Abstract
Cells have developed a variety of mechanisms to respond to heavy metal exposure. Here, we show that the yeast ubiquitin ligase SCFMet30 plays a central role in the response to two of the most toxic environmental heavy metal contaminants, namely, cadmium and arsenic. SCFMet30 inactivates the transcription factor Met4 by proteolysis-independent polyubiquitination. Exposure of yeast cells to heavy metals led to activation of Met4 as indicated by a complete loss of ubiquitinated Met4 species. The association of Met30 with Skp1 but not with its substrate Met4 was inhibited in cells treated with cadmium. Cadmium-activated Met4 induced glutathione biosynthesis as well as genes involved in sulfuramino acid synthesis. Met4 activation was important for the cellular response to cadmium because mutations in various components of the Met4-transcription complex were hypersensitive to cadmium. In addition, cell cycle analyses revealed that cadmium induced a delay in the transition from G1 to S phase of the cell cycle and slow progression through S phase. Both cadmium and arsenic induced phosphorylation of the cell cycle checkpoint protein Rad53. Genetic analyses demonstrated a complex effect of cadmium on cell cycle regulation that might be important to safeguard cellular and genetic integrity when cells are exposed to heavy metals.
INTRODUCTION
Heavy metals are a major environmental hazard and present a danger to human health. The cause of the cytotoxic effects of heavy metals is not completely understood, but it has been suggested that at least part of their toxicity is due to the formation of hydroxyl radicals, which lead to lipid, protein, and DNA damage (Stohs and Bagchi, 1995; Brennan and Schiestl, 1996; Halliwell and Gutteridge, 1984).
As with any cytotoxic and genotoxic insults, all organisms have developed strategies to respond to heavy metal exposure to maintain cellular and genetic integrity. These strategies include detoxification, repair, or removal of damaged molecules, and delay of cell division to prevent propagation of damaged cellular components (Jamieson, 1998).
The biological effects of cadmium are perhaps better studied than that of other heavy metals. High affinity for sulfhydryl groups, competition with Zn(II) in proteins, nonspecific interaction with DNA, generation of reactive oxygen species, and depletion of glutathione have been shown to contribute to the toxicity of cadmium (Stohs and Bagchi, 1995; Zalups and Ahmad, 2003; McMurray and Tainer, 2003). Recently, it has been demonstrated in yeast that the genotoxic effects of cadmium are indirect (Jin et al., 2003; McMurray and Tainer, 2003). Rather than by direct DNA damage, cadmium leads to genome instability by inhibition of the DNA mismatch repair system (Jin et al., 2003). Although the mechanism of how cadmium inhibits DNA repair is not clear, it has been suggested that damage of sulfhydryl groups containing components of the mismatch repair system might be responsible (Jin et al., 2003).
The damaging effect of cadmium on sulfhydryl groups containing proteins also is reflected in gene expression profiling experiments and proteome analyses in response to cadmium exposure (Vido et al., 2001; Fauchon et al., 2002). These experiments demonstrated up-regulation of proteins involved in the sulfur amino acid biosynthesis pathway, indicating the need to replace damaged proteins containing the sulfur amino acids methionine and cysteine. Interestingly, similar studies that analyzed the response to hydrogen peroxide-induced oxidative stress showed a striking difference between the cadmium and the oxidative stress response because a small decrease of sulfur amino acid pathway components was observed in response to hydrogen peroxide (Godon et al., 1998).
Cadmium exposure of yeast also induces expression of several isozymes of the carbohydrate metabolism such as pyruvate decarboxylase, enolase, and aldehyde dehydrogenase (Fauchon et al., 2002). All the cadmium-induced isozymes showed markedly reduced sulfur content, that is, less methionine and cysteine residues compared with the enzymes expressed under nonstress conditions. The physiological significance of this “sulfur sparing response” has not been tested rigorously, but it has been proposed that it allows cells to devote their sulfur resources to the synthesis of glutathione for cadmium detoxification. In addition, the isozyme switch may protect the carbohydrate metabolism because the sulfur-poor isozymes are predicted to be less cadmium sensitive than their sulfur-rich counterparts (Fauchon et al., 2002; Jamieson, 2002). Both isozyme switching and induction of sulfur amino acid synthesis pathway components in response to cadmium exposure have been shown to depend primarily on the yeast transcription factor Met4 (Fauchon et al., 2002). Microarray analysis showed that cadmium-dependent induction of >60 genes was severely affected in Δmet4 mutants (Fauchon et al., 2002). These results demonstrate that regulation of Met4 is an important component of the cellular response to cadmium exposure and perhaps heavy metal exposure in general.
Besides Met4 several other transcription factors are involved in regulation of genes of the sulfur assimilation pathway (Thomas and Surdin-Kerjan, 1997). These include Met28 (Kuras et al., 1996) and the DNA-binding factors Cbf1 (Kuras et al., 1997), Met31, and Met32 (Blaiseau et al., 1997). However, only Met4 has transactivating activity and seems to be the focus of regulation (Thomas and Surdin-Kerjan, 1997). Met4 is inactivated by ubiquitination, but interestingly by a direct regulatory role of ubiquitin, which is independent of proteolysis (Kaiser et al., 2000; Kuras et al., 2002; Flick et al., 2004; Hochstrasser, 2004). Activation of Met4 involves its deubiquitination by so far unknown ubiquitin hydrolase(s). The Met4 activity status correlates with the intracellular methionine level, but it is thought that S-adenosylmethionine (SAM) is the main effector molecule that regulates Met4 activity (Thomas and Surdin-Kerjan, 1997). SAM is the principal methyl-group donor in cells and is synthesized from methionine and ATP. Because the intracellular concentrations of free cysteine, free methionine and SAM are tightly linked, intracellular SAM levels are a good reflection of the sulfur amino acid content of a cell.
Normal methionine (or SAM) levels keep Met4 in its ubiquitinated form and thus inactive. Very low methionine levels shift Met4 into its deubiquitnated, active form and result in transcription of genes that are involved in sulfur assimilation and biosynthesis of sulfur amino acids (Kaiser et al., 2000; Kuras et al., 2002; Flick et al., 2004). In addition, full activation of Met4, for example, by blocking its ubiquitination, induces a cell cycle arrest that might be important to safeguard cellular integrity (Kaiser et al., 1998, 2000; Patton et al., 2000; Flick et al., 2004). How SAM regulates Met4 ubiquitination is not known, but it is clear that the ubiquitin ligase SCFMet30 together with the ubiquitin-conjugating enzyme Cdc34 catalyze Met4 ubiquitination (Kaiser et al., 2000; Patton et al., 2000). SCFMet30 and specifically Met30 are therefore the key regulators of Met4.
Met30 is essential for cell cycle progression because cell proliferation depends on Met4 ubiquitination. Accordingly, deletion of MET4 suppresses the essential function of Met30 and met4Δ met30Δ double mutants are viable (Kaiser et al., 2000; Patton et al., 2000). However, like Δmet4 mutants, met4Δ met30Δ double mutants are methionine auxotroph because of the importance of Met4 as a master regulator of the sulfur amino acid biosynthesis pathway (Patton et al., 2000).
