Abstract
Kinetochores are the proteinaceous complexes that assemble on centromeric DNA and direct eukaryotic chromosome segregation. The mechanisms by which higher eukaryotic cells define centromeres are poorly understood. Possible molecular contributors to centromere specification include the underlying DNA sequences and epigenetic factors such as binding of the centromeric histone centromere protein A (CENP-A). Frog egg extracts are an attractive system for studying centromere definition and kinetochore assembly. To facilitate such studies, we cloned a Xenopus laevis homologue of CENP-A (XCENP-A). We identified centromere-associated DNA sequences by cloning fragments of DNA that copurified with XCENP-A by chromatin immunoprecipitation. XCENP-A associates with frog centromeric repeat 1 (Fcr1), a 174-base pair repeat containing a possible CENP-B box. Southern blots of partially digested genomic DNA revealed large ordered arrays of Fcr1 in the genome. Fluorescent in situ hybridization with Fcr1 probes stained most centromeres in cultured cells. By staining lampbrush chromosomes, we specifically identified the 11 (of 18) chromosomes that stain consistently with Fcr1 probes.
INTRODUCTION
Kinetochores direct eukaryotic chromosome segregation (Biggins and Walczak, 2003; Cleveland et al., 2003; Mellone and Allshire, 2003). The proteins of the kinetochore assemble on the centromeric DNA and form the main site at which spindle microtubules attach to chromosomes. Interactions between kinetochores and microtubules direct much of the movement of chromosomes within the spindle. Kinetochores also act as signaling centers, at which the spindle assembly checkpoint monitors and responds to microtubule attachment and kinetochore tension.
The markers that cells use to identify the centromere remain unclear. Budding yeast is the exception: the “simple”, or “point,” centromere of Saccharomyces cerevisiae is completely specified by ∼125 base pairs of DNA (Carbon and Clarke, 1984; Panzeri et al., 1985). In animals, plants, and other fungi, centromeres are contained in sequences as long as several megabases and generally include arrays of repetitive DNA (Clarke, 1998; Csink and Henikoff, 1998; Houben and Schubert, 2003).
The role of these repetitive sequences in centromere specification remains controversial. The human centromeric repeat, alphoid DNA, exists in large arrays in all normal centromeres (Choo, 1997). Large, synthetic arrays of alphoid DNA occasionally support de novo centromere assembly on mammalian artificial chromosomes (Ohzeki et al., 2002). However, analysis of abnormal centromeres suggests that alphoid DNA is neither necessary nor sufficient to specify the site of kinetochore assembly. Although only ∼60 heritable neocentromeres have been described (Amor and Choo, 2002) and only a handful have been thoroughly characterized, at least two lack any detectable alphoid DNA (Lo et al., 2001; Alonso et al., 2003). Pseudodicentric chromosomes containing one active centromere and one inactive centromere provide further evidence against an exclusive role for alphoid DNA. The inactive centromere, defined as a locus that has lost centromere activity, sometimes retains large arrays of alphoid DNA (Sullivan and Willard, 1998; Amor et al., 2004). Current commentary thus favors both DNA sequence and epigenetic effects as factors in centromere definition.
Centromere protein A (CENP-A) is widely cited as a possible epigenetic marker of the centromere. CENP-A is a variant of histone H3 (Palmer et al., 1991) found only at the inner kinetochore (Warburton et al., 1997). CENP-A can replace H3 in the histone octamer in vitro (Yoda et al., 2000), and a variety of techniques strongly suggest the presence of CENP-A nucleosomes at the centromere (Ando et al., 2002; Smith, 2002). Several facts suggest CENP-A contributes to marking the centromere (Warburton, 2001). CENP-A localization correlates perfectly with centromere activity: the protein is found at normal centromeres and neocentromeres but not in inactive centromeres. CENP-A is one of only a few known constitutive components of the kinetochore; most kinetochore components localize there only during mitosis and/or meiosis. CENP-A seems to be required for localization of several other constitutive kinetochore components (Howman et al., 2000; Oegema et al., 2001). Finally, CENP-A homologues have been found in every closely examined eukaryote (Smith, 2002).
The ability to biochemically manipulate chromosome dynamics and the cell cycle makes Xenopus egg extracts an attractive system for studying centromeres and kinetochores. We report the identification and characterization of a X. laevis sequence homologue of CENP-A and a CENP-A–associated repetitive DNA element.
MATERIALS AND METHODS
CENP-A Cloning
Polymerase chain reaction (PCR) was performed on a Xenopus ovary cDNA library (Kinoshita et al., 1995) by using degenerate primers based on the conserved CENP-A peptides LQEAAEAFLVH and QLARRIRG (corresponding to residues 94–104 and 127–134 of hCENP-A, respectively). PCR products were cloned using the pGEM-T vector system (Promega, Madison, WI) and sequenced. Primers against the amplified sequence were paired with primers against plasmid backbone to amplify the 5′ end of the cDNA. PCR products were cloned, sequenced, and used to screen the same library by plaque hybridization (Sambrook et al., 1989), yielding several full-length cDNAs.
Sequence Analysis
Sequences were aligned using Clustal X (Thompson et al., 1997) and highlighted with Boxshade. Probable phylogenetic trees were generated using MrBayes (Ronquist and Huelsenbeck, 2003).
Antibodies
Rabbit antibodies were raised and purified against an amino-terminal peptide in XCENP-A: MRPGSTPPSRRKSRPPRRVSC-amide (BioSource International, Camarillo, CA). These antibodies were used for indirect immunofluorescence (detection with fluorescein-α-rabbit secondary antibodies; MP Biomedicals, Irvine, CA), Western blotting, and chromatin immunoprecipitation experiments. 594-α-XCENP-E and Alexa 488-α-XCENP-A (gifts from A. Straight, Stanford University) were used for direct immunofluorescent staining.
