Abstract
The expression of extracellular virulence determinants in Staphylococcus aureus is controlled by a 510-nucleotide RNA molecule (RNAIII) which is a part of the agr system. The agr operon, which encodes a multicomponent signal transduction system, is partially under the influence of an unlinked regulatory locus called sar. The sar locus is composed of three overlapping transcripts, designated sarA (0.56 kb), sarC (0.8 kb), and sarB (1.2 kb), originating from the P1, P3, and P2 promoters, respectively. In this study, we analyzed the differential expression of these promoters by using transcriptional fusion with the xylE reporter gene to study the activation of the sar locus. The data confirm the existence of three independent promoters with different promoter activities. Maximal promoter activity was observed with the combined fusion of P2-P3-P1 promoters. Expression studies with a sigB mutant revealed that the P3 promoter is SigB dependent. Analysis of these transcriptional fusions in a sarA mutant and in complemented strains with each of the sar transcriptional units revealed that the sar locus is autoregulatory, with SarA acting as a positive regulator. From various transcriptional fusion studies of the upstream region of the P1 promoter, we have localized a 34-bp sequence which seems to play a role in down-modulating P1 transcription. Using heparin-Sepharose and DNA-specific columns, we partially purified a 12-kDa protein, possibly a repressor, which binds to the promoter regions upstream of P2 and P1 and which also binds to the 34-bp sequence. These data indicated that the regulation of the sar locus is complex and may involve the sar gene product(s) and other regulatory protein(s).
Staphylococcus aureus is an important human pathogen (25). It causes a variety of infections in humans ranging from localized skin suppuration to life-threatening septicemia. S. aureus produces a plethora of exotoxins, including hemolysins, enterotoxins, and toxic shock syndrome toxin 1 (TSST-1). The latter toxins are the causative agents for food poisoning and toxic shock syndrome, respectively (24). The pathogenesis of S. aureus is complex and probably involves the synthesis of cell wall-associated adhesins and the secretion of extracellular toxins with damaging effect on host cells, including those within the immune system.
Many of these extracellular virulence determinants are regulated by pleiotropic regulatory elements such as sar and agr (2, 12, 13, 20). The agr locus has been shown to be an activator for the expression of extracellular virulence genes (i.e., α-toxin, β-hemolysin, TSST-1, enterotoxins, etc.), while it negatively regulates the synthesis of cell surface proteins, such as protein A and fibronectin-binding proteins (12, 13, 15). The agr locus is composed of two divergent transcripts, RNAII and RNAIII, with sizes of 3.0 and 0.5 kb, respectively. The transcript RNAII initiating from the P2 promoter contains agrA, -B, -C, and -D, all of which are required for the activation of P2 and the ensuing RNAIII transcription. RNAIII, which also contains the δ-hemolysin gene, is the agr effector molecule ultimately responsible for the control of extracellular and cell surface protein synthesis (13, 16, 19).
In addition to agr, staphylococcal accessory regulator (sar) and the exoprotein gene regulator (sae) have been recently identified as two distinct global regulatory elements that are also involved in the expression of extracellular and cell surface proteins (4, 5, 9, 24). The sar locus is composed of three overlapping transcripts, designated sarA (0.56 kb), sarC (0.8 kb), and sarB (1.2 kb) originating from three distinct promoters, P1, P3, and P2, respectively. The expression of each of the three transcripts varies during the growth cycle, with sarA and sarB being maximal at the exponential phase and sarC peaking during the postexponential phase (1). Sequence analysis revealed that the sarA transcript codes for a 124-amino-acid polypeptide (SarA), while the transcript sarC encodes SarA and a putative 39-amino-acid open reading frame (ORF3) (1). Molecular analysis indicated that the larger sarB transcript, encoding SarA, ORF3, and an additional 18-amino-acid ORF (ORF4), is essential for full expression of RNAII and RNAIII in S. aureus, while the shorter sarA and sarC transcripts only partially restored agr-related transcription. It is likely that agr activation is partially mediated by the binding of sar gene product(s) to the agr promoter (2, 7, 10). Accordingly, the mechanism by which sar is activated from its own promoter has bearing on agr expression.
In this paper, we examined the regulation of sar expression in a pair of isogenic sar strains of S. aureus by using transcriptional fusion with the xylE reporter gene. Expression studies suggested that the P1 promoter is the strongest promoter compared with the P2 and P3 promoters in the parental strain. In assaying transcriptional activity in an isogenic sigB mutant, we confirmed our previous speculation that the central promoter (P3) of sar is ςB dependent. Transcriptional fusion studies with the wild type and its isogenic sar mutant indicated that the expression of the sar locus is partially dependent on its own gene product. We have also identified a binding site for a putative repressor protein upstream of the P1 promoter. In gel shift assays, the partially purified 12-kDa protein binds to the sar P2 promoter region as well as to a 34-bp sequence upstream of P1. Therefore, we propose that the partially purified protein may act as a repressor for down-regulating sar expression, whereas sar gene product(s) may act as an activator of its own gene’s expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Phage φ11 was used as the transducing phage for S. aureus strains. CYGP, 0.3GL medium (17, 18), and tryptic soy broth were used for the growth of S. aureus strains, while Luria-Bertani medium was used for growing Escherichia coli. Antibiotics were used at the following concentrations: erythromycin, 5 μg/ml; chloramphenicol, 10 μg/ml; tetracycline, 5 μg/ml; and ampicillin, 50 μg/ml.
TABLE 1.
