Abstract
Hydrophobic membrane proteins are cotranslationally targeted to the endoplasmic reticulum (ER) membrane, mediated by hydrophobic signal sequence. Mitochondrial membrane proteins escape this mechanism despite their hydrophobic character. We examined sorting of membrane proteins into the mitochondria, by using mitochondrial ATP-binding cassette (ABC) transporter isoform (ABC-me). In the absence of 135-residue N-terminal hydrophilic segment (N135), the membrane domain was integrated into the ER membrane in COS7 cells. Other sequences that were sufficient to import soluble protein into mitochondria could not import the membrane domain. N135 imports other membrane proteins into mitochondria. N135 prevents cotranslational targeting of the membrane domain to ER and in turn achieves posttranslational import into mitochondria. In a cell-free system, N135 suppresses targeting to the ER membranes, although it does not affect recognition of hydrophobic segments by signal recognition particle. We conclude that the N135 segment blocks the ER targeting of membrane proteins even in the absence of mitochondria and switches the sorting mode from cotranslational ER integration to posttranslational mitochondrial import.
INTRODUCTION
The majority of membrane proteins in the secretory pathway of eukaryotic cells are synthesized on the endoplasmic reticulum (ER) and cotranslationally integrated into the membrane, and then sorted to their final destinations via vesicle transport. The ER targeting and membrane integration of nascent polypeptide chains are initiated by the signal sequence (Keenan et al., 2001), which is defined by a hydrophobic segment (Sakaguchi, 1997). The 54-kDa subunit of signal recognition particle (SRP) recognizes the hydrophobic segment just after it emerges from the ribosome, and then the SRP arrests polypeptide chain elongation on the ribosome. The SRP and ribosome-nascent chain complex is targeted to the ER membrane, where the hydrophobic segment is released from the SRP by the SRP receptor and is transferred into the protein translocation channel, the so-called translocon. The translocon is comprised of the Sec61 complex and forms an aqueous environment in the membrane (Johnson and van Waes, 1999). The translocon is responsible not only for protein translocation across the membrane but also for integration of various membrane proteins into the ER. The sequential processes of targeting and integration occur during protein synthesis.
In contrast to the cotranslational integration of membrane proteins into the ER membrane, most mitochondrial proteins are believed to be synthesized by free ribosomes in the cytosol. Once released into the cytoplasm as a preprotein bearing an N-terminal extension, the so-called presequence, the preprotein is imported into the mitochondria posttranslationally. The tight coupling of the targeting of hydrophobic signals to the ER and polypeptide chain elongation suggests that membrane proteins that possess hydrophobic segments are predominantly targeted to the ER. How do the various mitochondrial membrane proteins escape the cotranslational targeting mechanisms to the ER membrane?
Some membrane proteins destined for the mitochondria have less hydrophobic transmembrane (TM) segments than those destined for the ER, and it is possible that the lower hydrophobicity is critical determinant to escape the ER targeting mechanisms. Previous study demonstrated that the less hydrophobic characteristics in TM segments of Tom5 and Tom20 are essential for correct targeting to the mitochondrial outer membrane; when the hydrophobicity of the TM segments was increased by mutations, those mutants were targeted to the ER and were excluded from the mitochondria in cultured cells (Kanaji et al., 2000; Horie et al., 2002).
On the other hand, another group of mitochondrial membrane proteins possess highly hydrophobic TM segments. Mitochondrial isoforms of the ATP-binding cassette (ABC) transporter family are typical examples. The ABC transporters are multispanning membrane proteins involved in transmembrane transport of various materials (Schmitt and Tampe, 2002). Among them, many isoforms are integrated into the membrane via the cotranslational targeting mechanism to the ER and localized in the organelle membrane of the secretory pathway, whereas some isoforms are sorted to other organelles such as mitochondria and peroxisomes despite their high hydrophobicity. We examined, in this study, the sorting process of hydrophobic membrane proteins into mitochondria, by using a mitochondrial isoform of the ABC transporter, ABC-me (Shirihai et al., 2000), which is a homologue of the human ABCB10 isoform. It possesses a 135-residue N-terminal hydrophilic segment (termed N135 here), which is rich in positively charged residues and likely to be the presequence for mitochondrial targeting (Figure 1, A and B). The membrane domain after N135 is highly hydrophobic and has a similar hydropathy profile to that of its plasma membrane counterpart multidrug resistance protein 1 (MDR1). Based on its hydrophobicity, it should be targeted to the ER via an SRP-mediated mechanism; however, it localizes in the mitochondrial inner membrane. Recently, Graf et al. (2004) demonstrated the following on the import of ABC-me molecule into mitochondria: mouse ABC-me possesses an exceptionally long presequence of 105 residues. When it was deleted, the membrane domain was targeted to the ER instead of mitochondria and dimerized as the full-length molecule did on the mitochondrial inner membrane. They also examined structural requirements within the 105-residue presequence and demonstrated that the central one-third is essential for mitochondrial targeting and the N-terminal one-third is also critical for correct import. The details of the function of the unique presequence during membrane protein sorting, however, are still unclear.
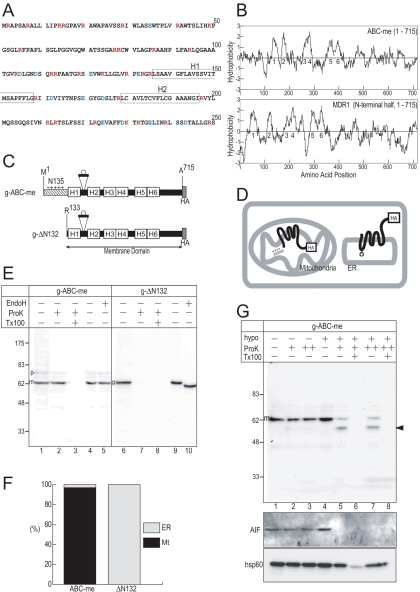
Figure 1.
Quantification of membrane topology and intracellular localization. (A) N-terminal sequence of mouse ABC-me. The hydrophobic segments (H1 and H2) are indicated by boxes. Positively and negatively charged residues are indicated by red and blue characters, respectively. (B) Hydrophobicity profiles of mouse ABC-me and human MDR1 were calculated according to the method of Kite and Doolittle (1982) (window set at 15 residues). Hydrophobic peaks predicted as TM are numbered. (C) A short 29-residue segment that includes a potential glycosylation site (open circle) was inserted between H1 and H2 of C-terminally HA-tagged constructs of ABC-me (g-ABC-me) and the membrane domain lacking N-terminal 132 residues (g-ΔN132). (D) Topology of the membrane domain on the mitochondrial inner membrane and the ER are illustrated. The N135-deleted mutant is integrated into the ER membrane and glycosylated between H1 and H2 (circle), whereas the full-length ABC-me is imported into the mitochondria, processed, and integrated into the inner membrane. The loop between H1 and H2 on the inner membrane is sensitive to proteases only under hypotonic conditions. (E) Postnuclear supernatant fractions (lanes 1–3 and 6–8) and membrane fractions (lanes 4–5 and 9–10) were treated with ProK and Endo H, respectively, and analyzed by immunoblotting. The precursor (p), processed mature (m), and glycosylated (g) forms are indicated. Where indicated, ProK treatment was performed in the presence of detergent (Tx100). (F) The localization percentages are calculated from the intensities of the ProK-resistant bands and the glycosylated precursor bands. (G) Membrane topology of ABC-me on the mitochondrial inner membrane. Postnuclear supernatant of COS7 cells were treated with ProK (+, 50 μg/ml; ++, 200 μg/ml) under either isotonic (hypo–) or hypotonic (hypo +) conditions. Arrowhead indicates inner membrane-protected C-terminal fragment. Apoptosis inducing factor (AIF) and heat shock protein 60 (hsp60) were used as a control for the intermembrane space and matrix proteins, respectively.
