Abstract
Phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] is a key second messenger that regulates actin and membrane dynamics, as well as other cellular processes. Many of the effects of PtdIns(4,5)P2 are mediated by binding to effector proteins that contain a pleckstrin homology (PH) domain. Here, we identify two novel effectors of PtdIns(4,5)P2 in the budding yeast Saccharomyces cerevisiae: the PH domain containing protein Slm1 and its homolog Slm2. Slm1 and Slm2 serve redundant roles essential for cell growth and actin cytoskeleton polarization. Slm1 and Slm2 bind PtdIns(4,5)P2 through their PH domains. In addition, Slm1 and Slm2 physically interact with Avo2 and Bit61, two components of the TORC2 signaling complex, which mediates Tor2 signaling to the actin cytoskeleton. Together, these interactions coordinately regulate Slm1 targeting to the plasma membrane. Our results thus identify two novel effectors of PtdIns(4,5)P2 regulating cell growth and actin organization and suggest that Slm1 and Slm2 integrate inputs from the PtdIns(4,5)P2 and TORC2 to modulate polarized actin assembly and growth.
INTRODUCTION
Proteins involved in the regulation of cell signaling and membrane trafficking are often targeted to specific cell membranes in response to the synthesis of phosphoinositide second messengers. Seven different phosphorylated derivatives of phosphatidylinositol (PI) are currently known to exist in mammalian cells (Odorizzi et al., 2000). These are phosphorylated at a single or multiple sites on the inositol head group of PI by an array of different phosphoinositide kinases (Fruman et al., 1998). Besides serving as intermediates for the synthesis of other lipid second messengers, most if not all of the seven phosphoinositide species can act directly as second messengers and control the activation and membrane recruitment of effector proteins through binding to specific domains such as the pleckstrin homology (PH), FYVE, Phox, and epsin N-terminal homology domains (Hurley and Meyer, 2001; Lemmon, 2003; Cozier et al., 2004).
From the various phosphoinositide second messengers, phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] occupies a central position in lipid signaling pathways. PtdIns(4,5)P2 is generated in all eukaryotic cells and regulates a diverse spectrum of cellular processes, including cytoskeletal reorganization, membrane trafficking, cell growth, apoptotic regulation, and ion channel activation (Martin, 2001; Takenawa and Itoh, 2001; Yin and Janmey, 2003; Itoh and Takenawa, 2004). Previously, PtdIns(4,5)P2 was considered to be only an intermediate in the generation of diacylglycerol, inositol-(1,4,5)-trisphosphate, and phosphatidylinositol-3,4,5-trisphosphate (Toker and Cantley, 1997; Toker, 1998). However, recent data show that PtdIns(4,5)P2 is an important regulatory molecule in its own right that controls the activation and membrane recruitment of diverse PtdIns(4,5)P2 binding proteins (Hurley and Meyer, 2001; Yin and Janmey, 2003). Many of these contain PtdIns(4,5)P2-binding modules, such as the PH domain, an ∼120-amino acid protein domain found in proteins involved in signal transduction, actin organization, and membrane trafficking (Lemmon, 2003; Cozier et al., 2004).
PtdIns(4,5)P2 is a relatively abundant molecule that is enriched in the plasma membrane of all cells tested, ranging from organisms as diverse as yeast and human. Although the number of identified proteins that can bind PtdIns(4,5)P2 in vitro has grown dramatically over the past years, it is still poorly understood how different pathways use this generic signaling molecule to specifically regulate many different cellular processes.
The phosphoinositide kinases and the pathway that generates PtdIns(4,5)P2 are evolutionarily conserved. Therefore, genetically tractable systems such as the budding yeast Saccharomyces cerevisiae offer powerful tools to investigate PtdIns(4,5)P2 signaling mechanisms on a genome-wide scale. In yeast, as in mammalian cells, PtdIns(4,5)P2 is synthesized via the sequential phosphorylation of PI to phosphatidylinositol 4-phosphate [PtdIns(4)P] and PtdIns(4,5)P2 through the action of PI 4-kinases and PtdIns(4)P 5-kinases, respectively (Fruman et al., 1998; Odorizzi et al., 2000). S. cerevisiae has a single and essential PtdIns(4)P 5-kinase, termed Mss4. Mss4p is localized to the plasma membrane and implicated in the regulation of cell cycle-dependent actin reorganization (Desrivieres et al., 1998; Homma et al., 1998).
PtdIns(4,5)P2 generated by Mss4 is thought to mediate many of its cellular functions by inducing the plasma membrane recruitment and activation of proteins containing PH domains. For example, PtdIns(4,5)P2 mediates the plasma membrane translocation of Rom2 by binding to its PH domain (Audhya and Emr, 2002). Rom2 is a GTP exchange factor that activates the partially redundant Rho1 and Rho2 GTPases, which in turn, regulate actin polarization, polarized secretion, endocytosis, and cell wall synthesis by signaling to multiple downstream effectors (Cabib et al., 1998; deHart et al., 2003; Dong et al., 2003; Valdivia and Schekman, 2003). Known Rho1 effectors include the protein kinase C (PKC) (Pkc1) (Kamada et al., 1996; Delley and Hall, 1999), the formin protein Bni1 (Kohno et al., 1996), the β-1,3 glucan synthase Fks1 (Qadota et al., 1996), the transcription factor Skn7 (Alberts et al., 1998; Ketela et al., 1999), and the exocyst complex component Sec3 (Guo et al., 2001). Similarly, PtdIns(4,5)P2 binds to the PH domains of the related Boi1 and Boi2 and thereby mediates their localization to the bud, which is important for their role in the regulation of polarized growth (Bender et al., 1996; Hallett et al., 2002). In addition, PtdIns(4,5)P2 modulates nuclear migration during mitosis via interaction with the Num1 PH domain (Farkasovsky and Kuntzel, 1995) and controls the membrane recruitment and activation of phospholipase D 1 (PLD1/Spo14), whose function is important for secretion and cellular differentiation during meiosis (Sciorra et al., 2002). Thus, diverse PH domain-containing proteins are critical effectors of PtdIns(4,5)P2 that mediate key aspects of PtdIns(4,5)P2 signaling.
To further our understanding of PtdIns(4,5)P2 signaling, we wished to identify novel in vivo targets of PtdIns(4,5)P2 and to elucidate the mechanism by which PtdIns(4,5)P2 regulates their cellular functions. We took a candidate gene approach and analyzed the phosphoinositide binding properties of yeast proteins containing a PH domain. Here, we report on the characterization of two novel effectors of PtdIns(4,5)P2, encoded by homologous genes termed Synthetic lethal with MSS4 (SLM)1 and SLM2. Slm1 and Slm2 function in a PtdIns(4,5)P2-regulated signaling branch that is required for cell growth and actin polarization. Slm1 localization to the plasma membrane is essential for its in vivo function and is modulated by interaction with PtdIns(4,5)P2 and components of the TORC2 signaling complex. We propose that Slm1 and Slm2 function in a signaling network that integrate inputs from both the Mss4 and Tor2 pathways to elicit downstream responses required for actin polarization and cell growth.
MATERIALS AND METHODS
Materials, Strains, and Plasmids
Aprotinin, chymostatin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride (PMSF) were obtained from Sigma-Aldrich (St. Louis, MO). Geneticin (G418) and Pfx polymerase were from Invitrogen (Carlsbad, CA). Glutathione (GSH)-conjugated or Ni2+-conjugated agarose beads were from BD Biosciences Clontech (Palo Alto, CA). Mouse monoclonal anti-glutathione S-transferase (GST) antibodies were from Sigma-Aldrich, anti-hemagglutinin (HA) antibodies were from Babco (Richmond, CA), Cy2-conjugated goat anti-mouse IgG were from Jackson ImmunoResearch Laboratories (West Grove, PA), and Alexa-594–conjugated phalloidin was from Molecular Probes (Eugene, OR). Molecular mass standards were from Bio-Rad (Hercules, CA).
S. cerevisiae strains and plasmids used are listed in Table 1 and Table 2, respectively. To construct temperature-sensitive strains, plasmids containing the temperature-sensitive pik1-83 and mss4-2 alleles were isolated from strains AAY104 and SD102 (Desrivieres et al., 1998; Foti et al., 2001), respectively, and transferred into heterozygous pik1Δ or mss4Δ deletion strains generated by transformation of the diploid W303 a/α strain with polymerase chain reaction (PCR) products containing pik1::KanMX (Open Biosystems, Huntsville, AL) and mss4::HIS3MX6 (from strain JK497) disruption alleles. Haploid segregants harboring the appropriate markers were obtained by sporulation and dissection of the diploid strains. For generation of double-mutant strains, single-mutant strains of opposite mating types were first mated and sporulated. Tetrads were then dissected, and segregants containing the appropriate markers were selected. To construct the haploid yeast strains harboring the slm1Δ slm2Δ and carrying plasmid-borne SLM1 or SLM2 genes, strains JK508 and JK509, respectively, were induced to sporulate, and tetrads were dissected onto YPG medium. Segregants were then selected for the presence of the appropriate markers. Yeast strains in the BY4741 strain background containing C-terminally green fluorescent protein (GFP)-tagged Slm1 or tandem affinity procedure (TAP)-tagged Avo2, Bit61 were purchased from Research Genetics (Invitrogen).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303a | MATa ade2-1 trp1-1 can1-100 leu2-3112 his3-11,15 ura3-1 GAL+ | Laboratory collection |
| W303α | MATa ade2-1 trp1-1 can1-100 leu2-3112 his3-11,15 ura3-1 GAL+ | Laboratory collection |
| W303a/α | MATa/MATα ade2-1/ade2-1 trp1-1/1trp1-1 can1-100/can1-100 leu2-3112/leu2-3112 his3-11,15/his3-11,15 ura3-1/ura3-1 GAL+/GAL+ | Laboratory collection |
| JK9-3da | MATa leu2-3112 ura3-52 rme1 trp1 his4 GAL HMLa | Laboratory collection |
| BY4741 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Laboratory collection |
| JK497 | Mata/MATα ade2-1 trp1 leu2-3112 his3-11,15 ura3 GAL+ mss4::HIS3MX6/MSS4 | This study |
| JK498 | W303α pik1::KanMX carrying pRS314pik1-83 | This study |
| JK499 | W303α mss4::HIS3MX6 carrying YCplac111::mss4-2ts | This study |
| JK502 | W303a except slm1::KanMX | This study |
| JK503 | W303α except slm1::KanMX | This study |
| JK504 | W303a except slm2::KanMX | This study |
| JK505 | W303α except slm2::KanMX | This study |
| JK506 | W303a except slm2::HIS3 | This study |
| JK507 | W303a/α except slm1::KanMX/SLM1 slm2::HIS3/SLM2 | This study |
| JK508 | W303a except slm1::KanMX/SLM1 slm2::HIS3/SLM2 carrying pJK702 | This study |
| JK509 | W303α except slm1::KanMX/SLM1 slm2::HIS3/SLM2 carrying pJK703 | This study |
| JK510 | W303a except SLM2-GFP::HIS3MX6 | This study |
| JK511 | MATa ade2-1 trp1 leu2-3112 his3-11,15 ura3 GAL+ mss4::HIS3MX6 ura3-52::mss4-2ts-URA3 | This study |
| JK512 | W303a except slm1::KanMX slm2::HIS3 carrying pJK702 | |
| JK513 | W303α except slm1::KanMX slm2::HIS3 carrying pJK703 | |
| JK514 | JK9-3da except fab1-2; obtained by backcrossing strain (Yamamoto et al., 1995) twice with JK9-3d | This study |
| JK515 | W303a except slm1::KanMX slm2::HIS3 carrying pSW4 | This study |
| JK516 | W303a except slm1::KanMX slm2::HIS3 carrying pSW5 | This study |
| JK517 | W303a except bit61::HIS3MX avo2::KanMX | This study |
| JK518 | JK515 except inp51::LEU2 | This study |
| YJC1426 | MATa ade2 ade3 ura3 leu2 trp1 lys2 stt4-7-LEU2 | Muhua et al. (1998) |
| 95700 | BY4741 except SLM1-GFP-URA3 | Research Genetics |
| 7502123 | BY4741 except AVO2-TAP-HIS3 | Open Biosystems |
| 7501161 | BY4741 except BIT61-TAP-HIS3 | Open Biosystems |
Table 2.