S. aureus strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| RN4220 | Mutant of strain 8325-4 that accepts foreign DNA | 17 |
| RN6390 | Parental strain | 17 |
| RUSA168 | sigB::Tn551 mutant of strain COL | 27 |
| ALC488 | sarA::ermC mutant of RN6390 | 3 |
| ALC812 | ALC488 with sarB (nt 1–1349 plus 180 bp of upstream sequence) integrated into the geh locus on the host chromosome | 2 |
| ALC996 | ALC488 with sarA (nt 530–1349) integrated into the chromosomal lipase gene (geh) via the integration vector pCL84 | 2 |
| ALC997 | ALC488 with sarC (nt 364–1349) integrated into the lipase gene (geh) | 2 |
| ALC1001 | sigB::Tn551 mutant of RN6390 | This study |
| CYL316 | Derivative of RN4220 supplying the integrase gene in trans (the recipient strain for pCL84) | 14 |
| COL | Methicillin-resistant strain | 27 |
| Plasmids | ||
| pCRII | E. coli cloning vector for direct cloning of PCR product | Invitrogen |
| pLC4 | Transcriptional fusion vector with a promoterless xylE gene | 21 |
| pSL24 | Derivative of pLC4 with a pUC18 multiple cloning site | 23 |
| pRN6735 | Derivative of pC194 containing pI258 bla promoter and 2/3 of the blaZ gene followed by the 1,566-bp MboI fragment of RNAIII lacking its promoter | 17 |
| pCL84 | S. aureus integration vector that inserts into the lipase gene (geh) of the host chromosome | 14 |
| pALC686 | pLC4 containing a 209-bp fragment upstream of sarA from nt 651–859; P11 | This study |
| pALC690 | pLC4 containing a 329-bp fragment upstream of sarA from nt 531–859; P14 | This study |
| pALC695 | pLC4 containing a 162-bp fragment upstream of sarC from nt 364–525; P3 | This study |
| pALC698 | pLC4 containing a 196-bp fragment upstream of sarB from nt 1–196; P22 | This study |
| ALC706 | pLC4 containing a 496-bp fragment upstream of sarA from nt 364–859; P3-P1 | This study |
| pALC707 | pLC4 containing a 1,039-bp fragment upstream of sarA from nt 1–859 plus 180 bp further upstream; P2-P3-P1 | This study |
| pALC926 | pUC18 containing a 49-bp fragment of the upstream P2 promoter region (nt 71–119) (1) at the BamHI site | This study |
| pALC932 | pLC4 containing a 148-bp fragment upstream of sarB from nt 49–196; P21 | This study |
| pALC936 | pLC4 containing a 376-bp fragment upstream of sarB from nt 1–196 plus 180 bp further upstream; P23 | This study |
| pALC1030 | pSL24 containing a 183-bp bla promoter fragment from pRN6735 | This study |
| pALC1050 | pLC4 containing a 258-bp fragment upstream of sarA from nt 601–859; P12 | This study |
| pALC1167 | pLC4 containing a 299-bp fragment upstream of sarA from nt 561–859; P13 | This study |
| pALC1227 | pLC4 containing a 277-bp fragment upstream of sarA from nt 567–859; P15 | This study |
| pALC1229 | pUC18 containing a 34-bp fragment of the upstream of P1 promoter from nt 567–600 (1) at the BamHI site | This study |
| pALC1362 | pCL84 containing the P14 promoter upstream of xylE reporter gene | This study |
Construction of transcriptional fusions.
DNA fragments encompassing various sar promoter regions (Fig. 1) were amplified by PCR by using genomic DNA of S. aureus RN6390 as the template and cloned into the TA cloning vector pCRII (Invitrogen, San Diego, Calif.). The EcoRI fragment containing the promoter region was cleaved from pCRII and cloned into plasmid pLC4 (21), generating transcriptional fusion to the xylE reporter gene. The orientation and authenticity of the cloned promoter fragments were confirmed by restriction analysis and DNA sequencing. As a positive control, a 183-bp HindIII fragment containing the bla promoter of S. aureus from plasmid pRN6735 (13) was cloned into plasmid pSL24 (23) to form a transcriptional fusion with the xylE reporter gene. For gel shift assays with the putative repressor protein, the 51-bp DNA fragment (nucleotides [nt] 71 to 119 with flanking BamHI sites) and the 34-bp region encompassing the repressor protein binding site (nt 567 to 600) (1) were synthesized chemically and cloned into the BamHI site of pUC18. All transcriptional fusions and relevant constructs in different mutants are described in Table 1.
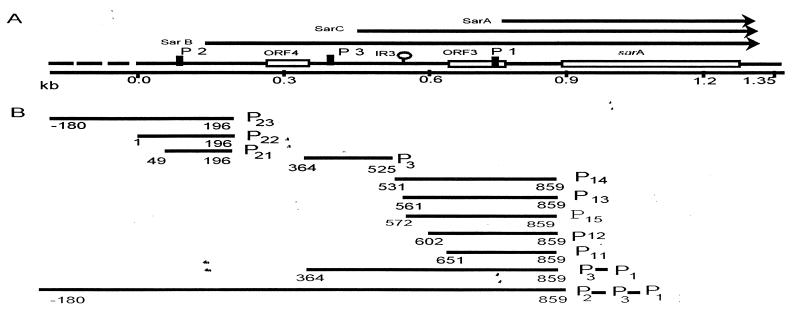
FIG. 1.
Organization of the sar locus. (A) Schematic representation of the sar locus showing various transcripts, sarA, sarC, and sarB that originate from the P1, P3, and P2 promoters (arrows), respectively. The ORFs are indicated by boxes, promoters are indicated by vertical solid bars, and an inverted repeat (IR3) is indicated by a hairpin loop structure. The DNA sequence of the 1.35-kb sar locus region has been published (1), as indicated by a solid line, whereas the broken line region (180 bp) is unpublished. (B) Different promoter fragments of the sar locus were used to construct the transcriptional fusions to the xylE reporter gene. The numbers are the nucleotide positions from the published sequence (1).
Genetic manipulations in S. aureus.
Different transcriptional fusions and other constructs were first transformed by electroporation to S. aureus RN4220, a restriction-deficient derivative of strain 8325-4 (17). Transformants were selected on NYE agar (14) containing 10 μg of chloramphenicol per ml. For transduction, phage φ11 was used to produce a phase lysate of strain RN4220 containing various sar transcriptional fusions. The phage lysate was then used to infect the recipient strain of S. aureus as described previously (4). The presence of the correct plasmids was confirmed by restriction mapping.
Single copies of specific sar fragments were introduced into the chromosome of sar mutant ALC488 as previously described (2). In brief, a specific sar fragment was cleaved from pCRII and cloned into the polyclonal site of the integration vector pCL84 (14). Upon transformation into strain CYL316, a derivative of RN4220 containing the integrase gene in trans, this vector inserts preferentially into the lipase gene (geh) of the host chromosome, resulting in tetracycline-resistant integrants with a loss of lipase activity. The integrated fragment was transduced into the sar mutant ALC488 as described previously (2, 3). Correct integration was verified by Southern blotting with lipase- and sar-specific probes.