Western Blotting
Nuclei were prepared from ∼5 × 105 Xenopus tissue culture (XTC) nuclei as described previously (Masumoto et al., 1989), resuspended in sample buffer, and separated by 7.5–15% gradient SDS-PAGE. Proteins were transferred to nitrocellulose for 1 h at 100V in 4°C CAPS transfer buffer (2.21 g of cyclohexylaminopropane sulfonic acid, 0.5 g of dithiothreitol [DTT], 150 ml of methanol, and 1 liter of double distilled H2O, pH 10.5). After blocking overnight in phosphate-buffered saline (PBS)-Tw (PBS plus 0.5% Tween 20) plus 4% milk, the membrane was incubated for 3 h at room temperature in ∼2 μg/ml anti-XCENP-A in PBS-Tw plus 4% milk. Subsequent washing, secondary antibody blotting, and electrochemical luminescent detection steps were performed by standard techniques.
Immunofluorescence
XTC cells were cultured at room temperature in 70% Leibovitz L-15 medium plus 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. For metaphase chromosome spreads, XTC cells were arrested in metaphase by overnight treatment with nocodozole (100 ng/ml), harvested by trypsinization, spun onto poly-l-lysine–coated slides in a cytocentrifuge (Shandon Instruments, Pittsburgh, PA), and fixed and stained as described previously (Van Hooser and Brinkley, 1999). For staining of cycling cells, XTC cells were cultured on poly-l-lysine–coated coverslips, fixed for 2 min in –20°C methanol, rehydrated in Tris-buffered saline (TBS), blocked with AbDil (TBS, 0.1% Triton X-100, and 2% bovine serum albumin [BSA]), and stained for 1 h with primary antibodies diluted in AbDil. After staining DNA with 2 μg/ml Hoechst 33342, coverslips were mounted in 90% glycerol, 1 mg/ml p-phenylenediamine, 20 mM Tris-Cl, pH 8.8. All images of XTC cells were acquired at room temperature on a Nikon E800 microscope, equipped with a PlanApo 60×/1.40 numerical aperture (NA) objective (XTC chromosome spreads) or a PlanApo 100×/1.40 NA objective (intact XTC cells), by using an Orca-ER digital camera (Hamamatsu, Hamamatsu City, Japan) and Meta-Morph acquisition software (Universal Imaging, Downingtown, PA). For costaining of XCENP-A and XCENP-E, images were taken at 0.2-μm z-steps. An approximation of the point-spread function was used to produce deconvolved reconstructions of the image stacks, which are presented as maximum intensity projections (MetaMorph).
Chromatin Immunoprecipitation
Isolation of Nuclei. Nuclei were prepared from ∼7 × 107 XTC cells as described previously (Masumoto et al., 1989). All steps after the first wash were performed on ice, using chilled buffers, unless otherwise noted. In brief, cells were harvested, washed with fresh medium, washed twice with PBS, washed twice with isolation buffer (3.75 mM Tris-HCl, pH 8.0, 0.05 mM spermine, 0.125 mM spermidine, 0.5 mM EDTA, 0.5 mM DTT, 20 mM KCl, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) and lysed by Dounce homogenization in isolation buffer plus 0.1% digitonin. Nuclei were collected by centrifugation, washed twice with isolation buffer plus 0.1% digitonin, and washed once with washing buffer (20 mM HEPES, pH 8.0, 20 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, and 0.1 mM PMSF). Nuclei were resuspended at 108 nuclei/ml in washing buffer plus 0.3 M NaCl and 10 μg/ml each leupeptin, pepstatin, and chymostatin (WB-NaCl) (Ando et al., 2002).
Fragmentation of Chromatin. CaCl2 was added to 2 mM, and chromatin was digested with micrococcal nuclease (200 U/ml) for 5 min at 37°C. Digestion was stopped by adding EDTA, pH 8.0, to 20 mM and chilling on ice. Insoluble material was collected by centrifugation at 4°C for 10 min at 8000g. After recovery of the supernatant to a fresh tube, the pellet was resuspended in one-half the original volume of the WB-NaCl and sonicated using the microtip attachment of a Sonifier 250 (Branson, Danbury, CT) for 10 s at setting 7, with a 70% duty cycle. After centrifugation, the supernatant was recovered and pooled with the first supernatant. NP-40 was added to 0.1%.
Immunoprecipitation. Fragmented chromatin was divided into two aliquots, and each was mixed with 50 μg of antibody (α-XCENP-A or preimmune IgG) for 6 h at 4°C. Reconstituted protein A-Sepharose CL4B beads (Sigma-Aldrich, St. Louis, MO) were washed three times in WB-NaCl. Immune complexes were recovered by mixing with 200 μl of beads for 1 h at 4°C. Beads were collected by centrifugation, washed three times with WB-NaCl plus 0.1% NP-40, and resuspended in WB-NaCl plus 0.1% NP-40 and 0.1% Tween 20.
DNA Extraction and Cloning. Bead-bound immunoprecipitates were digested with proteinase K (200 μg/ml) for 4 h at 56°C. DNA was extracted twice with phenol/chloroform, extracted once with chloroform, and precipitated with ethanol. A-overhangs were added by incubation with Taq polymerase and 200 μM dATP for 30 min at 70°C. Fragments were cloned into the TOPO-TA vector system (Invitrogen, Carlsbad, CA) by using product instructions. Ligation mixes were transformed into electrocompetent cells, and random clones were selected for sequencing. Initially, 24 anti-XCENP-A clones and 24 preimmune clones were sequenced. Later, 64 additional anti-XCENP-A clones were sequenced, for a total of 88. Finally, additional clones of Fcr1 were obtained by colony hybridization against the initial transformants (Sambrook et al., 1989).
Genomic Digests
One-microgram samples of genomic X. laevis DNA were digested with 25, 5, 1, 0.2, or 0.04 U of Nsi I for 16 h at 37°C. Samples were separated by 0.7% agarose gel electrophoresis and transferred to Hybond-N nylon membrane (Amersham Biosciences, Uppsala, Sweden) according to the manufacturer's instructions. Probes were constructed by nick translation of an ∼1-kb fragment of Fcr1 repeats by using an optimized DNA polymerase I/DNase I mixture (Invitrogen). Southern hybridization was performed in Rapid-hyb buffer (Amersham Biosciences) according to the manufacturer's instructions. Blots were exposed to Kodak (Eastman Kodak, Rochester, NY) BioMax MS film at –70°C.
