Abstract
The tumor suppressor protein p53 mediates stress-induced growth arrest or apoptosis and plays a major role in safeguarding genome integrity. In response to DNA damage, p53 can be modified at multiple sites by phosphorylation and acetylation. We report on the characterization of p53 C-terminal phosphorylation by CHK1 and CHK2, two serine/threonine (Ser/Thr) protein kinases, previously implicated in the phosphorylation of the p53 N terminus. Using tryptic phosphopeptide mapping, we have identified six additional CHK1 and CHK2 sites residing in the final 100 amino acids of p53. Phosphorylation of at least three of these sites, Ser366, Ser378, and Thr387, was induced by DNA damage, and the induction at Ser366 and Thr387 was abrogated by small interfering RNA targeting chk1 and chk2. Furthermore, mutation of these phosphorylation sites has a different impact on p53 C-terminal acetylation and on the activation of p53-targeted promoters. Our results demonstrate a possible interplay between p53 C-terminal phosphorylation and acetylation, and they provide an additional mechanism for the control of the activity of p53 by CHK1 and CHK2.
INTRODUCTION
The protein p53 is often referred to as a “tumor suppressor,” because it is frequently mutated in >50% of human cancers (Olivier et al., 2002). In response to stress, p53 undergoes extensive posttranslational modification, including phosphorylation and acetylation (Appella and Anderson, 2001; Brooks and Gu, 2003; Xu, 2003). Consequently, p53 is stabilized and activated (Vousden, 2002). As a transcription factor, p53 is composed of an N-terminal activation domain, a central specific DNA binding domain, and a C-terminal tetramerization domain, followed by a regulatory domain rich in basic amino acids (Ko and Prives, 1996). On activation, p53 binds to an array of gene promoters, some of which, such as p21/waf1, are responsible for stress-induced cell cycle arrest, whereas others, such as bax and puma, are responsible for driving cells into apoptosis (Vousden and Lu, 2002; Harms et al., 2004).
Phosphorylation of p53 mostly occurs in the N-terminal activation domain at the Ser6, Ser9, Ser15, Thr18, Ser20, Ser33, Ser37, Ser46, Thr55, and Thr81 residues, with some phosphorylation occurring in the C-terminal linker and basic regions at Ser315, Ser371, Ser376, Ser378, and Ser392. Phosphorylation on most of these sites is induced by DNA damage, with some, such as Thr55 and Ser376, being repressed upon genotoxic stress (Appella and Anderson, 2001; Holmberg et al., 2002). How these individual residues contribute to p53 stabilization and activation is still not fully understood, and, at times, has been the subject of debate. Many protein kinases have been implicated in phosphorylating p53 (Holmberg et al., 2002). Notably, phosphorylation at Ser15 by ATM/ATR, either directly or through CHK1/CHK2, or at Ser20 by CHK1/CHK2 has been shown to alleviate the inhibition or degradation of p53 by Mdm2, leading to p53 stabilization and activation (Shieh et al., 1997, 2000; Banin et al., 1998; Canman et al., 1998; Khanna et al., 1998; Tibbetts et al., 1999; Unger et al., 1999; Chehab et al., 2000).
Interestingly, introduction of Ala mutations at Ser18 or Ser23, the mouse equivalents of Ser15 and Ser20, respectively, into the endogenous p53 gene of murine embryonic fibroblasts showed no effect on p53 accumulation upon genotoxic assault (Wu et al., 2002; Sluss et al., 2004). Phosphorylation at Ser46 has been shown to be essential for the activation of apoptosis-related genes (Oda et al., 2000). Phosphorylation at Ser33, Ser46, and Thr81 also has been suggested to lead to p53 stabilization (Bulavin et al., 1999; Bushmann et al., 2001). It is very likely that phosphorylation-induced p53 stabilization and activation are mediated through multiple mechanisms and may vary according to the cellular context or microenvironment.
Another important modification of p53, acetylation, is also of great interest. p53 is specifically acetylated at Lys370, Lys372, Lys373, Lys381, and Lys382 by p300/CBP, and at Lys320 by p300/CREB-binding protein-associated factor (PCAF) (Gu and Roeder, 1997; Sakaguchi et al., 1998; Liu et al., 1999; Ito et al., 2001). Acetylation has been shown to augment p53 DNA binding, and to stimulate p53-mediated transactivation of its downstream target genes through the recruitment of coactivators (Gu and Roeder, 1997; Barlev et al., 2001). Acetylation also may regulate the stability of p53 by inhibiting its ubiquitination by MDM2 (Li et al., 2002). In vivo, acetylation at Lys320, Lys373, and Lys382 is induced by many genotoxic agents, including UV-radiation, ionizing radiation (IR), hypoxia, oxidative stress, and even depletion of ribonucleotide pools (Ito et al., 2001). The observation that both acetylation and phosphorylation are induced by DNA-damaging agents has led to the speculation that these modification events may be interdependent. Indeed, phosphorylation at the p53 N terminus has been shown to enhance its interaction with acetylase p300/CBP and to potentiate p53 acetylation (Lambert et al., 1998; Sakaguchi et al., 1998; Kar et al., 2002).
The mammalian cell cycle checkpoint kinases CHK1 and CHK2 are structurally distinct, but functionally overlapping, Ser/Thr kinases that are activated by a variety of genotoxic assaults (Rhind and Russell, 2000; Bartek and Lukas, 2003). Most notably, these kinases phosphorylate and inactivate Cdc25, leading to the inactivation of cyclin-dependent kinases (cdks) and cell cycle arrest (Rhind and Russell, 2000). On activation, both of these kinases also phosphorylate multiple sites in the p53 N-terminal domain. These include Ser15, Thr18, Ser20, and Ser37, which are all DNA-damage–inducible sites (Chehab et al., 2000; Hirao et al., 2000; Shieh et al., 2000).
In addition to the N terminus, the C terminus also has been shown to be heavily phosphorylated on unidentified sites by CHK1 and CHK2 without any clear functions being assigned (Shieh et al., 2000). Overexpression of CHK1 enhances p53 accumulation, whereas expression of a CHK1 antisense construct reduces it (Shieh et al., 2000). Similarly, overexpression of kinase-inactive CHK2 abrogates p53 response and Ser20 phosphorylation (Chehab et al., 2000). However, results from recent studies indicate that the regulation by CHK1 and CHK2 may be more complex than was previously expected. Disruption of CHK2 expression by homologous recombination has no effect on either p53 Ser20 phosphorylation or the p53 response (Jallepalli et al., 2003). Furthermore, down-regulation of CHK1 and CHK2 by small interfering RNA (siRNA) did not produce any discernible effect on the induction of p53 or its downstream targets (Ahn et al., 2003). To better define the roles of CHK1 and CHK2, we have attempted to characterize their phosphorylation sites on the p53 C-terminal domain. Our study identified six Ser/Thr residues in this region as potential targets of CHK1 and/or CHK2 phosphorylation. Using phospho-specific antibodies, we have demonstrated that at least three of these residues were DNA-damage–inducible sites. Phosphorylation of these sites was induced by various DNA-damaging agents having kinetics distinct from the N-terminal phosphorylation. Small interfering RNA-mediated knockdown of CHK1 and CHK2 abrogated phosphorylation on two of these sites and resulted in reduced induction of two p53 downstream targets, p21 and Bax, suggesting that these are the genuine in vivo CHK1 and CHK2 sites. Furthermore, mutation of these sites to nonphosphorylatable Ala, or to Asp, to mimic constitutive phosphorylation, had different impacts on p53 C-terminal acetylation, implying that interplay may exist between the two modifications.
MATERIALS AND METHODS
Cell Culture and DNA Damage Treatment
The human prostate carcinoma cell line LNCaP and human nonsmall-cell lung cancer cell line H1299 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) plus antibiotics. The human osteosarcoma cell lines U2OS (p53 wild type) and Saos2 (p53 null) were cultured in DMEM plus 10% FBS and antibiotics. A transformed human embryonic kidney cell line, 293T, was maintained in DMEM supplemented with 10% equine serum and antibiotics. The cells were irradiated at the exponential phase by using x-rays generated by a Torrex 150D inspection system (PerkinElmer Life and Analytical Sciences, Boston, MA) or with UV radiation in a UV cross-linker (Spectronics, Westbury, NY). Camptothecin, a DNA topoisomerase I inhibitor that generates DNA double-strand breaks was used at a final concentration of 0.5 μM.
