Abstract
The CAAX motif at the C terminus of most monomeric GTPases is required for membrane targeting because it signals for a series of three posttranslational modifications that include isoprenylation, endoproteolytic release of the C-terminal– AAX amino acids, and carboxyl methylation of the newly exposed isoprenylcysteine. The individual contributions of these modifications to protein trafficking and function are unknown. To address this issue, we performed a series of experiments with mouse embryonic fibroblasts (MEFs) lacking Rce1 (responsible for removal of the –AAX sequence) or Icmt (responsible for carboxyl methylation of the isoprenylcysteine). In MEFs lacking Rce1 or Icmt, farnesylated Ras proteins were mislocalized. In contrast, the intracellular localizations of geranylgeranylated Rho GTPases were not perturbed. Consistent with the latter finding, RhoGDI binding and actin remodeling were normal in Rce1- and Icmt-deficient cells. Swapping geranylgeranylation for farnesylation on Ras proteins or vice versa on Rho proteins reversed the differential sensitivities to Rce1 and Icmt deficiency. These results suggest that postprenylation CAAX processing is required for proper localization of farnesylated Ras but not geranygeranylated Rho proteins.
INTRODUCTION
The Ras and Rho families of monomeric GTPases regulate a wide array of cellular functions, including growth, differentiation, migration, polarity, and morphology (Matozaki et al., 2000). These disparate functions are regulated, in part, by numerous upstream and downstream regulatory elements that include guanine nucleotide exchange factors, GDP dissociation inhibitors (GDIs), GTPase activating proteins (GAPs), and effectors. However, distinct regulatory elements cannot explain entirely the diversity of function because subsets of these molecules are often shared among different pathways. Among the factors intrinsic to Ras and Rho family proteins that contribute to the diversity and specificity of function are their various subcellular localizations (Michaelson et al., 2001; Bivona et al., 2003).
Most functions of Ras and Rho family proteins require association of the GTPases with cellular membranes. The localization to membranes involves at least two signals within sequences at the C terminus of the proteins. The first of these is the C-terminal CAAX motif (where “A” refers to an aliphatic residue) that signals a series of posttranslational modifications. Nascent CAAX proteins are synthesized in the cytosol where they encounter one of two protein prenyltransferases that modify the CAAX cysteine by thioether linkage of a C-15 farnesyl or C-20 geranylgeranyl lipid (Clarke, 1992; Casey and Seabra, 1996). The substrate specificity for farnesyltransferase versus geranylgeranyltransferase type I (GGTase I) is determined by the residue in the X position of the CAAX motif: S or M specifies farnesylation, whereas L specifies geranylgeranylation (Moores et al., 1991; Reiss et al., 1991). Although Ras family proteins are farnesylated, most Rho family proteins are geranylgeranylated. Prenylated CAAX proteins have high affinity for the endoplasmic reticulum (ER) (Choy et al., 1999) where they encounter a prenyl-CAAX–specific protease, Rce1, that removes the –AAX amino acids (Boyartchuk et al., 1997; Schmidt et al., 1998). The newly exposed C-terminal isoprenylcysteine then becomes a substrate for an isoprenylcysteine-directed carboxyl methyltransferase (Icmt), also localized in the ER, that methylesterifies the α-carboxyl group of the isoprenylcysteine (Dai et al., 1998). The end product of this three-step posttranslational modification process is a hydrophobic domain at the C terminus of an otherwise hydrophilic protein.
CAAX processing alone is necessary but not sufficient to target CAAX proteins to the plasma membrane (PM). Transfer of processed CAAX proteins from the endomembrane to the PM requires a second C-terminal signal immediately upstream of the prenylcysteine—either a series of basic amino acids or cysteine residues that are sites of palmitoylation (Hancock et al., 1990, 1991b; Choy et al., 1999; Michaelson et al., 2001). Unlike Ras proteins, many Rho proteins are constitutively sequestered in the cytosol through interaction with RhoGDI. Targeting of these molecules to the PM or other cellular compartments requires release from this cytosolic chaperone as well as an appropriate second signal. Mutation of the second signal of Ras and Rho proteins leads to retention of the GTPase on the endomembrane (Choy et al., 1999). Some Rho family GTPases that lack second signals, such as Cdc42, reside on the endomembrane when dissociated from RhoGDI (Michaelson et al., 2001). Thus, the inherent membrane destination for any CAAX protein is determined by the nature of the second signal.
Although prenylation is necessary for targeting to the ER and the second signal is necessary for targeting to other membrane destinations, the specific functions of –AAX proteolysis and carboxyl methylation are not established. Nor is it clear why two different prenyltransferases, each using different prenyl groups, evolved. The majority of published studies that analyzed the requirements for CAAX processing in GTPase function did so by mutating the CAAX cysteine to another amino acid, thereby blocking all three CAAX modifications. These studies are therefore uninformative with regard to the specific roles of –AAX proteolysis and carboxyl methylation, both of which require prior prenylation. To investigate these issues, we have inactivated Rce1 and Icmt in the mouse with standard gene-targeting methods (Bergo et al., 2000, 2001, 2002). Both knockouts were lethal during embryonic development, but we had no difficulty in generating spontaneously immortalized fibroblast cell lines from Rce1–/– and Icmt–/– embryos. Preliminary studies with those cells suggested that both Rce1 and Icmt are required for efficient targeting of K-Ras4B to the PM (Bergo et al., 2000) and that Icmt is required for PM targeting of Gγ1 (Michaelson et al., 2002). Here, we use Rce1–/– and Icmt–/– mouse embryonic fibroblasts (MEFs) to examine the localization of all three Ras isoforms and four members of the Rho family of GTPases. All Ras proteins required postprenylation CAAX processing for efficient membrane association; but surprisingly, the Rho proteins did not. Moreover, although Rho proteins were substrates for Rce1 and Icmt, neither modification was required for actin remodeling. Finally, experiments involving swapping of prenyl groups revealed that the differences between the Ras and Rho proteins in their sensitivity to postprenylation processing could be attributed entirely to the type of prenyl modification. Farnesylated proteins required further processing, but geranylgeranylated proteins did not. Our findings demonstrate a biological difference between farnesylated versus geranylgeranylated proteins and suggest why two forms of prenylation evolved.
MATERIALS AND METHODS
Cell Culture and Transfection
Spontaneously immortalized MEFs were prepared from Icmt–/– and Rce1–/– embryos, along with control fibroblasts (Icmt+/+ and Rce1+/+) from littermate embryos (Bergo et al., 2001). The cells were cultured in DMEM with 15% calf serum (Colorado Serum, Denver, CO), nonessential amino acids, 0.1 mM β-mercaptoethanol, and l-glutamine in 5% CO2 at 37°C. For all microscopy, cells were plated, transfected, and imaged in the same 35-mm culture dish that incorporated a no. 1.5 glass coverslip-sealed 15-mm cut-out on the bottom (MatTek, Ashland, MA). Transfections were performed with LipofectAMINE Plus according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). In all cases, 2 μg of DNA total was used for each 35-mm dish. Transiently transfected cells were analyzed 1 d after transfection. When included, active Dbl was transfected at a 1:1 DNA ratio with the green fluorescent protein (GFP)-GTPase.