A sigB mutant of RN6390 was constructed as described previously (4) by transducing the parental strain with a phage lysate of strain RUSA168 carrying the sigB mutation (27).
Catechol 2,3-dioxygenase assays.
For enzymatic assays, overnight cultures were diluted 1:50 in 250 ml of tryptic soy broth containing the appropriate antibiotics and shaken at 37°C and 200 rpm. Starting after 3 h of growth, 10 to 50 ml of cell culture corresponding to different optical densities at 600 nm (OD600) was serially removed and centrifuged. The cells were washed twice with 1 ml of ice-cold 20 mM potassium phosphate buffer (pH 7.2). Pellets were resuspended in 500 μl of 100 mM potassium phosphate buffer (pH 8.0) containing 10% acetone and 25 μg of lysostaphin per ml and incubated for 15 min at 37°C and then iced for 5 min. Extracts were centrifuged at 20,000 × g for 50 min at 4°C to pellet cell debris. The XylE (catechol 2,3-dioxygenase) assays were determined spectrophotometrically at 30°C in a total volume of 3 ml of 100 mM potassium phosphate buffer (pH 8.0) containing 100 μl of cell extract and 0.2 mM catechol as described previously (28). The reactions were allowed to proceed for 25 min, with OD375 readings taken at the 2-, 5-, 15-, and 25-min time points. One milliunit is equivalent to the formation of 1.0 nmol of 2-hydroxymuconic semialdehyde per min at 30°C. Specific activity is defined as milliunits per milligram of cellular protein (28).
Purification of the 12-kDa protein.
The cell extract of S. aureus RN6390 was used to purify protein with binding activity to the sar promoter. The culture was grown overnight in 1 liter of CYGP medium and harvested by centrifugation. Cells were washed with 20 mM potassium phosphate (pH 7.5) and resuspended in buffer (100 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT]). The cells were lysed by incubation with lysostaphin (25 μg/ml) for 30 min at 10°C and then frozen at −70°C. After repeating freezing and thawing twice, the lysate was centrifuged at 35,000 rpm (TLA100.4 rotor in an Optima TL ultracentrifuge; Beckman Instruments, Fullerton, Calif.) for 40 min to remove cellular debris and dialyzed against buffer A (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol). The protein solution was applied to a 10-ml heparin-Sepharose column preequilibrated with buffer A. The column was then washed with five column volumes of buffer A and eluted with a continuous gradient of buffer A and buffer B (buffer A containing 1.5 M NaCl). Fractions were assayed for DNA binding activity by gel shift assays with an γ-32P-labeled 51-bp sar promoter fragment which encompasses the sequence upstream of the sar P2 promoter (nt 71 to 119) (1). This fragment (nt 71 to 119) (1) was originally synthesized and cloned into the BamHI site of pUC18. The 51-bp BamHI fragment for gel shift assays was gel purified. Fractions containing DNA binding activity were pooled, dialyzed against buffer A, and loaded onto a preequilibrated 5-ml DNA-specific column containing the 51-bp DNA fragment covalently linked to Sepharose as described by Hughes et al. (11). The column was washed with buffer A and eluted with a linear gradient of buffer A to buffer B. Fractions with DNA binding activity as determined by gel shift assays were pooled, dialyzed against buffer A with 50% glycerol, and stored at −70°C. Protein concentration was estimated with the Bio-Rad Protein Assay with bovine serum albumin as the standard. The apparent molecular weight of the putative protein was assessed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
Gel shift assays.
To determine if the 12-kDa DNA binding protein interacts with the sar promoter fragments, DNA fragments were end labeled with [γ-32P]ATP by using polynucleotide kinase. Labeled fragments were incubated at room temperature for 15 min with the indicated amount of purified protein in 25 μl of binding buffer (25 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 75 mM NaCl, 1 mM DTT, 10% glycerol) containing 0.5 μg of calf thymus DNA. The reaction mixtures were analyzed by nondenaturing polyacrylamide gel electrophoresis. The band shifts were detected by exposing dried gels to film.
RESULTS
Rationale for the construction of various transcriptional fusions of the sar locus.
In previous studies, we demonstrated that the expression of sar transcripts varies during the growth cycle, with sarA and sarB being most abundant in the exponential phase and with sarC being maximally expressed toward the postexponential phase (1). Our speculation is that these three transcripts reflect the activities of three different sar promoters and that one or more trans-acting regulatory elements may control the expression of different sar promoters by binding to the respective upstream region. To confirm the existence and the strength of these sar promoters and to detect possible regulatory regions within sar, a series of XylE transcriptional fusions with various lengths of different sar promoters were constructed (Table 1 and Fig. 1). Three fusions of the P2 promoter, P21, P22, and P23, containing 70, 120, and 300 bp upstream of the deduced −35 promoter box, respectively, were prepared. With the P3 promoter region relatively short, we constructed a single fusion with a 57-bp fragment upstream of the −35 promoter box. We speculated that an inverted repeat (IR3 [nt 553 to 593]) upstream of P1 may play an important role in down-regulating sarA transcription. To investigate the regulatory function of this region, we constructed five transcriptional fusions, P11, P12, P15, P13, and P14, to include sequences 35, 85, 119, 125, and 155 bp upstream of the P1 −35 promoter box, respectively (Fig. 1 and 2). All of these constructs were introduced into the wild type and an assortment of S. aureus mutant strains and assayed for the activity of catechol 2,3-dioxygenase, an enzyme which is the gene product of the xylE reporter gene.
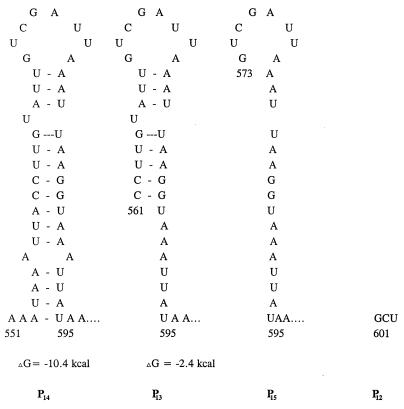
FIG. 2.