Generation of Phospho-specific Antibodies
The phospho-Ser366-specific, phospho-Ser378-specific, and phospho-Thr387-specific antisera were raised against chemically synthesized, eight-branch phosphopeptides GSRAH-S(PO3)-SHLKSK, KKGQST-S(PO3)-RHKKL, and KLMFK-T(PO3)-EGPDSD, respectively, by LTK Biolaboratories (Taipei, Taiwan). The antisera were further incubated with glutathione S-transferase (GST)-p53 (amino acid 361–393) binding to glutathione-Sepharose beads (Amersham Biosciences, Piscataway, NJ) to remove antibodies that could recognize unphosphorylated epitopes.
Cell Lysis, Immunoprecipitation with Anti-Phospho-specific Antibodies, and Western Blotting
Cell pellets were resuspended in TEGN buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 420 mM NaCl, 10% glycerol, and 0.5% NP-40) containing a cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN), 10 mM NaF, 10 mM β-glycerophosphate, 5 nM microcystin LR, and 1 mM dithiothreitol (DTT), incubated on ice for 15 min, and clarified using a 10-min spin in a Microfuge. The lysates were diluted with an equal volume of TEG buffer (10 mM Tris, pH 7.5, 1 mM EDTA, and 20% glycerol) and rocked with 15 μl of 50% protein G beads (Pierce Chemical, Rockford, IL) and the phospho-specific antibody for 2–3 h at 4°C. The proteins bound to the beads were washed three times in 10S buffer (50 mM HEPES, pH 7.9, 250 mM NaCl, 0.2% NP-40, 0.1% Triton X-100, and 0.01% SDS), boiled in protein sample buffer (2 M β-mercaptoethanol, 12% SDS, 0.5 M Tris, pH 6.8, 0.5 mg/ml bromphenol blue, and 30% glycerol), and then resolved using SDS-PAGE. For immunoblotting, the proteins were transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The p53 on the blots was detected using PAb1801 or PAb1801 in combination with DO1, and the signals were visualized with chemiluminescent reagents (Pierce Chemical). Other antibodies used were anti-CHK1 (Santa Cruz Biotechnology, Santa Crux, CA), anti-CHK2 (MBL), anti-p21 (Oncogene Science, Cambridge, MA), anti-hemagglutinin (HA) (Covance, Berkeley, CA), anti-His (Santa Cruz Biotechnology), anti-actin (Sigma-Aldrich, St. Louis, MO), anti-Acetyl-K382 (Oncogene Science), anti-Acetyl-K320 (Upstate Biotechnology, Lake Placid, NY), anti-Acetyl-K373 (Upstate Biotechnology), anti-phospho-p53 (Ser15) (Cell Signaling Technology, Beverly, MA), anti-p53 (pSer15) (U.S. Biochemical, Cleveland, OH), and anti-p53 (pSer20) (U.S. Biochemical).
Plasmids
To make His-tagged p53 protein, DNA encoding p53 was cloned into the pRSET B vector (Invitrogen, Carlsbad, CA) between the NcoI site and the HindIII site. All the p53 mutants were generated by polymerase chain reaction (PCR)-based site-directed mutagenesis and confirmed by DNA sequencing. These constructs were then expressed in bacteria, and the expressed proteins were purified using Ni-nitrilotriacetic acid agarose (QIAGEN, Valencia, CA).
For mammalian expression, the DNA encoding p53 was cut from the pRSET vectors with BamHI and HindIII and recloned into the pcDNA3.1 vector (Invitrogen) between the BamHI site and the EcoRV site.
To make the bax-luc reporter for the luciferase reporter assays, the oligonucleotide carrying one copy of the p53 binding site in the bax promoter was chemically synthesized and cloned into the SmaI site of the pGL3-promoter vector (Promega, Madison, WI). The p21-luc reporter has been described previously (Di Como et al., 1999). The 2 × AIP1-luc reporter was kindly provided by Dr. Y. Taya (National Cancer Center Research Institute, Tokyo, Japan).
In Vitro Kinase Assays and Phosphopeptide Mapping
Kinase assays were performed with purified His-p53, His-CHK2, and GST-CHK1 by using essentially the same method as described previously (Shieh et al., 2000). Phosphopeptide mapping was performed as described by Boyle et al. (1991). Briefly, the 32P-labeled phosphorylated products were gel purified, digested with N-tosyl-l-phenylalanine chloromethyl ketone-treated trypsin, and analyzed using electrophoresis on the first dimension in a pH 1.9 buffer, followed by ascending thin layer chromatography on the second dimension in phospho-chromatography buffer. The phosphopeptide was then detected using autoradiography.
Immunoprecipitation (IP)/Kinase Assay
For IP/kinase assays, CHK1 or CHK2 was first immunoprecipitated from LNCaP cell lysates with anti-CHK1 (sc-7898; Santa Cruz Biotechnology) or anti-CHK2 (Medical and Biological Laboratories, Nagoya, Japan), washed twice in 1:1 TEG/TEGN buffer, once in 1:1 TEG/TEGN buffer containing 0.7 M LiCl, and twice in kinase buffer. Kinase assay was then performed as described above by using GST-Cdc25C or His-p53 as substrate.
In Vitro Interaction of His-p53 with p300
To examine the interaction between p300 and p53, 293T cells were transiently transfected with a plasmid expressing HA-tagged p300. The HA-p300 protein was immunoprecipitated from the cell lysate with the monoclonal antibody (mAb) 12CA5. After three washes with the 10S buffer, the immunoprecipitates were then incubated with 400 ng of His-tagged, purified, recombinant p53 at 30°C for 30 min. The reaction was then washed three times with 10S and analyzed using SDS-PAGE followed by Western blotting by using anti-His (Santa Cruz Biotechnology) to detect the bound p53 proteins.
siRNA Transfection
Transfection of the siRNA was performed as described previously (Elbashir et al., 2001) by using Oligofectamine (Invitrogen) and siRNA at a final concentration of 0.15–0.2 μM. The cells were collected 1 d after transfection. The sequences targeted by chk1 and chk2 siRNA were 5′-ggtgcctatggagaagttc-3′ and 5′-aatgtgtgaatgacaactact-3′, respectively.
Northern Analysis
The total RNA was extracted from LNCaP and U2OS cells by using TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. Fifteen micrograms of total RNA was first denatured in 1.2 M glyoxal and then electrophoresed in 1× BTPE buffer (10 mM PIPES, 30 mM Bis-Tris, and 1 mM EDTA), blotted onto Nytran nylon membrane (Schleicher & Schuell). The p21 and bax cDNA probes were amplified from the LNCaP cDNA library by using the following primers: p21, 5′-ATGTCAGAACCGGCTGGGGA-3′ and 5′-TGCAGCAGAGCAGGTGAGGT-3′; and bax, 5′-GCCCTTTTGCTTCAGGGTTT-3′ and 5′-TCCAATGTCCAGCCCATGAT-3′. The purified PCR products and chk1 cDNA fragment were labeled with [α-32P]dCTP by using random priming (Promega), and the blots were hybridized with 32P-labeled probes at 42°C overnight. After hybridization, the blots were washed and then exposed to Kodak x-ray film with an intensifying screen at –80°C. The resulting autoradiographs were scanned and then quantified using densitometry.