The ubiquitin ligase SCFMet30 and its target Met4 coordinate cell cycle progression with the intracellular SAM and sulfur amino acid levels. Given the importance of sulfur amino acids for the response to cadmium and perhaps other heavy metals, we analyzed the effects of cadmium exposure on Met4. We report here that cadmium and arsenic prevent Met4 ubiquitination and consequently lead to Met4 activation. Met4 activation induces expression of genes that play important roles in the protection from cadmium damage and in the repair of damaged cellular macromolecules. Thus, Cdc34/SCFMet30 is an important component of the cellular defense system against heavy metals.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Yeast strains used in this study are isogenic to 15DaubΔ, a bar1Δ ura3Δns, a derivative of BF264-15D (Reed et al., 1985). Relevant genotypes of strains used in this study are listed in Table 1. All strains were grown in standard culture media, and standard yeast genetic methods were used (Guthrie and Fink, 1991). Heavy metals used were AsNaO2, CdCl2, CoCl2, NiSO4, and PbCl2.
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| 15Daub | a bar1Δ ura3Δns, ade1 his2 leu2-3112 trp1-1 | Reed et al. (1985) |
| PY23 | a bar1 cdc34-3 | S. Reed |
| PY175 | a bar1 cdc53-1 | S. Reed |
| PY187 | a bar1 cdc4-3 | S. Reed |
| PY236 | a bar1 pep4::URA3 | Kaiser et al. (2000) |
| PY283 | a bar1 met30-6::KAN | Kaiser et al. (1998) |
| PY589 | a bar1 met30::KAN met4::KAN | Kaiser et al. (2000) |
| PY640 | a bar1 cbf1::ZEO | This study |
| PY889 | a bar1 met32::KAN | This study |
| PY893 | a bar1 met30::KAN met32::KAN | This study |
| PY950 | a bar1 met28::HYG | This study |
| PY965 | a bar1 met31::HYG | This study |
| PY968 | a bar1 met32::KAN met31::HYG | This study |
| PY1073 | a bar1 12mycMET30::ZEO pep4::URA3 | I. Ouni |
| PY1117 | a bar1 rad53:: KAN <RNR1::TRP>2 μ | This study |
| PY1123 | a bar1 met4::KAN | This study |
| PY1161 | a bar1 tel1::HYG | This study |
| PY1164 | a bar1 mec1::HYG <RNR1::TRP>2 μ | This study |
| PY1197 | a bar1 mec1::HYG met32::KAN <RNR1::TRP>2 μ | This study |
Spotting Experiments
Strains were grown to logarithmic phase, sonicated, and counted. Serial dilutions were spotted onto YEPD plates by using a pin replicator (V&P Scientific, San Diego, CA). Plates were incubated at 30°C or either 25 or 28°C for temperature-sensitive strains as indicated.
Cell Survival Assay
Cells were cultured to logarithmic phase and incubated with designated amounts of cadmium for 30 min. Treated and untreated cells were then plated onto YEPD plates and incubated at 30 or 25°C for temperature-sensitive strains.
Cell Cycle Synchronization, Budding Index, and Flow Cytometry
Cells were synchronized in G1 with the mating pheromone α-factor (20 ng/ml final concentration). Cells were arrested at 30 or 25°C (temperature-sensitive strains) until ≥90% of cells were unbudded. Cells were washed in prewarmed media and incubated in fresh media with or without cadmium to release cells from the cell cycle block. Cells were prepared for flow cytometry and stained with SYTOX green (Molecular Probes, Eugene, OR) as described previously (Haase and Reed, 2002).
To determine the budding index, cells were collected and fixed with formaldehyde (3% final concentration). Two hundred cells for each time point were counted.
Protein Analyses
For immunoblot analysis, protein extracts were prepared in urea-buffer (8 M urea, 200 mM NaCl, 100 mM Tris, pH 7.5, 0.2% SDS, 10 mM Na-pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 0.1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 μg/ml each aprotenin, leupeptin, and pepstatin). Cells were broken with glass beads for 80 s at setting 4.5 in a FastPrep FP120 (Qbiogene, Carlsbad, CA), and cell debris was removed by centrifugation for 10 min at 13,000 rpm. Protein lysates were diluted to a final concentration of 4 M urea before separation by SDS-PAGE.
Separated proteins were transferred to a polyvinylidene difluoride membrane and the membrane was probed with the antibodies indicated. For immunopurifications, cells were broken with glass beads in Triton buffer (50 mM HEPES, pH 7.5, 0.2% Triton X-100, 200 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 10 mM Na-pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 0.1 mM orthovanadate, 1 mM PMSF, and 1 μg/ml each aprotenin, leupeptin, and pepstatin) for 3 × 20 s at setting 4.5 in a FastPrep FP120 with 1-min breaks. Cell debris was removed by centrifugation for 10 min at 13,000 rpm, and 2 mg of total protein lysates was used for immonopurification with anti-myc antibodies (SC-789-G; Santa Cruz Biotechnology, Santa Cruz, CA) and analyzed by immunoblotting.
Primary antibodies were used at the following dilutions: anti-Met4 (1: 20,000; gift from M. Tyers, Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada), anti-Rad53 (1:1000, SC-6749; Santa Cruz Biotechnology), and anti-Skp1(1:5000; gift from W. Harper, Harvard University, Cambridge, MA).
RNA Analyses and PCR
RNA was isolated with the RNeasy kit (QIAGEN, Germantown, MD) following the manufacturer's protocol with the following modification. Cell pellets (1–2 × 107 cells) were broken with glass beads in buffer RLT for four times 40 s at setting 4.5 in a FastPrep FP120 with 1-min breaks between the runs. Cell debris was removed by centrifugation for 2 min at 13,000 rpm. For RNA analysis by real-time reverse transcription-polymerase chain reaction (RT PCR), first strand cDNA synthesis was performed with SuperScriptII following the manufacturer's recommendations, with the exception that 1.5 μg of RNA was used in a 10-μl reaction with 0.3 μl of SuperScriptII. One-fiftieth of the cDNA preparation was used in real-time PCRs on an iCycler iQ (Bio-Rad, Hercules, CA) by using iQ SybrGreen Supermix (Bio-Rad). Primers for real-time PCR were designed to amplify 100- to 150-bp fragments by using Beacon Designer 2.1 software (Biosoft International, Palo Alto, CA). For each experiment, a standard curve was generated using fivefold dilutions of cDNA. The first dilution in the series was set arbitrarily to copy number 3000. Only when PCR products were falling within the range of the standard curve, the amount of cDNAs was calculated relative to the standard curve and normalized to the control (ACT1) samples. Samples were run in duplicates in a PCR program with an initial 3-min 95°C step, followed by 40 cycles of 10 s at 95°C and 45 s at 50°C. After each run a melting curve was run to ensure that no primer dimers or secondary products were formed. Primer sequences are available upon request from J.L.Y. (yenj@uci.edu)
RESULTS
Cdc34/SCFMet30 Is Required for the Cellular Response to Cadmium
The budding yeast transcription factor Met4 has been shown to play an important role in the cellular response to cadmium exposure (Fauchon et al., 2002). The major regulator of Met4 is the ubiquitin ligase SCFMet30, which together with the ubiquitin-conjugating enzyme Cdc34 controls Met4 activity in response to intracellular S-adenosylmethionine levels (Kaiser et al., 2000; Rouillon et al., 2000). To test whether Cdc34/SCFMet30 is involved in cadmium stress response, we tested growth of strains carrying temperature-sensitive mutations in components of SCFMet30 on cadmium-containing plates (Figure 1, A and B). Serial dilutions of the mutants were spotted onto plates and incubated at temperatures well below the restrictive temperatures of the mutants (Figure 1, A and B). met30 mutants carrying two different alleles were very sensitive to cadmium (Figure 1, A and B). In addition, cdc34 mutants and mutants in the SCF-core component Cdc53 were cadmium sensitive (Figure 1, A and B), suggesting that Met30 functions in the context of SCF in the cellular response to cadmium. Cdc4 is the closest homolog of Met30 and forms the ubiquitin ligase SCFCdc4, which has different substrate specificity than SCFMet30. cdc4 mutants were not cadmium sensitive (Figure 1, A and B). These results strongly suggest that SCFMet30 plays a role in the cellular response to cadmium exposure.