Plasmids
GFP fusion proteins of H-Ras, K-Ras4B, and N-Ras (Choy et al., 1999) as well as similar fusion proteins of Rac1, Cdc42hs (placental isoform), and RhoA and RhoB (Michaelson et al., 2001) have been described previously. GFP-RhoB-CAIM was the kind gift of Adrienne Cox (University of North Carolina School of Medicine, Chapel Hill, NC). Point mutations were generated by polymerase chain reaction with primers incorporating the desired mutation, and the resulting DNA was cloned into pEGFP-C3 (BD Biosciences Clontech, Palo Alto, CA). All constructs were verified by DNA sequencing. The pCMV6-Dbl construct was a kind gift from Richard Cerione (Cornell University, Ithaca, NY).
Fluorescence Microscopy
Cells were examined with a Zeiss Axioscope epifluorescence microscope (63× PlanApo 1.4 numerical aperture [NA] objective) equipped with a Princeton Scientific Instruments (Monmouth Junction, NJ) cooled charge-coupled device camera and MetaMorph digital imaging software (Universal Imaging, Downingtown, PA) or a Zeiss 510 laser scanning confocal microscope (100× PlanApo 1.4 NA objective). Digital images were processed with Adobe Photoshop 6.0.
Coimmunprecipitation of RhoGDI
Cells were transfected with GFP-Rac1 and lysed in Triton-radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100; 50 mM Tris, pH 7.4; 150 mM NaCl; 2 mM phenylmethylsulfonyl fluoride [PMSF]; 10 μg/ml each for chymotrypsin, pepstatin, and antipain; and 27 μg/ml aprotinin). GFP-Rac1 was immunoprecipitated from cell lysates with anti-GFP monoclonal antibodies (Roche Diagnostics, Indianapolis, IN) and analyzed by SDS-PAGE and 125I-protein A immunoblot for GFP (polyclonal antiserum; Molecular Probes, Eugene, OR) and for RhoGDI (Santa Cruz Biotechnology, Santa Cruz, CA) as described previously (Michaelson et al., 2002).
Subcellular Fractionation
Cells with or without overnight [35S]methionine labeling (20 μCi/ml) were scraped from 10-cm dishes into ice-cold phosphate-buffered saline (PBS) and lysed in hypotonic buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM dithiothreitol, 1 mM PMSF, and protease inhibitors). NaCl was added to 150 mM, and cells were then disrupted with 20 stokes of a Dounce homogenizer and separated into S100 and P100 fractions by centrifugation at 100,000 × g. Endogenous Ras was immunoprecipitated with monoclonal antibody Y13-259 (Santa Cruz Biotechnology) from RIPA extracts of [35S]methionine-labeled fractions. Endogenous RalA was analyzed by immunoblots of the unlabeled fractions by using a monoclonal anti-RalA antibody (BD Transduction Laboratories, Lexington, KY).
Methylation Assays
For in vitro methylation, human neutrophils were purified from peripheral blood of normal volunteers; disrupted by nitrogen cavitation; and separated into cytosol, light membrane, and granules as described previously (Philips et al., 1991). Endogenous Rac2, Cdc42, and RhoA were partially purified in the absence of detergent as 1:1 complexes with RhoGDI from the cytosol by DEAE ion-exchange chromatography and Sephacryl G200HR (Amersham Biosciences, Piscataway, NJ) gel filtration. Proteins were incubated for 30 min with 10 μg of human neutrophil light membranes containing endogenous Icmt and the methyl donor S-adenosyl-[3H]methyl-methionine [3H]AdoMet with or without guanosine 5′-O-(3-thio)triphosphate (GTPγS), and the reaction products were analyzed by SDS-PAGE and fluorography. In vivo methylation was performed on ECV304 cells stably expressing GFP-Cdc42, GFP-Rac1, or GFP-RhoA. Cells were transfected with pCMV or pCMV-Dbl (exchange factor for Rho proteins) and then metabolically labeled with [3H] methyl-methionine (precursor of AdoMet) and [35S]methionine. GFP-tagged proteins were immunoprecipitated with anti-GFP antibodies (Molecular Probes), analyzed by SDS-PAGE, and visualized as [35S]methionine-labeled proteins with a PhosphorImager. Immunoprecipitated proteins were excised from the gel and analyzed for carboxyl methylation by alkaline hydrolysis as described previously (Choy and Philips, 2000).
Phalloidin Staining
Cells transfected with GFP fusion construct of Rho proteins were serumstarved for 24 h followed by fixation in 4% paraformaldehyde for 20 min at room temperature (RT). Cells were permeablized with 0.5% saponin in PBS for 5 min at RT and then incubated for 20 min with 0.2 μg/ml Texas Red-phalloidin in PBS. Cells were washed 2 h in PBS and then imaged as described above.
RESULTS
Ras Proteins Require Full CAAX Processing for Proper Localization
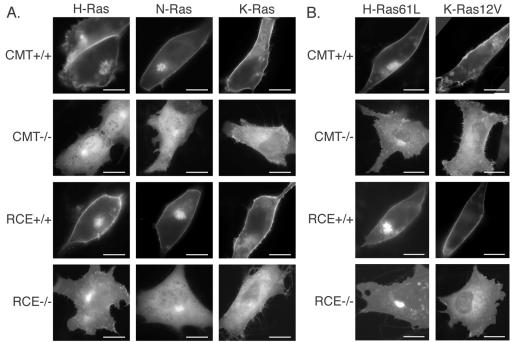
To determine the effects of postfarnesylation CAAX processing on the localization of Ras proteins we expressed GFP-tagged Ras isoforms in MEFs lacking Rce1 or Icmt. GFP-tagged Ras isoforms were localized in wild-type MEFs as we have reported in other cell types (Choy et al., 1999). GFP-N-Ras and GFP-H-Ras were expressed on PM, Golgi, and peri-Golgi vesicles, whereas GFP-K-Ras4B was localized predominantly on the PM (Figure 1A). In contrast, in both Rce1- and Icmt-deficient MEFs, significant portions of the GFP-tagged Ras proteins were mislocalized to the cytosol (Figure 1A). Nevertheless, some PM expression, particularly for GFP-K-Ras4B, was observed, albeit at much lower levels than in wild-type MEFs. These localization patterns were not dependent on the expression levels of the GFP fusion proteins. These data are consistent with our previous observations of GFP-K-Ras in processing-deficient MEFs (Bergo et al., 2000, 2002) and with mislocalization caused by the Icmt inhibitor N-acetyl-S-farnesylcysteine (Choy et al., 1999).
Figure 1.