Plasmids used in this study
| Plasmid | Characteristic | Source |
|---|---|---|
| pAS24 | YCplac111, CEN, LEU2, allows expression of proteins N-terminally tagged with a double HA-tag under the control of the GAL1 promoter | Schmidt et al. (1997) |
| pAS25 | YCplac33, CEN, URA3, allows expression of proteins N-terminally tagged with a 2 × HA-tag under the control of the GAL1 promoter | A. Schmidt |
| pJK701 | HA-SLM1 in pAS24 | This study |
| pJK702 | HA-SLM1 in pAS25 | This study |
| pSW1 | HA-SLM2 in pAS24 | This study |
| pJK703 | HA-SLM2 in pAS25 | This study |
| pJK704 | HA-SLM1ΔC in pAS25 | This study |
| pJK705 | HA-SLM2ΔC in pAS25 | This study |
| pSW2 | HA-SLM1ΔC in pAS24 | This study |
| PSW4 | slm1–3 in p416GPD, CEN, URA3 | This study |
| PSW5 | slm1–3 in p415GPD, CEN, LEU2 | This study |
| pAD41 | HA-SLM1M1 in pAS25 | This study |
| pAD42 | HA-SLM1M2 in pAS25 | This study |
| pAD43 | HA-SLM2M1 in pAS25 | This study |
| PJK706 | SLM1-PH in pGEX5 × –1 | This study |
| pAD44 | GST-SLM1-PHM1 in pGEX5 × –1 | This study |
| pAD45 | GST-SLM1-PHM2 in pGEX5 × –1 | This study |
| pJK708 | 6HIS-SLM1 in pET28a (Novagen) | This study |
| pJK709 | 6HIS-SLM2 in pET28a (Novagen) | This study |
| pJK710 | AVO2 in TNT521 (Udan et al., 2003) | This study |
| pJK711 | BIT61 in TNT521 | This study |
| pJK712 | YBR270C in TNT521 | This study |
| pC-186 | Genomic fragment containing the RHO1 gene, 2μ URA3 | Madaule et al. (1987) |
| pRHO2 | Genomic fragment containing the RHO2 gene in Yeplac195, 2μ ARS, URA3 | M. Hall |
| pPKC1 | Genomic fragment containing PKC1 in Yeplac195, 2μ ARS, URA3 | M. Hall |
| pPKC1(R398P) | Ycp50, URA3, CEN containing the dominant active allele PKC1(R398P) | Levin et al. (1990) |
| pBCK1 | Genomic fragment containing BCK1 in pRS314, TRP1, CEN | Lee and Levin (1992) |
| pBCK1-20 | Genomic fragment containing the dominant active allele BCK1–20 in pRS314, TRP1, CEN | Lee and Levin (1992) |
| pDLB824 | CEN HA-MKK1DD; constitutively activated MKK1-DD mutant, altering residues Ser-377 and Thr-381 to Asp | Harrison et al. (2004) |
| pMPK1 | Genomic fragment containing HA-MPK1 in Ycp50 (CEN, URA3) | M. Gustin |
| pHA-TOR2 | 2 × HA-tagged TOR2 in pAS25 | Kunz et al. (2000) |
| pHA-TOR2-KD | 2 × HA-tagged TOR2 kinase-dead mutant in pAS25 | Kunz et al. (2000) |
| PGAL1-SigD | SigD ORF under control of the GAL1 promoter in pTB227 (URA3, CEN; Kunz et al., 2000) | This study |
General Genetic Manipulations
Wild-type yeast strains were grown at 30°C, whereas temperature-sensitive mutants were maintained at 26°C unless otherwise indicated. Media were described previously (Sherman et al., 1983). Strain construction followed standard methods (Sherman et al., 1983). Yeast cells were transformed by the lithium acetate procedure (Ito et al., 1983), and transformants were selected on SD medium lacking the appropriate amino acid supplement for maintenance of the plasmid marker.
Cloning of Expression Constructs and Generation of Truncation and Point Mutants
SLM1, SLM2, AVO2, BIT61, and YBR270C open reading frames (ORFs) and ∼200-base pair flanking regions downstream of the stop codon were PCR amplified using Pfx polymerase from genomic DNA isolated from strain JK9-3D by using the following primers: SLM1-FOR 5′-CTACTACTCGAGATGTCGAAAAACAACACAATG and SLM1-REV 5′-CTACTACTCGAGCTTCCATGCATGGGACACA; SLM2-FOR 5′-CTACTACTCGAGATGTCTTACCAACGGAACAG SLM2-REV 5′-CTACTACTCGAGACCAGCAGTATTCATTGTATAA; AVO2-FOR: 5′-GTCGTAGGATCCATGTTGAAAGAGCCCTCAGTTC and AVO2-REV: 5′-GTAGTAGTCGACGAAGAACGCATCTCAGTGG; BIT61-FOR: 5′-GTAGTAAGATCTATGACAGCAGAAGATATACTCC and BIT61-REV: 5′-GTAGTA AGATCTGCTGTGTCCATCGCTGATTCC; and YBR270C-FOR: 5′-GTAGTA AGATCTATGGCAACAGACCTAAATCGTA and YBR270C-REV: 5′-GTAGTACTCGAGGTTCTTTCTGACTCTTTAGCAAG. The PCR reactions were digested with appropriate restriction enzymes (recognition site incorporated into the primer sequences; underlined in primer sequence) and cloned into various expression vectors. The SLM1ΔC and SLM2ΔC truncation mutants were generated by PCR amplification by using Pfx polymerase and SLM1 or SLM2 DNA as template with the following primers: SLM1-FOR and SLM1ΔC-REV (5′-CTACTACTCGAGTTATGTCCTCATCGGTAGATTAGG); SLM2-FOR and SLM2ΔC-REV (5′-CTACTACTCGAGTTAAGGATCGTTTTGATTGCTA). The Slm1 PH domain expression construct was generated using primers SLM1PH-FOR 5′-CTTCATTTCAAGGGATCCCAAC in combination with SLM1-REV. The resulting PCR fragment was cloned as a BamHI-XhoI fragment into the Escherichia coli expression vector pGEX5 × -1 (Pfizer, New York, NY). The SLM point mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Site-directed mutagenesis on SLM1 and SLM2 was performed according to the manufacturer's instructions by using gene-specific primers containing the desired codon changes incorporated. Primers were SLM1-M1FOR 5′-ATTTCTAAACTCCTATTCAAACGGGTATTATGTG; SLM1-M1REV 5′-CACATAATACCCGTTTGAATAGGAGTTTAGAAAT; SLM1-M2FOR 5′-ATCAGGGTTTTTAGAAAACAACTCAAAATTTCTAAA; SLM1-M2REV 5′-GGAGTTTAGAAATTTTGAGTTG T TTTCTAAAAACCC; SLM2-M1FOR 5′-AATTTTTAAACTCATACTCAAATGGGTTTTATGTCC; SLM2-M1REV 5′-GGACATAAAACCCATTTGAGTATGAGTTTAAAAATT; and SLM2-M2FOR 5′-ATCTGGGTTTCTGGAGAACAACTCAAAATTTTTAAA; SLM2-M2REV 5′-GAGTTTAAAAATTTTGAGTTGTTCTCCAGAAACCCA. The successful construction of all constructs and mutants with only the desired changes was confirmed by DNA sequence analysis.
Gene Disruptions
All gene disruptions, except when noted otherwise, were generated using DNA fragments containing KanMX2 disruption cassettes of each individual gene and 300-base pair upstream and downstream flanking regions. The disruption cassettes were PCR amplified from genomic DNA generated from strains in the S288C background and containing single genomic deletion disruptions (purchased from Open Biosystems). The PCR reactions were transformed into the diploid strain W303a/α for one-step gene replacement and G418-resistant transformants were selected. The disruptions were confirmed by PCR with the use of three different sets of primers. The slm2::HIS3 deletion allele was constructed by first cloning the 2.3-kb XhoI fragment containing the entire SLM2 ORF and 220 base pairs downstream of the stop codon into the SalI site of plasmid pGem5zf (Promega, Madison, WI) to generate pGem5zf-SLM2. The internal 0.6-kb BamHI-BglII fragment of pGem5zf-SLM2 was then replaced with a BamHI cassette containing the Schizosaccharamyces pombe HIS5 gene (complements a S. cerevisiae his3 mutant) PCR amplified from plasmid pFA6a-HIS3MX (Longtine et al., 1998). A NotI-SpeI fragment of pGem5zf-Slm2::HIS3 was transformed into strain W303a/α for one-step gene replacement, and His+ transformants were selected and confirmed by PCR. To construct the bit61::HIS3 mutant, the internal 1.6-kb BamHI fragment in vector pGem5zf-BIT61 was replaced by an BamHI restriction fragment containing HIS3MX. The resulting bit61::HIS3MX cassette was cut with SphI and SacI and transformed into yeast strain W303a for one-step gene replacement, and His+ transformants were selected and confirmed by PCR.
Synthetic Lethal Analysis
Doubly heterozygous diploids were generated by crossing haploid strains and selecting for the presence of appropriate markers. To obtain slm1Δ stt4-7 double mutants the slm1::KanMX strain JK503 was mated with strain YJ1426. The slm1::KanMX mutation was combined with the mss4-2 temperature-sensitive mutation by mating strains JK503 and JK511. The stt4-7-LEU2 and mss4-2-URA3 mutant strains JK503 and JK511 also were mated to strain JK505 containing a null mutation in SLM2 (slm2::KanMX). Diploids were sporulated at 26°C, and tetrads were dissected and allowed to germinate at 26°C on YEPD medium and scored for G418 resistance or the presence of auxotrophic markers. For slm1Δ stt4-7 genetic analysis, 53 tetrads were dissected (PD:TT: NPD = 9:36:8). Of the expected 52 double mutant segregants, none was recovered. For slm2Δ stt4-7 genetic analysis 22 tetrads were dissected (PD:TT: NPD = 6:12:4). Of the expected 20 double mutant segregants, 13 were recovered. For the analysis of slm1Δ mss4::HIS3 ura3-52::mss4-2:URA3 mutants 47 tetrads were dissected. Of the expected 23 G418R Ura+ His+ segregants, none was recovered, whereas 11 G418R Ura+ segregants and nine His+ Ura+ segregants were isolated. For slm2Δ mss4::HIS3 ura3-52::mss4-2:URA3, a total of 32 tetrads were dissected, of the expected 35 G418R Ura+ His+ segregants, 12 were recovered.
Expression and Purification of Proteins in E. coli
All proteins were expressed in E. coli BL21(λDE3) as GST or 6His fusion proteins and purified on GSH-conjugated and Ni2+-chelating agarose beads, respectively, following the manufacturer's instructions.
Protein Lipid Overlay Assay
Overlay assays were performed as described previously (Dowler et al., 2002) by using prespotted PIP Strip membranes (Echelon Biosciences, Salt Lake City, UT). Membranes were incubated overnight at 4°C with 500 ng/ml purified recombinant GST or GST fusion protein. Bound GST fusion proteins were detected by Western blot with anti-GST antibodies (1:2000 dilution) followed by horseradish peroxidase-coupled goat anti-rabbit antibody (1: 25,000 dilution) as described previously (Dowler et al., 2002).
Microscopy
To localize HA-Slm1 or HA-Slm2 proteins, plasmids containing wild-type SLM1, SLM2, or mutated variants thereof were transformed into the wild-type yeast strain W303a or strains containing temperature-sensitive mutations (together with their wild-type controls). These plasmids expressed the different SLM variants in yeast under the control of the GAL1 promoter as a fusion protein with a double HA epitope at the N terminus. The HA-tagged Slm1 and Slm2 proteins were both functional, because they complemented the growth defects of a slm1Δ slm2Δ double mutant. Liquid cultures were grown in selective raffinose medium at 25°C to early log phase, galactose (2%) was added to induce expression of HA-SLM1 and HA-SLM2 and incubation was continued at either 25 or 38°C for 1.5 h. Expression of HA-Slm1 was verified and quantitated by Western blot analysis of cell extracts. HA-Slm1 protein levels were comparable in wild-type and lipid kinase mutant strains assayed under the same condition, except in the fab1-2 strain background, which generally yielded somewhat lower levels of protein expression from the GAL1 promoter.
Cells were fixed for immunofluorescence experiments by addition of 1/10 1 M potassium phosphate buffer, pH 6.9, and formaldehyde solution (4% final concentration) directly to the media and incubation for 3–4 h shaking at room temperature (RT). Cells were then collected by centrifugation, washed, spheroblasted, permeabilized, and incubated with primary (mouse monoclonal HA-11 anti-HA) and secondary (Cy2-conjugated goat anti-rabbit IgG) as well as Alexa-594–conjugated phalloidin. Slm1-GFP experiments were performed using cells grown in selective media (selecting for maintenance of chromosomally encoded or plasmid-borne GFP fusions), diluted into SD complete medium for 2 h and mounted for viewing in 1% low-melting agarose.
Cdc42 was visualized essentially as described previously (Richman et al., 2002). Cells of strain JK515, transformed with plasmid p415MET-GFP-CDC42, were grown in SD-Leu medium to mid-log phase, diluted into SD-Met-Leu medium for expression of the methionine-repressible promoter, and incubated for 1 h at 25°C followed by 2 h at either RT or 38°C. To visualize Rho1, the plasmid pRB2138-GFP-RHO1was introduced into strain JK516. Transformants were grown in SD-Ura medium to mid-log phase at RT and for additional 2–3 h at either RT or 38°C. GFP fusion proteins were visualized by fluorescence microscopy in living cells, mounted for viewing in 1% low-melting agarose.