Figure 1.
SCFMet30 mutants are cadmium sensitive. (A) Serial dilutions of cells growing in YEPD medium in mid-log phase were spotted onto YEPD plates containing cadmium at the concentrations indicated. Plates were incubated at 28°C. (B) Same as in A, but cells were grown and plates were incubated at 25°C. Strains used were met30-6 (PY283), met30-9 (PY281), cdc34-3 (PY23), cdc53-1 (PY175), and cdc4-3 (PY187). (C) Wild-type cells and met30-6 (PY283) were grown in YEPD at the indicated temperatures and expression of GSH1 was measured by real-time RT-PCR. Expression levels were normalized to expression of the ACT1 gene. (D) Wild-type cells and met30-6 mutants were grown at 28°C in YEPD incubated with various concentrations of cadmium as indicated for 30 min. Dilutions of the cells were plated on YEPD plates and incubated at 25°C. Colonies were counted after 4 d. The number of colonies after no cadmium exposure was set to 100%. Error bars denote SE of the mean derived from four data sets.
The cadmium sensitivity of met30 mutants was unexpected, because microarray experiments showed increased GSH1 expression in met30 mutants (our unpublished data). GSH1 encodes γ-glutamylcysteine synthase the enzyme that catalyzes the first and rate-limiting step in glutathione biosynthesis (Grant et al., 1997). Increased Gsh1 expression should protect cells from cadmium stress because glutathione is important for cadmium detoxification (Perego and Howell, 1997). We first confirmed an increase in GSH1 expression in met30-6 mutants. Real-time RT-PCR showed significantly higher GSH1 levels in met30 mutants compared with wild-type cells (Figure 1C). Paradoxically, despite increased expression of GSH1, met30 mutants were hypersensitive to cadmium in the plate assay (Figure 1, A and B). However, the failure of met30 mutants to grow on plates containing cadmium can be explained by two scenarios. First, mutants in components of SCFMet30 cannot cope with the cadmium stress and die. Second, inactivation of SCFMet30 is a physiological response to cadmium exposure to achieve Met4 activation. Full activation of Met4 also induces a cell cycle arrest (Kaiser et al., 1998; Patton et al., 2000; Flick et al., 2004). The activity of the mutant Met30-6 protein is partially compromised; therefore, cadmium exposure would lead to further SCFMet30 inactivation. Because Met30 function is essential for cell cycle progression, continuous cadmium exposure could arrest met30-6 mutants as long as they are exposed to cadmium. To distinguish between these two scenarios, we analyzed survival of cells after a brief exposure to cadmium (Figure 1D). Cells were grown at permissive temperature and incubated with different concentrations of cadmium for 30 min, and various dilutions were plated to score the number of surviving cells. met30-6 mutants were more resistant than wild-type cells in this assay. This is consistent with the increased GSH1 expression we observed in met30 mutants (Figure 1C). This result supports the idea that continuous cadmium exposure induces a cell cycle arrest in met30 mutants and does not lead to cell death.
We next analyzed cell cycle progression in response to cadmium in wild-type cells and met30-6 mutants. Cells were synchronized in G1 with mating pheromone and released from the G1 arrest in medium containing cadmium (Figure 2A). Cadmium (500 μM) led to G1 arrest of wild-type cells over the entire time course of 3 h (our unpublished data). Lower cadmium concentrations of 200 μM induced a significant delay of cell cycle progression in wild-type cells, but cells eventually entered the cell cycle (Figure 2A). Cadmium seemed to affect both transition from G1 into S phase and progression through S phase (Figure 2A). In contrast to wild-type cells, cadmium exposed met30-6 mutants stayed arrested in G1 throughout the entire time course (Figure 2A), suggesting that met30 mutants are sensitized to the cadmium-induced cell cycle arrest.
Figure 2.
Cadmium induces a Met4 and Met32 dependent cell cycle arrest. (A) Wild-type cells and met30-6 mutants (PY283) were grown at 28°C in YEPD to early-log phase, arrested with α-factor until >90% of cells were unbudded and then released from the α-factor block into media with or without 200 μM cadmium. Cell cycle progression was followed by flow cytometry. (B and C) Serial dilutions of cells growing in YEPD medium in mid-log phase at 30°C were spotted onto YEPD plates containing cadmium at the concentrations indicated. Plates were incubated at 30°C. Strains used: met4Δ (PY1123), met4Δ met30Δ (PY589), met32Δ (PY889). met32Δ met30Δ (PY893), met28Δ (PY950), met31Δ (PY965), met32Δ (PY889), met31Δ met32Δ (PY968), and cbf1Δ (PY640).
The essential cell cycle function of SCFMet30 is inactivation of Met4. Deletion of MET4 or of its cofactor MET32 eliminate the cell cycle requirement for SCFMet30 (Kaiser et al., 2000; Patton et al., 2000; Flick et al., 2004). We therefore tested cadmium sensitivity of met30 met4 and met30 met32 double mutants (Figure 2B). Deletion of MET32 completely suppressed growth inhibition of met30 mutants that was induced by continuous exposure to cadmium (Figures 1, A and B, and 2B). Deletion of MET4 also suppressed the met30 growth defect on cadmium containing plates (compare 20 μM cadmium in Figures 1A and 2B). However, both met4 single and met30 met4 double mutants failed to grow on plates containing 50 μM cadmium (Figure 2B), confirming the importance of Met4 in the response to cadmium stress.
These results provide genetic support for the idea that yeast cells induce a cell cycle arrest in response to cadmium by affecting the SCFMet30-regulated cell cycle steps. In addition, cadmium sensitivity suggests an important role of Met4 activation in heavy metal detoxification.