Postprenylation CAAX processing is required for proper localization of all three Ras isoforms. (A) MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with GFP-tagged forms of H-, N- and K-Ras as indicated and imaged 24 h after transfection as described in Materials and Methods. (B) Constitutively active Ras mutants H-Ras61L and K-RasV12 were expressed and imaged as in A. Representative images are shown of >10 acquired on each of more than five independent experiments. Bars, 10 μm.
Expression on the Golgi of both GFP-N-Ras and GFP-H-Ras was retained in both Rce1- and Icmt-deficient cells. Although palmitoylation is required for the delivery of N-Ras and H-Ras to the PM (Choy et al., 1999), palmitoylation does not require CAAX processing (Cadwallader et al., 1994; Mitchell et al., 1994). Thus, the failure of GFP-N-Ras and GFP-H-Ras to progress beyond the Golgi in postprenylation processing deficient cells suggests that both acylation and carboxyl methylation are required for efficient delivery to and/or retention on the PM.
To determine whether the GTP binding state can influence the dependence on full CAAX processing for targeting to the PM, we studied the localization of GFP-tagged, constitutively active H-Ras61L and K-Ras12V in wild-type cells and in Icmt- and Rce1-deficient cells. The activated proteins localized like their proto-oncogenic counterparts in wild-type MEFs and were mislocalized to the cytosol in both Rce1- and Icmt-deficient MEFs (Figure 1B). We conclude that methylation of the C-terminal farnesylcysteine is required for efficient targeting to the PM of all three Ras isoforms and that this requirement is independent of GTP binding state.
Rho Proteins Bind RhoGDI and Localize Normally in Rce1- and Icmt-deficient Cells
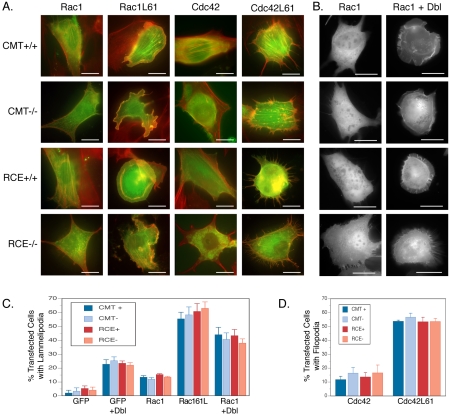
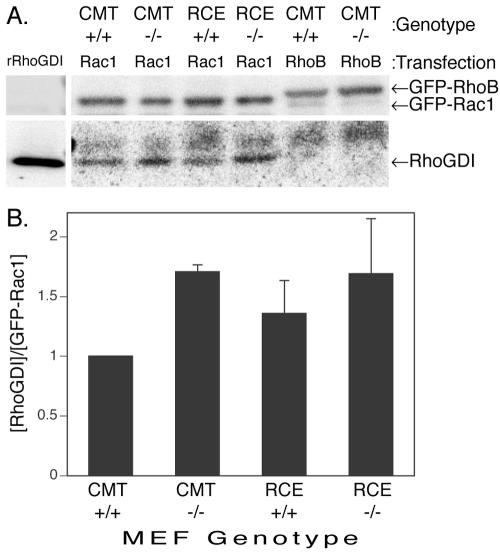
We next sought to determine whether Rho family Ras-related GTPases also require postprenylation processing for efficient membrane targeting. We expressed GFP-tagged members of the Rho family in wild-type MEFs and MEFs lacking Rce1 or Icmt. A subset of Rho family proteins, including Rac1, Cdc42, and RhoA, are normally sequestered in the cytosol in 1:1 complexes with RhoGDI. In COS-1 cells, low levels of expression of these GFP-tagged proteins result in their localization to the cytosol as a consequence of this interaction. However, when the levels of expression exceed the buffering capacity of RhoGDI, the native membrane targeting affinities intrinsic to the C termini of these proteins are revealed (Michaelson et al., 2001). GFP-tagged Rac1, Cdc42, or RhoA were observed only in the cytosol of wild-type, Rce1-, and Icmt-deficient MEFs (Rac1 and Cdc42, Figure 6; RhoA not shown). This was consistent with the lower levels of protein expressed in MEFs relative to COS-1 cells and suggested that these proteins complexed with RhoGDI in the absence of postprenylation processing. To confirm RhoGDI binding in the absence of –AAX proteolysis or carboxyl methylation, we tested directly the relevant protein–protein interactions. Endogenous RhoGDI was coimmunoprecipitated with GFP-Rac1 from the lysates of wild-type, Rce1-, and Icmt-deficient MEFs (Figure 2A). Interestingly, Rac1 bound RhoGDI more efficiently in Icmt-deficient MEFs than in wild-type MEFs (Figure 2B), suggesting that carboxyl methylation lowers the affinity of Rac1 for its chaperone. A trend toward increased binding in Rce1-deficient MEFs also was observed, although the difference was not significant.
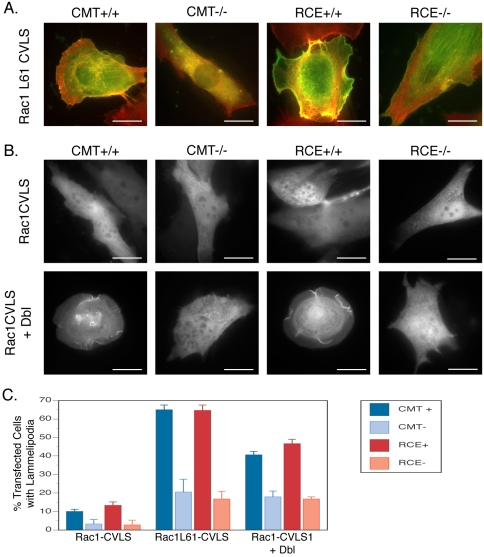
Figure 6.
Rac1 and Cdc42 do not require postprenylation processing to regulate actin remodeling. (A) MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with wild-type or constitutively active GFP-Rac1 or GFP-Cdc42. Twenty-four hours after transfection, cells were paraformaldehyde fixed, saponin permeabilized, and stained with Texas Red-phalloidin to image F-actin. Dual color images are shown (GFP in green and F-actin in red). Rac1-induced lamellipodia and Cdc42-induced fillopodia were equally evident in each cell type. (B) MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with wild-type GFP-Rac1 with or without oncogenic Dbl to induce ruffling. (C) Prevalence of ruffling shown in A and B among 100 randomly chosen cells transfected with the indicated constructs. (D) Prevalence of filopodia formation as shown in A among 50 randomly chosen cells transfected with the indicated constructs. Bars, 10 μm. Graphs show mean ± SEM, n = 3.
Figure 2.