Chromatin Immunoprecipitation and PCR Amplification
The procedure used was adapted from Szak et al. (2001). H1299 cells were seeded at 7.5 × 105 cells onto 10-cm dishes 1 d before transfection, and these were transfected with 2.5 μg of p53 wild-type or mutant p53 expression vectors by using 5 μl of LipofectAMINE 2000 (Invitrogen). Twenty-four hours after transfection, the cells were treated with 1% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature. Formaldehyde cross-linking was stopped by the addition of glycine at a final concentration of 0.125 M. Five minutes after the addition of glycine, the cells were washed twice with cold PBS, and lysed in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, 5 mM EDTA, and 1 mM DTT) containing the protease inhibitor cocktail (Roche Diagnostics) and phosphatase inhibitors (10 mM NaF, 1 mM sodium orthovanadate, and 10 mM β-glycerophosphate). The cell lysates were then sonicated to generate 500- to 1000-base pair fragments. After centrifugation for 10 min to remove cell debris, 2 mg of protein extract was precleared with mouse IgG binding to the protein G beads for 1 h with rocking at 4°C, followed by overnight incubation with 15 μl of protein G beads and p53 antibody PAb1801. The immune complexes were washed sequentially twice with RIPA buffer, four times with IP wash buffer (100 mM Tris, pH 8.5, 500 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride), and twice more with RIPA buffer. Each wash was performed with rocking for 5 min at 4°C. After washing, the immunoprecipitates were boiled in 300 μl of cross-linking reversal buffer (125 mM Tris, pH 6.8, 10% β-mercaptoethanol, and 4% SDS) for 30 min. The samples were vortexed for 10 s at 10-min intervals during the boiling stage. The proteins were removed by phenol/chloroform extraction, and the DNA was precipitated using ethanol. Finally, the DNA pellet was dried and dissolved in H2O.
All the PCRs were performed on a GeneAmp PCR System 2400 (PerkinElmer Life and Analytical Sciences) by using Thermoprime Plus DNA Polymerase (ABgene, Epsom, Surrey, United Kingdom). The following specific primer pairs were used to analyze the DNA from precipitated chromatin: p21, 5′-CATTGTTCCCAGCACTTCCTCTC-3′ and 5′-AGAAAGCCAATCAGAGCCACAG-3′; and AIP1, 5′-CACTTCAGGAGTCTCAAGTC-3′ and 5′-AGGCATCAGGAGCACTGGCC-3′. Two hundred nanograms of each primer was used in a 50-μl reaction containing 10% dimethyl sulfoxide and 1.5 mM MgCl2. The PCR products were resolved on either 8% polyacrylamide gels or 2% agarose gels.
Luciferase Reporter Assay
Saos2 cells were plated at a density of 1.2 × 105 per 35-mm dish 1 d before transfection. The transfection was performed using the calcium phosphate method, by using 200 ng of bax-luc, or 2 × AIP-luc reporter construct, or 100 ng of p21-luc reporter construct, together with wild-type p53, N2A, or C4A mutant. For normalization of the transfection efficiency, 100 ng of pCMV-β-gal expressing β-galactosidase was included as an internal control. Forty-eight hours after transfection, the cell lysates were prepared and assayed for their luciferase activity by using a Luciferase assay kit (Promega) and for the β-galactosidase activity as the normalization control.
RESULTS
Phosphorylation of p53 by CHK1 and CHK2 at the C Terminus
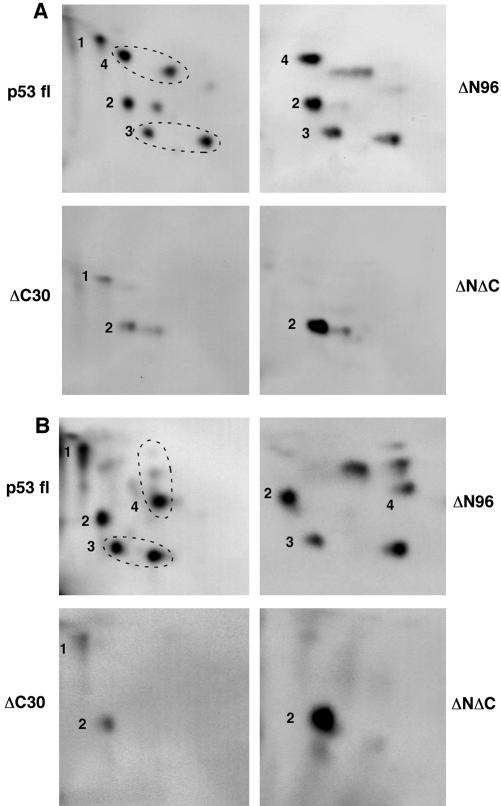
It has been previously demonstrated that CHK1 and CHK2 phosphorylate the p53 N terminus at Ser15, Thr18, Ser20, and Ser37 in vitro (Shieh et al., 2000). However, a construct with most of the N-terminal sequence deleted (ΔN96) still harbored a significant degree of phosphorylation (Shieh et al., 2000), suggesting that there are additional CHK1 and CHK2 sites in regions outside the N-terminal domain. To fully understand the regulation of p53 by CHK kinases, we sought to identify these additional sites by using two-dimensional phosphopeptide mapping. Three truncation mutants were generated to map the phosphorylated regions. In addition to the ΔN96 construct, which lacks the N-terminal 96 amino acids, ΔC30 (1-363) and ΔNΔC (97-363) truncated mutants also were compared. In vitro kinase assays were performed by incubating these truncated p53 proteins with either GST-CHK1 or His-CHK2. After phosphorylation, the proteins were digested using trypsin and initially analyzed using electrophoresis, followed by ascending chromatography on thin layer cellulose plates.
To our surprise, we observed that only one of the phosphopeptides was derived from the N terminus (Spot 1), because it was missing in the ΔN96 map (Figure 1). The majority of the phosphopeptides was derived from regions outside the N terminus: two from the C-terminal 30 amino acids (spots 3 and 4) and one from the region between amino acids 97 and 363 (spot 2). There were significant similarities between the maps resulting from CHK1 and CHK2 phosphorylation. Of the four major phosphopeptides detected, spots 1, 2, and 3 were shared by CHK1 and CHK2, whereas spot 4 was distinct in the two maps (Figure 1, A and B). To further locate the sites of phosphorylation, we created a series of S/T to A mutants in the backbone of ΔN96. As shown in Figure 2A, changes in both Ser313 and Ser314 to alanines altered the appearance of Spot 2 in the maps phosphorylated by CHK1 or CHK2, whereas simultaneous mutation of Thr377 and Ser378 eliminated or diminished spot 3. Among these four residues, the mutation of a single S or T to A site yielded maps indistinguishable from that of wild-type sites (our unpublished data). Two residues were found to be specifically phosphorylated by CHK1 or CHK2: the mutation of Ser366 to alanine completely removed spot 4 in the map phosphorylated by CHK2 (Figure 2B); and mutation of Thr387 abolished spot 4 in the map phosphorylated by CHK1 (Figure 2C). Therefore, we have identified at least six S/T residues in the C-terminal domain of p53 that are phosphorylated by CHK1 and/or CHK2 in vitro (summarized in Figure 2D). Among these sites, Ser313, Ser314, Thr377, and Ser378 were commonly phosphorylated by CHK1 and CHK2, whereas Ser366 and Thr387 seem to be specifically recognized by CHK2 and CHK1, respectively.
Figure 1.
CHK1 and CHK2 phosphorylate p53 on multiple sites spanning the N-terminal and the C-terminal domains. (A) Phosphopeptide mapping of p53 phosphorylated by GST-CHK1 in vitro. His-tagged, full-length, or truncated p53 was expressed in, and purified from, bacteria and incubated with GST-CHK1 expressed in, and purified from, insect cells in the presence of [γ-32P]ATP. Labeled p53 was tryptic digested and analyzed using two-dimensional separation on thin layer cellulose plate. (B) Phosphopeptide mapping of p53 phosphorylated by His-CHK2 in vitro. Reactions and analyses were carried out essentially the same as described in A, except that His-tagged CHK2 expressed in, and purified from, bacteria was used. p53 fl, amino acids 1–393; ΔN96, amino acids 97–393; ΔC30, amino acids 1–363; and ΔNΔC, amino acids 97–363.
Figure 2.