Met4 has been proposed to form different transcription complexes. The two suggested complexes consist of Met4, Met28, Cbf1, and Met4, Met31/Met32, Met28, respectively (Blaiseau and Thomas, 1998). Met4 is the sole transactivating factor in these complexes. Sequence-specific binding is thought to be mediated by the DNA-interacting factors Cbf1 and Met31/Met32, respectively. Met31 and Met32 are largely redundant because deletion of both factors is necessary to block transcription of known target genes and to produce a methionine auxotroph phenotype (Blaiseau et al., 1997). However, the Met4-induced cell cycle arrest is specifically dependent on Met32 and not Met31 because only deletion of MET32 can suppress the cell cycle defect of met30 mutants (Patton et al., 2000). We tested the requirement of components of the Met4 transcription complexes in the response to cadmium stress. Serial dilutions of wild-type and mutant cells were spotted on cadmium-containing plates, and growth was monitored (Figure 2C). Consistent with a redundancy of Met31 and Met32, either mutant was resistant to the cadmium concentrations tested. However, met31 met32 double mutants were very sensitive to cadmium (Figure 2C). Surprisingly, met31 met32 double mutants were more sensitive than met4 mutants (Figure 2C). Similarly, cadmium sensitivity of cbf1 mutants was greater than that of met4 mutants, suggesting that transactivating factors other than Met4 might form alternative transcription complexes with Cbf1 and Met31/Met32. Unexpectedly, met28 mutants were not cadmium sensitive, although they are methionine auxotroph and are required for robust expression of many Met4 target genes.
The cadmium sensitivity of cbf1 mutants is in contrast to previous results that demonstrated a cadmium hyper-resistant phenotype of cbf1 mutants (Dormer et al., 2000). We also tested the cbf1Δ mutant from the yeast deletion collection (S288C background) and found that it was not cadmium hyper-resistant (our unpublished data). The differences between our results and previously reported results (Dormer et al., 2000) might reflect a background-specific response to cadmium.
These results demonstrate that several components of the Met4 transcription complexes are involved in cadmium detoxification and that in addition to Met4, alternative transactivators might play a role in conjunction with Cbf1 and Met31/Met32.
Cadmium Prevents Met4 Ubiquitination by Inhibition of the Met30/Skp1 Interaction
We next tested whether cadmium exposure leads to Met4 activation. During normal growth conditions, Met4 is maintained in its inactive ubiquitinated form, and Met4 activation correlates with the appearance of deubiquitinated and phosphorylated species (Kaiser et al., 2000; Flick et al., 2004). The different modified Met4 species can be readily distinguished by immunoblotting (Kaiser et al., 2000; Flick et al., 2004). Exposure of cells to cadmium lead to the appearance of deubiquitinated, phosphorylated Met4 in a dose-dependent manner (Figure 3A). Exposure to a relatively low concentration of cadmium lead to a gradual shift from the ubiquitinated form to deubiquitinated and phosphorylated Met4 in a time-dependent manner (Figure 3B), suggesting that Met4 activation responds to the accumulating cadmium damage. Because ubiquitinated Met4 is very stable (Kaiser et al., 2000; Flick et al., 2004) the complete disappearance of ubiquitinated forms of Met4 is likely a reflection of cadmium-induced Met4 deubiquitination.
Figure 3.
Cadmium exposure activates Met4. (A) Wild-type cells were grown in YEPD to OD600 = 0.5, incubated with cadmium at the concentrations indicated for 30 min, and cell lysates were analyzed by immunoblotting (gel 7.5%) with polyclonal antibodies directed against Met4 and the proteasome subunit Rpt1 as a loading control. (B) Experimental procedures were as in A, but cells were incubated in YEPD containing 100 μM cadmium for the period indicated. (C) Wild-type cells were grown as in A, incubated in medium containing 1 mM cadmium for 30 min, and expression of Met4-target genes MET3, MET25, and GSH1 was analyzed by real-time RT-PCR. Expression levels were normalized to ACT1 expression. (D) Wild-type cells were grown in YEPD or YEPD supplemented with methionine (1 mM final concentration) to OD600 = 0.5, incubated with cadmium at the concentrations indicated for 30 min, and cell lysates were analyzed as described in A. (E) Cells expressing 12myc-Met30 under control of its own promoter (PY1073) were grown in YEPD to an OD600 = 0.5 and either exposed or not exposed to 1 mM cadmium for 30 min. Cells expressing untagged Met30 (PY236) were processed in parallel. Cell lysates were immunopurified with anti-myc antibodies and analyzed by immunoblotting with anti-Met4 and anti-Skp1 antibodies.
The appearance of deubiquitinated and phosphorylated Met4 correlated with Met4 activation, which is indicated by the induction of expression of Met4 target genes (Figure 3C). Cadmium induced activation of Met4 was not an indirect consequence of depletion of the intracellular methionine pool because supplementing the growth media with high concentrations of methionine did not overcome the effect of cadmium on Met4 modification (Figure 3D).
To address the mechanism of how cadmium prevents Met4 ubiquitination, we analyzed the interaction of Met30 with Met4 in response to cadmium. Myc-epitope tagged Met30 expressed under control of its own promoter was immunopurified from untreated cells or cells treated with 1 mM cadmium for 30 min. As expected, Met4 ubiquitination was inhibited by cadmium; however, the Met30/Met4 interaction seemed to be unaffected (Figure 3E). This was surprising because SCF-mediated ubiquitination is generally regulated at the level of F-box/substrate association. We next asked whether intact SCFMet30 complexes are bound to Met4 under these conditions. To this end, we probed for Skp1 in Met30 containing immunocomplexes because Skp1 forms a molecular bridge between Met30 and the other SCF components. Skp1 was present in Met30 containing immunocomplexes purified from untreated cells, but this interaction was dramatically reduced upon cadmium exposure (Figure 3E). Thus, cadmium specifically disrupts the Met30/Skp1 interaction to inhibit Met4 ubiquitination.
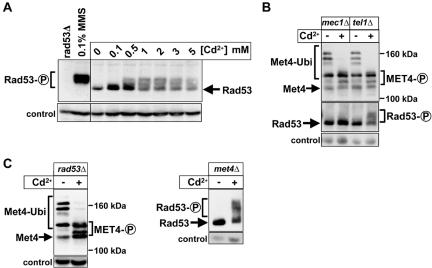
Cadmium Induces Rad53 Phosphorylation
Cadmium has been shown to induce DNA damage (Jin et al., 2003; Filipic and Hei, 2004). Therefore, we tested whether the DNA damage checkpoint is activated under the same conditions that we detected Met4 activation. Activation of the DNA-damage checkpoint leads to phosphorylation of Rad53, which can be monitored by immunoblotting (Sanchez et al., 1996). Similar to what we found for Met4 activation, cadmium induced Rad53 phosphorylation in a dose-dependent manner (Figure 4A). However, cadmium-induced Rad53 phosphorylation never reached the extent seen after treatment with the DNA-damaging agent methyl methanesulfonate (MMS) (Figure 4A). Consistent with cadmium-induced activation of the DNA-damage checkpoint, Rad53 phosphorylation was completely dependent on Mec1 but not Tel1 (Figure 4B). Given the strong correlation between activation of Met4 and activation of Rad53 in response to cadmium, we next asked whether these effects depend on each other. Appearance of deubiquitinated and phosphorylated Met4 was independent of Mec1, Rad53, and Tel1 (Figure 4, B and C). Conversely, Rad53 activation was independent of Met4 (Figure 4C), suggesting that cadmium activates both the Mec1/Rad53 pathway and Met4 independently of each other.
Figure 4.
