Abstract
Prolyl-phenylalanine-specific serine protease (dentilisin) is a major extracellular protease produced by Treponema denticola. The gene, prtP, coding for the protease was recently cloned and sequenced (K. Ishihara, T. Miura, H. K. Kuramitsu, and K. Okuda, Infect. Immun. 64:5178–5186, 1996). In order to determine the role of this protease in the physiology and virulence of T. denticola, a dentilisin-deficient mutant, K1, was constructed following electroporation with a prtP-inactivated DNA fragment. No chymotrypsin-like protease activity was detected in the dentilisin-deficient mutant. In addition, the high-molecular-mass oligomeric protein characteristic of the outer sheath of the organism decreased in the mutant. Furthermore, the hydrophobicity of the mutant was decreased, and coaggregation of the mutant with Fusobacterium nucleatum was enhanced compared to that of the wild-type organism. The results obtained with a mouse abscess model system indicated that the virulence of the mutant was attenuated relative to that of the wild-type organism. These results suggest that dentilisin activity plays a major role in the structural organization of the outer sheath of T. denticola. The loss of dentilsin activity and the structural change in the outer sheath affect the pathogenicity of T. denticola.
Treponema denticola is a helically shaped microorganism isolated from the human periodontal region (29, 30) and dermatitis lesions in cattle (4). Increased levels of the organism parallel the destruction of periodontal tissue. In addition, several potential virulence factors, such as an immunosuppressive factor (21, 48), proteolytic activity (32, 35, 43, 52, 53), and attachment factors (8, 19), are expressed by the organism. These observations suggested that this microorganism is potentially a pathogen involved in periodontitis.
Proteases are considered to be significant pathogenic factors in periodontal disease. Several proteases or peptidases of T. denticola have been described, and their pathogenic effects have been characterized (32–35, 43, 52, 53). Of these enzymes, a prolyl phenylalanine-specific protease (dentilisin; also called chymotrypsin-like protease) has a broad substrate specificity, including bioactive peptides (23, 34, 53). In addition, this enzyme is cytotoxic for human epithelial cells (52). These results suggested that the protease is a major pathogenic factor of T. denticola. The purified protease consists of three proteins (23, 37, 53), and activity was lost when the complex was dissociated (23). Several reports have also indicated that high-molecular-mass cell surface oligomeric proteins are expressed in T. denticola and were detected by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (6, 58). A 53-kDa major outer sheath protein (Msp) was also observed in an oligomeric form (11, 54). Recently, the DNA sequence of the major surface protease was determined (23). The protease is approximately 100 kDa under nonreducing conditions. However, it dissociated to 72-, 43-, and 38-kDa proteins on SDS-PAGE. The sequences of the 43- and 72-kDa proteins indicated that the open reading frames of the two proteins are tandemly oriented. The results of a homology search indicated that the 72-kDa protein is a protease with a molecular weight of 77,471. In the present study, we constructed a prtP-deficient mutant in order to determine the physiological role of the protease in the expression of cell surface proteins and its potential role in virulence.
MATERIALS AND METHODS
Microorganisms and plasmids.
The microorganisms and plasmids used in this study are listed in Table 1. T. denticola ATCC 35405 was propagated in TYGVS medium (43), while Porphyromonas gingivalis and Fusobacterium nucleatum were maintained on Tripticase soy agar (Becton Dickinson and Company, Cockeysville, Md.) containing 10% defribrinated horse blood, 5 μg of hemin per ml, and 0.5 μg of menadione per ml. The bacteria were incubated at 37°C under anaerobic conditions as described previously (22).