Proposed stem-loop structure for the inverted repeat region (IR3) upstream of the P1 promoter of the sar locus. The numbers are the nucleotide positions from the published sequence (1).
Transcriptional fusion studies of the wild type and sar mutant strains of S. aureus.
To determine the relative strength of these promoters and the regulatory region within IR3 upstream of the P1 promoter, the transcriptional activity of all three sar promoters was analyzed with the parental strain and an isogenic sarA mutant of S. aureus (Table 2). Based on XylE activity, the P1 promoter (P11 and P12) was the strongest, with ≈50-fold-more activity than the homologous P2 and P3 promoters in parental strain RN6390. The activities of the P21, P22, and P23 promoter fusions were similar, thus suggesting that a small region (i.e., beyond P21 from position 49 to −181 [Fig. 1]) upstream of the −35 promoter box is not critical to the activation of the P2 promoter. The P3 promoter was the weakest among these three promoters. Analysis of the expression of P11, P12, P15, P13, and P14 promoter fusions in parental strain RN6390 suggested that the activities of P11 and P12 were two of the strongest among all P1 promoter constructs. With P15, P13, and P14, XylE activities were approximately 2- to 3 1/2-fold lower than the corresponding P11 and P12 fusions. To assess the discrepancy in the expression of various P1 constructs, we analyzed the upstream region of the P1 promoter and found that P11 and P12 constructs lacked an inverted repeat region (IR3). Figure 2 shows the proposed secondary structure of the inverted repeat region (repeats at positions 553 to 569 and 578 to 593) among various constructs of the P1 promoter region. With both repeats, the P14 promoter fragment is capable of forming a stable stem-loop structure (ΔG = −10.4 kcal), whereas the P13 promoter fragment contained a partial stem-loop structure (ΔG = −2.4 kcal) (Fig. 2). The P15 construct retained half of the inverted repeats, thus disrupting the stem-loop structure. A lower level of expression for the P15 fusion compared with the P12 construct indicated that the region comprising approximately one-half of the inverted repeats (nt 567 to 593) likely encompasses the sequence necessary for down-regulating sarA transcription at the transcriptional level. Remarkably, the combined P3-P1 fusion activity was lower than that of the P11 or P12 fusion. We speculate that the P3-specific promoter sequence may be a binding site for a repressor protein (see below). Alternatively, the sequence itself may be part of secondary structure which interferes with transcription from the P1 promoter. Interestingly, maximum promoter activity was observed with the combined fusion of the P2-P3-P1 promoters, with the level of activity higher than that of any of the individual sar promoters. Using the bla promoter as a control, we found that it was stronger than the P2 and P3 promoters but about twofold weaker than the P1 promoter. In contrast, the vector control or the constructs with divergent orientation had little, if any, XylE activity (Table 2).
TABLE 2.
Expression of xylE fusion from different sar promoters in the wild type and an isogenic sarA mutant of S. aureus
| Construct | Orientation with respect to the xylE gene | XylE activity (mU/mg of protein)a
|
|
|---|---|---|---|
| RN6390 | ALC488 | ||
| P21 | Convergent | 2.9 | 0.9 |
| P22 | Convergent | 4.0 | 1.2 |
| P23 | Convergent | 5.5 | 0.8 |
| P3 | Convergent | 3.0 | 0.4 |
| P11 | Convergent | 313.8 | 27.6 |
| P12 | Convergent | 282.3 | 39.6 |
| P13 | Convergent | 91.8 | 13.8 |
| P15 | Convergent | 132.6 | 19.0 |
| P14 | Convergent | 137.2 | 18.4 |
| P3-P1 | Convergent | 74.6 | 35.4 |
| P2-P3-P1 | Convergent | 441.8 | 29.6 |
| Vector | 1.0 | 1.0 | |
| Pbla | Convergent | 69.8 | 61.1 |
| Pcb | Divergent | 1.1 | 0.8 |
Activity is defined as milliunits per milligram of total cellular protein. The value for each sample at a particular growth phase (OD600 = 1.7) is the mean value assayed at four different time points after the addition of the catechol.
Pc represents the average value of the divergent promoter fusions P21–23, P3, P11–15, P3-P1, P2-P3-P1, Pbla, with most values approaching the background level.
Similar expression studies were also performed with the isogenic sarA mutant ALC488 (3), in which the sarA gene was interrupted by an ermC gene inserted at nt 971 (1). Prior transcriptional analysis indicated that the transcription of sarA, -C, and -B was disrupted in this mutant (3). As shown in Table 2, a 2- to 15-fold reduction in promoter activity was observed in assorted transcriptional fusions during the early stationary phase in the mutant background. Similar reductions were also observed in the log and postexponential phases, thus indicating that the sar locus is involved in its own expression (data not shown). With the strongest promoter, P1, there was at least a sixfold decrease in promoter activity in all five fusion constructs in the sar-negative mutant background. The reduction was about 15-fold in the combined fusion of the P2-P3-P1 promoters. The lower transcriptional activities in the mutant background were not attributable to reduced plasmid copy number, because the plasmid yield was found to be consistent in quantity when isolated from various strains. In addition, we assayed for chloramphenicol acetyltransferase activities for the P2-P3-P1 fusion and found them to be equivalent in strains RN6390 and the sar mutant ALC488 harboring the fusion, thus implying similar plasmid copy numbers between the isogenic pair (data not shown). The relative constancy of the plasmid number was also supported by the observation that the control bla promoter as well as the divergent fusion constructs had similar promoter activities in the wild type and the isogenic sarA mutant. Taken together, these data support the notion that one or more of the sar gene products likely activates the expression of its own promoters.
To reconfirm the autoregulatory nature of the sar locus, we integrated a single copy of one of these promoter constructs, P14, into the lipase gene on the chromosome of parental strain RN6390 and isogenic sarA mutant ALC488 via the recombinant integration vector pCL84 containing the P14-xylE fusion in proper orientation. As with the multicopy counterpart, the transcriptional activity was reduced in the sar mutant ALC488 (2.24 mU/mg of cellular protein) compared with that in the parent (8.12 mU/mg of protein).
Growth-phase-dependent expression of different sar promoters.