Mec1-dependent activation of Rad53 in response to cadmium. (A) Experimental conditions were as in Figure 3A. Immunoblots were analyzed with polyclonal antibodies directed against Rad53. As a control, Rad53 from cells treated with 0.1% MMS for 30 min was analyzed. (B and C) Cells were grown as described in Figure 3A, incubated with 1 mM cadmium for 30 min, and analyzed by immunoblotting by using polyclonal antibodies to Met4 or Rad53. Strains used were mec1Δ (PY1164), tel1Δ (PY1161), rad53Δ (PY1117), and met4Δ (PY1123). mec1Δ and rad53Δ mutants were kept alive by a high-copy plasmid containing the RNR1 gene.
Cadmium Affects Cell Cycle Progression
Analysis of cell cycle progression during cadmium exposure demonstrated both G1- and S-phase delays (Figure 2A). The G1/S transition is primarily controlled by accumulation of G1-cyclins (Wittenberg and Flick, 2003). We therefore tested the effect of cadmium exposure on expression of the G1-cyclin Cln2. Cells were synchronized in G1 with mating pheromone and released from the pheromone block in the absence or presence of 200 μM cadmium (Figure 5A). Real-time RT-PCR analysis revealed strong repression of CLN2 transcription by cadmium (Figure 5A). The low level of CLN2 expression could possibly explain the cell cycle entry delay observed in cadmium-treated samples (Figure 2A). Both activation of Met4/Met32 and Mec1/Rad53 have been demonstrated to prevent accumulation of CLN2 transcripts (Sidorova and Breeden, 1997; Patton et al., 2000). Cadmium induced both the Met4/Met32 and the Mec1/Rad53 pathways (Figures 3 and 4) and could therefore be responsible for the cell cycle delay after cadmium treatment (Figure 2B). We therefore analyzed the cell cycle of met32Δ mec1Δ double mutants in response to cadmium exposure (Figure 5B). Mec1 is essential for viability. The essential function but not the cell cycle checkpoint function is suppressed by overexpression of the large subunit of ribonucleotide reductase (Rnr1) (Zhao et al., 1998). Therefore, all mec1Δ mutants used in this study carried a high-copy plasmid containing the RNR1 gene. Cells synchronized in G1 with mating pheromone were released from the pheromone block in the absence or presence of 200 μM cadmium, and cell cycle progression was monitored by flow cytometry. Wild-type cells showed a significant delay in the transition from G1 into S phase upon cadmium exposure (Figure 5, B and C). Consistent with the experiment shown in Figure 2B, after cadmium exposed cells had entered S phase, they progressed extremely slowly through S phase (Figure 5B). Surprisingly, cadmium affected the G1/S of met32Δ mec1Δ double mutant even stronger than that of wild-type cells (Figure 5B). This was confirmed when we monitored initiation of budding, which is an indicator of the G1/S transition (Figure 5C). Both met32Δ and mec1Δ mutants as well as met32Δ mec1Δ double mutants showed severely delayed budding when exposed to cadmium compared with wild-type cells, demonstrating that inactivation of Met32 and/or Mec1 enhanced the inhibitory effect of cadmium on G1/S transition (Figure 5C).
Figure 5.
Effects of cadmium exposure on cell cycle progression. (A) Wild-type cells were grown in YEPD at 30°C to early-log phase, arrested with α-factor until >90% of cells were unbudded, and then released from the α-factor block into media with or without 200 μM cadmium. Samples were taken at the time intervals indicated, and expression of CLN2 was analyzed by real-time RT-PCR. Expression levels were normalized to ACT1 expression. (B) Wild-type cells and mec1Δ met32Δ double mutants (PY1197) were synchronized and released as described in A, and cell cycle progression was monitored by flow cytometry. (C) Wild-type cells, met32Δ (PY889), mec1Δ (PY1164), and mec1Δ met32Δ double mutants (PY1197) were synchronized and released as in A, and the number of budded cells were counted at the time intervals indicated. (D) Wild-type cells and mec1Δ mutants were synchronized and released as in A, with the exception that 200 μM cadmium was added after cells had entered S phase (25 min after release). (E) The strains indicated were grown in YEPD medium to mid-log phase at 30°C, and serial dilutions were spotted onto YEPD plates containing no, 20 μM cadmium, or 70 μM cadmium. Plates were incubated at 30°C. Plates containing 70 μM cadmium were incubated longer because cell growth was significantly delayed. All strains carrying a MEC1 deletion were kept alive by a high-copy plasmid containing the RNR1 gene.
The slow progression through S phase we observed upon cadmium exposure (Figures 2A and 5B) could be caused by a checkpoint response or reflect a general effect of cadmium-induced cell damage on cell growth. To distinguish between these two possibilities, we synchronized wild-type cells and mec1Δ mutants in G1 with pheromone, released the cells, and added cadmium after cells had initiated S phase (Figure 5D, 25-min time point). Consistent with our previous results (Figures 2A and 5B), wild-type cells progressed through S phase very slowly (Figure 5D). Inactivation of the check-point kinase Mec1 eliminated the cadmium-induced S-phase delay (Figure 5D), indicating that cadmium activates a genuine checkpoint response.
We next tested the sensitivity of mec1Δ, met32Δ, and met32Δ mec1Δ double mutants to cadmium (Figure 5E). We had already shown that met32Δ mutants are insensitive to up to 50 μM cadmium due to the redundant function Met32 shares with Met31 (Figure 2C). However, met32Δ mutants were sensitive to higher concentrations of cadmium (70 μM; Figure 5E), indicating that despite the redundant Met31, Met32 probably plays some role in cadmium detoxification. This could explain the enhanced G1 cell cycle arrest of met32Δ and mec1Δ met32Δ double mutants. Due to less effective detoxification, theses mutants might accumulate more cadmium damage than wild-type cells. In contrast, mec1Δ mutants were not cadmium sensitive. The cadmium sensitivity of mec1Δ met32Δ double mutants was likely caused by the met32Δ mutation and did not show synergistic effects (Figure 5E).
The effects of mutations in MEC1 and MET32 on the cadmium-induced cell cycle delay were unexpected because both the Mec1/Rad53 and the Met4/Met32 pathways were clearly induced in response to cadmium exposure (Figures 3 and 4). Activation of either pathway can induce a cell cycle arrest, yet deletion of both pathways could not overcome the cadmium-induced cell cycle delay. This indicates that cadmium induces additional pathways that prevent cell cycle progression.