Identification of CHK1 and CHK2 phosphorylation sites in the C-terminal domain of p53. (A) Ser313, Ser314, Thr377, and Ser378 were phosphorylated by CHK1 and CHK2 in vitro. Phosphopeptide mapping was performed using His-tagged p53ΔN96 carrying wild-type residues or double Ala mutations at the indicated sites. After CHK1 or CHK2 phosphorylation, labeled p53 proteins were gel purified and analyzed as described in the legend to Figure 1. (B) Ser366 was specifically phosphorylated by CHK2 in vitro. His-tagged p53ΔN96 WT or the point mutant Ser366Ala (S366A) in the backbone of ΔN96 was phosphorylated with His-CHK2 and analyzed using two-dimensional separation. (C) Thr387 was specifically phosphorylated by CHK1 in vitro. Reaction and analyses were conducted as described in A by using ΔN96 WT or Thr387Ala (T387A) mutant and GST-CHK1. (D) Summary of the CHK1 and CHK2 phosphorylation sites in the p53 C-terminal domain between amino acids 301 and 393.
Phosphorylation of p53 at the Common Site Ser378, and the Specific Sites Ser366, Thr387, Is Inducible by DNA Damage
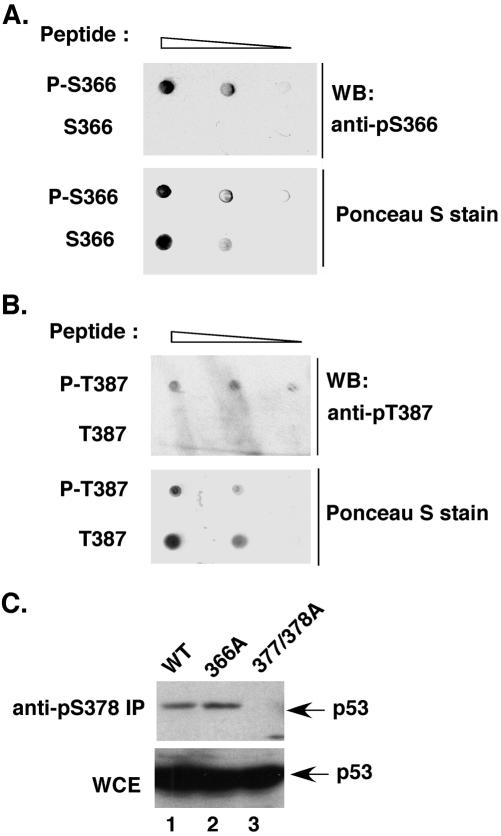
To investigate the physiological relevance of these identified in vitro phosphorylation sites, we generated anti-phospho-Ser366 –, phospho-Ser378 –, and phospho-Thr387–specific antibodies. The anti-phospho-Ser366 and anti-phospho-Thr387 antibodies recognized only the phosphopeptide but not the unphosphorylated peptide in a dot blot analysis (Figure 3, A and B). Furthermore, the anti-phospho-Ser378 antibody immunoprecipitated the transfected wild-type and S366A p53 but not the double mutant T377A/S378A, demonstrating the specificity of the antibody (Figure 3C). We then used these antibodies to determine whether the above-mentioned sites were phosphorylated in the cells and whether the phosphorylation status was altered in response to DNA damage. LNCaP cells, a human prostate carcinoma cell line expressing wild-type p53, were treated with either IR or UV, and cell lysates were prepared at different points in time after the treatment. In accordance with a previous report published by Waterman et al. (1998), the basal level of phosphorylation at Ser378 was readily detectable before damage (Figure 4B; our unpublished data). However, we found that it could be further induced by IR and UV with the maximum level of phosphorylation occurring at 3 and 6 h after irradiation, respectively. (Figure 4B, compare lanes 2–4 and lanes 7–9 in the top and bottom panels). In contrast to phosphorylation at Ser378, phosphorylation at Ser366 and Thr387 was barely detectable before irradiation, even when the basal level of p53 was increased using the proteasome inhibitor MG132 (Figure 4A, lane 12; and Figure 4C, lane 11). After irradiation, phosphorylation at these two sites was detected transiently at 3 h after IR (Figure 4, A and C) and at 6–9 h (for phospho-Ser366) or 15 h (for phospho-Thr387) after UV irradiation (Figure 4, A and C). Note that the kinetics of phosphorylation at Ser366 and Thr387 differed significantly after UV irradiation, which possibly indicates an intrinsic difference in the activation of CHK1 and CHK2 in phosphorylating p53. Both events also seemed much later than those for the p53 N terminus, which normally occurs within minutes after irradiation (Shieh et al., 1999).
Figure 3.
Specificity of the anti-phospho-Ser and anti-phospho-Thr antibodies. The phosphopeptides and unphosphorylated peptides were spotted on nitrocellulose membranes in a series of two-fold dilutions starting from 250 ng. After Ponceau S staining to confirm equal loading, the membrane was incubated with either anti-phospho-Ser366 antibody (A) or anti-phospho-Thr387 antibody (B), as for Western analysis. Both antibodies specifically recognized the corresponding phosphopeptide but not the unphosphorylated peptide. (C) The anti-phospho-Ser378 antibody specifically immunoprecipitates WT p53 and the 366A mutant but not the 377/378A mutant. H1299 cells were transfected with the indicated p53 constructs. p53 in the lysates was immunoprecipitated with the antiphospho-Ser378 antibody and detected by Western analysis by using the p53-specific antibodies PAb1801 and DO1.
Figure 4.
Phosphorylation of Ser366, Ser378, and Thr387 is induced by DNA damage in vivo. LNCaP (A–C) cells were irradiated with 8 Gy of x-rays or 50 J/m2 of UV-radiation and collected at the indicated times. p53 in the lysates was immunoprecipitated with anti-pSer366 (A), anti-pSer378 (B), or anti-pT387 (C), and then detected by Western analysis by using PAb1801. (D) LNCaP cells were treated with 0.5 μM CPT for the indicated length of time. The lysates were then analyzed for p53 phosphorylation, as described above. For controls, cells also were treated with 50 μM MG132 (MG), a proteasome inhibitor, to increase p53 in the absence of DNA damage. (E) 293T cells were irradiated with x-ray (8 Gy) or UV (40 J/m2) and analyzed as described above. (F and G) IP/kinase assays examining the CHK1 (F) or the CHK2 (G) activity at later time points after IR or UV. CHK1 or CHK2 was immunoprecipitated from the LNCaP lysates, and the kinase activities were determined by in vitro phosphorylation of GST-Cdc25C (F) or by CHK2 autophosphorylation (G) followed by autoradiography. (H) Induction of phosphorylation on Ser366 and Thr387 was abolished by caffeine. 293T cells were pretreated with 2 mM caffeine for 30 min before irradiation with x-ray or UV. Cell lysates were then analyzed as described above.
The possibility of induction of phosphorylation from other forms of DNA-damaging agents also was examined. As shown in Figure 4D, phosphorylation at Ser378, Ser366, and Thr387 also was induced by camptothecin (CPT), a DNA topoisomerase I inhibitor, which generates DNA double-strand breaks. The induction occurred hours after the addition of the drug, and again, was significant at a later period than the N-terminal phosphorylation events. Therefore, at least three of the sites identified in vitro (Ser378, Ser366, and Thr387) are phosphorylated in vivo in response to DNA damage.
Additionally, the induction of Ser366 and Thr387 phosphorylation also was detected in another cell line 293T that expresses constitutively high level of p53 (Figure 4E). Compared with the phosphorylation on Ser15, which was more persistent after the induction, phosphorylation on Ser366 and Thr387 seemed to be transient. In this cell, however, Ser378 phosphorylation was constitutive and was not significantly induced by either IR or UV (our unpublished data).
Because the phosphorylation at Ser366 and Thr387 occurred with delayed kinetics compared with the N-terminal modification, IP/kinase assays were carried out to examine whether CHK1 and CHK2 were still active at these late time points. As demonstrated in Figure 4F, CHK1 immunoprecipitated from LNCaP cells was clearly active in phosphorylating the substrate GST-Cda25C in vitro at 3 h after IR and 15 h after UV. Similarly, autophosphorylation of immunoprecipitated CHK2 was still detectable at 3 h after IR and 6 h after UV (Figure 4G).