GDI binding of Rac1 is not dependent on postprenylation CAAX processing. MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with GFP-Rac1. GFP-Rac1 was immunoprecipitated from cell lysates with anti-GFP antibodies and analyzed by SDS-PAGE and 125I-protein A immunoblot for GFP and for RhoGDI. (A) Representative coimmunoprecipitation with recombinant RhoGDI (rRhoGDI) as a control run on the same gel (shown as separate panel because of a much lower PhosphorImager exposure time). The identity of the nonspecific band running slightly higher than RhoGDI is not known. (B) Cumulative data where the ability of RhoGDI to interact with GFP-Rac1 is quantified by coimmunoprecipitation as the PhosphorImager volumes of [GDI]/[GFP-Rac1], and the data are normalized across experiments by assigning a value of 1 to ratios from Icmt +/+ cells. Bars shown are mean ± SEM, n = 6.
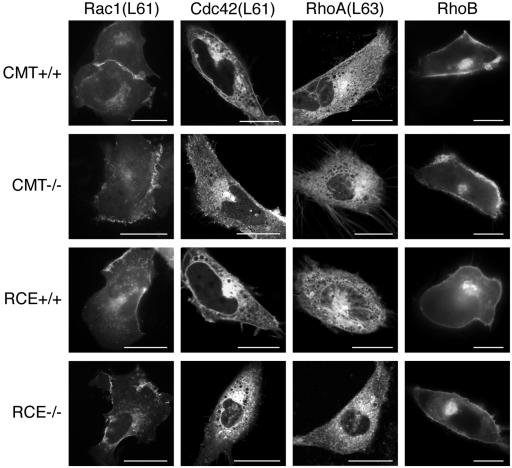
Because the GTP-bound forms of Rac1, Cdc42, and RhoA cannot bind to RhoGDI (Michaelson et al., 2001), we expressed GFP-tagged forms of the constitutively active proteins in MEFs to reveal their membrane targeting. In contrast to Ras proteins, the GFP-tagged, GTP-bound Rac1L61, Cdc42L61, and RhoAL63 proteins localized in Rce1- and Icmt-deficient MEFs in a pattern indistinguishable from that of wild-type MEFs (Figure 3). RhoB is an example of a Rho family GTPase that does not bind RhoGDI. GFP-tagged wild-type RhoB localized as in COS-1 cells (Michaelson et al., 2001) to the PM and Golgi in both wild-type MEFs and cells deficient in either Rce1 or Icmt (Figure 3). Thus, –AAX proteolysis and carboxyl methylation are required neither for the interaction of Rho proteins with RhoGDI nor for the proper membrane targeting of this family of proteins.
Figure 3.
GFP-Rho proteins do not require postprenylation CAAX processing for membrane localization. MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with constitutively active forms of GFP-Rac1, GFP-Cdc42, GFP-RhoA, or wild-type GFP-RhoB (which does not interact with RhoGDI). Constitutively active forms of RhoGDI binding proteins were used to block binding to RhoGDI and thus reveal intrinsic membrane targeting. Cells were imaged 24 h after transfection as described in Materials and Methods. Representative images are shown of >10 acquired on each of more than five independent experiments. Bars, 10 μm.
Carboxyl Methylation Is Required for Efficient Membrane Association of Farnesylated Ras but Not Geranylgeranylated RalA
Because Ras proteins are farnesylated and Rho proteins are geranylgeranylated, our localization results are consistent with the hypothesis that postprenylation processing is required for efficient membrane association only of farnesylated proteins. To test this idea with an independent method that would allow analysis of endogenous GTPases, we used subcellular fractionation. Because the expression of endogenous Ras in MEFs was below the level of detection by immunoblot, we used immunoprecipitation of [35S]methionine-labeled Ras for this analysis. We studied RalA (Figure 4) as a representative of a geranylgeranylated protein because this Ras-related GTPase is known to be strictly geranylgeranylated and does not bind to a cytosolic chaperone such as RhoGDI. Both Ras and RalA were >95% membrane associated in wild-type MEFs. In contrast, 30% of Ras was soluble in Icmt-deficient MEFs consistent with our previous results (Bergo et al., 2000). However, the soluble pool of RalA remained <5% in Icmt-deficient MEFs. Thus, carboxyl methylation is required for efficient membrane association of farnesylated Ras but not geranylgeranylated RalA.
Figure 4.
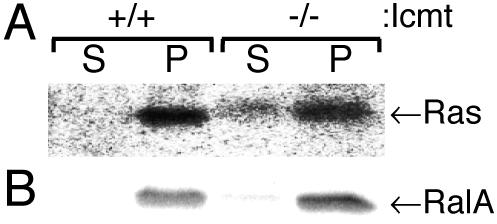
Subcellular fractionation confirms differential requirement for carboxyl methylation on membrane association of endogenous farnesylated versus geranylgeranylated proteins. (A) MEFs expressing (+/+) or deficient (–/–) in Icmt were metabolically labeled with [35S]methionine, disrupted with a Dounce homogenizer, and separated into 100,000 × g supernatants (S) and pellets (P). Fractions were extracted with RIPA buffer, immunoprecipitated with a pan-ras antibody, analyzed by SDS-PAGE, and imaged by PhosphorImager. (B) The same cells without metabolic labeling were used to generate identical S100 and P100 fractions that were analyzed for endogenous RalA by 125I-protein A immunoblots. RalA was chosen as a strictly geranylgeranylated Ras-related GTPase that lacks a cytosolic chaperon and is expressed at sufficient levels to be detected in MEFs. PhosphorImager analysis revealed 4.3 and 4.8% of total RalA and 2 and 30% of total Ras in the S100 fraction in +/+ versus –/– cells.
Rho Proteins Are Substrates for Icmt
We partially purified endogenous Rac2, Cdc42, and RhoA in 1:1 complexes with RhoGDI from the cytosol of human neutrophils and incubated the partially purified proteins with neutrophil membranes in the presence of [3H]AdoMet and in the absence or presence of GTPγS. Each Rho protein incorporated an [3H]methyl group in a GTP-dependent manner (Figure 5A). Thus, endogenous Rac2, Cdc42, and RhoA serve as substrates in vitro for Icmt. The increased efficiency of methylation induced by GTPγS can be attributed to dissociation of the GTP-loaded Rho protein from RhoGDI, thereby making the C terminus accessible for processing (Philips et al., 1993). To determine whether Rac1, Cdc42, and RhoA are methylated in vivo, ECV304 cells stably expressing GFP-tagged Rho family proteins were incubated with [3H]methyl-methionine to metabolically label endogenous pools of AdoMet. GFP-tagged proteins were immunoprecipitated from detergent lysates and analyzed by SDS-PAGE and fluorography: carboxyl methylation was determined by excising the labeled bands and quantifying base-releasable [3H]methanol. Each of the three fusion proteins was methylated in vivo, confirming that these proteins are substrates for Icmt (Figure 5B). To determine whether the GTP-binding state influences the level of methylation in vivo, we cotransfected cells with GFP-tagged Rho proteins and with oncogenic Dbl, a constitutively active exchange factor for Rac1, Cdc42, and RhoA. Although Dbl induced profound ruffling (Figure 6, B and C), confirming its biological activity as a stimulator of Rac1, it had no effect on the level of methylation of Rac1, Cdc42, or RhoA (Figure 5B), suggesting that exchange of GTP for GDP on these proteins does not lead to their methylation in vivo. Together, our data suggest that although Rho GTPases are substrates for Icmt, modification by this enzyme is not required for membrane targeting.
