins, Atg11ΔCC4 fused with a fluorescent protein would exhibit a punctate dot dependent on the wild-type Atg11. To check this possibility, we expressed CFP-Atg11ΔCC4 together with wild-type Atg11 in atg11Δ cells. As seen in Figure 3E, atg11Δ cells expressing CFP-Atg11ΔCC4 alone did not display an Atg11 dot. The presence of an additional empty vector did not change this distribution (Figure 8A). In contrast, the presence of wild-type Atg11 allowed CFP-Atg11ΔCC4 to assemble into a punctate structure (Figure 8A). An essentially identical result was obtained when GFP-Atg11ΔCC4 was expressed under the control of the authentic ATG11 promoter (our unpublished data). These results indicated that without the ability to interact with Atg19, Atg11ΔCC4 localized to a punctate dot dependent on the presence of another full-length Atg11 protein that could bind Atg19, supporting a model for Atg11 self-interaction.
Figure 8.
Atg11 forms a homo-oligomer at the PAS. (A) atg11Δ (AHY001) cells with a plasmid encoding CFP-Atg11ΔCC4 or wild-type Atg11 under control of the CUP1 promoter were grown to mid-log phase and observed by fluorescence microscopy. DIC, differential interference contrast. (B) Atg11 CC4 can act in trans for interaction with Atg19 and localization to the PAS. atg11Δ cells expressing CFP-Atg11ΔCC4 and wild-type Atg11 with YFP-Atg19 or YFP-Atg8 were grown to mid-log phase and examined by fluorescence microscopy. (C) Atg11 CC2 is unable to act in trans. atg11Δ or atg1Δ atg11Δ (YTS157) cells with plasmids containing CFP-Atg11ΔCC4 and Atg11ΔCC2 were grown to mid-log phase and observed by fluorescence microscopy. (D) Atg11ΔCC2 is defective in homo-oligomer formation. Detergent extracts of atg11Δ cells expressing wild-type HA-Atg11 or HA-Atg11ΔCC2 and myc-Atg11 were incubated with anti-HA antibody. Precipitated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-myc or anti-HA antibody.
Next, we examined whether the dot that formed from Atg11 and CFP-Atg11ΔCC4 hetero-oligomers localized at the PAS by examining colocalization with YFP-Atg19 or YFP-Atg8 (Figure 8B). The dot formed by CFP-Atg11ΔCC4 colocalized with both YFP-Atg19 and YFP-Atg8. This result suggested that Atg11ΔCC4 existed with the Cvt complex at the PAS. We also examined whether Atg11ΔCC4 could localize at the PAS when coexpressed with the Atg11ΔCC2 mutant, which did not transport the Cvt complex to the PAS and was defective in Atg11 self-interaction. atg11Δ cells expressing CFP-Atg11ΔCC4 and Atg11ΔCC2 were observed by fluorescence microscopy (Figure 8C). As seen in atg11Δ cells without wild-type Atg11, none of the cells displayed formation of a CFP-Atg11ΔCC4 dot. Similarly, PAS localization of CFP-Atg11ΔCC4 was not observed with coexpression of Atg11ΔCC3 even though the latter protein, like Atg11ΔCC2, retains the ability to bind Atg19 (our unpublished data). These results suggest that a single CC2 or CC3 domain is not sufficient for self-interaction.
As mentioned above, a yeast two-hybrid analysis revealed that Atg11ΔCC2 was not able to interact with wild-type Atg11 (Figure 6). To further examine the interaction between wild-type Atg11 and Atg11ΔCC2, we coexpressed myc-Atg11 and HA-tagged wild-type Atg11 or HA-Atg11ΔCC2 in atg11Δ cells and carried out immunoprecipitation with anti-HA antibody (Figure 8D). In the presence of wild-type HA-Atg11, myc-Atg11 was coprecipitated as shown previously (Kim et al., 2001b). In contrast, HA-Atg11ΔCC2 did not coprecipitate myc-Atg11. From these results, we concluded that Atg11ΔCC2 could not interact with Atg11 and as a consequence could not support the formation of a PAS dot by Atg11ΔCC4, even though Atg11ΔCC2 could bind Atg19. Together, these results suggest that Atg11 forms a homodimer or higher oligomer at the PAS and that both Atg11 proteins must retain the ability to self-interact to form a dimer.