Effects of MMS, H2O2, and Heavy Metals on Met4 and Rad53
Our results showed a specific effect of cadmium exposure on Met4 activity (Figure 3). Furthermore, genetic experiments demonstrated that Met4 activation plays an important role in the cellular defense against cadmium (Figure 2C). It has been suggested that at least part of the cytotoxic effects of cadmium are caused by induction of oxidative stress (Stohs and Bagchi, 1995; Brennan and Schiestl, 1996). We therefore tested whether induction of oxidative stress by hydrogen peroxide (H2O2) exposure has an effect on Met4 activity. In parallel, we tested the effects of other heavy metals as well as the DNA-damaging agent MMS. Cells were treated with three different concentrations of the chemicals for 30 min before protein lysates were analyzed by immunoblotting. To compare the biological effects of the concentrations used for the individual chemicals, the highest concentration was chosen such that cell viability after 30 min of treatment was between 50 and 70% (our unpublished data). Lead could not be used at concentrations that gave comparable biological effects because the lead solubility limit in YEPD medium was reached before any significant effects on cell viability were observed. The two lower concentrations of the different chemicals used in the experiment were 30 and 10% of the highest concentration, respectively. Immunoblot analyses revealed diverse effects of the different chemicals on both Met4 ubiquitination and Rad53 phosphorylation. MMS treatment had no effect on Met4 ubiquitination but, as expected, induced Rad53 phosphorylation (Figure 6). Hydrogen peroxide exposure led to a striking disappearance of all nonubiquitinated forms of Met4, suggesting that induction of oxidative stress activates Met4 ubiquitination or inactivates Met4 deubiquitination (Figure 6). Consistent with a previous report (Leroy et al., 2001), Rad53 phosphorylation was induced only at low concentrations of H2O2 (Figure 6). The effects of arsenic exposure were very similar to that of cadmium exposure (Figure 6). Met4 ubiquitination was effectively blocked and phosphorylated Met4 species appeared in response to arsenic (Figure 6). Consistent with the arsenic induced changes in Met4 modification, expression of Met4 target genes was induced (our unpublished data). Similar to cadmium, arsenic also induced Rad53 phosphorylation (Figure 6), emphasizing the similarities of the two highly toxic metals in respect to the cellular response they induce. Exposure of cells to lead, nickel, and cobalt had only small or no effects on Met4 and Rad53 modifications (Figure 6). Cobalt seemed to induce the appearance of phosphorylated Met4 species at the highest concentration, whereas nickel seemed to reduce the amount of deubiquitinated Met4 (Figure 6). The significance of these changes is not known.
Figure 6.
Effects of various chemicals on Met4 and Rad53. (A) Wild-type cells were grown to an OD600 = 0.5, incubated with different concentrations of the chemicals indicated for 30 min, and cell lysates were analyzed by immunoblotting with antibodies directed against Met4 or Rad53 (gel 7.5%, control anti-Rpt1). The concentrations of the chemicals were as follows: MMS, 0.01, 0.03, and 0.1%; H2O2, 1, 3, and 10 mM; Cd, 0.1, 0.3, and 1 mM; As, 1, 3, and 10 mM; Co and Ni, 5, 20, and 50 mM; and Pb, 0.5 and 1.5 mM. The first lane in each panel is an untreated sample.
Together, both arsenic and cadmium block Met4 ubiquitination and induce Rad53 phosphorylation, whereas induction of oxidative stress by hydrogen peroxide promotes Met4 ubiquitination.
DISCUSSION
To maintain cellular and genetic integrity, all organisms have developed strategies to respond to heavy metal exposure. These strategies include detoxification, repair or removal of damaged molecules, and delay of cell division to prevent propagation of damaged cellular components. Here we show that regulation of the yeast ubiquitin ligase SCFMet30 plays a central role in the response to heavy metal stress.
Cells exposed to cadmium showed a dramatic change in Met4 modification. Inactive ubiquitinated forms of Met4 rapidly disappeared and active phosphorylated forms of Met4 appeared (Figures 3 and 6). Similar to Met4 activation in response to methionine starvation, the total Met4 protein level remained constant indicating that proteolysis plays no role in these processes (Kaiser et al., 2000; Flick et al., 2004). The cadmium-induced Met4 activation was not an indirect consequence of depletion of intracellular methionine levels because addition of high concentrations of methionine could not prevent Met4 activation in response to cadmium exposure (Figure 3D).
Surprisingly, Met30 remained associated with Met4 in cadmium-treated cells, yet Met4 ubiquitination was blocked (Figure 3E). However, Skp1 binding to Met30 was severely reduced in response to cadmium treatment (Figure 3E), indicating that, under these conditions, Met4 ubiquitination is prevented by disassembly of SCFMet30. This regulation of the Met30/Skp1 interaction seems to be unique for the cadmium response because Rouillon et al. (2000) demonstrated that methionine starvation, which also blocks Met4 ubiquitination, did not disrupt the Met30/Skp1 complex. While this manuscript was in press, similar results on cadmium-mediated regulation of SCFMet30 were reported (Barbey et al., 2005). Moreover, using an in vitro Met4 ubiquitination system, the authors excluded a direct effect of cadmium on SCFMet30 activity, suggesting that cadmium induces modification of Met30 or association with a putative inhibitor (Barbey et al., 2005). Interestingly, studies with the mammalian Cul3-containing ubiquitin ligase SCF3Keap1 suggested that its substrate recognition subunit Keap1 dissociates from Cul3 in response to oxidative stress (Zhang et al., 2004). This in turn prevents ubiquitination of the transcription factor Nrf2, which coordinates the transcriptional response to oxidative stress (Jaiswal, 2004). Why cells have developed mechanisms to disrupt SCF-ligases in response to some signals rather than to modulate substrate recognition is not clear. One can speculate that by inactivation of the ubiquitin ligase complex, ubiquitination of a group of substrates can be prevented simultaneously, which might be desired under certain conditions.
Cadmium-mediated disruption of the Met30/Skp1 complex resulted in activation of Met4, which, consistent with proteomics studies and microarray experiments (Vido et al., 2001; Fauchon et al., 2002), led to induction of GSH1 expression and genes involved in the sulfur assimilation pathway (Figure 3C). Gsh1 encodes for γ-glutamylcysteine synthase the enzyme that catalyzes the first and rate-limiting step in glutathione biosynthesis and is therefore vital for heavy metal detoxification. GSH1 expression has previously been shown to depend on Met4 (Dormer et al., 2000; Wheeler et al., 2002, 2003). Induction of genes involved in sulfur amino acid biosynthesis is also an important aspect of the cellular response to cadmium exposure. The increased requirement for the cysteine containing glutathione requires induction of sulfur amino acid synthesis. In addition, the cytotoxicity of cadmium is thought to be at least in part due to its high affinity for sulfhydryl groups containing proteins (Stohs and Bagchi, 1995; McMurray and Tainer, 2003). Replacement of these cadmium-damaged proteins requires additional synthesis of sulfur amino acids.
Cadmium-induced Met4 activation has recently been implicated in a process called isozyme switching (Fauchon et al., 2002; Jamieson, 2002). It was noticed that cadmium exposure induced expression of isozymes of several carbohydrate metabolism genes and that all these cadmium-induced isozymes showed a markedly reduced content of the sulfur-containing amino acids methionine and cysteine compared with their normally expressed counterparts (Fauchon et al., 2002). Isozyme switching is at least in part achieved by activation of Met4 because expression of several sulfur-poor isozymes depends on Met4 (Fauchon et al., 2002). Although the physiological importance of this process is not clear, one can speculate that isozyme switching plays an important role in the cellular response to cadmium exposure because it protects the carbohydrate metabolism by expression of less cadmium-sensitive sulfur-poor isozymes (Fauchon et al., 2002; Jamieson, 2002).