Some mRNAs encoding mitochondrial proteins, such as ATP2 and ATM1 (mitochondrial inner membrane ABC transporter), are enriched on the mitochondrial membrane, and this localization is critical for mitochondrial biogenesis (Corral-Debrinski et al., 2000; Marc et al., 2002; Margeot et al., 2002). The enrichment of mRNA is suggested to be mediated by the 3′-untranslated region. These observations suggest that the targeting of mRNAs is involved in sorting of specific proteins into the mitochondria. It is not clear, however, whether the mRNA localization accounts for the escape of membrane proteins from the ER targeting and the localization into the mitochondria.
In this study, we extensively examined the membrane protein sorting mediated by the presequence, by using cultured cell system, in which membrane topology and localization were quantitatively assessed, and cell-free system, in which initial steps of the ER targeting can be examined. The membrane domain alone was integrated into the ER membrane in the same topology as MDR1. Other integral membrane proteins on the secretory organelles, synaptotagmin II (SytII) and MDR1, could be imported into mitochondria by the N135 segment. Another presequence of the mitochondrial matrix protein and some partially deleted N135 mutants cannot mediate the mitochondrial import of the membrane domain, although they can mediate efficient import of a soluble passenger protein, dihydrofolate reductase (DHFR). Furthermore, taking advantage of the DHFR domain, whose folding state can be regulated by the addition of its specific ligand, methotrexate (MTX), we distinguish the modes of intracellular sorting: MTX did not affect the ER membrane integration of the first hydrophobic segment of ABC-me (H1)-DHFR fusion protein, in which only H1 was fused to DHFR, whereas in the presence of the N135 segment, the mitochondrial import of the N135-H1-DHFR fusion protein was strictly inhibited by MTX. It is thus indicated that the H1-segment–mediated ER integration is cotranslational process, whereas the N135-H1–mediated mitochondrial import is posttranslational one. In a cell-free system, the N135 segment does not affect the recognition of the hydrophobic segments by the SRP but greatly suppresses targeting to the ER membrane, even in the absence of mitochondria. From the experimental data, we concluded that the cotranslational ER targeting mechanism for hydrophobic membrane protein is suppressed and switched to the sorting mechanism to posttranslational import by the presequence.
MATERIALS AND METHODS
Materials
Enzymes for DNA manipulation and in vitro transcription (Toyobo, Takara, Japan and New England Biolabs, Beverly, MA), proteinase K (ProK; Merck, Whitehouse Station, NJ), endoglycosidase H (Endo H; New England Biolabs), and peptide N-glycosidase F (PNGaseF; New England Biolabs) were obtained from the sources indicated. Rabbit reticulocyte lysate (Jackson and Hunt, 1983) and rough microsomal membrane (RM) from dog pancreas (Walter and Blobel, 1983) were prepared as described previously. RM were washed with 25 mM EDTA and treated with staphylococcal nuclease (Roche Diagnostics, Indianapolis, IN) to remove endogenous mRNA as described previously. Prestained molecular weight markers (New England Biolabs) were used to estimate the approximate size of the proteins. Anti-hemagglutinin (HA) monoclonal antibody (mAb) (16B12, CRP), rabbit antiserum specific for the SytII C-terminal domain (amino acids 139–267) (Fukuda et al., 1994), anti-DHFR mAb (BD Transduction Laboratories, Lexington, KY), rabbit antibody against the C-terminal segment of calnexin (StressGen Biotechnologies, San Diego, CA), MTX (Sigma-Aldrich, St. Louis, MO), and MitoTracker Orange (Molecular Probes, Eugene, OR) were obtained from the indicated sources. Rabbit antibody against SRP54 was raised to a synthetic peptide of the N-terminal 20 residues. cDNAs encoding human MDR1 and human ornithine transcarbamylase (OTC) were generous gifts from Dr. Ueda (Kyoto University, Kyoto, Japan) and Drs. Terada and Mori (Kumamoto University, Kumamoto, Japan), respectively.
Construction of Expression Plasmids
In the following procedures, the desired DNA fragments were obtained by polymerase chain reaction (PCR) with oligonucleotide primers containing the appropriate restriction enzyme sites, indicated in the parentheses. They were digested by the restriction enzymes and ligated with plasmid vectors that had been digested with the restriction enzymes. At the junction, the six bases of the restriction enzyme sites encoded two codons. Alternatively, two DNA fragments coding for the desired sequence were joined using an overlapextension procedure (Horton et al., 1993). For point mutations, mutagenesis was performed by the polymerase cycle reaction essentially as described previously (Weiner et al., 1994). The sequences of the oligonucleotides used are available upon request from the authors.
Open reading frames (ORFs) of mouse ABC-me were obtained by PCR amplification by using mouse cDNA, which was kindly provided by Dr. Yamazaki (Kyushu University, Fukuoka, Japan) and the following primers: ACTGGCGGCCGCCCACCATGCGCGCCCCTTCTGCTAGGGCG (sense for 5′ end of cDNA; NotI site, the Kozak sequence, and the initiation codon are underlined), and TACGTCTAGACGCCCGTGCAGGTTCCAGGAA (antisense for 3′ end; XbaI site is underlined). The full-length cDNA (Met1-Ala715) fragment (NotI/XbaI) and HA-tag sequence (XbaI/ApaI) were subcloned into the pRcCMV vector (NotI/ApaI) (Invitrogen, Carlsbad, CA). In all of the following constructs, N-terminal positions include initiation codon preceded by the restriction enzyme site and the Kozak sequence.
For domain deletion constructs (Figure 1), DNA fragments encoding either Arg133-Ala715 (NotI/XbaI) or Met1-Val140 (HindIII/XbaI) were joined with the HA-tag by using the XbaI site to obtain ΔN132 and N140. For insertion of the glycosylation loop (Figure 1), a DNA sequence coding for the glycosylation loop from band3 protein (Thr629-Ser657) was inserted between Ser169 and Glu170 to obtain g-ABC-me and g-ΔN132. For presequence replacements (Figure 2), DNA fragments encoding presequences of OTC (Met1-Gln36) were joined with the ABC-me membrane domain (Arg133-Ala715) or DHFR (Met1-Asp187) by using an overlap-extension procedure. For partial deletion mutants of the N135 segment (Figure 3), mutations were created by polymerase cycle reaction (Weiner et al., 1994). For MDR1 constructs (Figure 4), the cDNA encoding the N-terminal half of the human MDR1 (Asn20-Arg680; HindIII/XbaI) was subcloned into pRcCMV (HindIII/XbaI). The N-terminal sequence containing N135 (Met1-Val140, HindIII/XbaI) and the MDR1 sequence (Asn20-Arg680, XbaI/XbaI) were ligated on the pRcCMV-HA (HindIII/XbaI). For the SytII fusion construct (Figure 4), the endogenous glycosylation site of SytII was disrupted by the T34A mutation, and the glycosylation site was newly created by the mutations L50N, K51S, and E52T. The entire ORF (Met1-Lys422) of SytII was subcloned to pRcCMV-HA (HindIII/XbaI). The N140 sequence (Met1-Val140, HindIII/XbaI) and the glycosylation mutant of SytII (Asn50-Lys422, XbaI/XbaI) were subcloned into pRcCMV (HindIII/XbaI).