All cells were viewed using a 100× plan oil-immersion lens (numerical aperture 1.4). Images were acquired with the use of a DeltaVision deconvolution microscope (Applied Precision, Mississaugua, Ontario, Canada). Z series stacks were created by sequentially scanning green and red channels at 0.5-μm steps by using 2 × binning. For the quantitation of HA-Slm1 localization, Z series were collected from cells grown under the same growth conditions by using identical gain and exposure times. Images were deconvoluted using the Deltavision software and were then exported as Tiff files and processed with the use of Adobe Photoshop 6.0. Heightfield visualizations were performed on single z-sections of representative cells by using Amira 3.0 (Indeed Visual Concepts, Berlin, Germany). Single confocal sections were obtained from the middle of cells using a Zeiss LSM510 confocal laser scanning microscope. Density plots were generated using NIH Image 1.62. Pixel intensity was determined by a “raw average plot” from four to six cells.
Scoring of Cytoskeletal Polarization and Polarized Distribution of Marker Proteins
Actin polarization was examined in small- and medium-budded cells (∼100 cells analyzed in each case). The actin cytoskeleton was considered polarized if six or fewer actin patches were localized in the mother cells and patches were concentrated at the bud neck and actin cables were polarized. Cells with the majority of actin patches polarized to the bud and the bud neck and containing polarized actin cables were classified as partially polarized. Cells with more actin patches in the mother cell than in the bud were classified as depolarized. The confidence of the counts, which were done twice, was within ±3%.
The polarized localization of GFP-Cdc42 and GFP-Rho1 was assessed in ∼60 small-budded cells (bud <1/3 mother cell length) and visually scored based on whether the GFP or immunofluorescence signal was concentrated over background levels in the bud or was delocalized along the mother/daughter cortex or diffusely in the cytoplasm.
In Vitro Pull-Down Assays
In vitro transcription and translation and labeling of proteins with [35S] Easy Tag Express Protein Labeling Mix (PerkinElmer Life and Analytical Sciences, Boston, MA) was performed using the rabbit T7 in vitro transcription/translation kit from Promega. AVO2, BIT61, or YBR270C cloned into vector TNT521 (Udan et al., 2003) were used as DNA templates in the reactions. For pull-down assays, 6 μl of product from in vitro transcription and translation reactions was incubated for 1 h at 4°C with 1 μg of the different GST fusion proteins (or 6His-Slm1 and 6His-Slm2, purified as indicated above and bound to agarose beads) in binding buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.1% Tween 20, and 0.1% SDS, containing protease inhibitors [1 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A]). Agarose beads then were collected by centrifugation in a microcentrifuge at 500 × g for 2 min and washed five times with 1 ml of cold binding buffer. Bound proteins were eluted and denatured in SDS sample buffer by incubation at 68°C for 15 min. Proteins were separated by standard 10% SDS-PAGE gels. Gels were dried and exposed to BiomaxMR film (Eastman Kodak, Rochester, NY).
Coimmunoprecipitation Assays
SLM1 and SLM2 were expressed from the GAL1 promotor as a fusion protein with a double HA epitope at the N terminus (pAS24, CEN LEU2) or as untagged proteins (pAS23, CEN LEU2). Cells were cultivated in selective raffinose medium to an A600 of 0.5, followed by a 4- to 6-h galactose induction. To prepare whole cell extracts (typically from 2 liters of culture), yeast cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, and 10% glycerol, and 1 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A), lysed with glass beads in a bead-beater (6 × 30 s, 4°C; BioSpecs, Bartlesville, OK), and clarified by microcentrifugation (500 × g, 15 min, 4°C). TAP-tagged Avo2 or Bit61 was purified as described previous (Gould et al., 2004) from 3 mg of whole cell extract (diluted 1:5 with lysis buffer), by using an IgG-Sepharose column followed by washing steps and cleavage with tobacco etch virus protease. The eluted protein was then applied to calmodulin-conjugated beads for the second purification step. After washing five times with lysis buffer containing 500 mM NaCl and 0.5% Triton X-100 and once with phosphate-buffered saline (PBS), the purified complexes were resuspended in 1× SDS-PAGE sample buffer, denatured at 60°C for 10 min, and analyzed by SDS-PAGE and Western analysis by standard methods. To analyze Slm interaction with Tor2, a plasmid containing HA-TOR2 (pAS25, CEN, URA3 GAL1 containing 2 × HA-TOR2) (Kunz et al., 2000) was introduced into strain W303a, and expression of HA-TOR2 was induced by growing exponential yeast cultures for 4–6 h in the presence of 2% galactose. Yeast cell extracts (1 mg total protein diluted 1:5 in lysis buffer) were generated as described above and incubated with 1 μg of purified recombinant 6His-Slm1 and 6His-Slm2 bound to Ni2+-agarose beads or beads alone for1hat 4°C. Beads were then collected by centrifugation (500 × g), and washed four times with lysis buffer and once with PBS. The purified complexes were resuspended in 1 × SDS-PAGE sample buffer, denatured at 60°C for 10 min, and analyzed by SDS-PAGE and Western analysis.
Alkaline Phosphatase Treatment of Protein Extracts and In Vitro Kinase Assays
Twenty-five micrograms of whole yeast cell extract lacking or containing phosphatase inhibitor cocktails I and II (Novagen, Madison, WI) were incubated with 1 U of alkaline phosphatase (ALP) (Roche Diagnostics, Indianapolis, IN) in the presence of 2 mM MgCl2 for 30 min at 37°C. Samples were denatured at 60°C for 5 min and subjected to SDS-PAGE and Western analysis. In vitro protein kinase assays were performed as described previously (Inagaki et al., 1999) by using 1 μg of recombinant 6His-Slm1 and 6His-Slm2 and immunoprecipitated HA-Tor2 or HA-Tor2KD isolated from 0.5 mg of cell extract derived from W303a cells expressing HA-TOR2 or HA-TOR2KD.
RESULTS
Slm1 and Slm2 Are Related Proteins Containing PH Domains
To identify novel PtdIns(4,5)P2 effectors, we searched for yeast proteins that contain a PH domain. Thirty proteins predicted to contain a total of 33 PH domains were identified in the yeast genome by using the program SMART (http://www.smart.heidelberg.de). Among these were known effectors of PtdIns(4,5)P2 (Rom2, Boi1, Boi2, Num1, and Pld1; Farkasovsky and Kuntzel, 1995; Audhya and Emr, 2002; Hallett et al., 2002; Sciorra et al., 2002) and of PtdIns(4)P (the oxysterol-binding proteins Osh1 and Osh2 and the serine/threonine protein kinase Cla4; Levine and Munro, 2001; Levine and Munro, 2002; Roy and Levine, 2004; Wild et al., 2004). The remaining 21 PH domain-containing proteins listed in the SMART database had not been reported previously to bind phosphoinositides. Among these were a group of 10 proteins with unknown functions. In this study, we explored the functions and regulation by phosphoinositides of two of these proteins, encoded by ORFs YIL105C and YNL047C. Because YIL105C and YNL047C exhibit synthetic genetic interaction with MSS4 (see below), these genes were termed SLM1 and SLM2.
Slm1 (686 amino acids, YIL105C) and Slm2 (656 amino acids, YNL047C) are highly related and share an overall amino acid sequence identity of 59% (Figure 1A). Slm1 and Slm2 also exhibit lower sequence homology (38% identity) within their central regions to two other yeast proteins: Ask10, a component of the RNA polymerase II complex that is involved in the cellular response to heat and oxidative stress (Page et al., 1996; Cohen et al., 2003), and a protein of unknown function encoded by ORF YPR115W (Figure 1B). Sequences related to Slm1 and Slm2 are conserved throughout the fungal kingdom, but no clear homologues are present in higher eukaryotes.
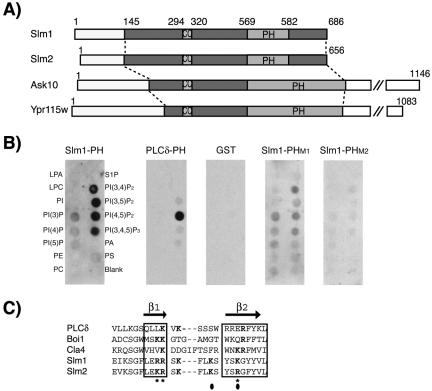
Figure 1.
Domain organization of Slm1 and Slm2 and lipid binding specificity. (A) Schematic diagram of Slm1, Slm2, Ask10, and Ypr115w structures showing the locations of PH (PH) and coiled-coil (CC) domains, respectively. Light and dark shaded boxes indicate the regions conserved among all four proteins. (B) Nitrocellulose-immobilized phospholipids (PIP Strip) were incubated with recombinant fusion proteins of GST to the C terminus of Slm1 (amino acids 441–686) containing either the wild-type PH domain (Slm1-PH) or the mutant variants PHM1, and PHM2. GST alone or a GST fusion to the PH domain of PLCδ, which specifically binds PtdIns(4,5)P2 in vitro were used as controls. Bound proteins were visualized by Western blot analysis with anti-GST antibodies. Spots contained lysophosphatidic acid (LPA), lysophosphacholine (LPC), PtdIns (PI), PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine-1-phosphate (S1P), PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, PtdIns(3,4,5)P3, phosphatic acid (PA), and phosphatidylserine (PS). (C) Sequence alignment of the β1/β2 regions of Slm1 and Slm2 PH domains with related PH domains from PLCδ, Boi1, and Cla4. Secondary structure is indicated above the sequence alignment. Bold letters indicate residues required for interaction with phosphoinositide ligands. Asterisks and closed circles indicate amino acid residues targeted for mutagenesis in SlmM1 and SlmM2 PH domain mutants, respectively.
Slm1, Slm2, Ask10, and Ypr115w are all predicted to contain C-terminal PH domains (Figure 1A). However, unlike the Slm PH domains, the PH domains of Ask10 and Ypr115w are interrupted by several, up to 70 amino acid-long insertions, indicating that they may bind different ligands. Indeed, Ask10 is localized to the nucleus rather than to a membrane compartment (Cohen et al., 2003). In addition to the PH domain, Slm1, Slm2, Ask10, and Ypr115w also contain a coiled-coil domain (Figure 1A). Coiled-coils are known to mediate protein-protein interactions and often promote homo- or heterodimerization. Thus, Slm1 and Slm2 are members of a novel protein family and have the potential to interact with both lipid and protein ligands.
The Slm1 PH Domain Is a Polyphosphoinositide Binding Module
To assess the lipid-binding activity of the Slm1 and Slm2 PH domains, we used a lipid overlay assay (Dowler et al., 2002). This assay has been used extensively to characterize the lipid binding specificity of proteins. A GST fusion protein containing the PH domain of Slm1 was used to probe a panel of lipids spotted onto nitrocellulose membranes (PIP Strip; Echelon Biosciences). This qualitative assay showed that the Slm1 PH domain bound strongly to all multiply phosphorylated phosphoinositides tested, including PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 (Figure 1B). The Slm1 PH domain also bound weakly to monophosphorylated forms of PI such as PtdIns(3)P, PtdIns(4)P, and PtdIns(5)P (Figure 1B). This contrasts with the highly specific lipid binding activity exhibited by the PH domain of PLCδ, which was tested as a control and exclusively bound to PtdIns(4,5)P2 (Figure 1B). As expected, GST alone showed no detectable binding (Figure 1B). We conclude that the PH domain of Slm1 is a functional phosphoinositide-binding domain that binds promiscuously to polyphosphoinositides in vitro.
Structural studies of PH domains have shown that phospholipid binding in all PH domains examined so far is dependent on conserved basic residues located in a region spanning β strands 1 and 2 (Ferguson et al., 1995, 2000; Figure 1C). An alignment of the Slm1 and Slm2 PH domains with a number of other PH domains showed that Slm1 and Slm2 contain basic residues in analogous positions (Figure 1C). Based upon this alignment, we constructed a panel of mutants in which several of these basic residues in the Slm1 PH domain were mutated in various combinations to alanine. These mutant PH domains were then tested in lipid overlay assays alongside the wild-type PH domain. We found that two mutations, K483A/K487A (designated PHM1) and R477A/R478A/K487A (designated PHM2), significantly reduced phosphoinositide binding to ∼20 and 5%, respectively, of wild-type levels. Thus, as in previously characterized PH domains, conserved basic residues in the Slm1 PH domain are required for lipid binding.
Slm1 and Slm2 Localize to the Plasma Membrane
Many PH domains mediate the recruitment of their host protein to cellular membranes. To determine whether the PH domains of Slm1 and Slm2 serve a similar function, we first analyzed the subcellular localization of Slm1 and Slm2. As shown in Figure 2A, a chromosomally expressed GFP fusion to the C terminus of Slm1 localized in a patch-like pattern around the cell periphery, suggesting that Slm1-GFP localizes to the plasma membrane. Weak diffuse GFP signal also was observed in the cytoplasm, indicating that a minor portion of Slm1-GFP is soluble. Similar to Slm1-GFP, a chromosomally expressed Slm2-GFP fusion protein was concentrated in clusters at the plasma membrane (our unpublished data), but produced a GFP signal of much lower intensity, possibly due to lower expression levels of Slm2 compared with Slm1 (Ghaemmaghami et al., 2003; our unpublished observations).