Figure 5.
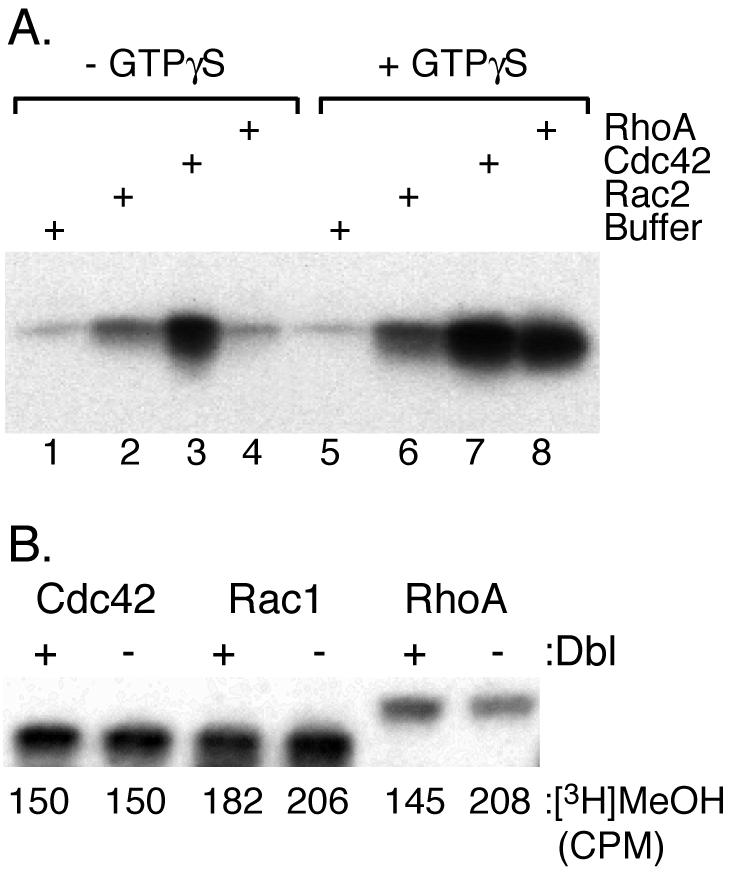
Rho family proteins are substrates for carboxyl methylation. (A) In vitro methylation: endogenous Rac2, Cdc42, and RhoA, partially purified as 1:1 complexes with RhoGDI from human neutrophil cytosol by ion exchange chromatography and gel filtration, were incubated for 30 min with 10 μg of human neutrophil light membranes containing endogenous Icmt and the methyl donor [3H]AdoMet with or without GTPγS, and the reaction products were analyzed by SDS-PAGE and fluorography. (B) In vivo methylation: ECV304 cells stably expressing GFP-Cdc42, GFP-Rac1, or GFP-RhoA were transfected with pCMV or pCMV-Dbl (exchange factor for Rho proteins) and then metabolically labeled with [3H]methyl-methionine (precursor of AdoMet) and [35S]methionine. GFP-tagged proteins were immunoprecipitated with anti-GFP antibodies, analyzed by SDS-PAGE, and visualized as [35S]methionine-labeled proteins by PhosphorImager. Immunoprecipitated proteins were excised from the gel and analyzed for carboxyl methylation by alkaline hydrolysis. Numbers below the bands indicate cpm for base-releasable [3H]methanol (background 25 cpm) The experiment shown is representative of four independent experiments.
Neither Rce1 nor Icmt Is Required for Rac1 and Cdc42 Function
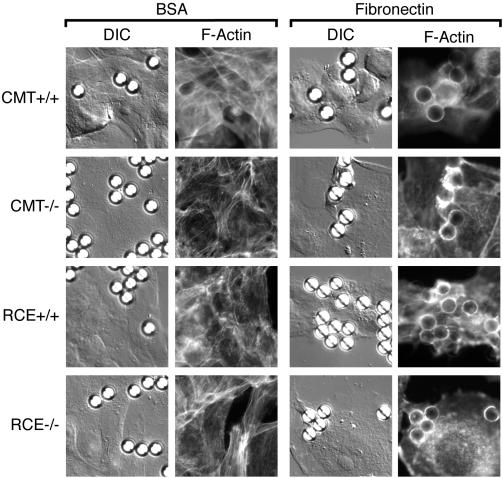
We studied the ability of constitutively active forms of Rac1 and Cdc42 to induce characteristic actin remodeling in wild-type MEFs and MEFs lacking Rce1 or Icmt (Figure 6A). GFP-Rac1L61 induced ruffling to similar degrees in wild-type, Rce1-, and Icmt-deficient cells. Similarly, GFP-Cdc42L61 stimulated filopodia in all three cell types. To confirm the capacity of wild-type Rac1 to regulate actin remodeling in the absence of postprenylation processing, we cotransfected MEFs with GFP-Rac1 and Dbl. In both wild-type MEFs and MEFs lacking Rce1 or Icmt, oncogenic Dbl induced profound membrane ruffling and translocation of GFP-Rac1 to the plasma membrane in the region of the ruffles (Figure 6B). To confirm stimulated actin remodeling in these cells in the absence of overexpression of Rho proteins, we used an assay in which fibronectin-coated beads stimulate local circumferential F-actin formation as a consequence of signaling through β1 integrins (Miyamoto et al., 1995). Bovine serum albumin (BSA)-coated control beads that settled on MEFs of each genotype induced no F-actin formation. In contrast, fibronectin-coated beads adherent to the cells induced characteristic rings of F-actin in each cell type (Figure 7). This result indicates that the control of actin polymerization downstream of integrin engagement, a process dependent on Rho proteins (Kjoller and Hall, 1999), requires neither –AAX proteolysis nor prenylcysteine carboxyl methylation.
Figure 7.
Postprenylation processing is not required for integrin-induced actin remodeling. MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were incubated with BSA- or fibronectin-coated 1-μm latex beads and then paraformaldehyde fixed, saponin permeabilized, and stained with Texas Red-phalloidin. Subplasmalemmal F-actin formation was visualized by wide-field epifluorescence microscopy and scored as a ring of fluorescence surrounding fibronectin- but not BSA-coated beads. The positions of the beads were revealed by differential interference contrast microscopy.