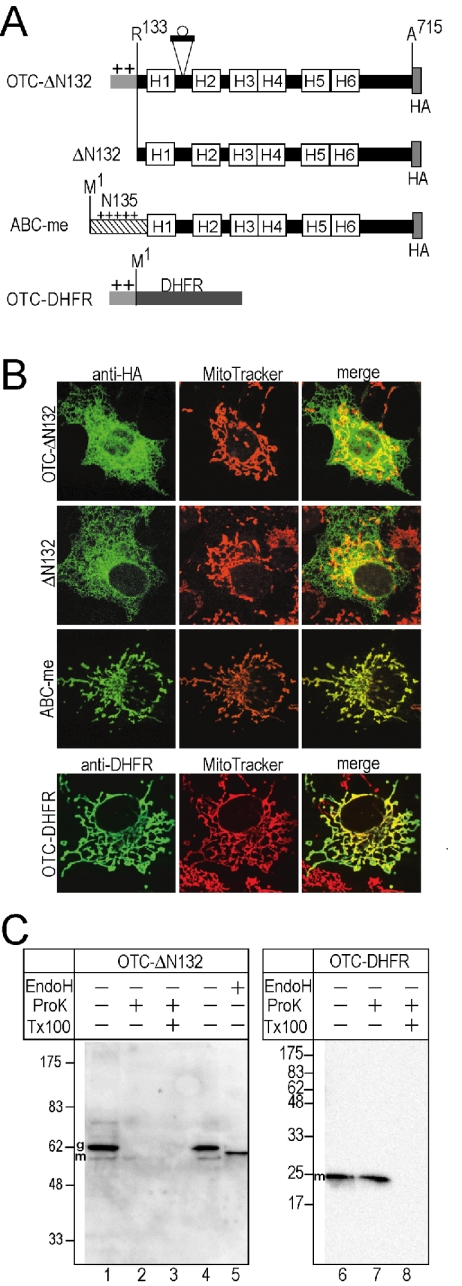
Figure 2.
Membrane domain cannot be localized in mitochondria by OTC presequence. (A) The presequence of OTC was fused either to Arg133 of g-ABC-me (OTC-ΔN132) or to N terminus of DHFR (OTC-DHFR). Original ABC-me and the membrane domain lacking N-terminal 132 residues (ΔN132) also were examined. (B) The constructs were expressed in the COS7 cells and detected by indirect immunofluorescent microscopy using anti-HA antibody and anti-DHFR antibody (anti-HA and anti-DHFR, green). Cells also were stained with a mitochondrial marker (MitoTracker Orange, red). The merged images also are shown (merge). (C) Localizations of the OTC-presequence fusion proteins were examined by immunoblotting as noted in Figure 1E.
Figure 3.
Difference of presequences required for membrane domain and for soluble protein. (A) Partial deletion mutants of N135 (left) were fused to the N-termini (dotted squares) of DHFR or the membrane domain of g-ABC-me (right). (B and C) Localizations of fusion proteins expressed in COS7 cells were examined by immunoblotting as described in Figure 1E. Import efficiency (percentage) of DHFR domain was calculated from the precursor (p) and ProK-resistant mature (m) forms. Localization (percentage) of ABC-me membrane domain into the mitochondria (Mt) and the ER (ER) were calculated from the ProK-resistant m and p form, respectively.
Figure 4.
N135 can import MDR1 and SytII into the mitochondria. (A) N-terminal 140-residues sequence was fused to the Asn21 of MDR1 (N140-MDR1). Three potential endogenous glycosylation sites between H1 and H2 are indicated by circles. (B and C) Localizations and intracellular topology were assessed as noted in Figure 2. Staining with ER marker (calnexin) also was performed. In lanes 5, 6 11, and 12, aliquots of membrane fractions were treated with either PNGaseF (PF) or Endo H (EH) to remove sugar chains. (D) N-terminal 140-residues sequence was fused to SytII at Asn50 (N140-SytII). Glycosylation sites are indicated by circles. (E and F) Localization and topology were assessed as noted in Figure 2. The di-, mono-, and unglycosylated forms are indicated (gg, g, and p, respectively).
For in vitro translation experiments (Figure 5), coding sequences of ABC-me (NcoI/XbaI) or ΔN132 (NcoI/XbaI) were inserted into the pCITE-2b vector (Novagen, Madison, WI) or pSPBP4 (Siegel and Walter, 1988) to improve translation efficiency. For the chemical cross-linking experiments, three Arg residues (Arg130, Arg133, and Arg135) were changed to Lys to improve the cross-linking efficiency. Furthermore, to improve the radioactivity of protein bands, three Ala residues (Ala261, Ala263, and Ala265) were exchanged with Met.
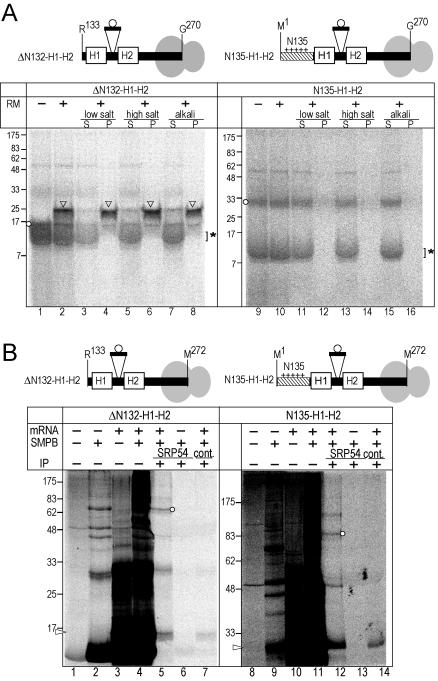
Figure 5.
N135 blocks ER targeting but does not affect interaction with SRP. (A) The truncated mRNAs encoding up-to Gly270 were translated. The nascent chains were attached to the ribosomes as peptidyl-tRNA. The circle indicates the inserted potential glycosylation site. After translation in the absence (–) or presence (+) of RM, aliquots were separated into membrane precipitate (P) and supernatant (S) fractions under low salt (150 mM KOAc), high salt (500 mM KOAc), and alkaline (0.1 M Na2CO3) conditions. The nonglycosylated product (circles), the glycosylated form (arrowheads), and globin (asterisk) are indicated in the panel. (B) The truncated mRNAs were translated in the absence of RM and then treated with heterobifunctional cross-linker (SMPB, + lanes). Two Arg (Arg133 and Arg135) of ΔN132-H1-H2 and three Arg (Arg130, Arg133, and Arg135) of N135-H1-H2 were changed to Lys to improve cross-linking efficiency. Three Ala (Ala261, Ala263, and Ala265) were changed to Met to improve band intensity. After the reactions, aliquots were immunoprecipitated with either anti-SRP54 antibody or control antibody (anti-yeast Bmh1p) (IP, +). The samples were analyzed by SDS-PAGE (13% acrylamide, left and 10%, right) and subsequent image analysis. Arrowhead and circles indicate the translation product and the product linked with the SRP-54-kDa subunit, respectively.