ATM and ATR function upstream of CHK1 and CHK2 in DNA damage checkpoint response. To determine whether the p53 C-terminal phosphorylation events we detected in vivo were related to this signaling pathway, 293T cells were pretreated with 2 mM caffeine to block the activation of ATM and ATR by DNA damage (Sarkaria et al., 1999). Under this condition, the induction of phosphorylation at Ser366 and Thr387 was essentially abolished (Figure 4H, lanes 5–8). Our results are consistent with, although do not fully prove, the involvement of ATM/ATR-CHK1/CHK2 signaling pathway in DNA damage-induced phosphorylation of Ser366 and Thr387. Note that the basal level of Ser366 phosphorylation was elevated upon caffeine pretreatment (Figure 4H, lane 5), suggesting that additional signaling pathways also may be involved in the regulation.
Disruption of CHK1 and CHK2 Expression by Small Interfering RNA Mitigates p53 C-Terminal Phosphorylation and Lys382 Acetylation
To further demonstrate unambiguously that CHK1 and CHK2 do indeed contribute to phosphorylation at these C-terminal sites in vivo, siRNA techniques were applied to knock down the expression of endogenous CHK1 and CHK2. As shown in Figure 5A, the levels of CHK1 and CHK2 were reduced by >98 and 80%, respectively, by their specific siRNAs. Although the overall p53 induction was not affected, phosphorylation at Ser20, a site reported to be targeted by CHK1 and CHK2 (Chehab et al., 2000; Hirao et al., 2000; Shieh et al., 2000), was reduced by CHK1- or CHK2-targeting siRNAs, demonstrating the effectiveness of this technique. Furthermore, phosphorylation at Ser15, a site targeted by ATM and ATR (Banin et al., 1998; Canman et al., 1998; Khanna et al., 1998; Tibbetts et al., 1999), also was slightly reduced, suggesting that CHK1 and CHK2 also may participate in the phosphorylation of Ser15 in response to CPT treatment.
Figure 5.
Down-regulation of CHK1 and CHK2 by RNA interference. (A) Abrogation of Ser366 and Thr387 phosphorylation by chk1 and chk2-targeting siRNA. LNCaP cells were transiently transfected with a scrambled control (C), chk1 siRNA, or chk2 siRNA for 20–24 h, and then treated with CPT (0.5 μM) for 9 h before harvest. The cell lysates were analyzed for Ser366 and Thr387 phosphorylation by IP/Western analysis, as described in the legend to Figure 4. In addition, Ser15 and Ser20 phosphorylation, Lys382 acetylation, and p21 induction also were examined by Western analysis. (B and C) p53 Lys382 acetylation was diminished in cells with knocked down CHK1 and CHK2. LNCaP (B) or U2OS (C) cells were irradiated with x-rays or UV radiation and then lysed to examine for Lys382 acetylation by Western analysis. (D and E) Impaired p21 and bax induction in chk1- and chk2-knockdown cells. LNCaP (D) or U2OS (E) cells were irradiated with x-ray or UV and allowed to recover for the indicated periods before the isolation of the total RNA for Northern analysis. The signals on the autoradiographs were quantified using densitometry.
More importantly, phosphorylation at Ser366 and Thr387 was markedly reduced in CHK1- or CHK2-knockdown cells. This result, in combination with our data from the phosphopeptide mapping, demonstrates that Ser366 and Thr387 are indeed two sites targeted by CHK1 and CHK2 in vivo. Unexpectedly, although Ser366 was identified as a CHK2 site in vitro, phosphorylation at this site also was diminished in CHK1-knockdown cells. Similarly, phosphorylation at Thr387, a CHK1-specific site in vitro, was markedly reduced in CHK2-knockdown cells. It is not clear at this time whether the phosphorylation at these two sites was interdependent in vivo.
C-terminal acetylation of p53 has been shown to increase the DNA binding activity of p53 and to facilitate the transcription of p53 downstream target genes (Gu and Roeder, 1997; Barlev et al., 2001). Because the phosphorylation sites we identified were in proximity to the C-terminal acetylation sites, we wondered whether the C-terminal phosphorylation by CHK1 or CHK2 would play any modulatory role in p53 acetylation. We therefore examined the status of Lys382 acetylation in CHK1- or CHK2-knockdown cells. Interestingly, as the C-terminal phosphorylation was abrogated, acetylation at the Lys382 site was accordingly diminished (Figure 5A). This did not occur only in cells treated with CPT, and CHK1- and CHK2-knockdown cells irradiated with x-rays or with UV similarly exhibited reduced Lys382 acetylation (Figure 5B). Furthermore, another cell line, U2OS, also displayed reduced acetylation at Lys382 when both CHK1 and CHK2 were ablated (Figure 5C). These results suggest that CHK1 and CHK2 may indirectly modulate the acetylation status of p53.
Because acetylation and phosphorylation at the C terminus of p53 have been shown to positively influence transcriptional activation by p53 (Appella and Anderson, 2001; Brooks and Gu, 2003), we also examined the induction of p53 downstream targets from DNA damage. Induction of p21 protein by CPT was reduced when either CHK1 or CHK2 were down-regulated by siRNA (Figure 5A). Of the RNA levels assessed using Northern analysis, p21 and bax were induced 19.8- and 4.2-fold, respectively, in control siRNA-transfected cells at 6 h after IR (Figure 5D, compare lanes 1 and 3). The magnitude of induction was reduced to 6.6 and 1.6 times for p21 and bax, respectively, in CHK1-ablated cells, mostly due to the reduced induction (p21) and the elevated basal expression (bax) when CHK1 was knocked down (Figure 5D, compare lanes 6 and 8). Similar effect also was observed after UV irradiation (Figure 5D, lanes 4, 5, 9, and 10). In U2OS cells (Figure 5E), induction of p21 was reduced from 10-fold in the control cells (compare lanes 1 and 3) to 1.9-fold in cells with both CHK1 and CHK2 ablation (compare lanes 4 and 6) at a period of 16 h after UV irradiation. Induction of bax was basically eliminated in CHK1 and CHK2 siRNA-transfected cells after UV (Figure 5E). Similar effect also was observed in the treatment with IR (Figure 5E, lanes 7–12). Note that the basal levels of bax expression were significantly elevated in most cases when CHK kinases were ablated, and the expression was not further enhanced by DNA damage in some cases, suggesting that the expression of bax was deregulated in the absence of CHK1 and CHK2. These results together indicate that CHK1 and CHK2 participate in the phosphorylation of p53 C terminus, the modulation of p53 acetylation, as well as the proper induction of its downstream targets by DNA damage.
C-Terminal Phosphorylation of p53 Differentially Regulates p53 Acetylation
Because CHK1 and CHK2 phosphorylate both the N terminus and the C terminus of p53, it is therefore impossible to compare the relative contribution of these two events for p53 acetylation in CHK1- and CHK2-knockdown cells. To determine the role of p53 C-terminal phosphorylation alone, we generated p53 mutants, in which either residues 377 and 378 or residues 366 and 387 were mutated to Asp, to mimic constitutive phosphorylation, or to nonphosphorylatable Ala. These p53 constructs were coexpressed with p300 or PCAF in p53-null H1299 cells. Interestingly, differences in the C-terminal acetylation were observed in these mutants (Figure 6A). Mutations of the 377 and 378 residues to Ala enhanced acetylation at Lys373 and Lys382, whereas substitution at the same positions with Asp reduced acetylation (Figure 6A, lanes 2 and 3). In contrast, mutations of the 366 and 387 sites to Asp greatly stimulated acetylation at Lys373 and Lys382 (Figure 6A, lane 6).
Figure 6.