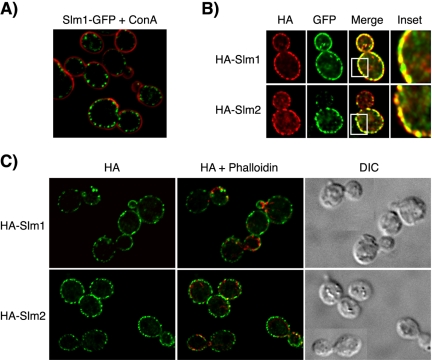
Figure 2.
Slm1 and Slm2 localize to the plasma membrane. (A) Cells containing chromosomally expressed Slm1-GFP fusion protein were mounted on glass slides and the localization of the GFP fusion protein (shown in green) was visualized by fluorescence microscopy, whereas cell walls were visualized by staining with Alexa-594–conjugated concanavalin A (ConA, shown in red). (B) HA-Slm1 and HA-Slm2 are concentrated in clusters along the plasma membrane that contain Pma1-GFP. HA-Slm1 (top, red), HA-Slm2 (bottom, red) expressed from the GAL1 promoter and chromosomally expressed Pma1-GFP (green) were visualized using antibodies directed against HA and GFP, respectively, followed by appropriate fluorescently labeled IgG. Yellow indicates signal overlap. The inset shows a magnification of the staining. (C) HA-Slm1 and HA-Slm2 do not localize to cortical actin patches. HA-Slm proteins were visualized as in B, whereas actin was stained with Alexa-594–conjugated phalloidin (shown in red). Differential interference images of the same cells are shown to the right.
To confirm that Slm1 and Slm2 localize to the plasma membrane, we performed double labeling experiments by using a yeast strain expressing plasmid-borne HA-Slm1 and HA-Slm2 in combination with a chromosomally encoded GFP fusion to Pma1, the plasma membrane H+-ATPase (Serrano et al., 1986). HA-tagged Slm1 and Slm2 fusion proteins expressed from a low-copy vector under the control of the GAL1 promoter colocalized with the endogenous Slm1-GFP signal in double labeling experiments (our unpublished data) and thus accurately reflected the localization of the endogenous Slm proteins. In agreement with earlier findings, Pma1-GFP exhibited a characteristic punctate fluorescence pattern along the plasma membrane (Figure 2B) that was often more pronounced in the mother cell (Malinska et al., 2003). The fluorescent patterns of HA-Slm1 and HA-Slm2 showed significant overlap with the Pma1-GFP signal (Figure 2B), demonstrating that Slm1 and Slm2 localize to the plasma membrane.
The Slm-containing patch-like clusters at the plasma membrane were reminiscent of cortical actin patches (Mulholland et al., 1999). However, neither Slm1-GFP nor HA-Slm1 colocalized with actin in double-labeling experiments by using phalloidin to visualize actin filaments and anti-HA or anti-GFP antibodies, respectively, to detect Slm1 (Figure 2C). Thus, both endogenous and plasmid-borne Slm1 and Slm2 proteins localize to a plasma membrane subcompartment that contains Pma1 but that is distinct from actin patches.
The PH Domains of Slm1 and Slm2 Mediate Plasma Membrane Localization
To determine whether the PH domains are required for targeting of Slm proteins to the plasma membrane, we investigated the subcellular localization of HA-tagged Slm truncation mutants Slm1ΔC (containing amino acids 1–453) and Slm2ΔC (containing amino acids 1–442), in which the C-termini containing the PH domains were deleted. Neither Slm1ΔC nor Slm2ΔC localized to the plasma membrane. Rather, they were distributed diffusely throughout the cytosol and to cytosolic speckles (Figure 3, A and B) demonstrating that the C-terminal domains of Slm1 and Slm2 mediate plasma membrane localization.
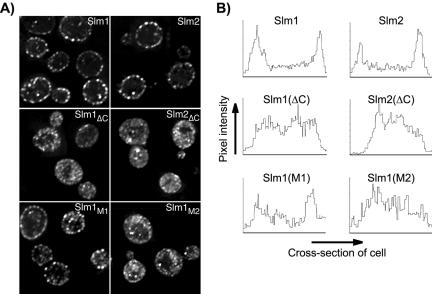
Figure 3.
The PH domain and phosphoinositide binding activity are necessary for plasma membrane association of Slm1 and Slm2. (A) Subcellular localization of HA-tagged wild-type and mutant variants of Slm1 and Slm2 expressed under the control of the GAL1 promoter and visualized by indirect immunofluorescence using antibodies against the HA-tag followed by Cy2-conjugated secondary antibodies. Shown are single z-sections of W303a cells expressing wild-type HA-tagged Slm1 and Slm2 (Slm1 and Slm2); C-terminally truncation mutants lacking the PH domain (Slm1ΔC and Slm2ΔC) and full-length Slm1 point mutants containing substitutions in the PH domain (Slm1M1 and Slm1M1). (B) Quantification of plasma membrane association of wild-type and mutant Slm1 and Slm2 variants. A density profile plot was generated using NIH Image 1.62. Pixel intensity was determined by a “raw average plot” of a cross section through the center of the cell from single confocal slices obtained from four to six cells.
A functional PH domain was necessary for Slm1 and Slm2 localization, because the ability of Slm proteins to associate with the plasma membrane in vivo correlated directly with their lipid-binding activity in vitro. Accordingly, membrane association was severely reduced in the Slm1M2 PH triple mutant, which had lost its lipid binding activity in vitro. This mutant accumulated in the cytosol and within cytosolic speckles (Figure 3, A and B), and only a small portion of the protein remained associated with the plasma membrane. By contrast, the Slm1M1 mutant that still retained some lipid binding in vitro was only partially delocalized from the plasma membrane (Figure 3, A and B). Together, our data demonstrate that Slm1 and Slm2 targeting to the plasma membrane is dependent on their PH domains and phosphoinositide binding activity.
Subcellular Localization of Slm Proteins Is Dependent on PtdIns(4,5)P2 Synthesis
The broad in vitro phosphoinositide binding specificity of the Slm1 PH domain raised the question of which lipid was recognized by Slm proteins in vivo. To assess the in vivo lipid binding specificity, we determined the consequences of manipulating specific phosphoinositide pools on Slm localization. To do this, we took advantage of a panel of yeast mutants defective in the lipid kinases responsible for the synthesis of the various phosphoinositide species in yeast.
Of the lipids bound by the Slm1 PH domain in vitro, yeast cells contain PtdIns(4,5)P2 and PtdIns(3,5)P2, whereas PtdIns(3,4)P2, and PtdIns(3,4,5)P3 have so far not been detected (Desrivieres et al., 1998; Gary et al., 1998; Homma et al., 1998; Odorizzi et al., 1998). Given the in vivo localization of Slm proteins to the cell periphery, we first investigated whether their localization is dependent on PtdIns(4,5)P2 synthesis at the plasma membrane mediated by Mss4. Because MSS4 is essential for growth, HA-Slm1 was expressed in cells containing the temperature-sensitive mss4-2ts allele. This temperature-sensitive mutation reduces cellular PtdIns(4,5)P2 levels by 90% at the nonpermissive temperature of 37°C (Desrivieres et al., 1998). Incubation of mss4-2ts cells at the nonpermissive temperature for 2 h caused a significant decrease of HA-Slm1 signal at the plasma membrane, accompanied by a concomitant increase in cytoplasmic signal (Figure 4, A and B). Under the same conditions, inactivation of MSS4 also led to the delocalization of a PtdIns(4,5)P2-specific GFP reporter protein containing the PH domain of PLCδ, indicating that cellular PtdIns(4,5)P2 levels were indeed down-regulated (our unpublished data). HA-Slm1 was already present in the cytoplasm in mss4-2 cells grown at the permissive temperature, whereas HA-Slm1 almost exclusively localized to the cell periphery in wild-type control cells grown at either 25 or 38°C (Figure 4, A and B). The increase in cytoplasmic HA-Slm1 in mss4-2 cells reflected the redistribution of the protein from the plasma membrane into the cytoplasm and was not due to increased de novo synthesis of HA-Slm1, because HA-Slm similarly relocalized into the cytosol in mss4-2 cells that were shifted back to glucose medium to repress HA-Slm1 expression before incubation at nonpermissive temperature (our unpublished data). Based on these results, we conclude that Slm1 plasma membrane recruitment is at least in part dependent on PtdIns(4,5)P2 synthesis.
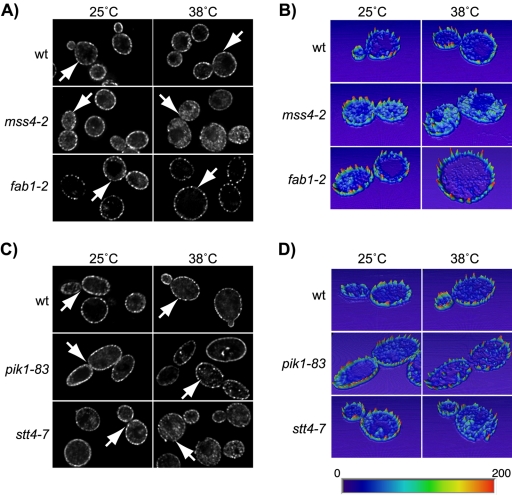
Figure 4.
Localization of Slm1 is dependent on PtdIns(4,5)P2 synthesis. The YCp-HA-SLM1 plasmid was introduced into the wild-type strains JK9-3Da (A) and W303a (C), and into isogenic strains containing mss4-2ts, fab1-2ts, pik1-83ts, and stt4-7ts temperature-sensitive mutations. Indirect immunofluorescence was performed on cells grown at 25°C or shifted to 38°C for 1.5 h by using antibodies against the HA-tag followed by Cy2-conjugated secondary antibodies. (B and D) Quantification of pixel intensity. Heightfield visualizations were performed on single z-sections of representative cells (marked with arrows) by using Amira 3.0. Maximum pixel intensity is indicated in red; minimum pixel intensity in dark blue in the color scheme.
The pool of PtdIns(4)P that serves as the substrate for Mss4 is generated by the essential PI 4-kinase Stt4, which also localizes primarily to the plasma membrane (Audhya and Emr, 2002). A separate pool of cellular PtdIns(4)P is generated on Golgi membranes by the second essential PI 4-kinase, Pik1 (Audhya et al., 2000; Hama et al., 1999). Inactivation of Pik1 by using the temperature-sensitive pik1-83ts allele (Audhya et al., 2000) had little effect on the intracellular distribution of HA-Slm1 (Figure 5, C and D). In contrast, a portion of HA-Slm1 was redistributed from the cell surface into the cytosol and onto intracellular speckles (Figure 4, C and D) in the temperature-sensitive stt4-7 mutant at 38°C (Muhua et al., 1998). Therefore, it seems that the Stt4-dependent plasma membrane pool of PtdIns(4)P modulates plasma membrane localization of Slm1, most likely by serving as the precursor for PtdIns(4,5)P2.
Figure 5.
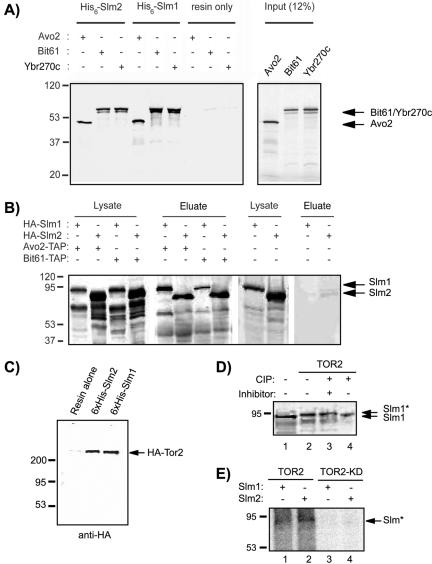
Slm1 and Slm2 physically interact with Tor2 and TORC2 components. (A) In vitro-transcribed and -translated 35S-labeled Avo2, Bit61, and Ybr270c were incubated with recombinant 6His-Slm1 and 6His-Slm2 fusion proteins bound to nickel beads or with resin alone. Bound 35S-labeled proteins that remained after washing were analyzed by SDS-PAGE and autoradiography. The molecular masses of protein markers are indicated to the left. The inferred positions of Avo2, Bit61, and Ybr270c are indicated by arrows. (B) Western blot analysis of proteins associated with Tap-Avo2 and TAP-Bit61 during TAP purification experiments. Cell extracts were prepared from a strain that expressed C-terminally TAP-tagged Avo2 or Bit61 and contained HA-Slm1 or HA-Slm2 expressed under the control of the GAL1 promoter and were then subjected to sequential purifications on IgG and calmodulin affinity resins. Bound proteins were eluted with SDS-PAGE buffer, separated on 10% SDS-PAGE gels, and immunoblotted with antibodies directed against HA. The inferred positions of HA-Slm1 and HA-Slm2 are indicated by arrows. (C) HA-Tor2 associate with Slm1 and Slm2. Clarified cell lysates from a wild-type strain expressing HA-TOR2 were incubated with nickel beads lacking or containing bound 6His-Slm1 or 6His-Slm2. Bound proteins were subjected to SDS-PAGE analysis and Western blotting with HA monoclonal antibody (mAb). The inferred position of HA-Tor2 is indicated by an arrow. D) Yeast cell extracts from strain JK9–3D transformed with vector alone (lane 1) or HA-TOR2 (lanes 2–4) were incubated with alkaline phosphatase (CIP) in the presence (+) or absence (-) of phosphatase inhibitor cocktail as indicated, before SDS-PAGE analysis and Western blotting with HA mAb. The asterisk indicates the phosphorylated form. (E) Recombinant 6His-Slm1 (lanes 1 and 3) and 6His-Slm2 (lanes 2 and 4) were incubated with HA-Tor2 or HA-Tor2KD immunoprecipitated from cell lysates in the presence of [γ-32P]ATP. After SDS-PAGE, 32P-labeled proteins (denoted with an asterisk) were visualized by autoradiography.