Fluorescent In Situ Hybridization on XTC Cells
Digoxigenin-labeled probes were synthesized by nick translation of an ∼1-kb fragment of Fcr1 repeats, by using an optimized DNA polymerase I/DNase I mixture (Invitrogen) and digoxigenin-11-dUTP (Roche Diagnostics, Indianapolis, IN). Probes were detected with rhodamine-α-digoxigenin antibodies (Roche Diagnostics). Metaphase spreads were prepared as described above. Hybridization and staining were performed as described previously (Van Hooser and Brinkley, 1999).
Lampbrush Chromosomes
Germinal vesicle spreads were prepared as described previously (Gall, 1998). Each of the 18 lampbrush chromosomes was identified by staining with antibodies against RNA polymerase III as described previously (Murphy et al., 2002), with secondary detection by Alexa 488-α-rabbit IgG (Molecular Probes, Eugene, OR). After antibody staining, slides were rinsed twice in 2× SSC and dehydrated through an ethanol series (from 30 to 100%). Slides were incubated in xylene overnight, followed by three xylene changes, for 1 h each. Slides were baked in a dry oven for 30 min at 80°C and cooked for 30 min in 2× SSC at 75–80°C. After three rinses in room temperature 2× SSC, slides were denatured by incubation in 0.07 N NaCl for 90 s at room temperature. After three rinses in room temperature 2× SSC, slides were dehydrated through an ethanol series.
Cy3-labeled RNA probes were constructed by in vitro transcription of an ∼1-kb fragment of Fcr1 repeats by described methods (Singer, 1998). Probes were resuspended in hybridization solution (10% dextran sulfate, 2 mM vanadyl-ribonucleoside, 0.02% RNase-free BSA, 40 μg of Escherichia coli tRNA, 2× SSC, and 50% formamide) and denatured for 1 min at 65°C. Probes were hybridized to slides overnight under sealed coverslips in a 37°C humid chamber. Coverslips were pried off under 2× SSC, and slides were washed twice in 2× SSC. Slides were washed three times for 15 min each in 2× SSC plus 50% formamide at 45°C and twice in room temperature 2× SSC. After staining DNA with 0.01 μg/ml 4,6-diamidino-2-phenylindole (DAPI), slides were mounted in 50% glycerol, 1 mg/ml p-phenylenediamine, and 20 mM Tris-Cl, pH 8.8. Images were acquired with a MicroMax charge-coupled device camera (Princeton Instruments, Trenton, NJ) on a Zeiss Axioplan microscope equipped with a 63×/1.25 NA objective. Images were acquired using IP Lab Spectrum (Scanalytics, Fairfax, VA), with additional analysis in MetaMorph. Specific chromosomes were identified by measuring positions of RNA polymerase III staining relative to total chromosome length. We mapped >50 chromosomes, including at least two examples of each of the 18 (with the exception of chromosome 3, of which we mapped only 1 example).
RESULTS
Identification of Xenopus CENP-A
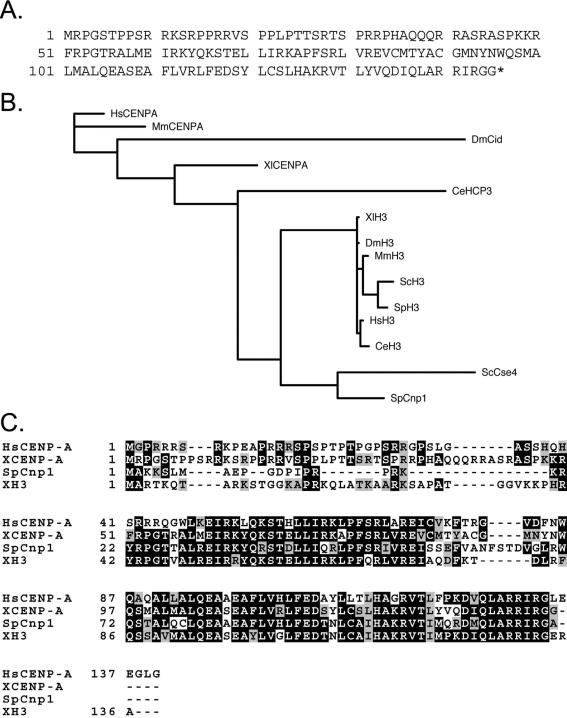
We used degenerate PCR to look for the frog homologue of CENP-A. We designed primers based on residues that are conserved among CENP-A homologues and, at some positions, distinguish CENP-A from normal histone H3. From a X. laevis cDNA library, we amplified a partial cDNA encoding a 40-amino acid sequence bearing significant identity to human CENP-A and distinct differences from X. laevis H3 (XH3). PCR between the identified fragment and the vector backbone of the library completed the 5′ end of the cDNA. Phage plaque hybridization by using a probe constructed from the 5′ portion of the open reading frame yielded a full-length cDNA encoding a 145-amino acid protein that we name XCENP-A (Figure 1A).
Figure 1.
Identification of a sequence homologue of CENP-A in X. laevis. (A) Predicted amino acid sequence of XCENP-A. The asterisk denotes the stop codon. (B) Phylogenetic tree depicting probable evolutionary relationships between CENP-A and H3 homologues from humans (HsCENP-A and HsH3, respectively), mice (MmCENP-A and MmH3), frogs (XCENP-A and XH3), fruit flies (DmCid and DmH3), worms (CeHCP-3 and CeH3), fission yeast (SpCnp1 and SpH3), and budding yeast (ScCse4 and ScH3). Branch lengths are proportional to predicted evolutionary distances. The tree was generated with MrBayes (Ronquist and Huelsenbeck, 2003). (C) Multiple sequence alignment of CENP-A homologues from fission yeast, humans, and frogs. The X. laevis H3 protein sequence is included in the alignment for reference. Dark shading indicates identical residues; light shading indicates similar residues.
Comparison with the sequences of CENP-A and H3 homologues from other organisms strongly suggests that XCENP-A is the X. laevis CENP-A homologue. Although the rapid evolution of CENP-A homologues (Malik and Henikoff, 2001) results in a phylogenetic tree that is somewhat distorted relative to species relationships, XCENP-A clearly clusters with animal CENP-A homologues (Figure 1B). Whereas H3 sequences from different species show near identity across the length of the protein, CENP-A homologues typically contain a divergent amino-terminal tail like that of XCENP-A (Figure 1C). XCENP-A also contains several CENP-A–specific residues in the more conserved histone core, such as lysine 63 and serine 78.