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Treponema denticola ATCC 35405 | Ems | 3 |
| Treponema denticola K1 | prtP::Emr | This study |
| Porphyromonas gingivalis ATCC 33277 | American Type Culture Collection | |
| Fusobacterium nucleatum ATCC 25586 | American Type Culture Collection | |
| Escherichia coli HB101 | ||
| Plasmids | ||
| pMCL191 | Cmr; pMCL19 (40) lacking EcoRI site | Y. Nakano |
| pCR II | Apr | Invitrogen |
| pTA2 | Apr; pCR II containing amplified prtP fragment | This study |
| pVA2198 | Emr | 13 |
| pDLCK3 | Cmr; pMCL191 containing amplified prtP fragment | This study |
| pKO3 | Cmr Emr | This study |
Escherichia coli HB101 was used to construct plasmids for isolation of the prtP mutant. Strain HB101 was grown on Luria-Bertani agar plates or in Luria-Bertani broth. Plasmids were maintained in cultures containing the following antibiotics: pMCL191, pDLCK3, and pKO3, 30 μg of chloramphenicol per ml; PCR II and pTA2, 100 μg of ampicillin per ml.
Construction of the prtP mutant.
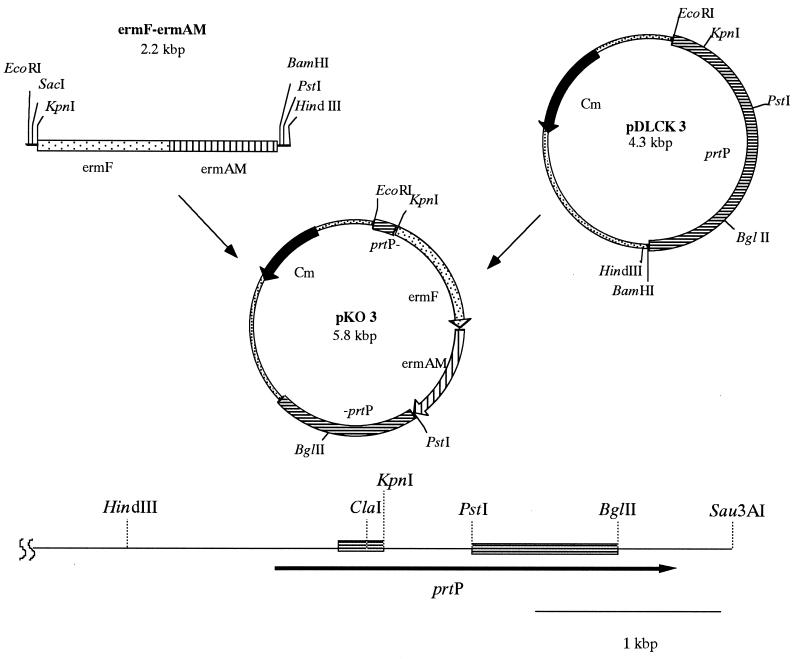
The process for construction of the prtP mutant is illustrated in Fig. 1. The sequence of mature prtP was amplified by the PCR with synthetic oligonucleotide primers (prtP forward primer, 5′-CGGTCTGACAGACGGTAATTATTTGG-3′; prtP reverse primer, 5′-ACGGATCCCCTGTAAACCGTAACTC-3′) as described previously (23). The amplified fragment was inserted into pCR II (Invitrogen, San Diego, Calif.) according to the supplier’s instructions. An EcoRI-BamHI fragment containing the prtP gene was isolated from the resulting plasmid and ligated to pMCL191 (41). The plasmid obtained was designated pDLCK3. An ermF-ermAM cassette (13) was isolated from plasmid pVA2198 following KpnI-PstI digestion and inserted into pDLCK3. The resulting plasmid, pKO3, was linearized following EcoRI and BglII digestion and used in electroporation (28). T. denticola ATCC 35405 was inoculated into 500 ml of TYGVS medium and incubated for 3 days as described above. Cells were placed on ice for 15 min, washed with 500 ml of ice-cold distilled water, and centrifuged at 4,000 × g for 10 min. Cells were resuspended in 250 ml of ice-cold distilled water, centrifuged, and resuspended in 10 ml of ice-cold distilled water containing 10% glycerol. After centrifugation, the cells were suspended in 1 ml of 10% glycerol. Eighty microliters of competent cells (approximately 5 × 1010 cells) was mixed with 10 μg of linearized pKO3. Competent cells were electroporated as previously described (28) and mixed with 2 ml of TYGVS medium. After the cells were incubated for 24 h under anaerobic conditions, 1 ml of the culture was mixed with 35 ml of TYGVS medium containing 0.8% agarose (TYGVS plates) and 40 μg of erythromycin per ml. The resulting plates were incubated for 4 to 8 days under anaerobic conditions. After incubation, individual colonies were isolated with a capillary pipette and reinoculated into TYGVS medium containing 40 μg of erythromycin per ml. One mutant, designated K1, was selected for further evaluation.