Rce1 and Icmt Are Required for Membrane Targeting of Farnesylated but Not Geranylgeranlyated GTPases
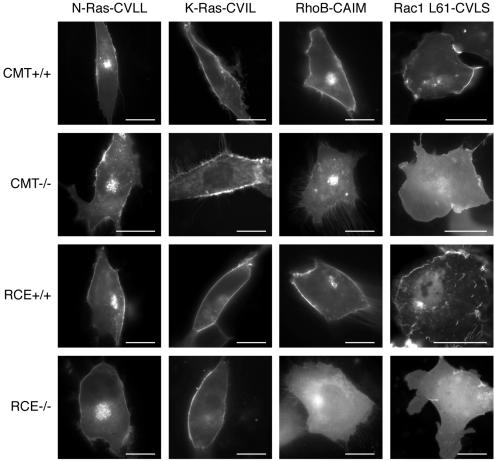
Ras proteins terminating in a CAAS/M sequence are farnesylated, whereas most Rho proteins terminate in a CAAL sequence and therefore are geranylgeranylated. The impact of postprenylation processing on the Ras and Rho proteins were very different, raising the possibility that the length of the isoprenyl lipid might determine whether endoproteolysis and carboxyl methylation are necessary for membrane targeting. To test this hypothesis, we constructed GFP-tagged Rac1, RhoB, N-Ras, and K-Ras proteins with CAAX motif mutations expected to switch the chain length of the isoprenyl modification (i.e., farnesylation for geranylgeranylation or vice versa) and then studied their subcellular localization in wild-type MEFs and MEFs lacking Rce1 or Icmt. Geranylgeranylated GFP-N-Ras-CVLL and GFP-K-Ras-CVIL localized in wild-type MEFs in a pattern indistinguishable from their farnesylated forms (Figures 1 and 8). However, whereas farnesylated GFP-N-Ras and GFP-K-Ras were mislocalized to the cytosol in the absence of endoproteolysis or methylation (Figure 1), geranylgeranylated GFP-N-Ras-CVLL and GFP-K-Ras-CVIL were not (Figure 8). Similarly, a switch in prenylation of Rac1L61 and RhoB did not affect subcellular localization in wild-type MEFs (Figure 8) or the ability of Rac1 to bind RhoGDI (our unpublished data). However, switching the isoprenyl chain length of Rac1 and RhoB had an effect opposite to that observed for Ras proteins: farnesylated GFP-Rac1L61-CVLS and GFP-RhoB-CAIM became sensitive to the absence of Rce1 and Icmt (Figure 8). Thus, –AAX proteolysis and carboxyl methylation are required for efficient membrane targeting but only for farnesylated GTPases.
Figure 8.
A prenyl swap reverses the differential sensitivity of Ras and Rho proteins to mislocalization in Icmt- and Rce1-deficient cells. MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with GFP-tagged Ras and Rho proteins with mutations in their CAAX sequences that direct alternative prenylation (i.e., geranylgeranylation of Ras proteins and farnesylation of Rho proteins). These constructs included GFP-N-Ras-CVLL, GFP-K-Ras-CVIL, GFP-RhoB-CAIM, and GFP-Rac1L61-CVLS.
Farnesylated but Not Geranylgeranylated Rac1 Requires –AAX Proteolysis and Carboxyl Methylation to Regulate Membrane Ruffling
To determine whether mislocalization of farnesylated Rac1 in cells lacking Icmt or Rce1 would result in impaired function, we assessed the ability of activated farnesyl-Rac1 to induce ruffles in wild-type MEFs and Rce1- and Icmt-deficient MEFs. GFP-Rac1L61-CVLS induced ruffling as revealed by phalloidin staining in wild-type MEFs but not in Rce1- or Icmt-deficient cells (Figure 9A). When cotransfected with oncogenic Dbl, GFP-Rac1-CVLS translocated to the ruffled plasma membrane in wild-type MEFs but not in Rce1- or Icmt-deficient MEFs (Figure 9B). In addition, whereas oncogenic Dbl increased significantly the extent of ruffling in wild-type MEFs expressing Rac1-CVLS, this effect was greatly diminished in Icmt- and Rce1-deficient cells expressing the same construct (Figure 9C). These data suggest that –AAX proteolysis and carboxyl methylation are required for Rac1-mediated lamellipodia formation only under conditions where the prenyl modification is a farnesyl lipid.
Figure 9.
Farnesylated Rac1 requires complete CAAX processing to regulate actin remodeling. (A) MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with GFP-Rac161L-CVLS, a constitutively active, farnesylated form of the GTPase. Twenty-four hours after expression, the cells were analyzed for lamellipodia formation as in Figure 6. (B) MEFs expressing (+/+) or deficient (–/–) in Icmt or Rce1 were transfected with GFP-Rac1 with or without oncogenic Dbl to induce ruffling. (C) Prevalence of ruffling shown in A and B among 50 randomly chosen, transfected cells. Bars, 10 μm. Graphs show mean ± SEM, n = 3.
DISCUSSION
The independent contributions to membrane targeting and protein function of each of the three sequential steps in CAAX processing have proven difficult to elucidate because of their interdependence. To address the importance of postprenylation processing, we examined the properties of specific CAAX proteins in Rce1–/– and Icmt–/– MEFs. We have previously found that K-Ras was mislocalized to the cytosol in both Rce1- (Kim et al., 1999; Bergo et al., 2002) and Icmt-deficient (Bergo et al., 2000) cells. In the case of the Rce1-deficient cells, the mislocalization of K-Ras was associated with reduced susceptibility to K-Ras–induced oncogenic transformation (Bergo et al., 2004). Similarly, we observed resistance to K-Ras–mediated transformation in Icmtflox/flox MEFs when the Icmt gene was inactivated with Cre expression (Bergo et al., 2004). Because K-Ras differs from N-Ras and H-Ras in lacking palmitoylation and because Ras proteins differ from many other small GTPases, including most Rho family proteins, in being farnesylated rather than geranylgeranylated, we considered it essential to determine whether the effects observed for K-Ras applied equally to other members of the Ras superfamily. In the present study, we have tested the role of postprenylation processing on the other Ras isoforms and on three geranylgeranylated members of the Rho family of GTPases. We found that although all three Ras isoforms (K-Ras, N-Ras, and H-Ras) required postprenylation processing for proper localization, this was not the case for four Rho family GTPases. Moreover, the ability of Rac1 to bind RhoGDI and of Rac1 and Cdc42 to regulate actin remodeling was preserved in cells deficient in Rce1 and Icmt. Prenyl swap experiments revealed that the resistance of Rho proteins to mislocalization and loss-of-function in cells lacking postprenylation processing could be attributed entirely to the nature of the polyisoprene modification. Thus, our data suggest that –AAX proteolysis and carboxyl methylation are required for farnesylated Ras proteins but not closely related Rho proteins that are geranylgeranylated.
Our results with small GTPases are consistent with an earlier study in which we analyzed the importance of postprenylation processing on heterotrimeric G proteins. In that study, we found that farnesylated Gγ1 was mislocalized in Icmt–/– MEFs, but geranylgeranylated Gγ2 was not (Michaelson et al., 2002). Our results also are consistent with studies of the ability of farnesylated peptides to partition into liposomes (Silvius and l'Heureux, 1994). In the latter studies, although carboxyl methylation enhanced slightly the lipid partition of geranylgeranylated peptides, the effect was trivial because unmethylated, geranylgeranylated peptides partitioned into the liposomes with high efficiency. In contrast, farnesylated peptides were dependent on carboxyl methylation for efficient partitioning into liposomes. Similarly, methylation has been reported to enhance the membrane affinity of in vitro-translated K-Ras (Hancock et al., 1991a).