For several fusion constructs (Figure 6), the ABC-me sequence coding for Met1-Arg178 (HindIII/XbaI) or Arg133-Arg178 (HindIII/XbaI), including the nine-residue glycosylation loop (KVSNSSARG) between Ser169 and Glu170, was fused to the N terminus of the DHFR (Met1-Asp187, XbaI/XbaI) or green fluorescent protein (GFP) domain (Met1-Lys238, XbaI/XbaI) on pRcCMV (HindIII/XbaI). In the SytII fusion constructs, the coding sequence for DHFR (Met1-Asp187, HindIII/EcoRI) that contained the mutation (Ile8Thr) for the glycosylation site was fused at the N terminus of SytII (EcoRI/XbaI; Arg2-Lys422). In this case, five extra amino acids (SRGSI) were inserted between Met1 and Arg2. The mutated DHFR domain (DHFRm), including point mutations (Cys7Ser, Ile8Thr, Ser42Cys, and Asp49Cys), also was used instead of DHFR.
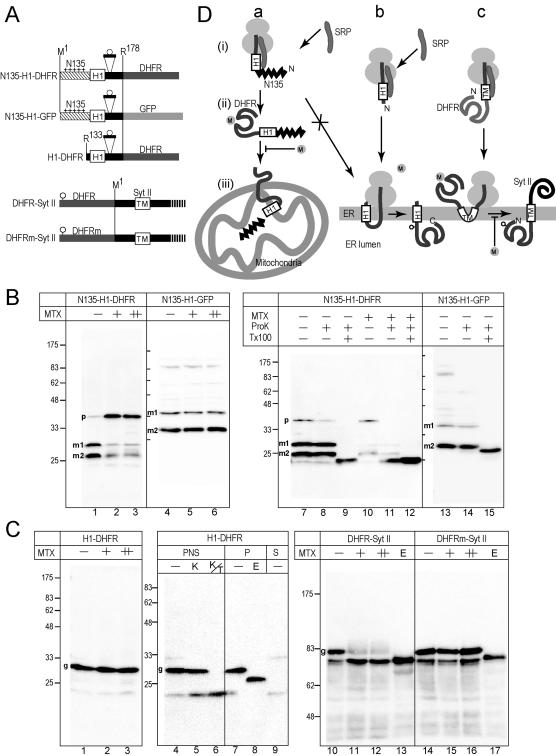
Figure 6.
Posttranslational translocation of the DHFR domain through the membrane is inhibited by MTX in COS7 cells. (A) The 178-residue N-terminal segment, which contains glycosylation sequence of nine residues, was fused to DHFR or GFP. In the third construct, the N-terminal 132 residues were deleted. In the last two constructs, the DHFR domains (DHFR and DHFRm) containing a glycosylation site (circle) were fused to SytII at Arg2. DHFRm denotes a DHFR mutant that does not bind MTX. (B) The constructs were expressed in COS7 cells in the presence of MTX (+, 10 μM; ++, 50 μM). As a mock control, the same volume of DMSO was added (–). Total cell lysate were subjected to immunoblotting analysis by using anti-DHFR or anti-GFP antibodies (lanes 1–6). Postnuclear supernatants were subjected to ProK treatment in the absence (–) or presence (+) of Tx100. Two processed mature forms (m1 and m2) and a precursor form (p) are indicated. (C) The constructs were expressed in COS7 cells in the presence or absence of MTX. Whole cell-lysate was analyzed by immunoblotting by using anti-SytII or anti-DHFR antibodies. Aliquots were treated with Endo H before the analysis (E lane). For ProK treatment, postnuclear supernatant was used (lanes 4–6). For Endo H treatment, the membrane precipitate fraction was used (lanes 7 and 8). Postmicrosomal supernatant fraction also was analyzed (S). In the case of SytII-related constructs, 12 and 36 μM MTX were used (+ and ++ lanes, respectively). (D) Schematic presentation of the effect of MTX. In the presence of N135, cotranslational integration of the H1 segments into the ER membrane was suppressed and posttranslational mitochondrial import was achieved (a). The cotranslational translocation of the DHFR domain across the ER membrane was not inhibited by MTX (b), whereas the translocation of the prefolded N-terminal DHFR domain was completely inhibited (c).
Cell Culture, Cell Fractionation, and Immunofluorescence Studies
COS7 cells were maintained in DMEM supplemented with 10% fetal calf serum under 10% CO2 atmosphere. Transfections using FuGENE 6 reagent (Roche Diagnostics) and indirect immunofluorescent microscopy were performed as described previously (Kanaji et al., 2000; Miyazaki et al., 2001).
For cell fractionation, COS7 cells were cultured on 10-cm culture dishes for 24 h after transfection. When indicated, MTX dissolved in dimethyl sulfoxide (DMSO) was added to the culture medium 16 h after transfection and further incubated for 8 h. Cells were harvested into 2 ml of phosphate-buffered saline and precipitated by centrifugation at 5000 rpm for 2 min. They were washed with HES buffer (10 mM HEPES-KOH, pH 7.5, 1 mM EDTA, and 0.25 M sucrose) and resuspended in 1 ml of HES buffer containing 20 μg/ml α2-macroglobulin. Cells were homogenized by aspirating 20 times through a 27-gauge needle. The homogenate was centrifuged at 2500 rpm for 5 min to obtain a postnuclear supernatant. Aliquots of the supernatant were centrifuged at 40,000 rpm for 10 min to obtain a membrane pellet, which was then resuspended in HES buffer and treated with Endo H or PNGaseF under denaturing conditions. Other aliquots were treated with ProK (50 μg/ml) on ice for 30 min either in the presence or absence of 0.5% Triton X-100 (Tx100). The enzyme reactions were terminated with 10% trichloroacetic acid (TCA), and the protein precipitates were subjected to SDS-PAGE and immunoblotting analysis.
For examination of the topology on the mitochondrial membranes, aliquots of postnuclear supernatant were treated by isotonic (HE containing 250 mM sucrose) or hypotonic (5 mM sucrose) conditions on ice for 1 h and then treated with ProK (50 μg/ml) on ice for 1 h in the presence or absence of 0.5% Triton X-100.
In Vitro Transcription and Translation
In vitro transcription and translation were performed essentially as described previously (Sakaguchi et al., 1992; Ota et al., 1998). For membrane binding assay, the DNA fragment was amplified by PCR by using the forward primer corresponding to the 220 base pairs upstream from T7 RNA polymerase promoter and the reverse primer at Gly270 of ABC-me, to produce truncated templates of pCITE-derived plasmid. The PCR fragment was purified by agarose gel electrophoresis and GFX DNA fragment purification kit (Amersham Biosciences, Piscataway, NJ) and was then subjected to transcription. For cross-linking assay, the pSPBP4-derived plasmids were lineralized by XbaI to produce templates truncated at Met272 and transcribed with SP6 RNA polymerase. The in vitro-synthesized mRNAs were translated at 30°C for 45 min in the presence or absence of RM in cell-free system containing 20% reticulocyte lysate. Salt conditions were optimized to be 100 mM potassium chloride and 1.5 mM magnesium acetate for pCITE-derived mRNAs and to be 100 mM potassium acetate and 1.5 mM magnesium acetate for pSPBP4 mRNAs.