Slm1 still localized to the plasma membrane in cells containing a temperature-sensitive mutation in FAB1 (fab1-2) encoding the single, nonessential PtdIns(3)P 5-kinase (Yamamoto et al., 1995; Gary et al., 1998; Odorizzi et al., 1998; Figure 4, A and B), thus demonstrating that Slm1 localization is independent on cellular PtdIns(3,5)P2 levels.
Another approach to demonstrate that Slm subcellular targeting is dependent on PtdIns(4,5)P2 is to manipulate cellular PtdIns(4,5)P2 levels by expression of a lipid phosphatase. The Salmonella phosphatase SigD (also known as SopB) was previously shown to promote the disappearance of PtdIns(4,5)P2 in mammalian cells and to displace PLCδ-PH-GFP from the plasma membrane (Terebiznik et al., 2002). To assess the effects of SigD expression on Slm1 targeting in yeast, we placed SigD under the control of the GAL1 promoter and introduced it into cells containing chromosomally encoded Slm1-GFP. As expected, Slm1-GFP was localized in a punctate pattern along the plasma membrane in cells grown in glucose medium where SigD expression is repressed (Supplementary Figure 1). By contrast, in cells grown in galactose medium, which induces SigD expression, the Slm1-GFP signal in plasma membrane clusters decreased in intensity and clusters often were not evident anymore (Supplementary Figure 1). SigD expression efficiently lowered plasma membrane PtdIns(4,5)P2 levels under these conditions, because it induced the detachment of the PLCδ-PH-GFP fusion protein from the membrane (our unpublished data). Together, these data demonstrate a requirement for PtdIns(4,5)P2 in recruiting Slm1 and Slm2 to the plasma membrane.
SLM1 and SLM2 Synthetically Interact with Mutations in STT4 and MSS4
To further investigate a relationship between Slm1/2 and the Stt4/Mss4 pathway, we looked for synthetic enhancement between mutations that enfeeble Stt4 or Mss4 activity and mutations in SLMs. Individual slm1Δ or slm2Δ deletion mutants conferred no growth defect. However, when the slm1Δ mutation was combined with the temperature-sensitive stt4-7 mutation, slm1Δ stt4-7 doubly mutant segregants were never recovered at the permissive temperature for stt4-7 after sporulation and tetrad analysis (n = 53; see Materials and Methods), indicating that the combination of these mutations is lethal. Likewise, when slm1Δ was combined with the mss4-2 temperature-sensitive mutation by crossing strains JK503 and JK511 (mss4::HIS3 ura3-52::mss4-2::URA3), no meiotic progeny were obtained from dissected tetrads (n = 47) that were Ura+ His+ G418R. Therefore, slm1Δ is synthetically lethal with stt4-7 and mss4-2.
Although slm2Δ stt4-7 and slm2Δ mss4-2 doubly mutant segregants were obtained from dissected tetrads (n = 22 and n = 32, respectively), they formed smaller colonies than the individual single mutants at 25°C and failed to grow at temperatures at or above 38°C (our unpublished data). Thus, slm2Δ confers synthetic growth defects when combined with stt4 and mss4 mutations. The much higher abundance of Slm1 protein compared with Slm2 (Ghaemmaghami et al., 2003; our unpublished observations) may explain why mutations in SLM1 confer more severe growth defects than mutations in SLM2.
The synthetic lethality and synthetic sickness phenotypes observed between slmΔ, and mutations in STT4 and MSS4 suggest that these genes function in a common process. Together with our previous results these data are thus consistent with Slm proteins being effectors of PtdIns(4,5)P2 and acting in the Stt4/Mss4 signaling pathway.
Slm1 and Slm2 Physically Interact with TORC2 Components
Unlike the isolated Slm1 PH domain or the PtdIns(4,5)P2-binding PLCδ-PH-GFP fusion protein, which localize uniformly along the plasma membrane (Yu et al., 2004), Slm1 and Slm2 target to distinctive plasma membrane clusters. This suggests that Slm subcellular targeting also is dictated by a PtdIns(4,5)P2-independent component and may involve protein-protein interaction(s). Recent genome-wide two-hybrid screens identified several candidate Slm1 or Slm2 interacting proteins (Uetz et al., 2000; Ito et al., 2001). Among these, Avo2 and Ybr270c interact with both Slm proteins (Uetz et al., 2000; Ito et al., 2001). Avo2 associates with the phosphatidylinositol kinase-related protein kinase Tor2 as part of the TORC2 signaling complex (Abraham, 2002; Loewith et al., 2002; Wedaman et al., 2003), one of two distinct protein complexes containing Tor2 (Crespo and Hall, 2002). TORC2 is composed of Tor2, Avo1, Avo2, Avo3, Lst8, and Bit61 (Loewith et al., 2002; Wedaman et al., 2003; Reinke et al., 2004) and mediates the essential Tor2-unique function required for the regulation of cell cycle-dependent actin polarization (Crespo and Hall, 2002). Ybr270c is highly related (45% amino acid identity and 61% similarity) to Bit61, and both proteins have been isolated in two-hybrid screens as binding partners of the TORC2 component Avo3 (Ito et al., 2001; Uetz et al., 2000). Notably, similar to Slm proteins, Tor2 and TORC2 components have been shown to localize to punctate clusters at or near the plasma membrane (Kunz et al., 2000; Wedaman et al., 2003). Together, these findings suggest that Slm1 and Slm2 are novel components of the TORC2 complex.
To confirm the physical association of Slm proteins with Avo2, Bit61, and Ybr270c, we performed in vitro pull-down assays. Avo2, Bit61, and Ybr270c were radioactively labeled using in vitro transcription/translation reactions in the presence of [35S]methionine and then incubated with recombinant purified 6His-Slm1 and 6His-Slm2 protein bound to agarose beads. After washing, the proteins retained on the beads were visualized by autoradiography. We found that beads containing 6His-Slm1 fusion protein, but not resin alone, efficiently and specifically retained Avo2, Bit61, and Ybr270c (Figure 5A). All three proteins also were precipitated on beads containing bound 6His-Slm2 (Figure 5A). We thus conclude that Slm1 and Slm2 bind Avo2, Bit61, and Ybr270c in vitro.
We next used the tandem affinity purification (TAP) method (Gould et al., 2004) to test whether Slm1 and Slm2 interact with Avo2 and Bit61 in vivo. We focused on Avo2 and Bit61, because these proteins had been previously characterized. Cell extracts of yeast cells containing chromosomally expressed TAP-tagged Avo2 or Bit61 and expressing plasmid-borne HA-Slm1 or HA-Slm2 were subjected to successive affinity purification on IgG-Sepharose (first step) and calmodulin-agarose (second step). The affinity-purified Avo2-TAP and Bit61-TAP complexes were then separated by SDS-PAGE and analyzed by Western blot analysis by using anti-HA antibodies to detect copurified HA-tagged Slm1 or Slm2.
Prominent bands of ∼70 kDa that correspond well with the predicted molecular masses of HA-Slm1 (78 kDa) and HA-Slm2 (75 kDa) were recovered from extracts containing HA-tagged Slm proteins in the Avo2-TAP and Bit61-TAP purification experiments, but they were absent from extracts containing untagged Slm1 or Slm2 proteins (Figure 5B). These 70-kDa protein bands also were not recovered in control experiments by using whole cell extracts from a yeast strain that expresses HA-Slm1 or HA-Slm2 but contains untagged Avo2 or Bit61 (Figure 5B). Thus, HA-Slm1 and HA-Slm2 are copurified only in the presence of Avo2-TAP or Bit61-TAP. We conclude that Slm1 and Slm2 specifically interact with both Avo2 and Bit61 in vitro and in vivo.
To further assess whether Slm1 and Slm2 are part of the TORC2 complex, we tested whether Slm1 and Slm2 also could be copurified with Tor2. Perhaps due to instability of the TORC2 complex, we were unable to demonstrate copurification of HA-tagged Tor2 with chromosomally expressed Slm1 and Slm2 TAP fusion proteins during TAP affinity purification. However, when purified recombinant 6His-tagged Slm1 and Slm2 proteins, immobilized on agarose beads, were incubated with whole yeast cell extracts containing HA-Tor2, a protein with the predicted molecular mass of Tor2 of ∼280 kDa was specifically retained on Slm1- and Slm2-containing beads, but not on resin alone (Figure 5C). Together, these results demonstrate that Slm1 and Slm2 physically associate with Tor2 and TORC2 components in a protein complex.
In the course of these experiments, we noticed an apparent shift of the Slm1-TAP (Figure 5D, lanes 2 and 3) and Slm2-TAP (our unpublished data) fusion proteins to a higher molecular mass when HA-TOR2 was overexpressed in these cells. This upshift in molecular mass likely reflects phosphorylation of Slm, because phosphatase treatment reverted the effect (Figure 5D, lane 4). Phosphorylation is either directly mediated by Tor2 or a Tor2-regulated protein kinase associated with TORC2, because recombinant 6His-Slm1 and -Slm2 proteins could be radioactively labeled in in vitro kinase assays in the presence of [γ-32P]ATP and immunoprecipitates containing HA-Tor2 (Figure 5E). In contrast, no phosphorylation was observed in kinase assays containing HA-Tor2 kinase-dead (Tor2KD) immunoprecipitates (Figure 5E). Thus, the TORC2 complex may regulate Slm function through phosphorylation.
Interaction with Bit61 and Avo2 Stabilizes Slm1 at the Plasma Membrane
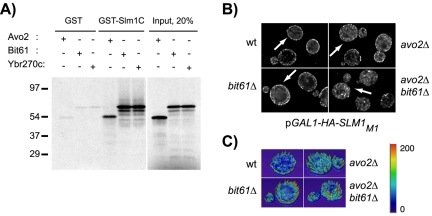
Because Slm1 and Slm2 physically interact with TORC2, we wanted to investigate its effect on Slm association with the plasma membrane. Our previous localization studies showed that the Slm1 deletion mutant Slm1ΔC, which contains amino acids 1–453 of Slm1, but lacks the C-terminal 233 amino acids that include the PH domain, was predominantly cytosolic. In contrast, a significant portion of Slm1 reproducibly remained associated with the plasma membrane when cellular PtdIns(4,5)P2 levels were down-regulated in the mss4-2 mutant. This suggested that the Slm C-terminus not only binds PtdIns(4,5)P2 but also mediates protein-protein interaction, potentially with TORC2 and that these interactions contribute to Slm localization. To test this, we performed pull-down assays with a GST fusion protein containing the Slm1 C-terminal domain (GST-Slm1C comprising amino acids 441–686 of Slm1) and 35S-labeled Avo2, Bit61, and Ybr270c proteins, generated by in vitro transcription/translation reactions. We found that all three proteins efficiently bound to GST-Slm1C, but not to GST alone (Figure 6A). Thus, the Slm1 C terminus contains binding sites for PtdIns(4,5)P2 and TORC2.
Figure 6.
Interaction with TORC2 stabilizes Slm1 association with the plasma membrane. (A) The Slm1 C terminus can mediate interaction with TORC2 components. In vitro-transcribed and -translated 35S-labeled Avo2, Bit61, and Ybr270c were incubated with resin alone or with resin containing GST fused inframe to the Slm1 C terminus (GST-Slm1C; containing amino acids 441–686 of Slm1). Bound 35S-labeled proteins that remained after washing were analyzed by SDS-PAGE and autoradiography. The molecular masses of protein markers are indicated to the left. (B) Plasmid pJK702 (GAL1-HA-SLM1) was introduced into wild-type cells (W303a), and isogenic avo2Δ, bit61Δ, ybr270cΔ single mutant, or avo2Δ bit61Δ double mutant cells. Cells were grown to early exponential phase in raffinose medium at 25°C and HA-Slm1 expression was induced by addition of galactose and further incubation for 2 h. HA-Slm1 was visualized in fixed cells as described in Figure 3. (C) Heightfield visualizations were performed on single z-sections of representative cells (marked with arrows) by using Amira 3.0. Maximum pixel intensity is indicated in red; minimum pixel intensity in dark blue in the color scheme.