The ability of RhoGDI to bind Rho GTPases in Rce1–/– and Icmt–/– cells has implications for the trafficking of nascent Rho proteins. GGTase I is cytosolic, whereas Rce1 and Icmt are associated with the ER. Thus, nascent geranylgeranylated Rho proteins must be transferred from the aqueous environment of the cytosol to the membrane environment of the ER for further processing. If the established cytosolic chaperone RhoGDI were unable to bind geranylgeranylated Rho proteins that were not processed beyond prenylation, then either the geranylgeranylated proteins would have to transit the aqueous environment of the cytosol with the isoprenoid exposed or an alternative chaperone would be required. Recent evidence has pointed to a surprising candidate for such a chaper-one—GGTase I itself. Structural analysis of this enzyme has revealed an unusual reaction mechanism wherein the product, the geranylgeranylated GTPase, remains associated with the transferase until the next cycle of substrate binding causes it to be dislodged (Taylor et al., 2003). Thus, if substrate binding were somehow linked to docking at the ER, GGTase I could serve as the required chaperone. However, our observation that RhoGDI binds geranylgeranylated Rho proteins that retain their –AAX sequence demonstrates that no alternative chaperone is required for nascent Rho proteins. GGTase I can transfer the geranylgeranylated protein directly to RhoGDI. Such a model is supported by the homology between RhoGDI and the Rab escort protein that serves such a function for geranylgeranyltransferase type II (Andres et al., 1993; Alexandrov et al., 1994). However, for Rho proteins that do not bind RhoGDI, such as RhoB, GGTase I as a cytosolic chaperone remains an attractive hypothesis.
If RhoGDI serves as the cytosolic carrier protein for nascent geranylgeranylated Rho proteins before –AAX proteolysis and carboxyl methylation, then what accounts for the release of the partially processed proteins at the ER? We have shown that Rho proteins that bind RhoGDI lose their affinity for the chaperone in the GTP-bound state (Michaelson et al., 2001). The increased carboxyl methylation of Rho GTPases that we observed in vitro in the presence of GTPγS is consistent with a model in which GTP exchange promotes dissociation from RhoGDI and delivery of the intermediate to Rce1 and then Icmt. The association of fully processed Rho GTPases with RhoGDI would then occur only after release of the proteins from the endomembrane after modification. Although extraction of Rho proteins from membranes is a well established function of RhoGDI, GDP-bound Rho proteins are preferred (Isomura et al., 1991). If it is the GTP-bound form of the nascent geranylgeranylated protein that is delivered to the endomembrane for –AAX proteolysis and carboxyl methylation, but a GDP-bound form that is extracted, then a GAP is likely to operate at the ER. The model thus predicts a round of GTP binding and hydrolysis associated with postprenylation processing of Rho proteins.
The observation that membrane targeting of Rho GTPases and their ability to regulate the actin cytoskeleton do not depend on postprenylation processing leads to the fundamental question of why geranylgeranylated proteins such as Rho GTPases serve as substrates for Rce1 and Icmt. Evidence for the processing state of endogenous Rho proteins is lacking. Our observation that endogenous RhoA, Rac2, and Cdc42 from human neutrophils serve as substrates for Icmt confirms that at least a portion of the pool of endogenous proteins are not fully processed. Carboxyl methylation is energetically costly, and its evolutionary conservation suggests that it serves a critical function. Our data support the hypothesis that its critical role involves farnesylated rather than geranygeranylated proteins. However, the biological processes controlled by Rho GTPases are numerous. Here, we have tested only the regulation of actin remodeling. Perhaps other functions of Rho proteins will be found to require postprenylation processing.
Why did two forms of protein prenylation evolve? Our data suggest that the answer lies in the relative reversibility of membrane targeting. The ability of the farnesyl modification to mediate membrane association is regulated by carboxyl methylation, but the membrane targeting capacity of a geranylgeranyl group is not. Carboxyl methylation, unlike prenylation, is readily reversible under physiological conditions. This suggests that farnesylation evolved as a relatively low affinity and rapidly reversible membrane-targeting modification, whereas geranylgeranylation evolved as a more permanent anchor requiring molecular chaperones for extraction from membranes. There is abundant evidence for cytosolic pools of farnesylated Ras proteins (Choy et al., 1999) and transducin (Fukada et al., 1994; Matsuda et al., 1994). One obvious function of the rapid reversal of membrane association of farnesylated proteins is the ability to rapidly attenuate signaling. Other functions of cytosolic farnesylated proteins may emerge.
Our observations also have practical application in the ongoing quest to develop anticancer drugs that target the pathway through which Ras gains access to membranes.
Although farnesyltransferase inhibitors (FTIs) are specific for farnesylated proteins such as Ras and therefore expected to avoid Rho-mediated toxicities, inhibitors of Rce1 and Icmt might be expected to have more toxicity because these enzymes can modify all CAAX proteins. However, our observations suggest that Rho proteins might function normally under conditions where Rce1 and/or Icmt are blocked, making these more attractive targets for drugs designed to work synergistically with FTIs. On the other hand, because the major limitation of FTIs is alternative prenylation (i.e., modification by GGTase I) of K-Ras and N-Ras under conditions of farnesyltransferase inhibition (Whyte et al., 1997), our data suggest that addition of an Rce1 or Icmt inhibitor might not overcome the effects of alternative prenylation.
Acknowledgments
This work is supported by grants from the National Institutes of Health (to M.R.P. and S. Y.), the Burroughs Wellcome Foundation (to M.R.P), the University of California Tobacco-Related Disease Research Program (to M. B and S. Y), the Swedish Cancer Foundation (to M. B), and a General Clinical Research Center grant from National Institutes of Health, National Center for Research Resources (M01RR00096).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–11–0960) on January 19, 2005.