Extraction of RM under Physiological, High Salt, and Alkaline Conditions
For physiological low salt extraction, the translation reaction mixture (15 μl) was diluted 3.5-fold with physiological buffer, and RM membranes were precipitated essentially as described previously (Ota et al., 1998; Ukaji et al., 2002). For high salt extraction, the translation mixture (15 μl) was diluted to 50 μl, which was finally adjusted to 500 mM KOAc, 5 mM Mg(OAc)2, and 30 mM HEPES, pH 7.4. The mixture was then layered onto a 100-μl cushion of 0.5 M sucrose in the same buffer. After centrifugation for 5 min at 50,000 rpm (Hitachi RP100AT2 rotor), the membrane pellet (P) was dissolved directly in SDS-PAGE sample buffer. Proteins in the supernatant (S) were precipitated with 5% TCA. For alkaline extraction, the translation mixture (15 μl) was added to 162 μl of 0.1 M Na2CO3 and incubated for 20 min on ice. The mixture was then layered onto a 100-μl cushion of 0.1 M Na2CO3 solution containing 0.5 M sucrose and centrifuged for 5 min at 70,000 rpm.
Cross-linking and Immunoprecipitation
Cross-linking reaction with the heterobifunctional cross-linker succinimidyl 4-[p-maleimidophenyl]butyrate (SMPB) (Pierce Chemical, Rockford, IL) was performed as follows: 1.6 μl of 50 mM SMPB was added to 14 μl of translation mixture and incubated on ice for 2 h. After quenching with 50 mM Tris-HCl and 50 mM dithiothreitol on ice for 10 min, the proteins were denatured by 1% SDS at 40°C for 10 min and then diluted to 1 ml with immunoprecipitation (IP) buffer (1% Triton X-100, 20 mM Tris-HCl, pH 7.5, and 110 mM NaCl). After insoluble materials were removed by centrifugation, the solution was incubated once for 30 min with protein A-Sepharose (Amersham Biosciences) alone and then the unbound fraction was incubated with 20 μl of antiserum and protein A-Sepharose at 4°C overnight. After the supernatant was removed, the protein A-Sepharose was washed once with 1 ml of IP buffer and then extracted with sample buffer at 97°C for SDS-PAGE.
RESULTS
Quantification of Membrane Topology and Intracellular Localization
To quantitatively assess the organelle localization of bulk molecules expressed in COS7 cells and to examine the membrane topology on the ER membrane, a well characterized loop segment with a glycosylation site was inserted between the H1 and H2 segments of ABC-me (Figure 1C). The loop sequence of 29 residues was derived from an extracellular loop between TM7 and TM8 of erythrocyte band3 protein, which is often used as a marker sequence for topology assessment (Popov et al., 1997; Ota et al., 1998, 2000). Because the corresponding lumenal loop between H1 and H2 of MDR1 is glycosylated, we assumed that the site would be glycosylated, if the membrane domain was correctly integrated into the ER membrane (Figure 1D). The HA-tagged constructs of full-length mouse ABC-me (Met1-Ala715) and the N135-deleted one (ΔN132: Arg133-Ala715) (the residue numbering used in the following text corresponds to the original amino acid numbers of the precursor proteins.) were expressed in COS7 cells (Figure 1E). The postnuclear supernatant was analyzed by immunoblotting. The g-ABC-me construct gave a single major band and trace amounts of a larger band (Figure 1E, lane 1). The major band was ProK resistant (Figure 1E, lane 2). In addition to the ProK-resistant processed mature from, we noted a trace but significant amount of ProK-resistant bands of the precursor form and intermediated form, which lost N-terminal 4.7-kDa segment (Figure 1E, lane 2). This suggested that the N135 segment contains another potential processing site in addition to the position 105. All of the resistant bands were degraded by ProK in the presence of detergent (Figure 1E, lane 3). Thus, g-ABC-me molecules expressed in COS7 cells were efficiently imported into mitochondria and processed to the mature form. When membrane precipitate obtained from the postnuclear supernatant was treated by Endo H, the major band was not affected (Figure 1E, lanes 4 and 5). In contrast, in the absence of N135, the single major band of the g-ΔN132 construct was sensitive to ProK (Figure 1E, lane 7) and had increased mobility after Endo H treatment (Figure 1 E, lane 10), indicating that the N135-deleted molecule was exclusively integrated into the ER membrane and the loop between H1 and H2 was glycosylated in the ER lumen, as in the case of MDR1. By quantifying the glycosylated and the imported ProK-resistant bands, the intracellular localization efficiency was determined (Figure 1E). The membrane domain alone was integrated into the ER membrane, showing the same topology as MDR1. Using this type of construct, we can quantitatively estimate localization and membrane topology of membrane proteins.
We then examined the membrane topology of ABC-me expressed in COS7 cells on mitochondrial inner membrane by using protease treatment (Figure 1G). When isolated mitochondrial fractions were treated with ProK in isotonic conditions, g-ABC-me was not degraded (Figure 1G, lanes 1–3). Under hypotonic conditions in which only the outer membrane was disrupted, g-ABC-me was partially degraded, but significant amounts of the ProK-resistant polypeptide fragment were observed (Figure 1G, lanes 5 and 7, arrowhead). The ProK-resistant fragment was similarly observed even when 4 times the amount of ProK was used (Figure 1G, lane 7). Based on the size and position of the HA-tag in the molecule, we concluded that the ABC-me molecule in the inner membrane was cleaved between H1 and H2, which was exposed on the intermembrane space (Figure 1D). The fates of apoptosis inducing factor and heat shock protein 60, which are marker proteins of the intermembrane space and matrix, respectively, indicated that under the hypotonic conditions, the intermembrane space was accessible to ProK, but the matrix was not. These data indicate that the g-ABC-me molecule was imported into mitochondria and integrated into the inner membrane in COS7 cells.
Difference of Presequences Required for Membrane Domain and for Soluble Protein
To examine whether the matrix-targeting presequence of soluble protein can import the membrane domain, the N-terminal 132-residue segment was replaced with the presequence of OTC (Yano et al., 1997) (Figure 2A, OTC-ΔN132). Localization was examined by indirect immunofluorescent microscopy with mAb against HA. The fusion construct (OTC-ΔN132) was localized in reticular structures, which are distinguished from mitochondria (Figure 2B) and was well merged with the ER marker (our unpublished data), as noted with membrane domain alone, ΔN132 (Figure 2B). In contrast, the full-size ABC-me was exclusively localized in the filamentous structures that were stained with the mitochondrial marker (Figure 2B). Almost all of the expressed molecules of OTC-ΔN132 were sensitive to ProK (Figure 2C, lane 2), and the major band was shifted down by the Endo H treatment (Figure 2C, lanes 4 and 5), indicating almost all of them were targeted to the ER but not in mitochondria. In contrast, the DHFR molecule, which was fused to the OTC presequence, was targeted to the mitochondria (Figure 2B) and resistant to ProK treatment (Figure 2C, lane 7), indicating that the OTC presequence directs the soluble DHFR molecule to the mitochondria. These results suggest that conventional presequences were insufficient for mitochondrial targeting of the ABC-me membrane domain.