Mutations at the p53 C-terminal CHK1 and CHK2 phosphorylation sites differentially regulate p53 C-terminal acetylation. (A) Asp substitutions at Thr377 and Ser378 inhibit acetylation at Lys373 and Lys382, whereas Asp substitutions at Ser366 and Thr387 potentiate acetylation. H1299 was transiently transfected with WT p53 or the phosphorylation site mutants together with either p300-HA (for observing acetylation at Lys373 and Lys382) or FLAG-PCAF (for observing Lys320 acetylation). The cell lysates were analyzed by Western analysis by using anti-acetyl-K382 antibodies or by initial immunoprecipitation with anti-acetyl-K373 or anti-acetyl-K320 antibodies followed by Western analysis by using p53-specific antibody PAb1801. (B) Comparison of Ser15 and Ser20 phosphorylation between WT and mutant p53. H1299 cells were transiently transfected with either WT p53 or the indicated phosphorylation site mutants, and the lysates were analyzed by Western analysis by using anti-phospho-Ser15 or anti-phospho-Ser20 antibodies. (C) Interaction of WT p53 or the phosphorylation site mutants with p300. HA-tagged p300 was first immunoprecipitated from transiently transfected 293T lysates by using the mAb 12CA5 and then incubated with His-tagged, purified recombinant p53 proteins. The bound p53 was detected by Western analysis by using anti-His antibody. (D) The 366/387D mutant binds better than the WT and the 366/387A mutant to the p21 and AIP1 promoters. Chromatin immunoprecipitation assays were performed by initially transfecting the H1299 cell with either WT or mutant p53. p53 was immunoprecipitated from the cross-linked lysates by using PAb1801, and a portion (1/15) of the immunoprecipitate was analyzed using Western analysis to monitor the amount of p53. The precipitated DNA was then amplified by PCR by using p21-specific or AIP1-specific primers.
Opposite to the effects observed above, acetylation at Lys320, as mediated by PCAF, was not affected by these mutations (Figure 5A, bottom). These results imply that phosphorylation of the p53 C terminus on these residues may differentially regulate p53 acetylation, specifically at Lys373 and Lys382.
Because N-terminal phosphorylation at Ser15 and Ser20 has been shown to augment p53 acetylation, possibly through an increased interaction with p300/CBP (Lambert et al., 1998; Sakaguchi et al., 1998; Kar et al., 2002), we therefore examined these mutants for their N-terminal phosphorylation status. Consistently, the 377/378A mutant displayed significantly higher Ser20 phosphorylation (Figure 6B, lane 5) and showed a better interaction with p300 (Figure 6C, lane 3) compared with the other mutants. However, this is not the case for mutations at Ser366 and Thr387. Despite the difference in their levels of acetylation, the 366/387A mutant and the 366/387D mutant interacted with p300 equally well (Figure 6C, lanes 5 and 6) and displayed similar degrees of Ser15 and Ser20 phosphorylation (Figure 6B, lanes 3 and 4). Therefore, although acetylation of the 377/378 mutants may be affected by their interaction with p300, acetylation of the 366/387 mutants may be governed by a different mechanism.
In addition to the interaction with p300, DNA binding by p53 also can affect its acetylation by p300, as implicated in the studies of Dornan et al. (2003). To assess this possibility, binding of the 366/387 mutants to p53-targeted promoters was examined using chromatin immunoprecipitation (ChIP) assays. As shown in Figure 6D, the D mutant bound better than the wild type (WT) or the A mutant to the p21 and AIP1 promoters. This result suggests that the enhanced acetylation observed on the 366/387 D mutant may be due to its increased ability to bind to p53-targeted promoters. These data also suggest that phosphorylation of the p53 C terminus by CHK1 and CHK2 at Ser366 and Thr387 may facilitate the expression of p53 downstream target genes.
Differential Regulation of p53-targeted Promoters by the N-Terminal and the C-Terminal Phosphorylation Sites
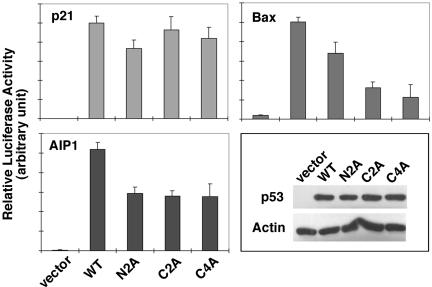
As we have demonstrated, CHK1 and CHK2 phosphorylate the N-terminal residues and the C-terminal residues. We therefore wondered which phosphorylation events contribute most to the activation of p53 downstream target genes. To address this proposition, we performed a series of luciferase reporter assays by transfecting p53 into p53-null Saos2 cells, together with reporters carrying various p53 targeted-response elements. In these assays, we compared the activity of WT p53 with that of mutants carrying Ala substitutions at Ser15, 20 (N2A), Ala substitutions at residues 366 and 387 (C2A), or Ala substitutions at residues 313, 314, 366, and 387 (C4A) on activation of the p21, bax, or AIP1 reporters. Mutation of Ser15 and Ser20 to Ala reduced the activation of all three reporters (Figure 7).
Figure 7.
Comparison of the transcriptional activity of p53 mutated at the N-terminal and the C-terminal phosphorylation sites. Saos2 cells were transiently transfected with constructs expressing p53 and a luciferase reporter driven by p53 binding sites derived from the p21, bax, or AIP1 promoter. N2A is a mutant p53 with Ala substitutions at Ser15 and Ser20. C2A represents a mutant p53 with Ala substitutions at Ser366 and Thr387, and C4A represents a mutant p53 with two additional Ala substitutions at Ser313 and 314. The expression levels of p53 were examined using Western analysis with PAb1801. The error bars represent the SD obtained from four independent, duplicated experiments.
The effect of C-terminal mutations seemed to be more selective. Although the activity of the p21 reporter was not apparently affected, the activity of the AIP1 reporter was reduced by 45% and that of the bax reporter was significantly abated by ∼70% (Figure 7). Therefore, unlike N-terminal phosphorylation, which is required for full activation of most p53-targeted promoters, C-terminal phosphorylation is possibly only required for a subset of p53 target genes.
In addition to the Ala mutants, we also generated a mutant carrying Asp substitutions at residues 366 and 387 (C2D). The activity of C2D was ∼20% higher than that of C2A, suggesting that negative charges can only partially mimic the effect of phosphorylation at these C-terminal sites (our unpublished data).
DISCUSSION
The cell cycle checkpoint kinases CHK1 and CHK2 have been shown to phosphorylate multiple sites in the N-terminal domain of p53, consequently leading to p53 stabilization and activation (Chehab et al., 2000; Hirao et al., 2000; Shieh et al., 2000). In this study, we have identified at least six additional CHK1 and CHK2 phosphorylation sites in the C-terminal domain of p53. Like their N-terminal counterparts, phosphorylation on at least three of these sites is inducible by DNA damage. Furthermore, phosphorylation in the C-terminal domain seems to differentially regulate C-terminal acetylation and is required for the activation of a subset of p53 downstream target genes.
One Kinase, Two Events
CHK1 and CHK2 phosphorylate Ser15, Thr18, Ser20, and Ser37 in the N-terminal domain of p53 in vitro. Phosphorylation at these sites is induced within minutes after the DNA damage treatment (Shieh et al., 1999), suggesting that preexisting kinases are responsible for the phosphorylation. Although it is possible that CHK1 or CHK2 is involved in these phosphorylation events in vivo, gene knockout or ablation by siRNA in certain cancer cell lines seriously questions this possibility (Jallepalli et al., 2003; Ahn et al., 2003). In our siRNA experiments using the prostate carcinoma cell line LNCaP (Figure 5A), chk1- or chk2-targeting siRNA did exhibit a reduction in, although it did not eliminate, the induction of phosphorylation at Ser15 and Ser20 by CPT without showing an effect on the overall p53 level. This result suggests that although CHK1 and CHK2 may contribute to the phosphorylation at Ser15 and Ser20, other kinases, possibly ATM and ATR, may have complementary roles in this event. Furthermore, the induction of p21 by DNA damage, at either the protein level or the steady-state RNA level, was found to be reduced when either CHK1 or CHK2 was knocked down (Figure 5, A and D). The diminishing induction of the p53 downstream target genes, p21 and bax, also was observed in U2OS cells, an osteosarcoma cell line (Figure 5E). Our data suggest that, at least in some cellular contexts, CHK1 and CHK2 participate in the phosphorylation of the p53 N terminus and are required for proper induction of p53 target genes after DNA damage.