XCENP-A Localization
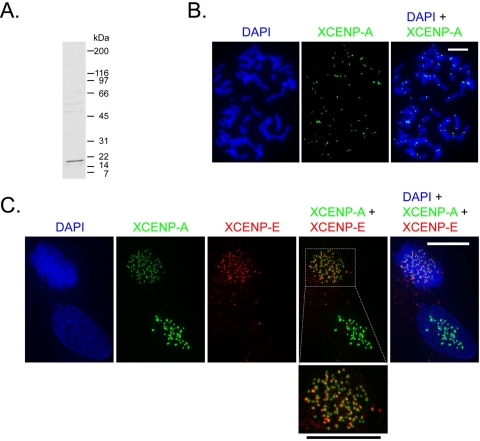
To confirm that XCENP-A encodes a centromeric variant of H3, we examined the localization of the protein by immunofluorescence (Figure 2). Antibodies against a peptide in the amino-terminal tail of XCENP-A stained the primary constriction of metaphase chromosomes (Figure 2B), providing cytological evidence of centromeric localization. In mitotic cells, XCENP-A antibodies stained foci resembling those stained by XCENP-E antibodies (Figure 2C). XCENP-E is a kinesin-like motor that associates with the outer region of the kinetochore during mitosis (Wood et al., 1997).
Figure 2.
XCENP-A localizes to kinetochores. (A) Anti-XCENP-A Western blot of nuclear proteins reveals a single prominent band with mobility consistent with the predicted molecular mass of XCENP-A (17 kDa). (B) Indirect immunofluorescence against XCENP-A (green) on DAPI-stained metaphase chromosome spreads (blue) from cultured cells. (C) Direct immunofluorescence on mitotic (top left) and interphase (bottom right) cells. Cells were stained with directly labeled antibodies against XCENP-A (green) and XCENP-E (red). DAPI-stained DNA is shown in blue. Boxed region is enlarged below. Bars, 10 μm. XCENP-A and XCENP-E foci seem to be adjacent rather than identical. This pattern might reflect the expected juxtaposition of inner and outer kinetochore components or the two images not being properly in register with each other.
Like other CENP-A homologues, XCENP-A is a constitutive component of the kinetochore. Unlike XCENP-E antibodies, which stained only mitotic chromosomes, XCENP-A antibodies also stained discrete foci in interphase nuclei (Figure 2C).
Identification of CENP-A–associated DNA Sequences in X. laevis
To find DNA sequences associated with the X. laevis centromere, we performed chromatin immunoprecipitation against XCENP-A. We purified chromatin from XTC cells and fragmented the material into oligonucleosomes by micrococcal nuclease digestion. After immunoprecipitation with peptide antibodies against XCENP-A or preimmune IgG, we extracted and cloned the coprecipitated DNA fragments. We initially sequenced 24 random clones from the anti-XCENP-A immunoprecipitates and 24 random clones from the preimmune IgG controls.
One sequence motif occurred in three of the 24 sequenced anti-XCENP-A clones and none of the sequenced control clones. We obtained additional clones of this motif both by sequencing an additional 64 random anti-XCENP-A clones (for a total of 88) and by colony hybridization against transformants from the anti-XCENP-A chromatin immunoprecipitation. We name this sequence element frog centromeric repeat 1 (Fcr1). Its defining characteristics are a unit length of 174 base pairs, occurrence as multimeric direct repeats, and a high degree of sequence homology between clones.
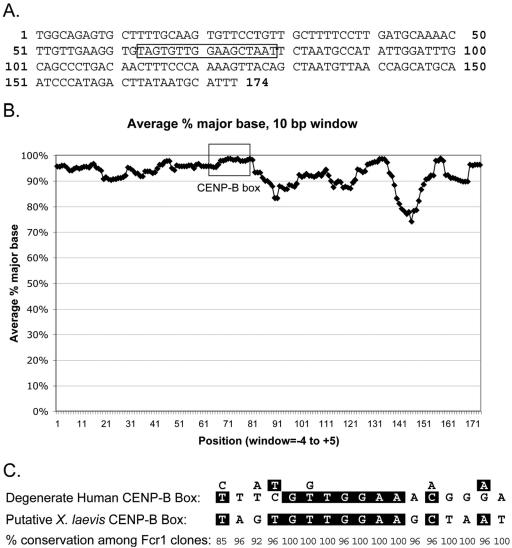
The consensus sequence of the Fcr1 monomer is shown in Figure 3A. The overall sequence is highly conserved from clone to clone (Figure 3B), with ∼90% identity between any pair of monomeric units. The consensus sequence of Fcr1 does not bear significant primary sequence similarity to any known sequences. The X. laevis sequence does show certain hallmarks, however, that are characteristic of centromere-associated repeats from other organisms. One such trait is high A/T content: the consensus Fcr1 sequence is 64% A/T.
Figure 3.
Identification of Fcr1, a 174-base pair centromere-associated repeat. (A) Consensus sequence of the monomer unit (with ends defined arbitrarily) of the DNA satellite cloned from α-CENP-A chromatin immunoprecipitates. The box indicates the putative CENP-B box (positions 63 through 79). (B) Conservation of Fcr1 sequences among 38 analyzed monomeric units and submonomeric fragments. The graph plots average (over a 10-base pair window) percentage of sequences containing the consensus nucleotide at each position. Boxed region indicates putative CENP-B box. (C) Similarity between a 17-base pair element in Fcr1 (“putative X. laevis CENP-B box”) and the degenerate human CENP-B box (Choo, 1997). The percentage of Fcr1 clones containing the consensus nucleotide is shown below each position.
Another widely conserved feature of centromeric sequences is repeat size: centromeric sequences in higher eukaryotes frequently contain repeats of roughly nucleosomal unit length, such as the 171-base pair α-satellite in primate centromeres. The consensus unit length of Fcr1 is 174 base pairs, although we observed three apparent deletions (1, 3, and 7 base pairs) and three apparent insertions (1, 1, and 16 base pairs) among the 38 analyzed monomeric units.