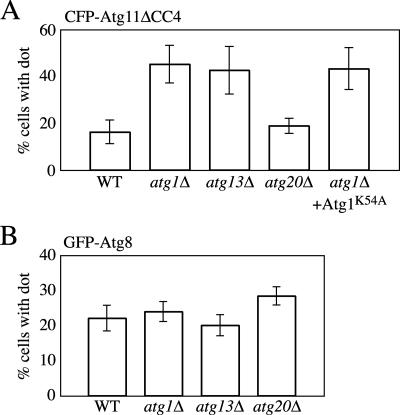
Atg1 is a kinase component and proposed to be able to form a complex with multiple proteins, including Atg11 (Kamada et al., 2000; Kim et al., 2001b). The role of Atg1 is not known, but in cells deleted for ATG1, the Cvt complex of Atg19 and prApe1 is blocked at the PAS and is not delivered to the vacuole (Shintani et al., 2002; Suzuki et al., 2002). Atg1 might be involved in some process after the recruitment of the Cvt complex to the PAS, and we wanted to determine whether it played a role in Atg11 oligomerization. To test the effect of deleting ATG1 on the Atg11 homo-oligomer at the PAS, we observed CFP-Atg11ΔCC4 with wild-type Atg11 in atg1Δ atg11Δ cells and quantified the number of cells containing a punctate dot of CFP-Atg11ΔCC4 (Figure 9A). This assay again examines the ability of CFP-Atg11ΔCC4, which cannot bind Atg19, to localize as a punctate dot at the PAS by virtue of its ability to homo-ologomerize with a second copy of Atg11 that can bind Atg19. CFP-Atg11ΔCC4 displayed a higher frequency of punctate localization in the absence of Atg1; 16% of the cells showed a punctate dot in the presence of Atg1, whereas ∼50% of the cells without Atg1 had the dot. This phenomenon also was observed with atg1Δ cells versus wild-type cells expressing GFP-Atg11ΔCC4 from the endogenous ATG11 promoter (our unpublished data). We also examined whether the absence of Atg1 would allow us to detect colocalization between the CFP-Atg11ΔCC4 and Atg11ΔCC2 constructs. When coexpressed with Atg11ΔCC2 (which can bind Atg19) in atg1Δ atg11Δ cells, however, CFP-Atg11ΔCC4 still did not show a punctate localization (Figure 8C). This indicates that Atg1 does not affect the stable PAS localization of CFP-Atg11ΔCC4 directly, but rather stabilizes localization of the wild-type copy of Atg11. It is possible that the increase in the number of cells displaying a CFP-Atg11ΔCC4 dot in the atg1Δ strain was due to an increase in the efficiency of PAS formation or stability. To check this, we observed the PAS marker GFP-Atg8 expressed in wild-type or atg1Δ cells by fluorescence microscopy and quantified the number of cells with a GFP-Atg8 dot (Figure 9B). Unlike the results seen with CFP-Atg11ΔCC4, there was no significant difference between the ratios of wild-type and atg1Δ cells containing a punctate dot (22 and 24%, respectively), suggesting that deletion of ATG1 did not affect the efficiency of PAS formation or its stability.
Figure 9.