References
- Alexandrov, K., Horiuchi, H., Steele-Mortimer, O., Seabra, M. C., and Zerial, M. (1994). Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated Rab proteins to their target membranes. EMBO J. 13, 5262–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, D. A., Seabra, M. C., Brown, M. S., Armstrong, S. A., Smeland, T. E., Cremers, F. P., and Goldstein, J. L. (1993). cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73, 1091–1099. [DOI] [PubMed] [Google Scholar]
- Bergo, M. O., Ambroziak, P., Gregory, C., George, A., Otto, J. C., Kim, E., Nagase, H., Casey, P. J., Balmain, A., and Young, S. G. (2002). Absence of the CAAX endoprotease Rce 1, effects on cell growth and transformation. Mol. Cell. Biol. 22, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo, M. O., Gavino, B. J., Hong, C., Beigneux, A. P., McMahon, M., Casey, P. J., and Young, S. G. (2004). Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J. Clin. Investig. 113, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo, M. O., Leung, G. K., Ambroziak, P., Otto, J. C., Casey, P. J., Gomes, A. Q., Seabra, M. C., and Young, S. G. (2001). Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 276, 5841–5845. [DOI] [PubMed] [Google Scholar]
- Bergo, M. O., Leung, G. K., Ambroziak, P., Otto, J. C., Casey, P. J., and Young, S. G. (2000). Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J. Biol. Chem. 275, 17605–17610. [DOI] [PubMed] [Google Scholar]
- Bivona, T. G., Perez De Castro, I., Ahearn, I. M., Grana, T. M., Chiu, V. K., Lockyer, P. J., Cullen, P. J., Pellicer, A., Cox, A. D., and Philips, M. R. (2003). Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424, 694–698. [DOI] [PubMed] [Google Scholar]
- Boyartchuk, V. L., Ashby, M. N., and Rine, J. (1997). Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275, 1796–1800. [DOI] [PubMed] [Google Scholar]
- Cadwallader, K. A., Paterson, H., Macdonald, S. G., and Hancock, J. F. (1994). N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol. Cell. Biol. 14, 4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, P. J., and Seabra, M. C. (1996). Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292. [DOI] [PubMed] [Google Scholar]
- Choy, E., Chiu, V. K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I. E., and Philips, M. R. (1999). Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98, 69–80. [DOI] [PubMed] [Google Scholar]
- Choy, E., and Philips, M. R. (2000). Expression and activity of human prenylcysteine-directed carboxyl methyltransferase. In: Regulators and Effectors of Small GTPases, Part D: Rho Family. Methods in Enzymology, vol. 325, ed. W. E. Balch, C. J. Der, and Alan Hall, San Diego, CA: Academic Press, 101–114. [DOI] [PubMed] [Google Scholar]
- Clarke, S. (1992). Protein isoprenylation and methylation at carboxyl terminal cysteine residues. Annu. Rev. Biochem. 61, 355–386. [DOI] [PubMed] [Google Scholar]
- Dai, Q., Choy, E., Chiu, V., Romano, J., Slivka, S., Steitz, S., Michaelis, S., and Philips, M. R. (1998). Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 273, 15030–15034. [DOI] [PubMed] [Google Scholar]
- Fukada, Y., Matsuda, T., Kokame, K., Takao, T., Shimonishi, Y., Akino, T., and Yoshizawa, T. (1994). Effects of carboxyl methylation of photoreceptor G protein g-subunit in visual transduction. J. Biol. Chem. 269, 5163–5170. [PubMed] [Google Scholar]
- Hancock, J. F., Cadwallader, K., and Marshall, C. J. (1991a). Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B). EMBO J. 10, 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J. F., Cadwallader, K., Paterson, H., and Marshall, C. J. (1991b). A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10, 4033–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J. F., Paterson, H., and Marshall, C. J. (1990). A polybasic domain or palmitoylation is required in addition to the CAAX motif to loacalize p21ras to the plasma membrane. Cell 63, 133–139. [DOI] [PubMed] [Google Scholar]
- Isomura, M., Kikuchi, A., Ohga, N., and Takai, Y. (1991). Regulation of binding of rhoB p20 to membranes by its specific regulatory protein, GDP dissociation inhibitor. Oncogene 6, 119–124. [PubMed] [Google Scholar]
- Kim, E., Ambroziak, P., Otto, J. C., Taylor, B., Ashby, M., Shannon, K., Casey, P. J., and Young, S. G. (1999). Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J. Biol. Chem. 274, 8383–8390. [DOI] [PubMed] [Google Scholar]
- Kjoller, L., and Hall, A. (1999). Signaling to Rho GTPases. Exp. Cell Res. 253, 166–179. [DOI] [PubMed] [Google Scholar]
- Matozaki, T., Nakanishi, H., and Takai, Y. (2000). Small G-protein networks: their crosstalk and signal cascades. Cell Signal. 12, 515–524. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., Takao, T., Shimonishi, Y., Murata, M., Asano, T., Yoshizawa, T., and Fukada, Y. (1994). Characterization of interactions between transducin alpha/beta gamma-subunits and lipid membranes. J. Biol. Chem. 269, 30358–30363. [PubMed] [Google Scholar]
- Michaelson, D., Ahearn, I., Bergo, M., Young, S., and Philips, M. (2002). Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol. Biol. Cell. 13, 3294–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson, D., Silletti, J., Murphy, G., D'Eustachio, P., Rush, M., and Philips, M. R. (2001). Differential localization of Rho GTPases in live cells. Regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. A., Farh, L., Marshall, T. K., and Deschenes, R. J. (1994). A polybasic domain allows nonprenylated Ras proteins to function in Saccharomyces cerevisiae. J. Biol. Chem. 269, 21540–21546. [PubMed] [Google Scholar]
- Miyamoto, S., Akiyama, S. K., and Yamada, K. M. (1995). Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science 267, 883–885. [DOI] [PubMed] [Google Scholar]
- Moores, S. L., Schaber, M. D., Mosser, S. D., Rands, E., O'Hara, M. B., Garsky, V. M., Marshall, M. S., Pompliano, D. L., and Gibbs, J. B. (1991). Sequence dependence of protein isoprenylation. J. Biol. Chem. 266, 14603–14610. [PubMed] [Google Scholar]
- Philips, M. R., Abramson, S. B., Kolasinski, S. L., Haines, K. A., Weissmann, G., and Rosenfeld, M. G. (1991). Low molecular weight GTP-binding proteins in human neutrophil granule membranes. J. Biol. Chem. 266, 1289–1298. [PubMed] [Google Scholar]
- Philips, M. R., Pillinger, M. H., Staud, R., Volker, C., Rosenfeld, M. G., Weissmann, G., and Stock, J. B. (1993). Carboxyl methylation of ras-related proteins during signal transduction in neutrophils. Science 259, 977–980. [DOI] [PubMed] [Google Scholar]
- Reiss, Y., Stradley, S. J., Gierasch, L. M., Brown, M. S., and Goldstein, J. L. (1991). Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc. Natl. Acad. Sci. USA 88, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, W. K., Tam, A., Fujimura-Kamada, K., and Michaelis, S. (1998). Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 95, 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius, J. R., and l'Heureux, F. (1994). Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33, 3014–3022. [DOI] [PubMed] [Google Scholar]
- Taylor, J. S., Reid, T. S., Terry, K. L., Casey, P. J., and Beese, L. S. (2003). Structure of mammalian protein geranylgeranyltransferase type-1. EMBO J. 22, 5963–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte, D. B., Kirschmeier, P., Hockenberry, T. N., Nunez-Oliva, I., James, L., Catino, J. J., Bishop, W. R., and Pai, J. K. (1997). K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272, 14459–14464. [DOI] [PubMed] [Google Scholar]