Aside from the N-terminal events, CHK1 and CHK2 also phosphorylate multiple sites spanning the C-terminal region of p53; four of which are commonly recognized by both kinases, whereas two of which, Ser366 and Thr387, are specifically targeted by CHK2 and CHK1, respectively, in vitro. Phosphorylation in vivo on at least three of these sites was induced by DNA damage (Figure 4). The induction was abrogated by CHK1- and CHK2-targeting siRNA (Figure 5A), suggesting that indeed, these are physiologically relevant CHK1 and CHK2 sites. What puzzles us most is that chk1 siRNA also abolished Ser366 phosphorylation, and chk2 siRNA disrupted Thr387 phosphorylation. The specificity we observed in vitro for CHK1 and CHK2 did not seem to recapitulate itself in vivo. It is possible that the kinases may have altered specificity in cells or that phosphorylation on these two sites may be cooperative in vivo. These possibilities, among others, await further investigation.
Interestingly, the kinetics of C-terminal phosphorylation, which occurred hours after the treatment, was significantly delayed compared with the N-terminal events. If indeed CHK1 and CHK2 impose phosphorylation on both the N terminus and C terminus, this would be difficult to reconcile with the apparently disparate kinetics. Several models can be envisaged. One possibility is that N-terminal phosphorylation is mainly contributed to by more upstream kinases, such as ATM and ATR, or similar kinases. These kinases could play a major role in stabilizing p53 and the initiation of the p53 response, which would subsequently be maintained by CHK1 and CHK2. This model would explain why in some studies blocking CHK1 or CHK2 did not have a major impact on the induction of p53 by DNA damage (Ahn et al., 2003; Jallepalli et al., 2003). However, this is somewhat inconsistent with the kinetics of CHK1 or CHK2 activation in the IP/kinase assay, showing that CHK2 is activated within 30 min after IR treatment (Falck et al., 2001) and that CHK1 is activated within 2 h after UV irradiation (Mailand et al., 2000). Among other possible explanations, it may be that although CHK1 and CHK2 are activated early, the availability of the p53 C terminus is dependent upon a conformational change, possibly elicited by N-terminal phosphorylation. Alternatively, phosphorylation of the C terminus by CHK1 and CHK2 may require a DNA damage-induced cofactor. This would explain the delayed kinetics and why p53 does not act as a good substrate in some CHK2 IP/kinase assays (Ahn et al., 2003). Based on the diverse conclusions derived from the different experimental settings in respect to phosphorylation of p53 by CHK1 and CHK2, it is probably not too bold to speculate that the actual occurrence would depend on the cellular microenvironment.
Role of p53 C-Terminal Phosphorylation by CHK1 and CHK2 in p53 Stabilization and p53-dependent Transactivation
If p53 N-terminal phosphorylation is responsible for its stabilization by DNA damage and the transactivation of its downstream target genes, then it is intriguing to know the role of CHK1- and CHK2-mediated p53 C-terminal phosphorylation. The C-terminal phosphorylation of p53 has long been known to enhance specific DNA binding by p53 (Meek, 1998). In fact, CHK1- or CHK2-phosphorylated p53 binds better than unphosphorylated p53 to its recognition site in vitro (Shieh and Prives, unpublished data). Consistently, the p53 366/387D mutant also binds better to the p21 and the AIP1 promoters in our ChIP assays (Figure 6D). This, however, does not necessarily lead to higher activation of all of the p53 target genes. The impact of Ser366 and Thr387 phosphorylation seems to be selective. Although mutation of these residues to Ala had limited effect on the activity of p21 and AIP1 reporters, it was detrimental to the activity of the bax reporter (Figure 7). It is likely that CHK1- and CHK2-mediated p53 C-terminal phosphorylation provides an additional layer of control for the differential regulation of p53 downstream targets.
How p53 is stabilized by DNA damage has been a subject of intense investigation. The current consensus is that DNA damage activates ATM and ATR, which phosphorylate p53 at Ser15 and Ser20 either directly, or through the activation of CHK1 and CHK2, leading to alleviation of p53 degradation by Mdm2 and accumulation of p53 (Shiloh, 2003). Concomitant phosphorylation of MDM2 by ATM also disrupts MDM2–p53 interaction and results in p53 stabilization (Appella and Anderson, 2001; Chène, 2003). Supporting evidence includes the delayed or diminished induction of p53 in AT cells (Kastan et al., 1992; Lu and Lane, 1993), reduced induction of p53 in chk2 null murine cells (Hirao et al., 2000), and the decreased stability observed for p53 mutated at the Ser20 site (Unger et al., 1999).
However, the results of recent studies also cast doubts on the role of N-terminal phosphorylation. To mention a few of these, the replacement of WT p53 with mutants carrying Ala substitution at Ser15 (Ser18 in mouse) or Ser20 (Ser 23 in mouse) had basically no effect on p53 stabilization in murine cells (Wu et al., 2002; Sluss et al., 2004). Furthermore, ablation of CHK1 and CHK2 by either recombination or siRNA had no impact on DNA damage-induced stabilization of p53 in certain cancer cell lines (Ahn et al., 2003; Jallepalli et al., 2003). In our siRNA studies, chk1 and chk2 siRNA slightly reduced the phosphorylation at Ser15 and Ser20 without grossly affecting the overall level of p53 induction (Figure 5A). These data point to the possibility that the major role of CHK1 and CHK2 lies in their phosphorylation of the p53 C terminus rather than the N terminus.
Based on the point in time at which phosphorylation at Ser366 and Thr387 is induced (Figure 4), it is not likely that these C-terminal events play any role in the initial stages of the p53 response. Whether they play a role in maintaining the response in the later stages remains to be determined. It is relatively more certain that without either CHK1 or CHK2, the induction of p53 downstream targets is significantly diminished (Figure 5, A, D, and E). This, in some way, is in agreement with what has been previously observed in chk2 null murine thymocytes or mouse embryonic fibroblast cells (Hirao et al., 2000, 2002; Takai et al., 2002) and may be a combinatorial effect resulting from phosphorylation on both the p53 N terminus and the C terminus.
The Interplay between C-Terminal Phosphorylation and C-Terminal Acetylation
p53 acetylation has been implicated in the regulation of its activity (Scolnick et al., 1997; Barlev et al., 2001), stability (Kawai et al., 2001; Li et al., 2002), and DNA binding (Gu and Roeder, 1997). Most of the acetylation events occurred within the C-terminal 100 amino acids and are contributed to by p300/CBP and PCAF (Sakaguchi et al., 1998; Liu et al., 1999; Ito et al., 2001). The functional interaction between acetylation and phosphorylation has been demonstrated previously; phosphorylation at the p53 N terminus potentiated acetylation at the C terminus (Lambert et al., 1998; Sakaguchi et al., 1998), possibly through an enhanced interaction between p53 and the acetylases as a result of phosphorylation (Lambert et al., 1998; Kar et al., 2002). In our study, we have demonstrated that p53 C-terminal phosphorylation by CHK1 and CHK2 may also modulate C-terminal acetylation.
Mutation of the Thr377 and Ser378 residues to Ala significantly raised the levels of acetylation at Lys373 and Lys382, whereas leaving Lys320 acetylation unchanged (Figure 6A). In contrast, mutation of the same residues to Asp suppressed the acetylation at Lys382 and Lys373. This may be the result of their different affinity for p300 acetylase, because the 377/378A mutant interacted with p300 better than did the 377/378D mutant (Figure 6C). Because phosphorylation at the Ser378 site is constitutive (Figure 4B), our data imply that under normal unstressed conditions, phosphorylation at Ser378, and possibly Thr377, suppresses p53 acetylation and keeps the activity of p53 in check. This suppression is relieved when combined with Ser366 and Thr387 phosphorylation, as when a stressed condition is encountered. This is supported by our observation that mutation of Ser366 and Thr387 to Asp greatly stimulates the acetylation of Lys373 and Lys382. In this case, this is due to enhanced promoter binding, rather than increased p300 interaction (Figure 6, C and D). Furthermore, siRNA knockdown of CHK1 and CHK2 attenuates the induction of acetylation (Figure 5, B and C), suggesting that the CHK kinase-mediated phosphorylation of p53 C terminus can modulate the degree of p53 acetylation. Together, these results reveal a novel mechanism by which CHK1 and CHK2 control the p53 activity through their compound effects on phosphorylation as well as acetylation.