The Atg1–Atg13 complex and Atg1 kinase activity affect Atg11 homo-oligomer disassembly. (A) Atg11 homo-oligomers accumulate in the absence of Atg1 or Atg13. atg11Δ (AHY001), atg1Δ (WHY1), atg13Δ (CWY233), or atg20Δ (D3Y109) cells with plasmids expressing CFP-Atg11ΔCC4 and wild-type Atg11, or atg1Δ cells with a plasmid expressing CFP-Atg11ΔCC4 and the Atg1K54A mutant and wild-type Atg11 were observed with a fluorescence microscope, and 200–250 cells containing fluorescent punctate dots were quantified. Error bars indicate the SD of three independent experiments. (B) The absence of Atg1 or Atg13 does not increase the stability of the PAS. Wild-type (SEY6210), atg1Δ (WHY1), atg13Δ (CWY233), or atg20Δ (D3Y109) cells transformed with a plasmid expressing GFP-Atg8 were observed and cells containing punctate dots were quantified. Error bars indicate the SD of three independent experiments.
Because Atg1 interacts with Atg13, we decided to quantify the PAS localization of CFP-Atg11ΔCC4 in an atg13Δ strain. Forty-three percent of ATG13-deleted cells showed the CFP-Atg11ΔCC4 dot, which is a comparable level with that in atg1Δ cells. This result suggests that the Atg1–Atg13 complex is involved in the efficiency of localization or stability of the Atg11 homo-oligomer at the PAS. We have found that Atg11 can physically interact with Atg20 (Figure 6). Accordingly, we checked the effect of deleting Atg20 on the formation of Atg11 homo-oligomers (Figure 9A). In contrast to the cases of atg1Δ and atg13Δ cells, only 19% of atg20Δ cells had a CFP-Atg11ΔCC4 dot. This result suggests that Atg20 has no effect on Atg11 homo-oligomerization. We also examined the role of Atg13 and Atg20 in PAS formation/stability by observing atg13Δ or atg20Δ cells containing GFP-Atg8. Both strains exhibited the same level of GFP-Atg8 punctate staining as seen in the wild-type and atg1Δ cells. This result suggests that Atg13 and Atg20 do not affect PAS formation or stability.
Finally, to check whether the enhancement of CFP-Atg11ΔCC4 localization at the PAS requires the kinase activity of Atg1, which is controlled by Atg13, we examined atg1Δ cells expressing the Atg1K54A mutant, which is defective for kinase activity (Kamada et al., 2000). Cells expressing Atg1K54A showed the same frequency of CFP-Atg11ΔCC4 punctate staining as seen in the atg1Δ or atg13Δ strains (Figure 9A). This result suggests that the kinase activity of Atg1 is involved in the efficient disassembly of Atg11 proteins, because without kinase-active Atg1 a higher level of oligomeric Atg11 accumulated at the PAS.
DISCUSSION
For precise delivery, cargo proteins need to be selectively recruited and incorporated into the appropriate transport vesicles. For example, the resident vacuolar hydrolases Ape1 and Ams1, which are synthesized in the cytosol, are selectively packaged into Cvt vesicles and delivered to the vacuole via the Cvt pathway. This selective incorporation of cargo is thought to occur at the PAS, so it requires transport to this site. Recently we carried out a temporal analysis of the mechanism of cargo selection in the Cvt pathway (Shintani et al., 2002). After homo-oligomerization, an Ape1 complex directly binds to a receptor protein, Atg19, to form the Cvt complex; the complex is transported to the PAS by the mediation of Atg11 (Scott et al., 2001; Shintani et al., 2002). The function of Atg19 in this process was examined by mapping its interaction domains; however, it was not clear how Atg11 functions in the Cvt pathway.
An important question then is what is the role of Atg11 in specific autophagy? To gain insight to this question, we decided to explore one of the most notable features of this protein, the fact that it interacts with several other Atg proteins that seem to function at discrete steps of the import process. By mapping the interacting domains and mutating them, we have started to resolve one of the major hurdles in understanding the mechanism of autophagy, defining a temporal order of action to the >20 Atg-specific proteins. Our previous studies showed that Atg11 localizes with other Atg components, including prApe1 and Atg19 at the PAS (Kim et al., 2002), and interacts with Atg19, whereas ATG11 deletion prevents localization of the prApe1–Atg19 complex at the PAS (Shintani et al., 2002). There might be two possibilities for how Atg11 recruits the Cvt complex to the PAS through the interaction with Atg19. One is that Atg11 first localizes at the PAS and then recruits the Cvt complex to the PAS; the other is that Atg11 is first targeted to the Cvt complex and then this complex is transported to the PAS. Because it was shown that Atg19 localization in a punctate complex is defective in the absence of prApe1 (Shintani et al., 2002), we decided to observe the localization of Atg11 by fluorescence microscopy. We found that Atg11 is no longer seen at the PAS in the absence of the prApe1–Atg19 complex (Figure 2). This result supports the latter idea that Atg11 assembles with the prApe1–Ag19 complex before targeting to the PAS. The finding that Atg11 deleted for CC1–3 colocalized with the Cvt complex but could not guide it to the PAS (Figure 3) also supports this model.