Although centromeric sequences from different higher eukaryotic species generally lack any universal homology at the primary sequence level, many plant and animal centromeric repeats contain short motifs similar to the 17 base pair “CENP-B box” bound by the human CENP-B protein (Masumoto et al., 1989). Fcr1 contains a 17-base pair motif (the boxed region in Figure 3A) that is identical to the degenerate human CENP-B box at 11 of 17 positions (Figure 3C), including an eight-base pair stretch of perfect identity near the middle of the repeat. The putative X. laevis CENP-B box is highly conserved among all analyzed clones of Fcr1 (Figure 3C).
Genomic Organization and Localization of the Fcr1 Repeat
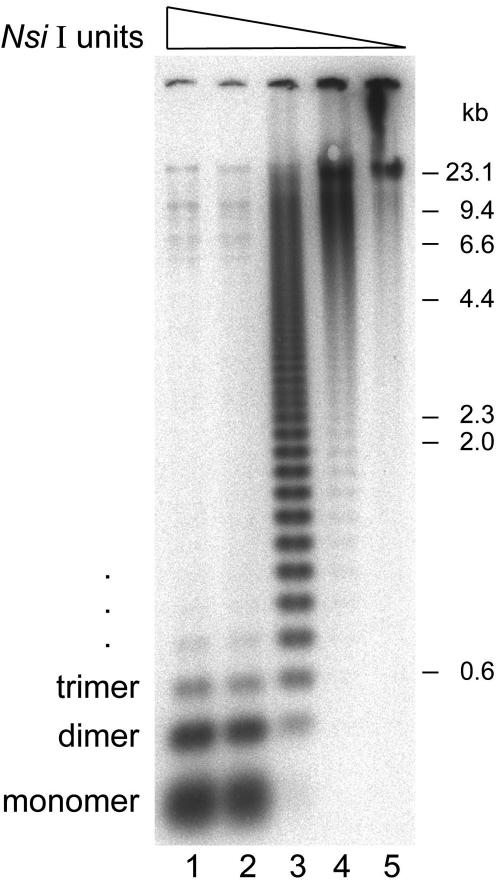
To examine the organization of Fcr1 repeats in the X. laevis genome, we digested genomic DNA with varying quantities of the restriction endonuclease Nsi I, which is predicted to cut the consensus monomeric unit at a single site. Southern blotting with radioactive Fcr1 probe revealed a large ladder of bands, with step sizes consistent with the 174-base pair monomeric unit length (Figure 4). The size and spacing of this ladder suggests the presence of large, homogeneous arrays of directly repeated units of Fcr1 in the genome.
Figure 4.
Fcr1 is present in large ordered arrays in the frog genome. Southern blot of Nsi I-digested genomic DNA with radioactive probe for Fcr1. Genomic DNA (1 μg per lane) was digested with 25 U (lane 1), 5 U (lane 2), 1 U (lane 3), 0.2 U (lane 4), or 0.04 U (lane 5) of Nsi I.
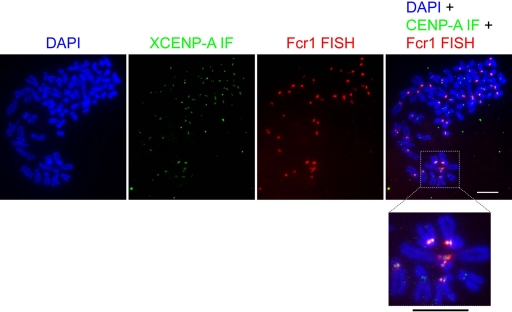
We examined the localization of Fcr1 arrays in the genome by fluorescent in situ hybridization against metaphase chromosome spreads from XTC cells (Figure 5). Probes constructed by nick translation of an ∼1-kb fragment of Fcr1 repeats stained most X. laevis chromosomes. Costaining with antibodies against XCENP-A confirmed the centromeric localization of the sites of Fcr1 hybridization. Although persistent aneuploidy in XTC cells made precise quantification difficult due to the varying total numbers of chromosomes per cell, Fcr1 probes consistently stained 60–70% of the centromeres in each metaphase spread.
Figure 5.
In situ hybridization against Fcr1 stains most centromeres. Fluorescent in situ hybridization of digoxigenin-labeled Fcr1 probe (red) against DAPI-stained metaphase chromosome spreads (blue) from cultured cells. Indirect immunofluorescence against XCENP-A is shown in green. Boxed region is enlarged below. Bars, 10 μm.
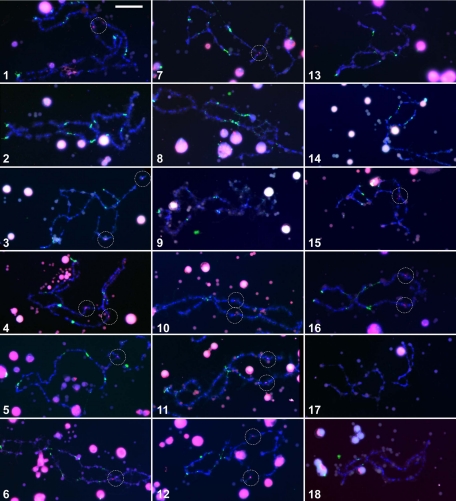
To determine whether the same chromosomes consistently stain (or fail to stain) with Fcr1 probes, we performed in situ hybridization on lampbrush chromosomes. Murphy et al. (2002) have shown that antibodies against RNA polymerase III (pol III) stain a distinct and identifiable pattern of loci on each X. laevis lampbrush chromosome (one chromosome, 17, is distinct in its lack of any loci of significant pol III staining). By costaining with fluorescence in situ hybridization (FISH) against Fcr1 and immunofluorescence against pol III, we were able to map the location of Fcr1 hybridization (if any) on each of the 18 bivalent lampbrush chromosomes. We analyzed an average of three examples of each chromosome (Supplementary Table 1). We find that Fcr1 stains 11 chromosomes (1, 3, 4, 5, 6, 7, 10, 11, 12, 15, and 16) and consistently fails to stain seven chromosomes (2, 8, 9, 13, 14, 17, and 18). A chi-square test rejects the null hypothesis that each chromosome is equally likely to stain (p = 0.00023). Representative examples of each chromosome are shown in Figure 6. Hybridization results for each chromosome were identical in every replicate analyzed (with the exception of chromosome 5, of which we identified two Fcr1+ examples and one Fcr1– example).