To determine whether TORC2 interaction is required for stabilizing Slm1 at the plasma membrane, we investigated whether the localization of the Slm1M1 mutant, which has reduced PtdIns(4,5)P2 binding activity, was affected in yeast cells lacking individual TORC2 components. HA-Slm1M1 localized predominantly to the plasma membrane in wild-type cells but became more cytosolic in yeast cells containing single deletion mutants in AVO2, BIT61, or YBR270C (Figure 6, B and C; our unpublished data). These effects were additive, as HA-Slm1M1 became significantly more delocalized from the plasma membrane in avo2Δ bit61Δ double mutant cells as compared with the single mutants (Figure 6, B and C). Thus, the presence or absence of TORC2 components that physically interact with Slm1 influences Slm1 association with the plasma membrane. Together, our data suggest that Slm localization is coordinately regulated by binding to PtdIns(4,5)P2 and TORC2.
Slm1 and Slm2 Have Redundant Functions and Are Essential for Growth
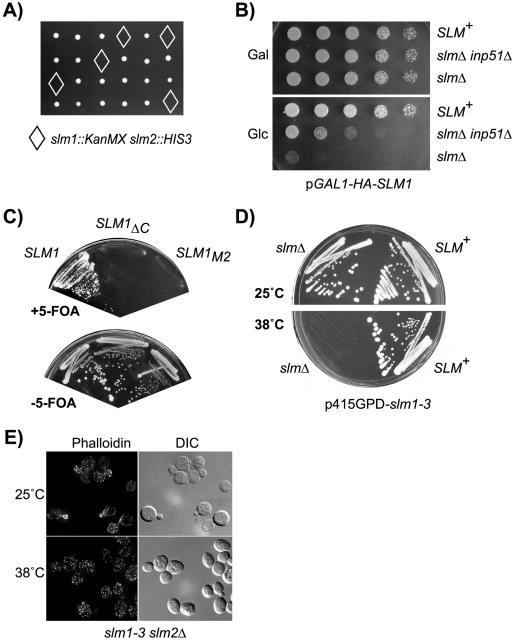
To examine the phenotypic effects associated with loss of SLM function, we constructed a diploid yeast strain heterozygous for deletion alleles of SLM1 and SLM2. Sporulation and tetrad analysis revealed that deletion of both genes led to inviability (Figure 7A). Microscopic examination of doubly mutant spores showed that the spores germinated, but arrested growth in various stages of the cell cycle within one generation (our unpublished data). Slm function is therefore required for cell growth after spore germination. Conditional expression of wild-type SLM1 (Figure 7B) or SLM2 (our unpublished data) from the GAL1 promoter restored growth to slm1Δ slm2Δ double mutant cells on medium containing galactose as the carbon source, demonstrating that the lethality of double mutant cells is due to loss of Slm functions. Together, these data demonstrate that Slm1 and Slm2 are functionally redundant and perform an essential function required for cell growth.
Figure 7.
Slm1 and Slm2 are essential for growth and actin cytoskeleton polarization. (A) Viable meiotic progeny of the SLM1/slm1::KanMX SLM2/slm2::HIS3 heterozygous diploid strain JK507 were recovered after microdissection of spores on YPD plates that were either G418-resistant (indicative of slm1::KanMX) or histidine prototroph (indicative of slm2::HIS3), but not both (marked by diamonds). B) The lethality of slm null mutants is suppressed by conditional expression of SLM1 or by disruption of INP51. Serial dilutions of cultures of wild-type (W303a) or of slm1-3 slm2Δ (JK516) and slm1-3 slm2Δ inp51Δ (JK518) mutant cells containing galactose-inducible SLM1 (pGAL1-HA-SLM1) were spotted onto rich medium containing glucose (Glc) or galactose (Gal) as carbon source. Shown are plates incubated for 3 d. (C) Only a LEU2 plasmid containing wild-type SLM1 but not the SLM1ΔC and SLM1M2 mutant variants can support growth of strain JK513 (slm1::KanMX slm2::HIS3 YCpURA3Gal1-HA-SLM2) on medium containing 5-fluoroorotic acid (+5-FOA), which counterselects the SLM2 URA3 plasmid. (D) Wild-type and slm1-3 slm2Δ mutant cells were streaked on solid SD-Ura medium and incubated at 25 or 38°C for 3 d. (E) Loss of Slm function causes actin cytoskeletal defects. Exponentially growing slm1-3 slm2Δ yeast cells were grown at 25°C or shifted to 38°C for 2 h. Cells were fixed and stained with Alexa-594-phalloidin to visualize the actin cytoskeleton.
To confirm a role of PtdIns(4,5)P2 in the regulation of Slm activity, we next asked whether up-regulation of cellular PtdIns(4,5)P2 levels could suppress the lethality of slmΔ mutant cells. Yeast cells lacking the nonessential PtdIns(4,5)P2 5-phosphatase INP51 have a two- to fourfold increase in cellular levels of PtdIns(4,5)P2 (Stolz et al., 1998a,b). We found that disruption of the INP51 gene in a slmΔ null mutant strain containing the glucose-repressible SLM1 gene was able to suppress the lethality of this strain on glucose medium (Figure 7B). This result suggests that PtdIns(4,5)P2 positively regulates Slm function and likely activates the residual Slm1 protein that is present in these cells due to leaky expression from the GAL1 promoter. In addition, this result implicates Inp51 as a negative regulator of the Slm pathway.
PtdIns(4,5)2 Binding Is Essential for Slm In Vivo Function
We next wanted to determine whether the lipid binding activity of Slm1 and Slm2 is important for their function in vivo. We used a plasmid shuffle assay to assess the ability of Slm mutants lacking a functional PH domain to provide Slm function in vivo. slm1Δ slm2Δ double mutant cells carrying a SLM2 URA3 plasmid were transformed with a second plasmid containing LEU2 as a marker and expressing either wild-type SLM1, or the mutant versions SLM1ΔC and SLM1M2. Transformants were then streaked on solid medium containing 5-fluoroorotic acid (5-FOA) to counterselect the SLM2 URA3 plasmid. We found that the lethality of slm doubly mutant cells was complemented on 5-FOA–containing medium only by wild-type SLM1, but not by the C-terminal truncation mutant SLM1ΔC or the SLM1M2 mutant, which is defective for phosphoinositide binding (Figure 7C). We thus conclude that Slm function is dependent on an intact PH domain that can bind PtdIns(4,5)P2.
Slm Proteins Are Required for Polarized Actin Assembly
The TORC2 signaling complex and the Stt4/Mss4 pathway have both been linked to the regulation of actin cytoskeleton polarization (Desrivieres et al., 1998; Homma et al., 1998). To determine whether Slm proteins, like Tor2, Stt4, and Mss4, are required for proper actin organization, we examined the actin cytoskeleton in cells containing the temperature-sensitive slm1-3 allele (slm1-3 slm2Δ; strain JK515). The slm1-3 allele was generated in the course of our mutational analysis of PH domain residues and contains two point mutations in the PH domain (K501A, R505A) that reduce PtdIns(4,5)P2 binding activity by ∼50% (our unpublished data). This enfeebled Slm1 mutant is able to support growth at 26°C, but not at temperatures ≥38°C (Figure 7D) when expressed from a low-copy vector in slm1Δ slm2Δ double mutant cells.
Incubation of slm1-3 slm2Δ cells at 38°C for 2 h resulted in severe cytoskeletal defects. More than 80–90% of small- to medium-budded cells displayed cortical actin patches that were distributed randomly between mother and daughter cell, rather than being concentrated in the emerging bud (Figure 7E). Furthermore, actin cables were either faint or completely absent. In contrast, the actin cytoskeleton in slm1-3 slm2Δ cells grown at the permissive temperature was similar to wild-type cells, with actin cables traversing from mother to bud and actin patches polarized toward the bud tip (Figure 7E). The actin and growth phenotypes of slm1-3 slm2Δ mutants were reversible after shorter periods of incubation (4–6 h) at 38°C (our unpublished data). However, after overnight incubation at the nonpermissive temperature, ghost cells indicative of dead or lysed cells accumulated, and cells had morphological defects, including aberrations in cell shape and size. In addition, these cells often grew as chains of cells suggesting defects in cell separation (our unpublished data). Together, our data suggest that Slm1 and Slm2 play redundant roles required for cell growth and for the proper assembly of a polarized actin cytoskeleton.
The Polarized Distribution of Cdc42 and Rho1 Is Perturbed in slm Mutant Cells
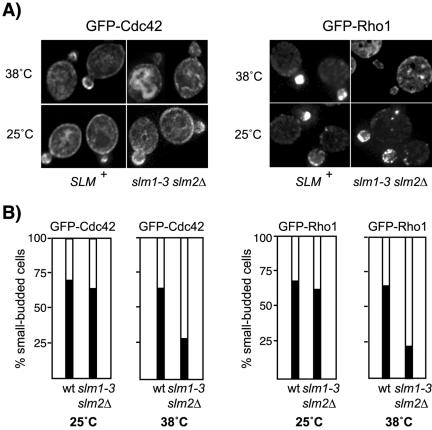
The two Rho GTPases Cdc42 and Rho1 both depend on actin cables to concentrate at sites of polarized growth, such as the growing bud (Abe et al., 2003; Wedlich-Soldner et al., 2003; Pruyne et al., 2004). To confirm that actin cable assembly and polarization is defective in slm mutant cells, we examined the localization of GFP-Cdc42 and GFP-Rho1 fusion proteins in wild-type and slm1-3 slm2Δ mutant cells. As reported previously (Richman et al., 2002), GFP-Cdc42 localized to the plasma membrane and internal membranes and was clustered at the presumptive bud site and in small buds in wild-type and slm1-3 slm2Δ cells grown at 25°C (Figure 8, A and B). GFP-Rho1 also was polarized and localized almost exclusively in the growing bud under these conditions (Figure 8, A and B). After2hat 38°C, the fraction of slm1-3 slm2Δ cells that retained the polarized distribution of GFP-Cdc42 and GFP-Rho1 was significantly reduced, whereas it remained relatively constant in wild-type cells (Figure 8, A and B). Because the retention of Cdc42 and Rho1 at the bud requires actin cables, our results thus support a role of Slm proteins in actin cable organization.
Figure 8.
Polarized localization of Cdc42 and Rho1 is perturbed in slmΔ mutant cells. (A) The distribution of GFP-Rho1 and GFP-Cdc42 in wild-type and slm1-3 slm2Δ mutant cells is shown. Exponential cultures were grown in selective medium at 25°C followed by growth at 38°C for additional 2 h. Expression of GFP-Cdc42 was induced on SD-Leu-Met medium for 1 h at 25°C before shift to nonpermissive temperature. GFP-Cdc42 and GFP-Rho1 localization was visualized by immunofluorescence in living cells mounted in 1% agarose. (B) Approximately 60 small-budded cells expressing GFP-Cdc42 and GFP-Rho1 were scored for polarized distribution of the GFP signal to the bud (black bars) or depolarized localization to the cortex or the cytoplasm (white bars).
PKC1 Is a Dosage Suppressor of slm Null Mutants
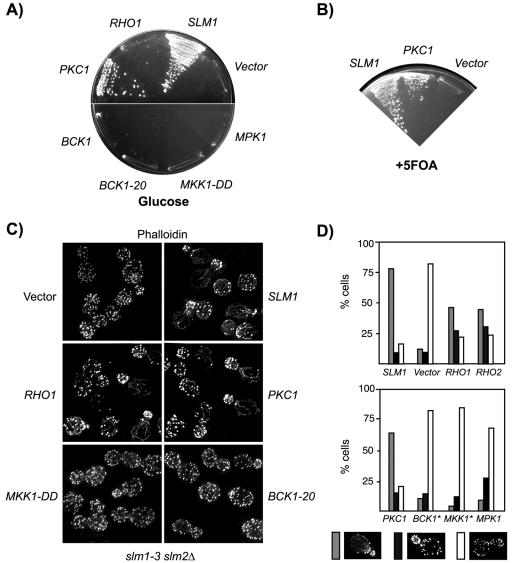
Previous genetic studies suggested that TORC2 acts upstream or in coordination with the Stt4/Mss4 pathway to control actin polarization through the cell integrity pathway (Schmidt et al., 1997; Helliwell et al., 1998a,b; Loewith et al., 2002). Accordingly, genes encoding Mss4 or components of the cell integrity pathway, including the GTP exchange factor Rom2; the partially redundant GTPases Rho1 and Rho2; Pkc1; and a mitogen-activated protein kinase (MAPK) cascade module, including Bck1, the redundant Mkk1 and Mkk2, and Mpk1, act as high-copy suppressors of tor2ts or TORC2 mutants (Schmidt et al., 1997; Helliwell et al., 1998a,b; Loewith et al., 2002). To test whether Slm1 and Slm2 control cell growth and actin polarization through the cell integrity pathway, we examined whether overexpression of cell integrity pathway components can suppress the phenotypes of slmΔ null mutants. We found that only overexpression of PKC1 suppressed the lethality of Slm-depleted cells on glucose (Figure 9A) or of slm1-3 slm2Δ cells at 38°C (our unpublished data). Plasmid shuffling experiments further demonstrated that an activated PKC1 allele, PKC1R398P, present on a TRP1 plasmid, can bypass the complete loss of SLM function, because it allowed slmΔ mutant cells to grow in the absence of a SLM1 URA3 plasmid on medium containing 5-FOA (Figure 9B). In contrast, wild-type or activated forms of the MAPK components that function downstream of Pkc1, including BCK1 (BCK1-20), MKK1 (MKK1-DD), and MPK1 were unable to suppress the growth defects of Slm-depleted or of slm1-3 slm2Δ mutant cells grown at 38°C (Figure 9A; our unpublished data).