FIG. 1.
Construction of a prtP mutant with the ermF-ermAM cassette.
Growth rate of the mutant.
Cultures of T. denticola ATCC 35405 and mutant K1 were adjusted to an absorbance of 0.2 at 660 nm in TYGVS medium, and 1.0 ml of each was inoculated into 100 ml of TYGVS medium and incubated at 37°C under anaerobic conditions. Growth rates were determined by measuring the absorbance at 660 nm and by use of a Petroff-Hauser bacterial counting chamber.
Southern blot analysis.
Genomic DNAs from T. denticola ATCC 35405 and K1 were isolated and hybridization was performed as described previously (22). Briefly, chromosomal DNA from T. denticola was digested with HindIII, electrophoresed through 1.0% agarose gels, denatured, and transferred to Hybond-N+ paper by capillary transfer (49). DNA probes (571-bp KpnI-PstI frag ment of the prtP gene and KpnI-BamHI fragment from pVA2198; Fig. 1) were labeled with digoxigenin-dUTP by use of a DIG DNA labeling system (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) according to the manufacturer’s protocol. Hybridization was performed at 42°C for 18 h in an aqueous buffer containing 50% formamide, 5 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% blocking buffer (Boehringer), 0.1% sarcosine, and 0.01% SDS. Hybridized membranes were washed as specified by the supplier, and hybridizing bands were detected on Hybond-N+ paper by use of a DIG DNA detection kit (Boehringer).
Assays of dentilisin activity of the mutant.
Proline-phenylalanine-specific protease activity was measured with the synthetic substrate N-succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanine p-nitroanilide (SAAPNA; Sigma Chemical Company, St. Louis, Mo.). The cells were disrupted by sonication (Branson, Danbury, Conn.) at 100 W for 5 min on ice. Insoluble material was removed by centrifugation at 8,000 × g for 20 min. The protein concentration of the sonicate was determined by the DC protein assay (Bio-Rad Laboratories, Hercules, Calif.). A 5-μl aliquot of sonicate was mixed with 150 μl of 100 mM Tris-HCl buffer (pH 8.0) containing 1.0 mM SAAPNA. The mixture was incubated at 37°C for 15 min, and the reaction was stopped by the addition of 50 μl of 20% acetic acid. The release of p-nitroaniline was determined by measuring its absorbance at 405 nm. One unit of enzyme was defined as the amount required to release 1.0 μmol of p-nitroaniline in 1 min at 37°C under these conditions.
Hydrolysis of fibronectin (Sigma) was performed as described before (23). Samples containing 10 μg of fibronectin were incubated with 2 μg of the sonicate of T. denticola for 6 h at 37°C. Reaction mixtures were subjected to SDS-PAGE analysis, and the protein bands were stained with Coomassie brilliant blue R-250.
Antisera.
Rabbit antiserum against T. denticola ATCC 35405 whole cells and rabbit antiserum against dentilisin were prepared as described previously (23). Rabbit antiserum against T. denticola ATCC 35404 Msp was kindly provided by T. Umemoto (Asahi University, Gifu, Japan).
SDS-PAGE and immunoblot analyses.