Figure 6.
Fcr1 hybridization against lampbrush chromosomes confirms consistent staining of 11 of the 18 frog chromosomes. Each of the 18 X. laevis lampbrush chromosomes (stained with DAPI, blue) can be identified by the distinct patterns of loci stained by antibodies against RNA polymerase III (green) (Murphy et al., 2002). Fluorescent in situ hybridization with Cy3-labeled Fcr1 probe is shown in red. Chromosomes are numbered in descending order according to length (Murphy et al., 2002). This composite combines representative images from an average of three analyzed examples of each of the 18 lampbrush chromosomes. Cross-hybridization with extrachromosomal nucleoli (large DAPI- and Fcr1-stained spheres) is believed to be caused by polylinker sequences at the ends of the Fcr1 probe, which can hybridize with rRNA (Witkiewicz et al., 1993). Dashed circles highlight chromosomal sites of Fcr1 hybridization. Bar, 20 μm.
DISCUSSION
Identification of a Putative CENP-A Homologue in X. laevis
We have identified a cDNA encoding a predicted protein we name XCENP-A. Sequence characteristics and protein localization support the conclusion that this open reading frame encodes the X. laevis homologue of CENP-A. The 145-amino acid predicted sequence is similar to other CENP-A homologues and to Xenopus H3, but it is clearly distinct from the latter. Localization studies place XCENP-A near the outer kinetochore component XCENP-E, consistent with the expected inner kinetochore localization of a CENP-A homologue. Punctate nuclear staining for XCENP-A is preserved throughout the cell cycle. The fact that all other reported frog kinetochore components localize there only during cell division precludes any definitive characterization of the interphase localization of XCENP-A. However, the observed staining supports the conclusion that XCENP-A is the first identified constitutive component of the X. laevis kinetochore.
Use of Lampbrush Chromosomes to Identify Fcr1+ Chromosomes
We used chromatin immunoprecipitation to isolate XCENP-A associated DNA sequences, identifying a 174-base pair repetitive element we term Fcr1. Fcr1 is present in large, directly ordered arrays in the genome and colocalizes with most, but not all, centromeres by FISH. Interpretation of this failure to stain all X. laevis centromeres was complicated by hyperploidy in XTC cells and the difficulty of identifying specific chromosomes in metaphase spreads of cultured cells.
Lampbrush chromosomes provided an elegant solution to these problems. Isolated from the germinal vesicle (giant nucleus) of the oocyte, lampbrush chromosomes are significantly condensed yet still transcriptionally active (Callan, 1986). Murphy et al. (2002) have shown that antibodies that recognize RNA pol III yield a distinct pattern of stained loci on each lampbrush chromosome from X. laevis oocytes. These pol III “bar codes” can be used to reproducibly identify each of the 18 bivalent lampbrush chromosomes. By hybridizing Fcr1 probes to lampbrush chromosomes and costaining with pol III antibodies, we determined that 11 chromosomes reproducibly stain positively for Fcr1. Seven of the lampbrush chromosomes consistently fail to show significant staining for Fcr1. These results, which correspond to 61% of chromosomes staining positively for Fcr1, are consistent with the 66 ± 3% of centromeres that stain positively in hyperploid spreads from cultured cells (Figure 5; our unpublished data).
To date, we have been unable to directly confirm that the localization of the Fcr1 hybridization signals on lampbrush chromosomes corresponds to the positions of the centromeres (our XCENP-A antibodies fail to stain lampbrush chromosomes). However, the physical location of each centromere relative to loci of RNA pol III staining has been mapped by localization of myc-tagged human CENP-C (Wu and Gall, personal communication). The mapped locations of Fcr1 hybridization correspond very closely to those of hCENP-C-GFP staining. The colocalization of Fcr1 hybridization and XCENP-A staining on metaphase spreads from XTC cells also provides indirect evidence that the lampbrush hybridization is centromeric.
Evidence That Fcr1 Is a Centromeric Satellite
Although Fcr1 probes fail to stain some chromosomes, a variety of factors support the conclusion that Fcr1 is a component of centromeric DNA in X. laevis. Fcr1 was identified by coprecipitation of small, homogeneous arrays of the repeat with XCENP-A in chromatin immunoprecipitations, suggesting that centromeric XCENP-A nucleosomes contain Fcr1 DNA. The signals from in situ hybridizations with Fcr1 also colocalize with anti-XCENP-A immunofluorescence.
Sequence analysis of Fcr1 reveals several hallmarks of centromeric satellites, providing further support for the characterization of Fcr1 as a member of this class. Centromeres often contain repetitive elements with monomer lengths that are very similar to the observed length of DNA in a single nucleosome. In humans and other primates, the length of the alphoid DNA monomer is 171 base pairs. Similar lengths are found in the centromeric repeats in some insects: a centromeric repeat in Chironomus pallidivittatus is 155 base pairs (Rovira et al., 1993). Several plants also have centromeric repeats with lengths in this range: the Arabidopsis CENP-A homologue, for example, associates with arrays of a 180-base pair repeat (Nagaki et al., 2003). The unit length of Fcr1 lies in the heart of this range at 174 base pairs. Fcr1 also contains a 17-base pair motif resembling the CENP-B box, another widely conserved feature of centromeric sequences. CENP-B box-like motifs are found in the centromeric satellites of a variety of animals and some plants. The putative Xenopus CENP-B box is among the best conserved sequences in the Fcr1 monomer. Finally, Fcr1 is A/T rich, a characteristic of centromere sequences in species ranging from humans—alphoid DNA is 64% A/T (Choo, 1997)—to budding yeast: Cse4, the budding yeast homologue of CENP-A, interacts with the centromeric CDEII domain, which is 90% A/T (Keith and Fitzgerald-Hayes, 2000).