To further characterize requirements for import of membrane domain, we examined the effect of deletions within the N135 sequence on the mitochondrial import function (Figure 3A). All the deletion mutants supported the mitochondrial import of DHFR (Figure 3B). The fusion constructs expressed in COS7 cells gave processed mature forms, which were resistant to ProK treatment (Figure 3B, lanes 2, 5, 8, and 11). Although some precursor forms might be observed, the precursor forms became negligible, when less expression plasmids were used to make the expression level lower (our unpublished data). Unexpectedly, even the Δ81–132 construct, which did not contain the processing site at position 105, also was processed to mature form. This processing likely occurred in the former position as observed with full-length ABC-me molecule (Figure 1E, lanes 1 and 2). On ProK treatment in the presence of Tx100, the processed mature form of this construct gave less ProK-resistant core DHFR domain than the other constructs did (e.g., Figure 3B, lane 6 vs. 9). It is likely that the different segment within the N termini of the DHFR domain caused the different proteinase K sensitivity. In contrast, import of the membrane domain was greatly affected by the deletions of 2–60 and 81–132, and in turn those mutants were integrated into the ER (Figure 3C); less ProK-resistant processed mature forms were detected with these constructs (Figure 3C, lanes 7 and 12), and the major bands had increased mobility after Endo H treatment (Figure 3C, lanes 10 and 15). Even when the expression level was limited, the patterns were essentially the same (our unpublished data). Thus, these deletion mutations affected the import efficiency of the membrane domain, whereas they could import soluble DHFR domain into the mitochondria. Therefore, the N135 segment contains some extra functions specific for membrane protein import, in addition to the conventional feature required for import of soluble protein.
N135 Directs Authentic ER Membrane Proteins into the Mitochondria
We also examined whether membrane proteins destined for the ER can be imported into the mitochondria by N135 (Figure 4). First, we examined the effect on the location of the N-terminal half of MDR1. When expressed in COS7 cells, the MDR1 protein was localized mainly in the ER and partly in a reticular pattern wider than the calnexin stain, but not in the mitochondria (Figure 4B), indicating that it is localized in the secretory pathway. The product produced a single major band upon immunoblotting analysis, which was sensitive to ProK treatment (Figure 4C, lane 2) and was shifted down by Endo H and PNGaseF treatments (Figure 4C, lanes 5 and 6). These data indicated that the MDR1 molecule was integrated into the ER and glycosylated. In contrast, when N135 was fused to the N-terminal half of the MDR1 molecule, the apparent localization was shifted to mitochondria (Figure 4B). A significant amount of ProK-resistant mature form was observed (Figure 4C, lane 8, m), although some was glycosylated (Figure 4C, lane 10–12, g). These results indicate that N135 can direct significant amounts of the N-terminal half of the MDR1 molecule into the mitochondria by suppressing the cotranslational ER targeting mechanism. The targeting efficiency of MDR1 was not complete. This fact might suggest that the membrane domain of ABC-me contains additional information that supports the mitochondrial targeting function of N135 sequence, although it does not contain targeting functions by itself. This would be agreement with the observation by Graf et al. (2004) that mutated presequence of ABC-me targeted ABC-me membrane domain more efficiently than GFP. The targeting-supporting information might be related higher structure, by which the N135 segment is freely exposed to molecular surface.
Second, N135 was fused to mouse SytII, which is a single spanning membrane protein with an Nexo/Ccyt topology (Kida et al., 2000) (Figure 4D). SytII is targeted to the translocon on the ER immediately after the TM segment emerges from the ribosome (Kida et al., 2000). When expressed in COS7 cells, the SytII molecule was located on the cell surface (Figure 4E). This pattern was clearly different from the mitochondrial pattern (Figure 4E, top). When the N140 sequence was fused to the N terminus, the location was dramatically changed into a mitochondrial pattern, although a weak cell surface pattern also was observed (Figure 4E, bottom). Immunoblotting analysis revealed a major band that was resistant to ProK (Figure 4F, lane 2, m) and trace amounts of three larger bands (p, g, and gg). Because the N135-fusion construct possessed two potential glycosylation sites (Figure 4D) and the larger two bands were shifted down by PNGaseF and Endo H treatment (Figure 4F, lanes 5 and 6), the larger three bands corresponded to none-, mono- and diglycosylated precursor forms. It should be noted that original SytII expressed in COS7 cells was efficiently glycosylated (Kida et al., 2000). Thus, >75% of the N140-SytII-fusion protein was targeted to the mitochondria, despite the strong ER targeting signal.
N135 Prevents ER Targeting but Not Recognition by SRP
To examine the effect of N135 on targeting and insertion of H1 and H2 segments of ABC-me into the ER in the cell-free system, truncated mRNA encoding the sequence from either Met1 or Arg133 up-to Gly270 was translated in a reticulocyte lysate cell-free system in the presence or absence of RM (Figure 5). In this system, the mRNAs possess no in-frame termination codon, so that the nascent polypeptides are retained on ribosomes as peptidyl-tRNA. In the presence of RM, the N135-deleted construct was glycosylated (Figure 5A, lane 2, arrowhead). Most of the glycosylated form was recovered in the membrane precipitates, even under alkali conditions (Figure 5A, lane 8), whereas the trace amounts of the nonglycosylated form was in the supernatant, indicating that the hydrophobic segments emerging from the ribosome were integrated into the ER membrane. In contrast, in the presence of N135, the nascent polypeptide was not glycosylated (Figure 5A, lanes 9 and 10, circles). Furthermore, all of the products were recovered in the supernatant fraction even under the low salt conditions (Figure 5A, lanes 11 and 12). Thus, the H1-H2 segments were correctly targeted to RM as a ribosome nascent chain complex in the absence of the N135 segment, whereas the ER targeting was suppressed by N135. Because the reticulocyte lysate cell-free system does not contain mitochondria, N135 suppresses the ER targeting process in the absence of mitochondria. The switching from ER integration to mitochondrial import is not a competitive mechanism between ER and mitochondria, but the cotranslational process to the ER was suppressed by N135.
Next, we addressed recognition of the hydrophobic segments by SRP by using a chemical cross-linking (Figure 5B). To improve cross-linking efficiency, two Arg residues of the ΔN132-construct and three Arg residues of the N135-construct were replaced with Lys. To improve radioactivity of the nascent chain, three Ala residues (Ala261, Ala263, and Ala265) were exchanged with Met residues. Although both of the constructs gave heterogeneous bands after the cross-linking reaction (Figure 5B, lanes 4 and 11), immunoprecipitation with anti-SRP 54-kDa subunit antibodies revealed that both of constructs were cross-linked with the SRP subunit (Figure 5B, lanes 5 and 12). The immunoreactive cross-linked bands were not detected, when unrelated antibodies were used (anti-yeast bmh1 antibodies; Figure 5B, lanes 7 and 14) or when no mRNA was used for the translation reaction (Figure 5B, lanes 6 and 13). The molecular masses of the cross-linked products were 70 and 86 kDa, which were consistent with the sum of the probe (16 and 30 kDa) and SRP54 subunit (54 kDa). These results suggest that N135 does not affect recognition of the hydrophobic segments by SRP.
N135 Switches the Sorting Mode from Cotranslational ER Integration to Posttranslational Mitochondrial Import
We then examined the targeting modes to the ER and mitochondria in cultured cells by using the DHFR domain as a reporter (Figure 6). The binding of a specific ligand, MTX, stabilizes the DHFR domain and inhibits unfolding-dependent translocation, as discovered by Eilers and Schatz (1986); they demonstrated using a cell-free system that mitochondrial import of a DHFR passenger is inhibited by MTX.