Wild-type cells and cells of the prtP mutant were examined by SDS-PAGE with or without 1 μM serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) by use of the discontinuous system of Laemmli (26). Proteins (10 μg) were separated on a 10 to 20% gradient resolving gel (Daiichi-Kagaku, Tokyo, Japan). Cells or sonicates were treated either at 100°C for 5 min or at 4°C overnight in the presence of β-mercaptoethanol. Protein bands were visualized by staining with Coomassie brilliant blue R-250.
For zymography, the cells were incubated overnight with SDS sample buffer at 4°C and the mixtures were separated by SDS–6% PAGE with gels containing 200 μg of gelatin per ml. After electrophoresis, the gels were incubated in 100 mM Tris-HCl buffer (pH 8.0) for 1 h and stained with Coomassie brilliant blue R-250.
For immunoblot analysis, 2 μg of proteins from treponemal cells was electrophoresed as described above and transferred by the method of Towbin et al. (51) with a Transblot cell (Bio-Rad). The blotted membranes were immunostained with rabbit antidentilisin serum (23), anti-T. denticola ATCC 35405 whole-cell serum, or anti-T. denticola ATCC 35404 Msp serum. Antibody bound to protein immobilized on the membranes was detected with peroxidase-conjugated goat anti-rabbit immunoglobulins (Bio-Rad).
Coaggregation assays.
Coaggregation was determined by a modification of the method of Cisar et al. (5). Briefly, cells were washed with phosphate-buffered saline (pH 7.2) and suspended in coaggregation buffer (1 mM Tris-HCl buffer containing 0.1 mM CaCl2, 0.1 mM MgCl2, 0.02% NaN3, and 0.15 M NaCl). All suspensions were adjusted to an optical density of 2.0 at 660 nm with a U2000 spectrophotometer (Hitachi, Tokyo, Japan). Aliquots of 500 μl from each bacterial suspension and coaggregation partner suspension were vortexed for 10 s, allowed to stand at room temperature for 1 h, and visually scored for coaggregation. Tubes containing each cell suspension alone were included as controls. When the bacterial suspensions autoagglutinated, the tubes were mixed again, and the existence of coaggregated cells was reexamined. Coaggregation was confirmed by phase-contrast microscopy. Briefly, the mixtures were allowed to stand at room temperature overnight, and an aliquot was gently vortexed and placed between a glass slide and coverslip for visual confirmation of coaggregation.
Determination of hydrophobicity.
Evaluation of cell hydrophobicity was carried out as described by Rosenberg et al. (46) and Gibbons et al. (15). Bacterial suspensions in PUM buffer (K2HPO4 · H2O, 22.2 g; KH2PO4, 7.3 g; urea, 1.8 g; MgSO4 · 7H2O, 0.2 g) were adjusted to an optical density of approximately 0.5 at 400 nm. Duplicate samples of bacterial suspensions (1.2 ml) in PUM buffer were placed in 10- by 70-mm glass tubes, and 600 μl of hexadecane (Sigma) was added. The tubes were vigorously vortexed for 60 s and allowed to stand for 15 min, after which the A400 of the aqueous phase was measured. The percent hydrophobicity was calculated as follows: [(A400 before mixing − A400 after mixing)/A400 before mixing)] × 100. Each isolate was assayed twice, and the values obtained were averaged.
Evaluation of pathogenicity of the mutant.
Virulence was assessed with a mouse abscess model described by Kesavalu et al. (24). Briefly, T. denticola ATCC 35405 and K1 were grown for 72 h under anaerobic conditions as described above and harvested. Cells were resuspended in phosphate-buffered saline (pH 7.2) and quantitated with a Petroff-Hauser bacterial counting chamber. Twenty-two BALB/c mice (6 to 8 weeks old) were separated into two groups and challenged subcutaneously (s.c.) on the posterior dorsolateral surface with 200 μl (3 × 109 cells) of ATCC 35405 or K1 cell suspension. Following challenge, the animals were examined at least once daily for 14 days for lesion formation, and the size of each lesion was measured with a caliper gauge. At 3 and 5 days after s.c. challenge, the mice were euthanatized by CO2 asphyxiation, and the contents of each abscess were aspirated with a syringe after disinfection of the lesion skin with ethyl alcohol. The viability of the spirochetes was evaluated by dark-field microscopy and inoculation on TYGVS plates under anaerobic conditions.