Regulation of SCFMet30-dependent Met4 ubiquitination by cadmium is therefore important for the defense against cadmium, the replacement of damaged proteins, and isozyme switching. The importance of Met4 regulation in response to cadmium is underlined by the cadmium hypersensitivity of met4 mutants and mutants in components of the Met4 transcription complexes (Figure 2C). Surprisingly, met28 mutants were not sensitive to cadmium, even though Met28 is important for expression of several genes involved in sulfur amino acid biosynthesis in response to methionine starvation (Kuras et al., 1996). It is possible that a protein with redundant function is induced in response to cadmium exposure that can replace Met28. Remarkably, both cbf1 mutants and met31 met32 double mutants were significantly more sensitive to cadmium than met4 mutants. Cbf1 and the redundant Met31 and Met32 proteins are DNA binding factors lacking transactivating activity (Thomas and Surdin-Kerjan, 1997). In contrast, devoid of DNA binding activity the transactivating factor Met4 relies on Cbf1, Met31, or Met32 for DNA binding (Blaiseau and Thomas, 1998). The greater cadmium sensitivity caused by mutations in the DNA binding factors compared with the transactivating factor Met4 (Figure 2C) suggests that alternative transcription factors can bind Cbf1, Met31, and Met32 and partially compensate for loss of Met4. Even though the genetic results imply the existence of additional components that are involved in coordination of the transcriptional response to cadmium exposure, our results demonstrate the importance of Met4, Cbf1, Met31, and Met32 in this process.
Cells have developed complex pathways that delay or halt cell cycle progression under stress situations to allow repair of damaged cellular components before cell division occurs (Hartwell and Weinert, 1989). These pathways are referred to as cell cycle checkpoints and are important to maintain cellular and genetic integrity. Cell cycle checkpoints in response to DNA damage (Sancar et al., 2004), DNA replication block (Boddy and Russell, 2001), problems in spindle assembly (Amon, 1999), and morphogenesis (Lew and Reed, 1995) have been described in all eukaryotes. To the best of our knowledge, the effects of heavy metal exposure on cell cycle regulation have so far not been analyzed. We monitored cell cycle progression of G1-synchronized cells in response to cadmium exposure. Higher concentrations of cadmium (500 μM) completely blocked entry into S phase (our unpublished data), whereas lower concentrations (200 μM) delayed S-phase entry and budding (Figure 5, B and C). Once cadmium-exposed cells initiated S phase, DNA replication was very slow (Figures 2A and 5B). The transition from G1 into S phase is initiated by a burst of expression of G1 cyclins (Wittenberg and Flick, 2003). Expression of the G1-cyclin Cln2 was strongly repressed by cadmium exposure (Figure 5A), suggesting that insufficient G1-cyclin activity contributes to the cadmium-induced delay in S-phase entry. It has been shown that inhibition of Met4 ubiquitination induces a cell cycle arrest in G1 and to repress expression of the G1-cyclin Cln2 (Patton et al., 2000). Cadmium exposure led to a complete block of Met4 ubiquitination (Figures 3 and 6). This suggests an attractive model where Met4 activation in response to cadmium on one hand regulates the defense against cadmium toxicity by induction of glutathione and sulfur amino acid synthesis, and on the other hand induces a cell cycle checkpoint in G1. Accordingly, deletion of either MET4 or MET32, a transducer of the Met4-induced cell cycle arrest, should prevent induction of the G1 arrest. However, both met4Δ and met32Δ mutants arrested in G1 in response to cadmium exposure (our unpublished data; Figure 5, B and C). Interpretation of these results is complicated by the fact that Met4 also is required for detoxification of cadmium and consequently met4Δ mutants might accumulate more cadmium damage. Met32 is less important for cadmium detoxification than Met4 probably because of the redundant Met31 (Figure 2C). However, the Met4-induced G1 cell cycle arrest depends on Met32 and not Met31 (Patton et al., 2000). Nonetheless, met32Δ mutants arrested in G1 in response to cadmium exposure, and surprisingly the arrest was even more pronounced than that of wild-type cells (Figure 5C; our unpublished data). This suggests that in addition to the Met4/Met32 pathway cadmium induces other pathways that block the G1/S transition. One such pathway is activation of the Mec1/Rad53 cell cycle checkpoint pathway. Our results demonstrated that cadmium induced Mec1-dependent phosphorylation of the checkpoint kinase Rad53. Among other effects, the Mec1/Rad53 pathway has been shown to down-regulate Cln2 expression in response to MMS exposure and consequently to delay S-phase initiation (Sidorova and Breeden, 1997). Furthermore, it has been demonstrated that Mec1-dependent activation of Rad53 inhibits firing of late replicating origins and thus to slow S-phase progression (Santocanale and Diffley, 1998; Shirahige et al., 1998). It is therefore feasible that activation of the Mec1/Rad53 pathway contributes to the delay in the G1/S transition and slow S-phase progression that we observed in response to cadmium exposure (Figures 2B and 5, B and C). The slow progression through S phase was dependent on Mec1 (Figure 5D) and is probably a manifestation of inhibition of late replication origins. Whether the Mec1/Rad53 checkpoint pathway contributes to the delay in G1/S transition could not be experimentally determined because similar to met32Δ mutants, mec1Δ mutants showed an enhanced G1 cell cycle arrest in response to cadmium exposure compared with wild-type cells (Figure 5C). Remarkably, cadmium induced a robust G1 arrest even in mec1Δ met32Δ double mutants, suggesting that cadmium activates a so far unknown pathway that blocks S-phase entry (Figure 7). It is possible that the G1/S transition defect we observed is not a consequence of activation of a cell cycle checkpoint but caused by direct cadmium-mediated damage of proteins important for cell cycle progression. However, suppression of the S-phase defect by deletion of Mec1 indicates a genuine checkpoint effect of cadmium at least in S phase (Figure 5D). This argues against major unspecific, cadmium-mediated protein damage as a cause for the cell cycle response.
Figure 7.
Model of the cellular response to cadmium stress. Cadmium blocks Met4 ubiquitination, which leads to Met4 activation. Active Met4 induces glutathione synthesis, biosynthesis of sulfur amino acids, and a cell cycle arrest. Cadmium also activates the cell cycle checkpoint kinase Rad53 as well as a so far unidentified cell cycle arrest pathway.
Together our results demonstrate a complex regulation of cell cycle progression by cadmium. Activation of both the Met4/Met32 and the Mec1/Rad53 pathways in response to cadmium as well as arsenic exposure was strongly suggested, for example, by changes in Met4 and Rad53 modifications that are consistent with their activation. Genetic analyses revealed a third, so far unknown, cadmium-induced pathway that blocks the transition from G1 to S phase (Figure 7).
Cadmium and arsenic are two of the most toxic environmental contaminants. Surprisingly little is known about how cells sense and respond to cadmium and arsenic exposure. Our results suggest the ubiquitin ligase SCFMet30 as a central regulator of the cellular heavy metal response.
Acknowledgments
We are grateful to Duncan Clarke for materials used in this study and Miguel Rodriguez for suggestions on experiments with arsenic. We thank Wade Harper for anti-Skp1 antibodies and Ikram Ouni for yeast strains and valuable advice on immunoprecipitations. We express thanks to Karin Flick for helpful suggestions and comments on the manuscript and members of the Kaiser and the Nomura laboratories for helpful discussions. This work was supported by National Institutes of Health grant GM-66164 to P. K.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1130) on February 2, 2005.