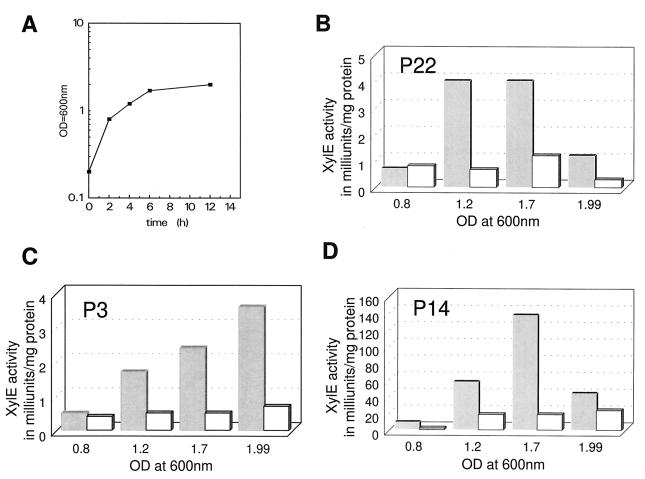
Samples from S. aureus strains containing different fusions were assayed for XylE activity during the growth cycle (Fig. 3). With P22 and P14 promoters, XylE activity generally increased toward the early stationary phase and then tapered off in the late stationary phase (overnight growth). In contrast, the P3 promoter revealed a steady increase in promoter activity, even toward the late stationary phase (3.6 versus 0.5 mU/mg of protein for overnight culture and mid-log phase, respectively). This result is akin to our prior transcriptional data (1), in which we found that the sarC transcript that originated from the P3 promoter was maximally transcribed during the postexponential phase. These findings support the notion that the expression of sar from different promoters is growth phase dependent.
FIG. 3.
XylE activity in the P22, P3, and P14 constructs (B, C, and D) during the growth cycle. Cells obtained at four sample points during the growth cycle (A [semilog scale]) were lysed with lysostaphin and assayed for catechol 2,3-dioxygenase activity as described previously. The average values for the constructs (n = 4) are presented in milliunits per milligram of cellular protein. The open and shaded bars represent data derived from RN6390 and ALC488 (sar mutant), respectively.
The P3 promoter is sigB dependent.
In previous studies, we showed that the P1 promoter shares homology with the ς70-dependent consensus sequences, while the P3 promoter closely resembles the consensus sequence of sigB-dependent promoters (1). To investigate whether the P3 promoter is truly sigB dependent in vivo, a sigB mutant, ALC1001, of S. aureus RN6390 was constructed by transducing the mutation from mutant RUSA168 (27). Transcriptional fusions of the sar promoters were introduced into the mutant strain ALC1001 and assayed for XylE activity (Table 3). With the exception of the P3 promoter, the pattern of activities for most fusions remained similar to those found in the wild type. Notably, the P3 promoter activity became negligible in this background. As with those found in the parental background, the combined P3-P1 fusion activity was also substantially lower than the activity of the P11 promoter in the sigB mutant background. These results suggest that the central P3 promoter of the sar locus likely utilizes a ςB-dependent form of RNA polymerase to initiate transcription in S. aureus. As an additional confirmation of these data, transcriptional analysis of the sar locus in the sigB mutant ALC1001 revealed that the sarC transcript initiated from the P3 promoter was absent (data not shown). A recent in vitro study with purified SigB and RNA polymerase from S. aureus has also shown that the P3 promoter of sar is dependent on SigB (8).
TABLE 3.
Activities of different sar promoters in a sigB mutant of S. aureus
| Constructa | XylE activity (mU/mg of protein)b
|
|
|---|---|---|
| Wild-type RN6390 | ALC1001 | |
| P21 | 7.4 | 8.2 |
| P3 | 6.1 | 0.6 |
| P11 | 302.0 | 256.0 |
| P3-P1 | 73.1 | 38.0 |
| P2-P3-P1 | 486.0 | 273.0 |
| Vector | 0.6 | 0.8 |
Note that all constructs (excluding vector) were convergent in terms of their orientation with respect to the xylE gene.
Activity is defined as milliunits per milligram of total cellular protein. The value for each sample at a particular growth phase (OD600 ≈1.9 from overnight cultures) is the mean value assayed at two different time points after the addition of catechol.
Transcriptional fusion studies in complemented sar mutant strains.
In previous studies, we found that the introduction of a sarC fragment into a sar mutant was sufficient for complementation; however, complete restoration of the sar-related phenotypes required the presence of a sarB fragment in the mutant (2). In promoter fusion studies involving the parental strain and the sar mutant ALC488 (sarA::ermC), our data clearly indicated that sar gene products partially regulate promoter activation from sar promoters (Table 2). To determine whether the observed reduction in promoter activity in the sar mutant is due to a loss of sarA function or to those proteins encoded by the sequence upstream of sarA (e.g., ORF3 and ORF4), single copies of sarA, sarC (sarA with ORF3), and sarB (sarA with ORF3 and ORF4) were introduced into sar mutant ALC488 to form ALC996, ALC997, and ALC812 via the integration vector pCL84, which preferentially integrated into the lipase gene (geh) on the host chromosome (Tables 1 and 4).
TABLE 4.
Expression of xylE fusion from different sar promoters in sarA mutant strain ALC488 complemented with various sar fragments
| Construct | Orientation with respect to xylE | XylE activity (mU/mg of protein)b
|
||||
|---|---|---|---|---|---|---|
| RN6390a | ALC488a | ALC812 (sarB) | ALC996 (sarA) | ALC997 (sarC) | ||
| P22 | Convergent | 4.00 | 1.20 | 4.63 | 2.94 | 8.3 |
| P3 | Convergent | 3.00 | 0.4 | 13.42 | 4.0 | 5.0 |
| P11 | Convergent | 313.8 | 27.61 | 407.50 | 272.4 | 177.8 |
| Vector | 1.02 | 0.9 | 0.73 | 0.55 | 0.61 | |
| Pcc | Divergent | 1.07 | 0.83 | 0.79 | 0.60 | 1.08 |
The data for RN6390 and ALC488 are taken from Table 2.
Activity is defined as milliunits per milligram of total cellular protein. The value for each sample at a particular growth phase (OD600 = 1.7) is the mean value assayed at four different time points after the addition of the catechol.