The 178-residue N-terminal sequence of ABC-me that contains N135 and H1 segment was fused to DHFR (Figure 6A, N135-H1-DHFR). When expressed in COS7 cells, two major bands were detected by the anti-DHFR mAb (Figure 6B, lane1, m1 and m2). These two bands were not degraded by ProK treatment (Figure 6B, lanes 7 and 8), whereas they were degraded in the presence of detergent (Figure 6B, lane 9) and not affected by Endo H treatment (our unpublished data), indicating that the two forms were imported into the mitochondria, processed at one of two different sites, and protected by the mitochondrial membranes. When MTX was added to the culture medium, import of the DHFR domain was greatly affected; the amount of the two processed mature bands decreased and the precursor form (p) increased (Figure 6B, lanes 2 and 3). The precursor form observed in the presence of MTX is sensitive to ProK treatment (Figure 6B, lane 11). In contrast to the DHFR constructs, import of the corresponding GFP-fusion construct was not affected by MTX (Figure 6B, lanes 4–6); the two processed bands were similarly observed (m1 and m2), both of which were resistant to ProK treatment (Figure 6B, lanes 13 and 14). Therefore, import inhibition by MTX was due to the DHFR domain, which was more tightly folded in the presence of MTX, and MTX had no effect on the general import mechanism. We estimated the processed mature forms (m1 and m2) of both constructs and found that the m1 forms correspond to the products that had been processed at the position 105, which is the authentic processing site of the N135 segment. The m2 forms should be products cleaved at the following positions. The nature of the cleavage is unknown but should be caused by the absence of the following hydrophobic segments. These data demonstrated that the DHFR domain can fold once in the cytoplasm to bind MTX before being imported and the MTX binding inhibits mitochondrial import even in cultured cells (Figure 6D, pathway a).
When the H1 segment alone was fused to the N terminus of the DHFR domain (Figure 6A, H1-DHFR), the construct was targeted to the ER instead of mitochondria and translocated through the membrane irrespective of MTX (Figure 6C, lanes 1–3); the single major product (Figure 6C, lane 4) was resistant to ProK treatment (Figure 6C, lane 5) and accessible to the protease only in the presence of detergent (Figure 6C, lane 6). When membranes were recovered by ultracentrifugation, the major band was in the membrane fraction (Figure 6C, lane 7), and it was shifted down by Endo H treatment (Figure 6C, lane 8). Thus, the H1 segment mediated cotranslational translocation of the following DHFR domain through the ER membrane, allowing no time for folding of the DHFR domain (Figure 6D, pathway b).
As a control, we examined the effect of MTX on the translocation of the DHFR domain through the ER membrane by a type I signal-anchor sequence, which mediates translocation of the preceding region (Kida et al., 2000). The DHFR domain and mutated DHFR (DHFRm) domain, which cannot bind MTX, were fused to the N terminus of SytII. The major bands synthesized in COS7 cells (Figure 6C, lanes 10 and 14) were shifted down by Endo H treatment (Figure 6C, lanes 13 and 17), indicating that the DHFR domain was translocated through the membrane and glycosylated by a lumenal enzyme. The translocation and glycosylation of the wild-type DHFR domain was clearly inhibited by MTX (Figure 6C, lanes 11 and 12), whereas that of DHFRm was not affected (Figure 6C, lanes 15 and 16). It is clear that the N-terminal domain can fold once before translocation (Figure 6D, pathway c). Thus, the inhibition by MTX is a clear indication of polypeptide chain folding before membrane translocation in both cases of the ER and mitochondria.
Together, the cultured cell experiments indicated that N135 suppresses the cotranslational integration into the ER membrane mediated by the H1 segment and achieves post-translational import into the mitochondria.
DISCUSSION
In this study, to explore the targeting and integration of the hydrophobic membrane protein to mitochondria, we focused on the functional detail of the exceptionally long mitochondria-targeting presequence of ABC transporter. The requirements for import of membrane domain are clearly different form those for soluble protein. The cell-free experiments demonstrated that the N135 segment suppresses initial targeting mechanism for the ER membrane even in the absence of mitochondria. The cultured cell experiments proved that targeting mechanism for hydrophobic membrane proteins is switched by the N135 segment from cotranslational ER targeting to the posttranslational mitochondrial import.
The DHFR-fusion experiments provide clear evidence that the N135-mediated mitochondria import of membrane protein proceeds posttranslationally in living cells. In the absence of N135, the H1 segment mediates translocation of the following DHFR domain through the ER membrane. The cotranslational ER translocation is not affected by MTX, consistent with the elongating polypeptide chain not binding MTX. In the presence of N135, however, the N135-H1-DHFR construct was excluded from the ER targeting mechanism and imported into the mitochondria. This import was inhibited by MTX. Because the crystal structure of the DHFR molecule indicates that the C-terminal region is involved in overall folding, the entire sequence is required for the MTX binding. Therefore, the entire molecule remains in the cytoplasm before it is imported into the mitochondria and the import of the DHFR domain should proceed in an unfolded state. We also demonstrated that translocation through the ER membrane of the N-terminal DHFR domain before the type I signal-anchor sequence is inhibited by MTX. This is a decisive indication that the N-terminal domain of the type I signal-anchor sequence can once fold in cytoplasmic side in cultured cells, so that the DHFR domain can bind MTX. The translocation of the N-terminal DHFR domain also has to cross the ER membrane in an unfolded state. Thus, the inhibition by MTX of the translocation of the DHFR domain through the membrane is a clear demonstration of that the entire DHFR domain remains in cytosol before translocation and can bind MTX.
The hydrophobic character of the ABC-me molecule is in contrast to that of the other group of mitochondrial membrane proteins, which possesses less hydrophobic TM segments. For example, by increasing the hydrophobic character by several mutations, the mitochondrial outer membrane proteins Tom20 (Kanaji et al., 2000), Tom5 (Horie et al., 2002), and Tom22 (Nakamura et al., 2004) completely changed the localization from the mitochondria to ER. It seems in these cases that a higher hydrophobic segment is the dominant signal for translocation to the ER membrane. This group of mitochondrial membrane proteins possesses no presequence, and their TM segments are generally less hydrophobic than those on the secretory membranes. The less hydrophobic segment possesses weak ER targeting function by itself and becomes a mitochondrial signal when several positive charges exist in the C-terminal flanking region (Kanaji et al., 2000). It is likely that the low hydrophobicity is critical for escaping the ER targeting mechanism. In contrast to the group of membrane proteins, highly hydrophobic membrane proteins destined for the mitochondria, such as ABCme, should require extra targeting sequences for escaping the ER targeting mechanism.
Cell-free experiments demonstrated that the initial step of ER targeting of the SRP-ribosome-nascent chain complex to the ER membrane is suppressed by N135, although the recognition of the H1 segment by the 54-kDa subunit of the SRP is not affected by N135. It is possible that when the hydrophobic segment of the nascent chain is recognized by SRP, the N135 itself and/or additional factor(s) interacting with the sequence prevent the following interaction on the ER membrane. We initially hypothesized that the long presequence in the N135 segment might be a strong targeting sequence that mediates direct targeting of the nascent polypeptide chain on the ribosome to the mitochondria and thus might be able to achieve cotranslational import of the membrane proteins into the mitochondria. This mechanism could explain the suppression of the ER targeting mechanism. The N135, however, achieves posttranslational import of protein that possesses an ER targeting signal sequence and it blocks RM targeting in vitro, even in the absence of mitochondria. This finding does not rule out the cotranslational targeting of nascent polypeptide and mRNAs to the mitochondrial surface and is consistent with the enrichment of mRNAs of some mitochondrial proteins (Corral-Debrinski et al., 2000; Margeot et al., 2002). It should be noted, however, that blocking of the ER targeting of hydrophobic proteins occurs even in the absence of mitochondria. Thus, the inhibition of the ER targeting is not a competitive mechanism between targeting routes to the ER and the mitochondria.