Statistical analysis.
Statistical differences in lesion area were determined by the Mann-Whitney U test.
RESULTS
Construction of the dentilisin-deficient mutant.
To determine the role of dentilisin in the physiology of T. denticola, an isogenic mutant defective in the prtP gene was constructed by allelic exchange mutagenesis (Fig. 1). The 2.1-kbp ermF-ermAM cassette was cloned into the KpnI-PstI site of plasmid pDLCK3. The recombinant plasmid was then linearized with EcoRI and BglII and electroporated into T. denticola ATCC 35405. Since the plasmid was linearized, erythromycin-resistant transformants could arise as a result of a double-crossover event between the regions flanking the erm cassette and the wild-type gene on the chromosome. This event would result in the replacement of the central Kpn-PstI fragment of the prtP gene with a fragment conferring erythromycin resistance.
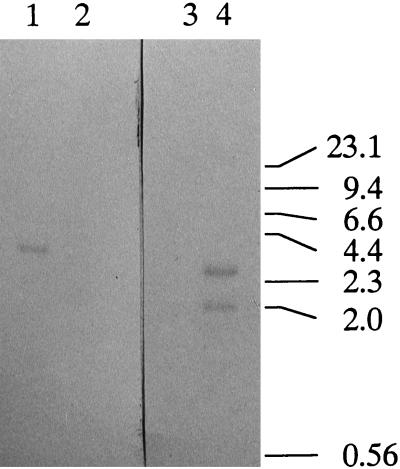
We obtained 24 Emr colonies following 7 days of incubation. The efficiency of the recombination event was approximately 1.2 colonies per μg of DNA. We isolated seven of the putative mutants and designated them K1 to K7. The growth rate of the mutants was the same as that of the wild type in TYGVS medium. The mutants also exhibited pronounced autoaggregation activity in TYGVS medium at the stationary phase. To confirm the predicted recombination event, Southern blot analysis was carried out (Fig. 2). A 3.3-kbp band was observed for wild-type strain ATCC 35405 when the KpnI-PstI fragment from the prtP gene was used as a probe (Fig. 2, lane 1). Since the KpnI-PstI fragment was replaced with the ermF-ermAM cassette in the transformants, no positive band was observed in Emr transformant K1 when the KpnI-PstI fragment was used as a probe (Fig. 2, lane 2). When the ermF-ermAM cassette was used as a probe, no band was observed for the wild-type cells (Fig. 2, lane 3). Since the ermF-ermAM cassette contains a single HindIII site, two bands (2.0 and 2.5 kbp) were observed in the Emr mutant (Fig. 2, lane 4) when the ermF-ermAM cassette was used as a probe. Likewise, the sizes of the amplified fragments of wild-type T. denticola and mutant K1 in PCR with the prtP forward and reverse primers were approximately 2 and 4 kbp, respectively (data not shown). These data suggested that the predicted recombination event had occurred, resulting in the interruption of the wild-type protease gene by the antibiotic resistance gene cassette. Identical results were observed for the other mutants, and one mutant, K1, was chosen for further analysis.
FIG. 2.
Southern blot analysis of T. denticola ATCC 35405 and prtP mutant K1. Chromosomal DNAs from T. denticola ATCC 35405 (lanes 1 and 3) and prtP mutant K1 (lanes 2 and 4) were digested with HindIII and hybridized with a digoxigenin-labeled KpnI-PstI fragment from the prtP gene (lanes 1 and 2) or the KpnI-BamHI fragment from pVA2198 (lanes 3 and 4). Numbers at right are kilobase pairs.