Pc represents the average value of the divergent promoter fusions P22, P3, and P11.
As shown in Table 4, both P2 (P22) and P3 promoter activities in sarA-complemented strains (2.94 and 4.0 mU/mg of protein, respectively, for P2 and P3 in ALC996) were generally comparable to those found in parental strain RN6390 (4.0 and 3.0 mU/mg of protein). However, in comparison to the sar mutant ALC488, P2 and P3 promoter activities were higher as the size of the complemented sar fragments increased from sarA to sarB (4.63 and 13.42 mU/mg for P2 and P3 activities, respectively [Table 4]). The restoration of P2 and P3 promoter activities in the sar mutant ALC488 to parental levels by the sarA transcript fragment itself (ALC996) revealed that the SarA protein was probably responsible for modulating these promoter activities in the mutant, since we have previously shown that the effector molecule within sar is the protein rather than the sarA transcript (2). As with the P2 and P3 promoters, the P1 promoter activity was significantly enhanced in complemented strains (e.g., 272.4 mU/mg for P11 in sarA-complemented strain ALC996) compared with the mutant ALC488 alone (27.6 mU/mg of protein for P11). The augmentation in XylE activity in P11 was higher in the sarB-complemented strain (ALC812) than in the sarA-complemented strain (ALC996). This finding argues for additional factors other than sarA but that are encoded by sarB and that may serve to augment transcriptional activity of the P1 promoter of the sar locus in S. aureus.
Purification of a putative 12-kDa protein.
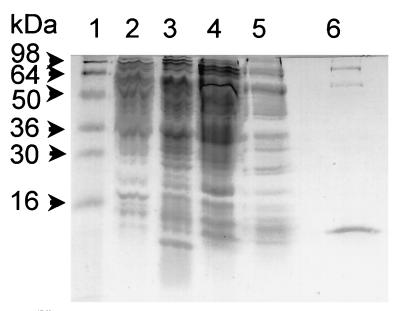
Two experimental observations led us to consider that some factor(s) or protein(s) may bind the upstream region to down-modulate the expression from sar promoters. First, transcriptional analysis of the sar locus revealed a gradual decrease in sarA and sarB transcription and an increase in sarC transcription as bacterial cells make the transition between the late log and stationary phases of growth (Fig. 3). Second, the differential promoter activity as expressed in the complemented sar mutant strain implies that a factor or factors other than SarA protein may bind to the sar promoter region (Table 4). A close inspection of the upstream region, including the inverted repeat (IR3) and the minimum 34-bp sequence (nt 567 to 600 in P15 [Fig. 2]) required for down-regulating the P1 promoter activity, reveals the presence of a 7- to 8-bp sequence (TAAATTAA) which is repeated 11 times within the P1-P3-P2 promoter region. It seems reasonable to surmise that this sequence may be involved in presenting the binding site for a common regulatory protein. Accordingly, we synthesized a 49-bp DNA sequence of the P2 promoter region (from nt 71 to 119) (1) which encompassed the AT-rich UP box of the P2 promoter region as well as the 8-bp repeat. This DNA fragment was then conjugated to CNBr-activated Sepharose 4B (Pharmacia) to produce an affinity column. Putative DNA protein was purified from lysates of S. aureus RN6390 by first using a heparin-Sepharose column followed by the DNA-specific column. The details for purification of the putative protein were described in Materials and Methods. Gel shift analysis of putative fractions obtained during the purification was done with a labeled 51-bp fragment encompassing the 49-bp sequence (data not shown). By this technique, we purified a 12-kDa protein which was analyzed by SDS gels (Fig. 4, lane 6) to be ∼90% pure.
FIG. 4.
SDS-polyacrylamide gel showing different stages of purification and the purity of the purified protein. Fractions were subjected to SDS–12% polyacrylamide gel electrophoresis and stained with Coomassie blue dye. Lanes 1, molecular mass marker (Novex, San Diego, Calif.); 2, 10 μg of total cell extract proteins; 3, 10 μg of heparin-Sepharose column-bound fraction; 4, 10 μg of nonbound heparin-Sepharose column fraction; 4, 10 μg of flowthrough fraction of the DNA-specific column; 6, 1 μg of the purified protein fraction from the affinity column.
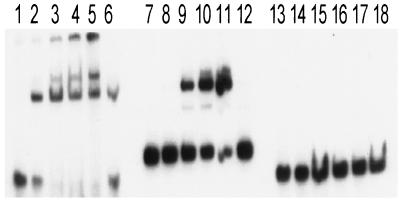
In gel shift studies of the labeled 51-bp probe with the purified 12-kDa protein, retardation of the probe was observed with increasing amounts of the purified protein, suggesting binding specificity (Fig. 5, lanes 1 to 5). As expected, unlabeled P2 promoter fragment (P21) competed successfully as an inhibitor (lane 6). The retarded bands in the gel shift assay indicate that the 51-bp fragment possibly contains three binding sites for this protein. An alternative explanation will be the binding of 12-kDa multimers to a single binding site. Interestingly, when a labeled 80-bp fragment containing the 34-bp sequence (nt 567 to 600 [Fig. 2]) upstream of P1 and flanking polylinker sequence of pUC18 was used for gel shift assays, a single retarded band was observed (Fig. 5, lanes 7 to 11). In contrast, a labeled 53-bp fragment containing only the polylinker region from pUC18 did not result in any gel shift activity with equivalent amounts of the purified protein (Fig. 5, lanes 13 to 18). Thus, the 34-bp sequence (nt 567 to 600) (Fig. 2) likely constitutes one binding site for this protein. Additional gel shift experiments with overexpressed and highly purified protein suggested that the partially purified protein behaves similarly and thus is specific to its substrates. Because the 34-bp sequence is responsible for down-modulating P1 transcription (see P15 versus P12), we speculate that the 12-kDa protein may act as a repressor for sar expression. Clearly, more extensive work needs to be done to confirm the functional and physical characterization of this 12-kDa protein. Preliminary analysis suggests that the purified protein is not SarA nor histone (HU)-like protein, but does have limited homology to the SarA protein of S. aureus.
FIG. 5.