Figure 9.
Overexpression of PKC1 suppresses the growth and actin defects of slm mutants. (A) Vector alone or 2μ plasmids containing SLM1, PKC1, BCK1, MPK1, or the activated alleles BCK1-20 and MKK1-DD were transformed into strain JK513 (slm1::KanMX slm2::HIS3/ YCpURA3::GAL1-HA-SLM2). Growth of transformants was tested on SD-Ura-Leu medium containing glucose as carbon source incubated at 30°C for 3 d. (B) Overexpression of PKC1 bypasses Slm function and supports growth of strain JK512 (slm1::KanMX slm2::HIS3/ YCpURA3::GAL1-HA-SLM1) on medium containing 5-fluoroorotic acid (+FOA). (C) Actin cytoskeleton polarization in slm1-3 slm2Δ cells (strain JK515) transformed with plasmids expressing the indicated genes. Cells were grown in SD-Ura-Leu medium, shifted to the nonpermissive temperature of 38°C for 3 h, fixed, and stained for F-actin by using Alexa-594-phalloidin. (D) Quantitation of the actin polarization defect of slm1-3 slm2Δ cells transformed with the indicated plasmids from C. Small- to medium-budded cells (∼100) were scored for their actin polarization state. Cells were classified as having a polarized (gray bars), partially polarized (black bars), or depolarized (white bars) actin cytoskeleton.
Overexpression of PKC1, but not of MAPK components also restored proper actin cable assembly and actin polarization in slm1-3 slm2Δ cells at 38°C (Figure 9, C and D). Overexpression of RHO1 or expression of a constitutively activated form (our unpublished data) did not restore growth, but partially restored the proper polarization of the actin cytoskeleton in slm1-3 slm2Δ cells at 38°C (Figure 9, A, C, and D). Because RHO1 reduced the growth rate in wild-type cells when overexpressed (our unpublished data), we investigated the possibility that any suppression by RHO1 may be masked by its toxicity. However, overexpression of the related RHO2, which is not toxic, conferred similar effects and partially corrected the actin defects of slm1-3 slm2Δ mutants at 38°C, but not the growth defects of Slm-depleted cells (Figure 9C; our unpublished data). Together, our data argue that activation of Pkc1, but not the Pkc1-activated MAPK signaling cascade, is sufficient to restore growth and proper actin polarization in slm null mutant cells.
DISCUSSION
In this study, we show that the PH domain-containing proteins Slm1 and Slm2 are functionally redundant and essential for cell growth and PtdIns(4,5)P2-dependent signaling to the actin cytoskeleton. We further demonstrate that Slm1 and Slm2 physically interact with Avo2 and Bit61 and are novel components of the TORC2 signaling complex. Slm1 (and Slm2) localize to the plasma membrane via interaction with PtdIns(4,5)P2 and TORC2. In addition, Tor2 positively and possibly directly regulates the phosphorylation of Slm1 and Slm2 suggesting that Slm function is modulated in response to signals that control Tor2. Collectively, our data suggest that Slm1 and Slm2 are novel PtdIns(4,5)P2 effectors that integrate inputs from the PtdIns(4,5)P2 and Tor2 signaling pathways to modulate polarized actin assembly and growth. We propose that the synergistic interaction with lipid and protein ligands ensures the proper localization and regulation of Slm1 and Slm2 and is important for the compartmentalization and differential effects of PtdIns(4,5)P2-dependent signaling events.
Our data suggest that Slm1 and Slm2 respond specifically to changes in PtdIns(4,5)P2 levels and thus constitute novel PtdIns(4,5)P2 effectors. The lipid binding activity of Slm1 and Slm2 resides in their C-terminal PH domains, and our analysis of Slm truncation and point mutants suggests that these domains serve as membrane anchors that direct the two proteins to sites enriched in their ligand PtdIns(4,5)P2. This model is supported by a recent survey of yeast PH domains, which found that the isolated Slm1 and Slm2 PH domains localized to the plasma membrane in a manner that was dependent on PtdIns(4,5)P2 synthesis (Yu et al., 2004). Although the PH domain of Slm1, like other PH domains, binds to many polyphosphoinositides with no apparent selectivity in vitro, our data further argue that in vivo, Slm proteins specifically bind PtdIns(4,5)P2 and that PtdIns(4,5)P2 binding is essential for Slm localization and function. First, targeting of Slm1 and Slm2 to the plasma membrane is dependent upon the integrity of their PH domains and requires lipid binding activity. Second, plasma membrane association of Slm1 and Slm2 is affected by changes in PtdIns(4,5)P2 levels such as in stt4ts and mss4ts mutants, and in cells that express the phosphatase SigD, which hydrolyzes PtdIns(4,5)P2. Third, manipulating cellular PtdIns(4,5)P2 levels by disruption of the PtdIns(4,5)P2 phosphatase INP51 suppresses the lethality of slmΔ null mutant cells. Fourth, slm1Δ mutations are synthetically lethal with hypomorphic mutations in STT4 or MSS4. Last, we demonstrate that the ability to bind PtdIns(4,5)P2 is essential for Slm function, because Slm mutants that fail to bind phosphoinositides in vitro, also lack the ability to complement the lethality of slmΔ null mutant cells in vivo.
Although the PH domains of Slm1 and Slm2 are necessary and sufficient to direct plasma membrane targeting (Yu et al., 2004; our unpublished observations), our data suggest that Slm proteins interact with a second, PtdIns(4,5)P2-independent factor at the plasma membrane that directs Slm localization to specific plasma membrane subdomains. This notion is based on the finding that Slm proteins do not uniformly localize along the plasma membrane, as do isolated PH domains that bind PtdIns(4,5)P2, but concentrate in discrete clusters. Our studies identify the TORC2 complex as a candidate PtdIns(4,5)P2-independent localization component, although we cannot rule out contribution by additional factors. Slm1 and Slm2 physically interact with the TORC2 components Avo2, Bit61, and Ybr270c, and our findings from biochemical and immunofluorescence studies are consistent with a role of these proteins in stabilizing and possibly refining Slm localization at the plasma membrane. Such combinatorial control of protein targeting by both lipid and protein determinants may be a common mechanism to regulate the localization and function of PH domain-containing proteins. For example, the targeting of the mammalian four-phosphate adaptor proteins 1 and 2 (Fapp1 and Fapp2) to the Golgi is mediated by the simultaneous binding of their PH domains to PtdIns(4)P and the GTPase Arf1 (Godi et al., 2004). PtdIns(4)P and a Golgi-localized factor also determine the targeting of the related PH domain-containing oxysterol binding proteins (Fang et al., 1996; Levine and Munro, 2002). Similarly, Cla4, Boi1, and Boi2 are targeted to sites of polarized growth by both lipid-dependent and -independent mechanisms (Bender et al., 1996; Hallett et al., 2002; Wild et al., 2004). Such a synergistic control mechanism may explain why many PH domain-containing proteins in vitro show little specificity in phosphoinositide recognition and bind with only moderate affinity, but still target in a specific manner in vivo.
Like mutants in STT4, MSS4, and TOR2, slm deletion and temperature-sensitive mutants exhibit defects in actin cable assembly and actin patch polarization. Together with our biochemical data demonstrating a direct physical interaction between Slm proteins, PtdIns(4,5)P2, and TORC2 components, these findings suggest that Slm proteins integrate inputs from Tor2 and PtdIns(4,5)P2 to modulate actin cytoskeleton polarization. Consistent with the observed cytoskeletal defects, we found that two proteins, Cdc42 and Rho1, which depend on actin cables for their polarized transport to and retention at sites of growth failed to efficiently concentrate in the growing bud upon loss of Slm activity. How do Slm proteins affect actin cytoskeleton dynamics? Because the precise molecular function of Slm1 and Slm2 is at present unknown, there are multiple ways of how a loss of Slm function might lead to the observed cytoskeletal defects. Two likely possibilities are that Slm proteins either directly affect actin cable assembly or that they play a role in vesicular trafficking to the plasma membrane. Rho1 and Cdc42 are key regulators of actin cytoskeletal dynamics that promote formin-dependent actin cable assembly (Tolliday et al., 2002; Dong et al., 2003) and their transport to the bud is mediated by vesicular transport along actin cables (Abe et al., 2003; Wedlich-Soldner et al., 2003; Pruyne et al., 2004). Thus, a failure or decrease in their secretion may indirectly cause the observed cytoskeletal defects in slmΔ mutants. Consistent with such a hypothesis, a block in secretion in several sec mutants also causes actin cable and actin polarization defects (Mulholland et al., 1997; Pruyne et al., 2004). Regardless of the precise mechanism, the phenotypic suppression of slm null mutants by overexpression of the cell integrity pathway component PKC1 suggests that Slm1 and Slm2 modulate Pkc1 activity or localization. Whether Slm1 and Slm2 signaling to Pkc1 involves the upstream activator Rho1 remains to be determined. Contrary to Rho1 and Cdc42, the localization of Pkc1 to the cell cortex is independent of vesicular trafficking (Andrews and Stark, 2000), which may explain why PKC1 is a more effective suppressor of slm null mutant phenotypes compared with RHO1.
In yeast, signaling from Tor2p to the actin cytoskeleton bifurcates at the level of Pkc1p (Delley and Hall, 1999). Interestingly, overexpression or mutational activation of components of the Pkc1-activated mitogen-activated protein (MAP) kinase cascade failed to suppress the phenotypes of slmΔ null mutant cells, and no synthetic lethality was observed between slm1Δ mutants and mutants in the MAP kinase module (our unpublished data). Thus, Slm signaling likely involves the MAP kinase independent Pkc1 signaling branch. This branch controls the cell cycle-dependent polarization of the actin cytoskeleton through yet to be defined effectors (Delley and Hall, 1999). Alternatively, and equally consistent with our data, Slm1 and Slm2 may act in a pathway that has a function overlapping with Pkc1.
Future studies also need to address the precise roles of Slm proteins and the TORC2 components Avo2, Bit61, and Ybr270c in the Tor2 signaling pathway. Preliminary studies indicate that SLM overexpression does not suppress the lethality of tor2ts mutants and only marginally restores actin cable assembly in these mutants (our unpublished observations). However, suppression is not necessarily to be expected because most components of the TORC2 complex (except Avo2) are essential proteins (Loewith et al., 2002), and mutants therein also exhibit actin defects (Loewith et al., 2002). It is thus likely that Slm1 and Slm2 regulate a subset of TORC2 functions activated in response to specific environmental signals. Future identification of downstream effectors of Slm1 and Slm2 and characterization of the role of TORC2 in Slm1 and Slm2 regulation will likely provide insight into the exact molecular function of Slm proteins and reveal how they signal to the actin cytoskeleton.
Interestingly, the TORC2 complex and its modulation of PKC function may be conserved in mammalian cells. Two recent publications reported that the mammalian mTOR protein is part of a protein complex that contains the Avo3 homolog rictor (Jacinto et al., 2004; Sarbassov dos et al., 2004). This complex mediates rapamycin-insensitive mTOR signaling to the actin cytoskeleton (Jacinto et al., 2004; Sarbassov dos et al., 2004) through modulation of protein kinase Cα (Sarbassov dos et al., 2004), suggesting that at least some aspects of TORC2 signaling are conserved between yeast and mammals.
In summary our studies identify two novel effectors of PtdIns(4,5)P2 that mediate an essential function required for growth and actin cytoskeletal organization. The yeast actin cytoskeleton is highly dynamic and quickly adapts to changes in various environmental conditions and in response to the cell cycle phase. Thus, actin dynamics is likely controlled and modulated by a network of signaling pathways that sense and integrate different stimuli. Slm1 and Slm2 contain lipid and protein binding modules and seem to function as part of such a signaling network to integrate signals from the Stt4/Mss4 and Tor2 pathways. In that way, PtdIns(4,5)P2 signaling is coordinated with and dependent on other signaling pathways such as the Tor2 pathway. This, in turn, may allow the differential regulation of PtdIns(4,5)P2-dependent signaling processes in response to various environmental and intracellular stimuli.