The presequence of OTC and the partially deleted mutants of N135 are insufficient for import of ABC-me, although they can import the DHFR domain. This fact strongly suggested that N135 possesses some features specific for hydrophobic membrane proteins in addition to the usual mitochondrial targeting function; N135 is not only rich in positively charged residues, Ser, and Thr residues but also is abnormally long. Among human ABC transporter isoforms that are reported to be in mitochondria, ABCB7, ABCB8, and ABCB10 possess long N-terminal hydrophilic sequences. The number of positive charges and the length are similar to that of ABC-me, whereas the amino acid sequence and the hydropathy profiles are very different. The structural bases for posttranslational import of the hydrophobic polypeptide chain remain to be determined. The mitochondrial targeting function specific for membrane proteins might be related to the length, number of positive charges, or specific higher structure. This sequence might directly mask the hydrophobic segments and prevent ER targeting by SRP. Alternatively, some factor(s) might recognize the N135 sequence and suppress cotranslational targeting to the ER membrane.
The present findings indicate that the mitochondrial targeting of hydrophobic membrane proteins is as follows (Figure 6D, pathway a): 1) In the early phase of protein synthesis, when N135 and the following H1 segment emerge from the ribosome, the cotranslational targeting of ribosome-nascent chain complex to the ER is suppressed, although the hydrophobic segment is recognized by SRP even in the presence of N135 presequence. 2) The nascent chain remains on the cytoplasmic side where the following passenger domain can fold once into a functional conformation. 3) Then the precursor polypeptide is posttranslationally imported into the mitochondria.
Acknowledgments
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, the Protein 3000 program, and the Takeda Science Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0707) on January 26, 2005.
Abbreviations used: DHFR, dihydrofolate reductase; Endo H, endoglycosidase H; MTX, methotrexate; N135, N-terminal 135 residue hydrophilic segment of ABC-me; PNGaseF, peptide N-glycosidase F; ProK, proteinase K; OTC, ornithine transcarbamylase; SMPB, succinimidyl 4-[p-maleimidophenyl]butyrate; SRP, signal recognition particle; SytII, synaptotagmin II; TM, transmembrane segment.
References
- Corral-Debrinski, M., Blugeon, C., and Jacq, C. (2000). In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20, 7881–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers, M., and Schatz, G. (1986). Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322, 228–232. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Aruga, J., Niinobe, M., Aimoto, S., and Mikoshiba, K. (1994). Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J. Biol. Chem. 269, 29206–29211. [PubMed] [Google Scholar]
- Graf, S. A., Haigh, S. E., Corson, E. D., and Shirihai, O. S. (2004). Targeting, import, and dimerization of a mammalian mitochondrial ATP binding cassette (ABC) transporter, ABCB10 (ABCme). J. Biol. Chem. 279, 42954–42963. [DOI] [PubMed] [Google Scholar]
- Horie, C., Suzuki, H., Sakaguchi, M., and Mihara, K. (2002). Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13, 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R. M., Ho, S. N., Pullen, J. K., Hunt, H. D., Cai, Z., and Pease, L. R. (1993). Gene splicing by overlap extension. Methods Enzymol. 217, 270–279. [DOI] [PubMed] [Google Scholar]
- Jackson, R. J., and Hunt, T. (1983). Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96, 50–74. [DOI] [PubMed] [Google Scholar]
- Johnson, A. E., and van Waes, M. A. (1999). The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15, 799–842. [DOI] [PubMed] [Google Scholar]
- Kanaji, S., Iwahashi, J., Kida, Y., Sakaguchi, M., and Mihara, K. (2000). Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, R. J., Freymann, D. M., Stroud, R. M., and Walter, P. (2001). The signal recognition particle. Annu. Rev. Biochem. 70, 755–775. [DOI] [PubMed] [Google Scholar]
- Kida, Y., Sakaguchi, M., Fukuda, M., Mikoshiba, K., and Mihara, K. (2000). Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 150, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Marc, P., Margeot, A., Devaux, F., Blugeon, C., Corral-Debrinski, M., and Jacq, C. (2002). Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot, A., Blugeon, C., Sylvestre, J., Vialette, S., Jacq, C., and Corral-Debrinski, M. (2002). In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J 21, 6893–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, E., Sakaguchi, M., Wakabayashi, S., Shigekawa, M., and Mihara, K. (2001). NHE6 protein possesses a signal peptide destined for endoplasmic reticulum membrane and localizes in secretory organelles of the cell. J. Biol. Chem. 276, 49221–49227. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Suzuki, H., Sakaguchi, M., and Mihara, K. (2004). Targeting and assembly of rat mitochondrial translocase of outer membrane 22 (TOM22) into the TOM complex. J. Biol. Chem. 279, 21223–21232. [DOI] [PubMed] [Google Scholar]
- Ota, K., Sakaguchi, M., Hamasaki, N., and Mihara, K. (2000). Membrane integration of the second transmembrane segment of band 3 requires a closely apposed preceding signal-anchor sequence. J. Biol. Chem. 275, 29743–29748. [DOI] [PubMed] [Google Scholar]
- Ota, K., Sakaguchi, M., von Heijne, G., Hamasaki, N., and Mihara, K. (1998). Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell 2, 495–503. [DOI] [PubMed] [Google Scholar]
- Popov, M., Tam, L. Y., Li, J., and Reithmeier, R.A.F. (1997). Mapping the ends of transmembrane segments in a polytopic membrane protein. J. Biol. Chem. 272, 18325–18332. [DOI] [PubMed] [Google Scholar]
- Sakaguchi, M. (1997). Mutational analysis of signal-anchor and stop-transfer sequences in membrane proteins. In: Membrane protein assembly, ed. G. von Heijne, Austin, TX: R.G. Landes Company, 135–150.
- Sakaguchi, M., Hachiya, N., Mihara, K., and Omura, T. (1992). Mitochondrial porin can be translocated across both endoplasmic reticulum and mitochondrial membranes. J. Biochem. 112, 243–248. [DOI] [PubMed] [Google Scholar]
- Schmitt, L., and Tampe, R. (2002). Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12, 754–760. [DOI] [PubMed] [Google Scholar]
- Shirihai, O. S., Gregory, T., Yu, C., Orkin, S. H., and Weiss, M. J. (2000). ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 19, 2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, V., and Walter, P. (1988). Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 52, 39–49. [DOI] [PubMed] [Google Scholar]
- Ukaji, K., Ariyoshi, N., Sakaguchi, M., Hamasaki, N., and Mihara, K. (2002). Membrane topogenesis of the three amino-terminal transmembrane segments of glucose-6-phosphatase on endoplasmic reticulum. Biochem. Biophys. Res. Commun. 292, 153–160. [DOI] [PubMed] [Google Scholar]
- Walter, P., and Blobel, G. (1983). Preparation of microsomal membranes for co-translational protein translocation. Methods Enzymol. 96, 84–93. [DOI] [PubMed] [Google Scholar]
- Weiner, M. P., Costa, G. L., Schoettlin, W., Cline, J., Mathur, E., and Bauer, J. C. (1994). Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151, 119–123. [DOI] [PubMed] [Google Scholar]
- Yano, M., Kanazawa, M., Terada, K., Namchai, C., Yamaizumi, M., Hanson, B., Hoogenraad, N., and Mori, M. (1997). Visualization of mitochondrial protein import in cultured mammalian cells with green fluorescent protein and effects of overexpression of the human import receptor Tom20. J. Biol. Chem. 272, 8459–8465. [DOI] [PubMed] [Google Scholar]