References
- Amon, A. (1999). The spindle checkpoint. Curr. Opin. Genet. Dev. 9, 69-75. [DOI] [PubMed] [Google Scholar]
- Barbey, R., Baudouin-Cornu, P., Lee, T. A., Rouillon, A., Zarzov, P., Tyers, M., and Thomas, D. (2005). Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. Embo J. (in press). [DOI] [PMC free article] [PubMed]
- Blaiseau, P. L., Isnard, A. D., Surdin-Kerjan, Y., and Thomas, D. (1997). Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell. Biol. 17, 3640-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau, P. L., and Thomas, D. (1998). Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 17, 6327-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., and Russell, P. (2001). DNA replication checkpoint. Curr. Biol. 11, R953-956. [DOI] [PubMed] [Google Scholar]
- Brennan, R. J., and Schiestl, R. H. (1996). Cadmium is an inducer of oxidative stress in yeast. Mutat. Res. 356, 171-178. [DOI] [PubMed] [Google Scholar]
- Dormer, U. H., Westwater, J., McLaren, N. F., Kent, N. A., Mellor, J., and Jamieson, D. J. (2000). Cadmium-inducible expression of the yeast GSH1 gene requires a functional sulfur-amino acid regulatory network. J. Biol. Chem. 275, 32611-32616. [DOI] [PubMed] [Google Scholar]
- Fauchon, M., Lagniel, G., Aude, J. C., Lombardia, L., Soularue, P., Petat, C., Marguerie, G., Sentenac, A., Werner, M., and Labarre, J. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell. 9, 713-723. [DOI] [PubMed] [Google Scholar]
- Filipic, M., and Hei, T. K. (2004). Mutagenicity of cadmium in mammalian cells: implication of oxidative DNA damage. Mutat. Res. 546, 81-91. [DOI] [PubMed] [Google Scholar]
- Flick, K., Ouni, I., Wohlschlegel, J. A., Capati, C., McDonald, W. H., Yates, J. R., and Kaiser, P. (2004). Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell. Biol. 6, 634-641. [DOI] [PubMed] [Google Scholar]
- Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., Boucherie, H., Toledano, M. B., and Labarre, J. (1998). The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273, 22480-22489. [DOI] [PubMed] [Google Scholar]
- Grant, C. M., MacIver, F. H., and Dawes, I. W. (1997). Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol. Biol. Cell 8, 1699-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. R. (1991). Guide to Yeast Genetics and Molecular Biology, San Diego: Academic Press.
- Haase, S. B., and Reed, S. I. (2002). Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1, 132-136. [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J. M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., and Weinert, T. A. (1989). Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629-634. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2004). Ubiquitin signalling: what's in a chain? Nat. Cell. Biol. 6, 571-572. [DOI] [PubMed] [Google Scholar]
- Jaiswal, A. K. (2004). Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 36, 1199-1207. [DOI] [PubMed] [Google Scholar]
- Jamieson, D. (2002). Saving sulfur. Nat. Genet. 31, 228-230. [DOI] [PubMed] [Google Scholar]
- Jamieson, D. J. (1998). Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511-1527. [DOI] [PubMed] [Google Scholar]
- Jin, Y. H., Clark, A. B., Slebos, R. J., Al-Refai, H., Taylor, J. A., Kunkel, T. A., Resnick, M. A., and Gordenin, D. A. (2003). Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34, 326-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, P., Flick, K., Wittenberg, C., and Reed, S. I. (2000). Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102, 303-314. [DOI] [PubMed] [Google Scholar]
- Kaiser, P., Sia, R. A., Bardes, E. G., Lew, D. J., and Reed, S. I. (1998). Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 12, 2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., Barbey, R., and Thomas, D. (1997). Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 16, 2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., Cherest, H., Surdin-Kerjan, Y., and Thomas, D. (1996). A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15, 2519-2529. [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., Rouillon, A., Lee, T., Barbey, R., Tyers, M., and Thomas, D. (2002). Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell. 10, 69-80. [DOI] [PubMed] [Google Scholar]
- Leroy, C., Mann, C., and Marsolier, M. C. (2001). Silent repair accounts for cell cycle specificity in the signaling of oxidative DNA lesions. EMBO J. 20, 2896-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D. J., and Reed, S. I. (1995). A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129, 739-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, C. T., and Tainer, J. A. (2003). Cancer, cadmium and genome integrity. Nat. Genet. 34, 239-241. [DOI] [PubMed] [Google Scholar]
- Patton, E. E., Peyraud, C., Rouillon, A., Surdin, K. Y., Tyers, M., and Thomas, D. (2000). SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 19, 1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego, P., and Howell, S. B. (1997). Molecular mechanisms controlling sensitivity to toxic metal ions in yeast. Toxicol. Appl. Pharmacol. 147, 312-318. [DOI] [PubMed] [Google Scholar]
- Reed, S. I., Hadwiger, J. A., and Lorincz, A. T. (1985). Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82, 4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon, A., Barbey, R., Patton, E. E., Tyers, M., and Thomas, D. (2000). Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 19, 282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar, A., Lindsey-Boltz, L. A., Unsal-Kaccmaz, K., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39-85. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B. A., Jones, W. J., Liu, Q., Wang, B., and Elledge, S. J. (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357-360. [DOI] [PubMed] [Google Scholar]
- Santocanale, C., and Diffley, J. F. (1998). A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615-618. [DOI] [PubMed] [Google Scholar]
- Shirahige, K., Hori, Y., Shiraishi, K., Yamashita, M., Takahashi, K., Obuse, C., Tsurimoto, T., and Yoshikawa, H. (1998). Regulation of DNA-replication origins during cell-cycle progression. Nature 395, 618-621. [DOI] [PubMed] [Google Scholar]
- Sidorova, J. M., and Breeden, L. L. (1997). Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes. Dev. 11, 3032-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohs, S. J., and Bagchi, D. (1995). Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18, 321-336. [DOI] [PubMed] [Google Scholar]
- Thomas, D., and Surdin-Kerjan, Y. (1997). Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61, 503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vido, K., Spector, D., Lagniel, G., Lopez, S., Toledano, M. B., and Labarre, J. (2001). A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276, 8469-8474. [DOI] [PubMed] [Google Scholar]
- Wheeler, G. L., Quinn, K. A., Perrone, G., Dawes, I. W., and Grant, C. M. (2002). Glutathione regulates the expression of gamma-glutamylcysteine synthetase via the Met4 transcription factor. Mol. Microbiol. 46, 545-556. [DOI] [PubMed] [Google Scholar]
- Wheeler, G. L., Trotter, E. W., Dawes, I. W., and Grant, C. M. (2003). Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J. Biol. Chem. 278, 49920-49928. [DOI] [PubMed] [Google Scholar]
- Wittenberg, C., and Flick, K. (2003). Cell cycle regulation during G1 phase in yeast: decisions, decisions, decisions. In: G1 Phase Progression, ed. J. Boonstra, Georgetown, TX: Landes Biosciences, 14-39.
- Zalups, R. K., and Ahmad, S. (2003). Molecular handling of cadmium in transporting epithelia. Toxicol. Appl. Pharmacol. 186, 163-188. [DOI] [PubMed] [Google Scholar]
- Zhang, D. D., Lo, S. C., Cross, J. V., Templeton, D. J., and Hannink, M. (2004). Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Muller, E. G., and Rothstein, R. (1998). A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 2, 329-340. [DOI] [PubMed] [Google Scholar]