Acknowledgments
We are grateful to Drs. Yoichi Taya for the AIP1-luc reporter constructs, Hsiu-Ming Shih for the pCMVβp300-CHA plasmid, and Yoshihiro Nakatani for pCI-FLAG-PCAF. This work was supported by National Science Council grants awarded to S.-Y.S.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0689) on January 19, 2005.
References
- Ahn, J., Urist, M., and Prives, C. (2003). Questioning the role of checkpoint kinase 2 in the p53 DNA damage response. J. Biol. Chem. 278, 20480–20489. [DOI] [PubMed] [Google Scholar]
- Appella, E., and Anderson, C. W. (2001). Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268, 2764–2772. [DOI] [PubMed] [Google Scholar]
- Banin, S., et al. (1998). Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Barlev, N. A., Liu, L., Chehab, N. H., Mansfield, K., Harris, K. G., Halazonetis, T. D., and Berger, S. L. (2001). Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Bartek, J., and Lukas, J. (2003). chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3, 421–429. [DOI] [PubMed] [Google Scholar]
- Boyle, W. J., van der Geer, P., and Hunter, T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–152. [DOI] [PubMed] [Google Scholar]
- Brooks, C. L., and Gu, W. (2003). Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 activation. Curr. Opin. Cell Biol. 15, 164–171. [DOI] [PubMed] [Google Scholar]
- Bulavin, D. V., Saito, S., Hollander, M. C., Sakaguchi, K., Anderson, C. W., Appella, E., and Fornace, A. J., Jr. (1999). Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18, 6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmann, T., et al. (2001). Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol. Cell. Biol. 21, 2743–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman, C. E., Lim, D.-S., Cimprich, K. A., Taya, Y., Tamai, K., Sakaguchi, K., Appella, E., Kastan, M. B., and Siliciano, J. D. (1998). Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- Chehab, N. H., Malikzay, A., Appel, M., and Halazonetis, T. D. (2000). Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14, 278–288. [PMC free article] [PubMed] [Google Scholar]
- Chène, P. (2003). Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 3, 102–109. [DOI] [PubMed] [Google Scholar]
- Di Como, C. J., Gaiddon, C., and Prives, C. (1999). p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan, D., Shimizu, H., Perkins, N. D., and Hupp, T. R. (2003). DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 278, 13431–13441. [DOI] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Falck, J., Mailand, N., Suljuasen, R. G., Bartek, J., and Lukas, J. (2001). The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410, 842–847. [DOI] [PubMed] [Google Scholar]
- Gu, W., and Roeder, R. G. (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Harms, K., Nozell, S., and Chen, X. (2004). The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61, 822–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao, A., et al. (2002). Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22, 6521–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao, A., Kong, Y. Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S. J., and Mak, T. W. (2000). DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287, 1824–1827. [DOI] [PubMed] [Google Scholar]
- Rhind, N., and Russell, P. (2000) Chk1 and Cds 1, linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113, 3889–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, C. I., Tran, S.E.F., Eriksson, J. E., and Sistonen, L. (2002). Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 27, 619–627. [DOI] [PubMed] [Google Scholar]
- Ito, A., Lai, C.-H., Zhao, X., Saito, S., Hamilton, M. H., Appella, E., and Yao, T.-P. (2001). p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli, P. V., Lengauer, C., Vogelstein, B., and Bunz, F. (2003). The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278, 20475–20479. [DOI] [PubMed] [Google Scholar]
- Kar, S., Sakaguchi, K., Shimohigashi, Y., Samaddar, S., Banerjee, R., Basu, G., Swaminathan, V., Kundu, T. K., and Roy, S. (2002). Effect of phosphorylation on the structure and fold of transactivation domain of p53. J. Biol. Chem. 277, 15579–15585. [DOI] [PubMed] [Google Scholar]
- Kastan, M. B., Zhan, Q., El-Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B., and Fornace, A. J., Jr. (1992). A mammalian cell cycle checkpoint pathway utilizing p53 and Gadd45 is defective in ataxia-telangiectasia. Cell 71, 587–597. [DOI] [PubMed] [Google Scholar]
- Kawai, H., Nie, L., Wiederschain, D., and Yuan, Z.-M. (2001). Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 276, 45928–45932. [DOI] [PubMed] [Google Scholar]
- Khanna, K. K., Keating, K. E., Kozlov, S., Scott, S., Gatei, M., Hobson, K., Taya, Y., Gabrielli, B., Chan, D., Lees-Miller, S. P., and Lavin, M. F. (1998). ATM associates with and phosphorylates p 53, Mapping the region of interaction. Nat. Genet. 20, 398–400. [DOI] [PubMed] [Google Scholar]
- Ko, L. J., and Prives, C. (1996). p 53, puzzle and paradigm. Genes Dev. 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- Lambert, P. F., Kashanchi, F., Radonovich, M. F., Shiekhattar, R., and Brady, J. N. (1998). Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- Li, M., Luo, J., Brooks, C. L., and Gu, W. (2002). Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277, 50607–50611. [DOI] [PubMed] [Google Scholar]
- Liu, L., Scolnick, M., Trievel, R. C., Zhang, H. B., Marmorstein, R., Halazonetis, T. D., and Berger, S. L. (1999). p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., and Lane, D. P. (1993). Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell 75, 765–778. [DOI] [PubMed] [Google Scholar]
- Mailand, N., Falck, J., Lukas, C., Syljuaswn, R. G., Welcker, M., Bartek, J., and Lukas, J. (2000). Rapid destruction of human Cdc25A in response to DNA damage. Science 288, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Meek, D. W. (1998). Multisite phosphorylation and the integration of stress signals at p53. Cell. Signal. 10, 159–166. [DOI] [PubMed] [Google Scholar]
- Oda, K., et al. (2000). p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser46-phosphorylated p53. Cell 102, 849–862. [DOI] [PubMed] [Google Scholar]
- Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C., and Hainaut, P. (2002). The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19, 607–614. [DOI] [PubMed] [Google Scholar]
- Sakaguchi, K., Herrera, J. E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C. W., and Appella, E. (1998). DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M., and Abraham, R. T. (1999). Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59, 4375–4382. [PubMed] [Google Scholar]
- Scolnick, D. M., Chehab, N. H., stavridi, E. S., Lien, M. C., Caruso, L., Moran, E., Berger, S. L., and Halazonetis, T. D. (1997). CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57, 3693–3696. [PubMed] [Google Scholar]
- Shieh, S.-Y., Ahn, J., Tamai, K., Taya, Y., and Prives, C. (2000). The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14, 289–300. [PMC free article] [PubMed] [Google Scholar]
- Shieh, S.-Y., Ideda, M., Taya, Y., and Prives, C. (1997). DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334. [DOI] [PubMed] [Google Scholar]
- Shieh, S.-Y., Taya, Y., and Prives, C. (1999). DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh, Y. (2003). ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3, 155–168. [DOI] [PubMed] [Google Scholar]
- Sluss, H. K., Armata, H., Gallant, J., and Jones, S. N. (2004). Phosphorylation of Serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak, S. T., Mays, D., and Peitenpol, J. A. (2001). Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, H., et al. (2002). Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21, 5195–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts, R. S., Brunbaugh, K. M., Williams, J. M., Sarkaria, J. N., Cliby, W. A., Shieh, S.-Y., Prives, C., and Abraham, R. T. (1999). A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger, T., Juven-Gershon, T., Moallem, I., Berger, M., Sionov, R. V., Lozano, G. Oren, M., and Haupt, Y. (1999). Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, K. H. (2002). Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602, 47–59. [DOI] [PubMed] [Google Scholar]
- Vousden, K. H., and Lu, X. (2002). Live or let die: the cell's response to p53. Nat. Rev. Cancer 2, 594–604. [DOI] [PubMed] [Google Scholar]
- Waterman, M.J.F., Stavridi, E. S., Waterman, J.L.F., and Halazonetis, T. D. (1998). ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19, 175–178. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Earle, J., Saito, S., Anderson, C. W., Appella, E., and Xu, Y. (2002). Mutation of mouse p53 Ser23 and the response to DNA damage. Mol. Cell. Biol. 22, 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. (2003). Regulation of p53 responses by post-translational modifications. Cell Death Differ. 10, 400–403. [DOI] [PubMed] [Google Scholar]