In the next step of cargo packaging, the prApe1–Atg19–Atg11 complex is targeted to the PAS. The Cvt complex containing an Atg19 mutant that loses the interaction with Atg11 was not transported to the PAS (Shintani et al., 2002), suggesting that the Atg19–Atg11 interaction was important for the PAS transport of the cargo. The present results with Atg11ΔCC1–3, however, show that the interaction between Atg11 and Atg19 is not enough for the Cvt complex to be transported to the PAS (Figure 3). Accordingly, we have examined whether the N-terminal or central regions of Atg11 have some role in this process. No conserved domains are found in Atg11, except for four coiled-coil domains. Among them, deletion of the second and third coiled-coil domains (Δ536–576 and Δ627–858, respectively) resulted in defects in the transport of the Cvt complex to the PAS. Our mapping of Atg11 shows that this protein forms a complex with multiple Atg components, and these interactions are affected by deletion of the coiled-coil domains. Although it remains to be determined how the prApe1–Atg19 complex is attached to the PAS by Atg11, it is possible that there is an acceptor protein at the PAS that links the Cvt complex and the PAS through an interaction with Atg11, probably through the coiled-coil segments. One possible candidate may be Atg12 because it was reported that Atg12 localizes at the PAS and physically interacts with Atg11 (Ho et al., 2002; Kim et al., 2002), although we did not detect an interaction between these two proteins in the present study (our unpublished data).
Another possible mechanism for transport of the Cvt complex to the PAS would involve homo-oligomerization of Atg11. It has been observed that Atg11 interacts with itself (Kim et al., 2001b) and yeast two-hybrid analyses suggested that Atg11 forms an oligomer. Indeed, Atg11 deletions (Atg11ΔCC2 and ΔCC3) that could not self-assemble were not able to transport the Cvt complex to the PAS (Figures 4 and 6). Atg11 might be assembled with the Cvt complex in a homo-oligomer form to transport it to the PAS. In other trafficking pathways, it has been reported that incorporation of the cargo complex into the vesicles requires a homo-oligomeric state for the transport machinery. For example, Emp47 is a receptor for Emp46, and they are both transported from the endoplasmic reticulum to the Golgi via COPII vesicles (Sato and Nakano, 2003). Incorporation of these proteins into COPII vesicles is dependent on Emp47 oligomerization. It was shown that an Emp47 mutant lacking one coiled-coil motif was not able to form a homo-oligomer and was defective in vesicular incorporation of not only Emp46 but also Emp47 itself. In comparison with this example, however, Atg11 oligomerization seems to have a different role. For efficient incorporation of cargoes into COPII vesicles, the formation of the homo-oligomeric cargoes is required for their concentration. On the other hand, prApe1 proteins are assembled with themselves and form a concentrated homo-oligomeric cargo independent of Atg11 and Atg19. So, we also may speculate that Atg11 homo-oligomerization may be required for more efficient attachment to the putative acceptor on the PAS through increased affinity, possibly resulting in highly selective sorting of the cargo in the Cvt pathway.