Autoradiogram of a nondenaturing 10% polyacrylamide gel showing the binding of the 12-kDa purified protein to a 51-bp radiolabeled DNA fragment [GATCC (nt 73) TTAAGACC-TAAATTAA-TGTTATTTTTTAA-TAATTTA-CACC-AAATTAA-(nt 119) G (the nucleotides in the repeated 7- or 8-bp sequence in the proper or complementary orientation are underlined)] (1) of the upstream P2 promoter region, an 80-bp labeled fragment containing the 34-bp sequence with the polylinker region of pUC18, and a 53-bp polylinker fragment from pUC18. Lanes: 1 to 5, mobility of the 51-bp DNA fragment of the upstream P2 promoter in the presence of 0, 30, 100, 200, and 300 ng of the purified protein, respectively; lane 6, mobility of the same fragment in the presence of 300 ng of the purified protein and 40-fold (molar ratio) of unlabeled P21 fragment as the specific competitor. Lanes 7 to 11 and 13 to 17 represent mobility shifts of the 80-bp DNA fragment containing the 34-bp binding site for the putative repressor together with flanking polylinker sequence and equivalent amounts of 53-bp polylinker fragment of pUC18, respectively. Lanes 12 and 18 are competition assays with unlabeled P21 and 53-bp pUC18 fragments, respectively.
DISCUSSION
The sar locus is a complex regulatory system with a 372-bp sarA gene preceded by an 800-bp extended triple-promoter region. Within the promoter region are multiple repeats and inverted repeats as well as potential peptide coding regions that may form a complex network for sar promoter activation (1). To confirm the existence of three distinct promoters and to ascertain the role of these secondary structures and its mode of regulation, we cloned various lengths of putative sar promoters, both singly and in combinations, into the transcriptional vector pLC4 by using xylE as a reporter gene. The results of our reporter fusion studies (Table 2) clearly confirm our previous transcriptional data (1) that the smaller transcripts do not arise from degradation of the larger sarB transcript because these three promoters can exist as distinct entities in transcriptional fusions. Among these, P1 is the strongest individual promoter compared with P2 and P3. Because the region immediately upstream of the P1 promoter including a 16-bp inverted repeat (IR3 in Fig. 2) and a putative ORF (ORF3) may modulate sarA transcription from P1 (1), we cloned various lengths of the P1 promoter into the fusion vector and found that shorter promoter fragments (P11 and P12) were stronger than their lengthy counterparts (i.e., P15, P13, and P14 in Table 2). Recognizing that P15, P14, and P13 encompass a promoterless ORF3, it becomes unlikely that ORF3 plays a major role in down-modulating sarA transcription from P1. In retaining the entire or partial 16-bp inverted repeats in P14 and P13 and also preserving only one-half of the repeat in P15, we showed that it is the sequence within the inverted repeat (IR3) rather than the repeat itself (1) that is critical to the down-modulation of sarA transcription from the P1 promoter, since comparable promoter activities were found among the P13, P14, and P15 constructs (Table 2).
The P2 promoter is at least 20-fold weaker than P1 but comparable to P3 (Table 2). Conceivably, differences in the promoter structure between P1 and P2 may have accounted for the disparate promoter activities. Both the P1 and P2 promoters have features that resemble a ς70-dependent promoter; a comparative analysis revealed that the −10 and −35 consensus sequence of the P1 promoter (TTTACT-N18-TATAAT), like that of the blaZ promoter (TTGACA-N18-TATTAT), closely resembles the E. coli canonical consensus sequence (TTGACA-N14–21-TATAAT) (22), whereas the P2 promoter (TAGCAAA-N17-TAATAT) is less conserved. However, the direct reliance of the P2 promoter on the ςA-dependent form of RNA polymerase has not been confirmed in vitro. The effect in varying the extent of the P2 promoter is not significant compared with that of P1, thus implying that the structure 70 bp upstream of P2 (i.e., beyond P12) is not critical to the regulation of P2 transcription. Alternatively, another factor or factors may bind to this region to suppress promoter activity of the transcriptional fusion. Remarkably, when the P2 promoter was fused to P3 and P1, the activity became more potent than that of P11. However, we observed that the P3-P1 promoter fusion had lower activity than P1 alone. A plausible explanation for this divergent finding may be that the P3 promoter region may be a binding site for a repressor protein that down-modulates P1 transcription while the region upstream of P2 may be involved in either positive regulation of P1 or negative regulation of P3 promoter. Clearly, more extensive work needs to be done to understand the mode of P2 and P3 promoter activation.
In contrast to P1 and P2, P3 has a putative sigB-dependent promoter with a typical −10 promoter box (GGGTAT) (1). Because sigB promoters in gram-positive bacteria (e.g., Bacillus subtilis) are typically activated during periods of stress, including the postexponential phase (26), our results also showed that P3 was most active in late stationary or overnight cultures (Fig. 3), while the ςA-dependent promoters of P1 and P2 had lower activities compared to those during the late-log or early stationary growth phases. Wherein P1 and P2 remained active in a sigB mutant, the P3 promoter activity became silent in this background (Table 3). These findings coincided with our observation that sarC transcription is absent in a sigB mutant (unpublished data). In addition, recent in vitro transcription assays with purified SigB protein and core RNA polymerase of S. aureus also supported the notion that P3 promoter is sigB dependent (8). Taken together, our data confirmed that the P3 promoter utilizes the SigB-dependent form of RNA polymerase as found in other stress-response promoters of gram-positive bacteria.
Previously, the transcription of RNAII and RNAIII was found to be dramatically reduced in an agr mutant (15), thus revealing the autoregulatory feature of the agr system. Similar to agr, we discovered that activation of sar promoters, either singly or in combination, is dependent on the expression of sar gene products (see sar mutant ALC488 in Table 2). In contrast, the Pbla promoter activity remained unaffected in the sar mutant background. To minimize the issue arising from plasmid copy number, we have also introduced single copy of the promoter construct P14 to a pair of isogenic sar strains. As anticipated, the P14-XylE activity in the sarA mutant was reduced approximately sevenfold compared to that of the parental strain when measured with the multicopy fusion and approximately threefold when measured with the single-copy fusion. To further confirm the autoregulatory nature of sar, we conducted complementation studies of sar mutant strains (ALC488) carrying single copies of sarA, sarC, or sarB. As predicted, promoter activities of P1, P2, and P3 were restored to near-parental levels with the strain containing the largest transcript, sarB, while those containing sarA and sarC were at slightly lower levels. Collectively, our data strongly support the notion that sar gene products, consisting of SarA and possibly other encoded elements upstream (2), contribute directly or indirectly to sar’s own regulation. However, factors other than sar gene products must also play a role in modulating sar promoter activity, because significant residual promoter activities (P1, P3-P1, and P2-P3-P1) were retained in the sar mutant ALC488 (Tables 2 and 4).