Explanations for the Hybridization of Fcr1 to Only a Subset of Centromeres
There are several possible explanations for the observation that in situ hybridization against Fcr1 stains only a subset of the chromosomes. One possibility is that the nonstaining centromeres contain no copies of the Fcr1 repeat and thus comprise one or more alternative centromeric sequences. There are precedents in other systems for centromeres on different chromosomes consisting of completely different DNA sequences. The lack of any universal centromeric sequence in Drosophila is a prominent example (Sun et al., 1997), as are rare human neocentromeres that lack any detectable alphoid DNA (Lo et al., 2001; Alonso et al., 2003). There is even a precedent within Amphibia: a centromeric satellite in Rana ridibunda is found in only a subset of centromeres (Ragghianti et al., 1995).
The possibility that there are multiple types of centromere sequence in X. laevis is particularly intriguing in light of the genomic composition of this frog. X. laevis is pseudotetraploid (Kobel and Du Pasquier, 1986). Such tetraploids can arise as a result of endoreduplication, the occurrence of two rounds of DNA replication without an intervening mitosis. Such events are selected for in the hybrids formed by mating between two species (Greig et al., 2002). If the two parental species show >1% sequence divergence, meiotic recombination fails, and no useful gametes are produced. Viable gametes are produced, however, after endoreduplication because the resulting two copies of a chromosome from one of the parental species can pair with and recombine with each other. This scenario may explain the origin of X. laevis (Kobel and Du Pasquier, 1986), and if the centromeric repeats in the two species were sufficiently different, only one of the ancestral sets of chromosomes would hybridize with Fcr1. Although it might seem unlikely that species that were sufficiently closely related to form fertile hybrids could have different centromere-associated repeat sequences, centromeric satellites are among the most rapidly evolving eukaryotic DNA sequences (Henikoff et al., 2001). Indeed, homologous chromosomes in closely related species of Drosophila sometimes contain completely divergent centromeric satellites (Csink and Henikoff, 1998).
The hypothesis that the tetraploid origin of X. laevis produced multiple types of centromere sequences in this frog raises the question of whether the Fcr1– centromeres might be bound by a distinct CENP-A protein. Although the absence of a complete genome sequence prevents us from excluding this possibility, our results suggest that X. laevis has only one CENP-A homologue. Degenerate PCR and hybridization studies against a X. laevis cDNA library identified only the single XCENP-A sequence reported here. Furthermore, peptide antibodies against this protein stain all X. laevis kinetochores by immunofluorescence (Figure 2). In principle, the immunofluorescent signal on the Fcr1– centromeres could correspond to a closely related but distinct CENP-A protein, but any protein similar enough to be recognized by anti-XCENP-A antibodies should have been identified in the colony hybridization experiment. Thus, although the existence of one or more additional CENP-A homologues remains a formal possibility, we consider it an unlikely one.
Because anti-XCENP-A antibodies stained all X. laevis kinetochores by immunofluorescence, the sequences of all 18 centromeres should have been enriched in the XCENP-A chromatin immunoprecipitation. However, the clones we have sequenced offer few clues about the structure and composition of the seven apparently Fcr1– centromeres. Analysis of the Fcr1– clones revealed two general classes of sequences, each represented by nearly equal numbers of clones. Roughly one-half have no significant homology to any known sequence. The remaining clones are each similar to a large number of otherwise unrelated Xenopus sequences and thus seem to be dispersed (and possibly mobile) elements. Indeed, several clones contain sequences that are highly similar to known transposons in the Xenopus genome. Known X. laevis satellite sequences REM 1 and satellite I are also represented. Neither of these known satellites are particularly attractive candidates for a second centromeric repeat, because previous in situ hybridization studies indicate that both are dispersed in a large number of loci throughout the genome (Jamrich et al., 1983; Hummel et al., 1984). However, in light of earlier anecdotal reports that the multitude of satellite I loci include positions near roughly one-half of the centromeres (Lam and Carroll, 1983), we performed two-color fluorescent in situ hybridization by using Fcr1 and satellite 1 probes. Our analysis confirmed Jamrich and colleagues' conclusion that satellite I loci are scattered widely across the chromosomes and do not cluster near the centromeres.
A more prosaic explanation of our results is that the seven nonstaining centromeres simply contain fewer copies of the Fcr1 repeat and are thus below the level of detection in these experiments. Although we know of no a priori reason to expect centromere size to correlate with chromosome length, we note that the positively staining chromosomes are among the longest in the X. laevis genome. Across evolution, average chromosome size in a species correlates roughly with the average number of microtubules bound to each kinetochore (Bloom, 1993). It seems plausible that larger chromosomes evolved larger arrays of centromeric satellite to support larger numbers of bound microtubules. Finally, there are precedents for variation in the size of centromeric satellite arrays. The size of human centromeric arrays varies over an order of magnitude (Choo, 1997).
Another interpretation of the hybridization pattern is that all X. laevis centromeres contain comparable arrays of Fcr1, but the staining protocol is inconsistent or inefficient. By this theory, all of the chromosomes have the same potential to stain positively, but a fraction fail to stain in any given experiment for technical reasons. The consistent staining of a distinct subset of lampbrush chromosomes suggests that such an explanation is insufficient. We also note that the two fluorescent in situ hybridization experiments (XTC chromosome spreads and lampbrush chromosomes) differed significantly in a variety of parameters, such as probe composition and detection, denaturation conditions, and hybridization medium. The consistency of the results between the two systems, despite large differences in the protocols, provides further evidence against a strictly technical explanation for the failure of some centromeres to stain with Fcr1 probes.
Implications for Future Experiments
The significance of our results resides in the matched set of CENP-A homologue and cognate centromeric satellite DNA and the advantages of Xenopus as an experimental system. Kinetochore assembly on sperm chromatin in egg extracts recapitulates the establishment of kinetochores on sperm chromosomes after fertilization, which is the closest approximation to de novo centromere assembly in a normal life cycle. Reconstitution experiments in egg extracts may thus represent the ideal system to test the centromere-defining capacity of repetitive centromeric DNA sequences and potential epigenetic markers such as CENP-A.