Note added in proof. While this article was under review, S. Emr and colleagues (Audhya et al., 2004) reported on the identification of Slm1 and Slm2 in a synthetic lethal screen with mss4ts. Their and our findings are in good agreement.
Supplementary Material
Acknowledgments
We thank John A. Cooper, Scott Emr, Michael Gustin, Michael Hall, Dan Johnson, David Levin, Tim Levine, Sergio Grinstein, and Daniel J. Lew for gifts of strains and plasmids; Richard Atkinson, Robin Hiesinger, and Wei-Ting Chao for help with the Deltavision Deconvolution microscope and image quantification; and Georg Halder for comments on the manuscript. This work was supported by grants GM-068098 from the National Institutes of Health and Q-1536 from the Welch Foundation (to J. K.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0564) on February 2, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abe, M., Qadota, H., Hirata, A., and Ohya, Y. (2003). Lack of GTP-bound Rho1p in secretory vesicles of Saccharomyces cerevisiae. J. Cell Biol. 162, 85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, R. T. (2002). Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell 111, 9-12. [DOI] [PubMed] [Google Scholar]
- Alberts, A. S., Bouquin, N., Johnston, L. H., and Treisman, R. (1998). Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein β subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273, 8616-8622. [DOI] [PubMed] [Google Scholar]
- Andrews, P. D., and Stark, M. J. (2000). Dynamic, Rho1p-dependent localization of Pkc1p to sites of polarized growth. J. Cell Sci. 113, 2685-2693. [DOI] [PubMed] [Google Scholar]
- Audhya, A., and Emr, S. D. (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2, 593-605. [DOI] [PubMed] [Google Scholar]
- Audhya, A., Foti, M., and Emr, S. D. (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., Loewith, R., Parsons, A. B., Gao, L., Tabuchi, M., Zhou, H., Boone, C., Hall, M. N., and Emr, S. D. (2004). Genome-wide lethality screen identifies new PI4,5P(2) effectors that regulate the actin cytoskeleton. EMBO J. 23, 3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, L., Lo, H. S., Lee, H., Kokojan, V., Peterson, V., and Bender, A. (1996). Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J. Cell Biol. 133, 879-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., Drgonova, J., and Drgon, T. (1998). Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67, 307-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, T. J., Lee, K., Rutkowski, L. H., and Strich, R. (2003). Ask10p mediates the oxidative stress-induced destruction of the Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p. Eukaryot. Cell 2, 962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozier, G. E., Carlton, J., Bouyoucef, D., and Cullen, P. J. (2004). Membrane targeting by pleckstrin homology domains. Curr. Top. Microbiol. Immunol. 282, 49-88. [DOI] [PubMed] [Google Scholar]
- Crespo, J. L., and Hall, M. N. (2002). Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66, 579-591, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart, A. K., Schnell, J. D., Allen, D. A., Tsai, J. Y., and Hicke, L. (2003). Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell 14, 4676-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley, P. A., and Hall, M. N. (1999). Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147, 163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres, S., Cooke, F. T., Parker, P. J., and Hall, M. N. (1998). MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273, 15787-15793. [DOI] [PubMed] [Google Scholar]
- Dong, Y., Pruyne, D., and Bretscher, A. (2003). Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161, 1081-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler, S., Kular, G., and Alessi, D. R. (2002). Protein lipid overlay assay. Sci STKE 2002, PL6. [DOI] [PubMed] [Google Scholar]
- Fang, M., Kearns, B. G., Gedvilaite, A., Kagiwada, S., Kearns, M., Fung, M. K., and Bankaitis, V. A. (1996). Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15, 6447-6459. [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky, M., and Kuntzel, H. (1995). Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131, 1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, K. M., Kavran, J. M., Sankaran, V. G., Fournier, E., Isakoff, S. J., Skolnik, E. Y., and Lemmon, M. A. (2000). Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373-384. [DOI] [PubMed] [Google Scholar]
- Ferguson, K. M., Lemmon, M. A., Schlessinger, J., and Sigler, P. B. (1995). Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037-1046. [DOI] [PubMed] [Google Scholar]
- Foti, M., Audhya, A., and Emr, S. D. (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998). Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481-507. [DOI] [PubMed] [Google Scholar]
- Gary, J. D., Wurmser, A. E., Bonangelino, C. J., Weisman, L. S., and Emr, S. D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003). Global analysis of protein expression in yeast. Nature 425, 737-741. [DOI] [PubMed] [Google Scholar]
- Godi, A., Di Campli, A., Konstantakopoulos, A., Di Tullio, G., Alessi, D. R., Kular, G. S., Daniele, T., Marra, P., Lucocq, J. M., and De Matteis, M. A. (2004). FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6, 393-404. [DOI] [PubMed] [Google Scholar]
- Gould, K. L., Ren, L., Feoktistova, A. S., Jennings, J. L., and Link, A. J. (2004). Tandem affinity purification and identification of protein complex components. Methods 33, 239-244. [DOI] [PubMed] [Google Scholar]
- Guo, W., Tamanoi, F., and Novick, P. (2001). Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3, 353-360. [DOI] [PubMed] [Google Scholar]
- Hallett, M. A., Lo, H. S., and Bender, A. (2002). Probing the importance and potential roles of the binding of the PH-domain protein Boi1 to acidic phospholipids. BMC Cell Biol. 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, H., Schnieders, E. A., Thorner, J., Takemoto, J. Y., and DeWald, D. B. (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294-34300. [DOI] [PubMed] [Google Scholar]
- Harrison, J. C., Zyla, T. R., Bardes, E. S., and Lew, D. J. (2004). Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 279, 2616-2622. [DOI] [PubMed] [Google Scholar]
- Helliwell, S. B., Howald, I., Barbet, N., and Hall, M. N. (1998a). TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148, 99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, S. B., Schmidt, A., Ohya, Y., and Hall, M. N. (1998b). The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8, 1211-1214. [DOI] [PubMed] [Google Scholar]
- Homma, K., Terui, S., Minemura, M., Qadota, H., Anraku, Y., Kanaho, Y., and Ohya, Y. (1998). Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J. Biol. Chem. 273, 15779-15786. [DOI] [PubMed] [Google Scholar]
- Hurley, J. H., and Meyer, T. (2001). Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 13, 146-152. [DOI] [PubMed] [Google Scholar]
- Inagaki, M., Schmelzle, T., Yamaguchi, K., Irie, K., Hall, M. N., and Matsumoto, K. (1999). PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol. Cell. Biol. 19, 8344-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., and Takenawa, T. (2004). Regulation of endocytosis by phosphatidylinositol 4,5-bisphosphate and ENTH proteins. Curr. Top. Microbiol. Immunol. 282, 31-47. [DOI] [PubMed] [Google Scholar]
- Jacinto, E., Loewith, R., Schmidt, A., Lin, S., Ruegg, M. A., Hall, A., and Hall, M. N. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122-1128. [DOI] [PubMed] [Google Scholar]
- Kamada, Y., Qadota, H., Python, C. P., Anraku, Y., Ohya, Y., and Levin, D. E. (1996). Activation of yeast PKC by Rho1 GTPase. J. Biol. Chem. 271, 9193-9196. [DOI] [PubMed] [Google Scholar]
- Ketela, T., Green, R., and Bussey, H. (1999). Saccharomyces cerevisiae mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181, 3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, H., et al. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 6060-6068. [PMC free article] [PubMed] [Google Scholar]
- Kunz, J., Schneider, U., Howald, I., Schmidt, A., and Hall, M. N. (2000). HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275, 37011-37020. [DOI] [PubMed] [Google Scholar]
- Lee, K. S., and Levin, D. E. (1992). Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae PKC homolog. Mol. Cell. Biol. 12, 172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M. A. (2003). Phosphoinositide recognition domains. Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- Levin, D. E., Fields, F. O., Kunisawa, R., Bishop, J. M., and Thorner, J. (1990). A candidate PKC gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62, 213-224. [DOI] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (2001). Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 12, 1633-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (2002). Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12, 695-704. [DOI] [PubMed] [Google Scholar]
- Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., Oppliger, W., Jenoe, P., and Hall, M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10, 457-468. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Madaule, P., Axel, R., and Myers, A. M. (1987). Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84, 779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska, K., Malinsky, J., Opekarova, M., and Tanner, W. (2003). Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14, 4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, T. F. (2001). PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493-499. [DOI] [PubMed] [Google Scholar]
- Muhua, L., Adames, N. R., Murphy, M. D., Shields, C. R., and Cooper, J. A. (1998). A cytokinesis checkpoint requiring the yeast homologue of an APC-binding protein. Nature 393, 487-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland, J., Konopka, J., Singer-Kruger, B., Zerial, M., and Botstein, D. (1999). Visualization of receptor-mediated endocytosis in yeast. Mol. Biol. Cell 10, 799-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland, J., Wesp, A., Riezman, H., and Botstein, D. (1997). Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol. Biol. Cell 8, 1481-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S. D. (1998). Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95, 847-858. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S. D. (2000). Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25, 229-235. [DOI] [PubMed] [Google Scholar]
- Page, N., Sheraton, J., Brown, J. L., Stewart, R. C., and Bussey, H. (1996). Identification of ASK10 as a multicopy activator of Skn7p-dependent transcription of a HIS3 reporter gene. Yeast 12, 267-272. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Gao, L., Bi, E., and Bretscher, A. (2004). Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., Python, C. P., Inoue, S. B., Arisawa, M., Anraku, Y., Zheng, Y., Watanabe, T., Levin, D. E., and Ohya, Y. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272, 279-281. [DOI] [PubMed] [Google Scholar]
- Reinke, A., Anderson, S., McCaffery, J. M., Yates, J., 3rd, Aronova, S., Chu, S., Fairclough, S., Iverson, C., Wedaman, K. P., and Powers, T. (2004). TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752-14762. [DOI] [PubMed] [Google Scholar]
- Richman, T. J., Sawyer, M. M., and Johnson, D. I. (2002). Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1, 458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A., and Levine, T. P. (2004). Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279, 44683-44689. [DOI] [PubMed] [Google Scholar]
- Sarbassov dos, D., Ali, S. M., Kim, D. H., Guertin, D. A., Latek, R. R., Erdjument-Bromage, H., Tempst, P., and Sabatini, D. M. (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296-1302. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., Bickle, M., Beck, T., and Hall, M. N. (1997). The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531-542. [DOI] [PubMed] [Google Scholar]
- Sciorra, V. A., Rudge, S. A., Wang, J., McLaughlin, S., Engebrecht, J., and Morris, A. J. (2002). Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 159, 1039-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, R., Kielland-Brandt, M. C., and Fink, G. R. (1986). Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 319, 689-693. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Hicks, J. B. (1983). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Stolz, L. E., Huynh, C. V., Thorner, J., and York, J. D. (1998a). Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148, 1715-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz, L. E., Kuo, W. J., Longchamps, J., Sekhon, M. K., and York, J. D. (1998b). INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J. Biol. Chem. 273, 11852-11861. [DOI] [PubMed] [Google Scholar]
- Takenawa, T., and Itoh, T. (2001). Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190-206. [DOI] [PubMed] [Google Scholar]
- Terebiznik, M. R., Vieira, O. V., Marcus, S. L., Slade, A., Yip, C. M., Trimble, W. S., Meyer, T., Finlay, B. B., and Grinstein, S. (2002). Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 4, 766-773. [DOI] [PubMed] [Google Scholar]
- Toker, A. (1998). The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr. Opin. Cell Biol. 10, 254-261. [DOI] [PubMed] [Google Scholar]
- Toker, A., and Cantley, L. C. (1997). Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387, 673-676.9192891 [Google Scholar]
- Tolliday, N., VerPlank, L., and Li, R. (2002). Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12, 1864-1870. [DOI] [PubMed] [Google Scholar]
- Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C., and Halder, G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell. Biol. 5, 914-920. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A. comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- Valdivia, R. H., and Schekman, R. (2003). The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100, 10287-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman, K. P., Reinke, A., Anderson, S., Yates, J., 3rd, McCaffery, J. M., and Powers, T. (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner, R., Altschuler, S., Wu, L., and Li, R. (2003). Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231-1235. [DOI] [PubMed] [Google Scholar]
- Wild, A. C., Yu, J. W., Lemmon, M. A., and Blumer, K. J. (2004). The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 279, 17101-17110. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., DeWald, D. B., Boronenkov, I. V., Anderson, R. A., Emr, S. D., and Koshland, D. (1995). Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell 6, 525-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H. L., and Janmey, P. A. (2003). Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65, 761-789. [DOI] [PubMed] [Google Scholar]
- Yu, J. W., Mendrola, J. M., Audhya, A., Singh, S., Keleti, D., DeWald, D. B., Murray, D., Emr, S. D., and Lemmon, M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 13, 677-688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.