After arrival of the Cvt complex with Atg11 at the PAS, other Atg components might be recruited to the PAS to form the vesicle that enwraps the Cvt complex (Shintani and Klionsky, 2004). In this model, the Cvt complex might itself function as the framework for the vesicle-forming machinery. Atg11 could mediate the assembly of the machinery around the cargo because of its interaction with multiple proteins. This mechanism might explain the high selectivity of cargoes in the Cvt vesicles and the exclusion of cytosolic materials from these vesicles (Baba et al., 1997). The prApe1–Atg19 complex is incorporated into the Cvt vesicles at the PAS and transported into the vacuole, whereas during the formation of the Cvt vesicles, Atg11 is thought to be released from the complex because Atg11 is not transported to the vacuole along with the Cvt complex, although it is not clear at which step the release occurs (Kim et al., 2001b). We observed an increase in the percentage of cells with an Atg11ΔCC4 dot in the absence of the Atg1–Atg13 kinase component, suggesting more efficient or stable homo-oligomerization of Atg11. One possible interpretation of this result is that the Atg11 homo-oligomer might be dissociated in a process dependent upon Atg1 kinase activity, and then Atg11 proteins might be released from the Cvt complex at the PAS before Cvt vesicle completion. Further study is needed to address this issue and to determine whether there is a role for Atg11 at the PAS after the delivery of the Cvt complex.
The relationship among Atg proteins is not completely understood. It has been reported that Atg11 has multiple interacting partners, including Atg1, Atg11 itself, and Atg19 (Kamada et al., 2000; Kim et al., 2001b; Ho et al., 2002; Shintani et al., 2002). Here, we mapped the binding domains of some of these Atg components within Atg11 (summarized in Figure 1B) and examined the defects resulting from the loss of these interactions. Deletion of the CC2 domain affected the interactions of Atg11 with four of the proteins that we examined. One question now under investigation is whether these binding proteins share this coiled-coil domain to interact with Atg11 and whether there is a temporal order for the interactions. We also identified two new binding partners, Atg17 and Atg20 (Figures 6 and 7). The multiple binding partners of Atg11 present an intricate network of interactions; it remains to be determined whether a complex of these proteins is formed individually or at the same time and whether the interactions are direct or indirect. Our current hypothesis is that Atg11 connects the cargo (via a direct interaction with Atg19) to the vesicle forming machinery (through a direct interaction with Atg1) at the PAS in a process that depends on Atg11 homo-oligomerization, although we still do not know how Atg11 itself is targeted to the PAS. We think that a more detailed understanding of Atg11 will provide further insight into specific autophagy.
Acknowledgments
We thank Dr. Takahiro Shintani (Tohoku University, Sendai, Japan) for supplying plasmids and strains and members of the Klionsky laboratory for helpful discussions. This work was supported by Public Health Service grant GM-53396 from the National Institutes of Health (to D.J.K.)
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–11–1035) on January 19, 2005.
References
- Abeliovich, H., Zhang, C., Dunn, W. A., Jr., Shokat, K. M., and Klionsky, D. J. (2003). Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema, E. H., Lewis, M. J., Black, M. W., and Pelham, H.R.B. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Hutchins, M. U., and Klionsky, D. J. (2001). Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 276, 20491–20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., and Klionsky, D. J. (2001a). Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002). Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001b). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Scott, S. V., Oda, M. N., and Klionsky, D. J. (1997). Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Klionsky, D. J., Cueva, R., and Yaver, D. S. (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and Ohsumi, Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Nice, D. C., Sato, T. K., Stromhaug, P. E., Emr, S. D., and Klionsky, D. J. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, M. N., Scott, S. V., Hefner-Gravink, A., Caffarelli, A. D., and Klionsky, D. J. (1996). Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 132, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S., Klionsky, D. J., Banta, L. M., and Emr, S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., and Nakano, A. (2003). Oligomerization of a cargo receptor directs protein sorting into COPII-coated transport vesicles. Mol. Biol. Cell 14, 3055–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Baba, M., Ohsumi, Y., and Klionsky, D. J. (1997). Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Guan, J., Hutchins, M. U., Kim, J., and Klionsky, D. J. (2001). Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and Klionsky, D. J. (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kamada, Y., and Ohsumi, Y. (2002). Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell 3, 815–824. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]