Supplementary Material
Acknowledgments
We are especially grateful to Joe Gall, Christine Murphy, and the entire Gall laboratory for invaluable help with lampbrush chromosomes. We thank Aaron Straight for antibodies, Len Hook from Scanalytics for help with image conversion, Nicholas Ingolia for help with phylogenetic analysis, and Margaret Saha for X. laevis satellite I DNA. We thank members of the Murray laboratory for encouragement and constructive criticism. This work was supported by grants from National Institutes of Health (to A.W.M.) and a fellowship from National Science Foundation (to N.S.E.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–09–0788) on January 26, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P. E. (2003). Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12, 2711–2721. [DOI] [PubMed] [Google Scholar]
- Amor, D. J., Bentley, K., Ryan, J., Perry, J., Wong, L., Slater, H., and Choo, K. H. (2004). Human centromere repositioning “in progress.” Proc. Natl. Acad. Sci. USA 101, 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D. J., and Choo, K. H. (2002). Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71, 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, S., Yang, H., Nozaki, N., Okazaki, T., and Yoda, K. (2002). CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22, 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Walczak, C. E. (2003). Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13, R449–R460. [DOI] [PubMed] [Google Scholar]
- Bloom, K. (1993). The centromere frontier: kinetochore components, microtubule-based motility, and the CEN-value paradox. Cell. 73, 621–624. [DOI] [PubMed] [Google Scholar]
- Callan, H. G. (1986). Lampbrush Chromosomes, Berlin: Springer.
- Carbon, J., and Clarke, L. (1984). Structural and functional analysis of a yeast centromere (CEN3). J. Cell Sci. Suppl. 1, 43–58. [DOI] [PubMed] [Google Scholar]
- Choo, K. H. (1997). The Centromere, Oxford, United Kingdom: Oxford University Press.
- Clarke, L. (1998). Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 8, 212–218. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Mao, Y., and Sullivan, K. F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Csink, A. K., and Henikoff, S. (1998). Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Gall, J. G. (1998). Spread preparation of Xenopus germinal vesicle contents. In Cells: A Laboratory Manual, ed. D. Spector, R. Goldman, and L. Leinwand, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 52.1–52.4.
- Greig, D., Borts, R. H., Louis, E. J., and Travisano, M. (2002). Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. B. Biol. Sci. 269, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H. S. (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., and Schubert, I. (2003). DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6, 554–560. [DOI] [PubMed] [Google Scholar]
- Howman, E. V., Fowler, K. J., Newson, A. J., Redward, S., MacDonald, A. C., Kalitsis, P., and Choo, K. H. (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel, S., Meyerhof, W., Korge, E., and Knochel, W. (1984). Characterization of highly and moderately repetitive 500 bp Eco RI fragments from Xenopus laevis DNA. Nucleic Acids Res. 12, 4921–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrich, M., Warrior, R., Steele, R., and Gall, J. G. (1983). Transcription of repetitive sequences on Xenopus lampbrush chromosomes. Proc. Natl. Acad. Sci. USA 80, 3364–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. C., and Fitzgerald-Hayes, M. (2000). CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, N., Minshull, J., and Kirschner, M. W. (1995). The identification of two novel ligands of the FGF receptor by a yeast screening method and their activity in Xenopus development. Cell. 83, 621–630. [DOI] [PubMed] [Google Scholar]
- Kobel, H. R., and Du Pasquier, L. (1986). Genetics of polyploid Xenopus. Trends in Genet. 2, 310–315. [Google Scholar]
- Lam, B. S., and Carroll, D. (1983). Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J. Mol. Biol. 165, 567–585. [DOI] [PubMed] [Google Scholar]
- Lo, A. W., Magliano, D. J., Sibson, M. C., Kalitsis, P., Craig, J. M., and Choo, K. H. (2001). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H. S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto, H., Masukata, H., Muro, Y., Nozaki, N., and Okazaki, T. (1989). A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone, B. G., and Allshire, R. C. (2003). Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13, 191198. [DOI] [PubMed] [Google Scholar]
- Murphy, C., Wang, Z., Roeder, R. G., and Gall, J. G. (2002). RNA polymerase III in Cajal bodies and lampbrush chromosomes of the Xenopus oocyte nucleus. Mol. Biol. Cell. 13, 3466–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P. B., Zhong, C. X., Dawe, R. K., Henikoff, S., and Jiang, J. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A. A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki, J., Nakano, M., Okada, T., and Masumoto, H. (2002). CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D. K., O'Day, K., Trong, H. L., Charbonneau, H., and Margolis, R. L. (1991). Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88, 3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri, L., Landonio, L., Stotz, A., and Philippsen, P. (1985). Role of conserved sequence elements in yeast centromere DNA. EMBO J. 4, 1867–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragghianti, M., Guerrini, F., Bucci, S., Mancino, G., Hotz, H., Uzzell, T., and Guex, G. D. (1995). Molecular characterization of a centromeric satellite DNA in the hemiclonal hybrid frog Rana esculenta and its parental species. Chromosome Res. 3, 497–506. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rovira, C., Beermann, W., and Edstrom, J. E. (1993). A repetitive DNA sequence associated with the centromeres of Chironomus pallidivittatus. Nucleic Acids Res. 21, 1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Volume 1, ed. N. Ford, Cold Spring Harbor, NY: Cold Spring harbor Laboratory Press.
- Singer, R. H. (1998). Preparation of probes for in situ hybridization. www.singerlab.org/protocols.
- Smith, M. M. (2002). Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14, 279–285. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., and Willard, H. F. (1998). Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 20, 227–228. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wahlstrom, J., and Karpen, G. (1997). Molecular structure of a functional Drosophila centromere. Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser, A., and Brinkley, W. R. (1999). Methods for in situ localization of proteins and DNA in the centromere-kinetochore complex. Methods Cell Biol. 61, 57–80. [DOI] [PubMed] [Google Scholar]
- Warburton, P. E. (2001). Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 17, 243–247. [DOI] [PubMed] [Google Scholar]
- Warburton, P. E., et al. (1997). Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Witkiewicz, H., Bolander, M. E., and Edwards, D. R. (1993). Improved design of riboprobes from pBluescript and related vectors for in situ hybridization. Biotechniques 14, 458–463. [PubMed] [Google Scholar]
- Wood, K. W., Sakowicz, R., Goldstein, L. S., and Cleveland, D. W. (1997). CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 91, 357–366. [DOI] [PubMed] [Google Scholar]
- Yoda, K., Ando, S., Morishita, S., Houmura, K., Hashimoto, K., Takeyasu, K., and Okazaki, T. (2000). Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.