In scanning the UP element in P2 and P3 promoters, as well as the 34-bp binding site (Fig. 2) for a putative repressor protein, we found a conserved 7- to 8-bp sequence (TAAATTAA) which was repeated 11 times in the 800-bp sar promoter region, including several repeats in the region upstream of the P1 and P2 −35 promoter boxes. This 7- to 8-bp sequence is also part of the inverted repeat (nt 553 to 593) found within the P15, P13, and P14 promoter constructs. Although S. aureus chromosomal DNA is 70% AT rich, this 8-bp sequence is rarely found, as demonstrated by the fact that this motif was found only once in a 6-kb agr sequence. We thus speculate that this may be a possible binding site for a DNA binding protein involved in regulating sar gene expression.
Remarkably, we were able to purify a 12-kDa protein with a DNA-specific column to which a 49-bp sequence (nt 71 to 119) comprising the UP box and this repeat was covalently linked via CNBr-activated Sepharose. Although this sequence was derived from the P2 promoter region, gel shift assays with the purified 12-kDa protein revealed that it also bound to a 34-bp fragment (nt 567 to 600) (Fig. 2) upstream of the P1 promoter in a dose-dependent manner (Fig. 5). Notably, this fragment yielded only one retarded band, whereas the 51-bp fragment resulted in three shifted bands in gel retardation assays (Fig. 5). In comparing these two sequences, we found that the 51-bp fragment contained three 7- to 8-bp repeats (see legend to Fig. 5), while the 34-bp fragment (TGTCGATTAAATTAA-GG-TAAATTA-TAA) encompassed two repeats. We speculate that proper conformation of the conserved 7- to 8-bp sequence, as influenced by the adjoining sequence, may serve as a binding site for the putative 12-kDa protein in sar regulation. Whether the formation of three shifted bands with the 51-bp fragment is a result of multiple binding sites or binding of protein multimers with increasing number of repeats is not clear. Because the 34-bp sequence constituting half of the 16-bp inverted repeat plays a role in down-regulating sarA expression from the P1 promoter (i.e., P15 versus P12 activity in Table 2), it is likely that the 12-kDa protein is involved in repressing promoter activity from the P1 promoter by binding to the 34-bp sequence in the wild-type strain. Nevertheless, the functional sequelae as a result of the binding of this protein to the 51-bp sequence upstream of the P2 promoter region are largely undefined but are currently under investigation.
In the araC family of regulatory proteins, it is known that environmental parameters may affect DNA topology, which, in turn, alters the transcriptional activity of the promoter. For instance, the invasive gene virB of Shigella flexneri is activated by an AraC-like regulator called virF but negatively regulated by virR. Although VirF has a limited sequence similarity to SarA (2, 6), it remains to be determined if the 12-kDa protein is an analogous repressor while SarA serves as an its own activator. Preliminary gel shift assays with purified SarA protein suggest that it also binds to the sar promoter fragments in a dose-dependent manner. However, the binding sequence appears to differ from the repressor site (data not shown). Nevertheless, this speculation is preliminary and will require additional confirmatory experiments. With additional functional characterization, including binding studies of P1 and P2 promoter fragments with the 12-kDa protein and SarA and analogous in vitro transcription assays, we will be able to dissect the regulatory mechanism in the expression of sar in S. aureus.
ACKNOWLEDGMENTS
We thank Alexander Tomasz for providing the sigB mutant strain RUSA168, Chia Lee for providing pCL84, and Y.-T. Chien and Kelly Eberhardt for helpful discussion.
This work was supported in part by grants-in-aid from the American Heart Association and the New York Heart Association and by NIH grants AI30061 and AI37142. M. G. Bayer was supported by a Norman and Rosita Winston Fellowship. A. L. Cheung is a recipient of the Irma T. Hirshl Career Scientist Award as well as the AHA-Genentech Established Investigator Award for the American Heart Association.
REFERENCES
- 1.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Projan S J, Edelstein R E, Fischetti V A. Cloning, expression, and nucleotide sequencing of a Staphylococcus aureus gene (fbpA) encoding a fibrinogen-binding protein. Infect Immun. 1995;63:1914–1920. doi: 10.1128/iai.63.5.1914-1920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien Y T, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 8.Deora R, Tseng T, Misra T K. Alternative transcription factor ς5B of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes M J, Jost J P, Jiricny J. Purification of sequence specific DNA-binding proteins by affinity chromatography. Biomethods. 1991;5:221–231. [Google Scholar]
- 12.Janzon L, Arvidson S. The role of the δ-hemolysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornblum J, Kreiswirth B, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 14.Lee C Y, Buranen S L, Ye Z H. Construction of single copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 15.Morfeldt E, Janzon L, Arvidson S, Löfdahl S. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol Gen Genet. 1988;211:435–440. doi: 10.1007/BF00425697. [DOI] [PubMed] [Google Scholar]
- 16.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 17.Novick R P. The staphylococcus as a molecular genetic system. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 1–40. [Google Scholar]
- 18.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 19.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3977. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray C, Hay R E, Carter H L, Moran C P., Jr Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J Bacteriol. 1985;163:610–614. doi: 10.1128/jb.163.2.610-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 23.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smeltzer M S, Hart M E, Iandolo J J. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect Immun. 1993;61:919–925. doi: 10.1128/iai.61.3.919-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldvogel F A. Staphylococcus aureus. In: Mandell G L, Douglas R G J, Bennett J E, editors. Principles and practice of infectious diseases. New York, N.Y: John Wiley & Sons; 1985. pp. 1097–1116. [Google Scholar]
- 26.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zukowski M M, Gaffney D G, Speck D, Kauffman M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]