Proteolytic activity of mutant K1.
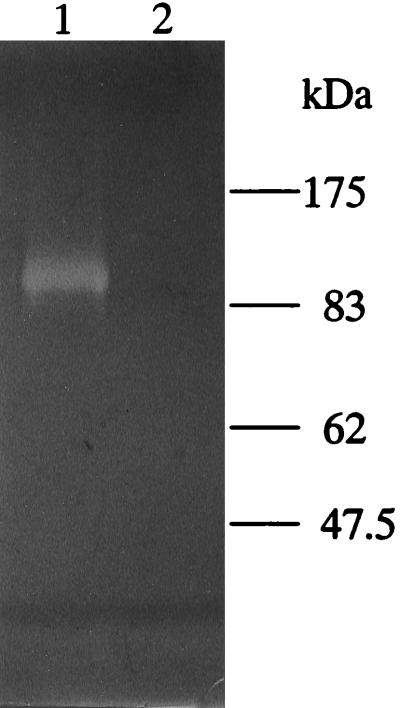
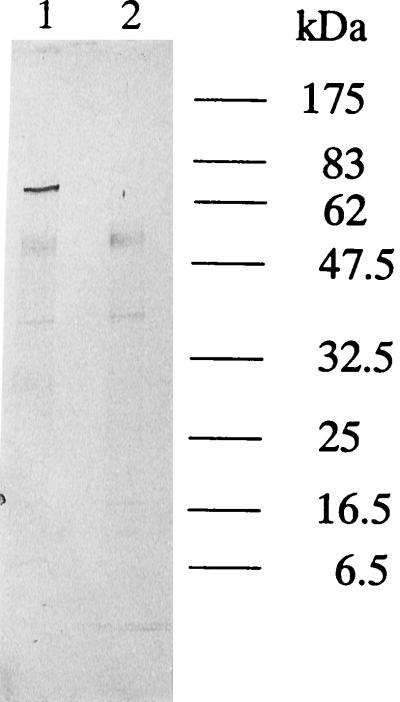
The proteolytic activities of whole cells and sonic extracts from T. denticola ATCC 35405 and K1 were assayed with the synthetic chymotrypsin substrate SAAPNA. Wild-type T. denticola ATCC 35405 exhibited SAAPNA-hydrolyzing activity (1.0 × 10−4 ± 0.020 × 10−4 U/1.1 × 109 cells) whereas K1 displayed little SAAPNA-hydrolyzing activity (0.010 × 10−4 ± 0.012 × 10−4 U/1.1 × 109 cells) in whole cells. Zymography with gelatin as a substrate (Fig. 3) indicated that a proteolytic band of approximately 100 kDa could be readily detected for wild-type strain ATCC 35405 but that this band was absent in mutant K1. The wild-type strain also hydrolyzed fibronectin, while mutant K1 did not (data not shown). The zymography data indicated that K1 was missing the predicted proteolytic band of approximately 100 kDa corresponding to the chymotrypsin-like protease activity of T. denticola (53).
FIG. 3.
Gelatin zymography of sonicates of T. denticola ATCC 35405 (lane 1) and K1 (lane 2).
SDS-PAGE and immunoblot analyses of the mutant.
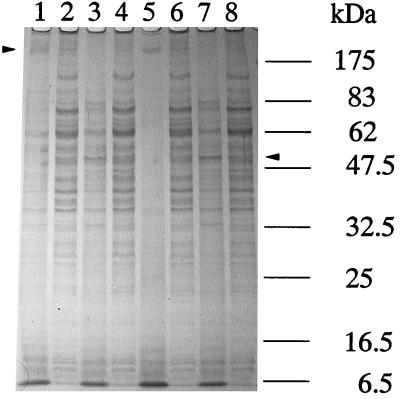
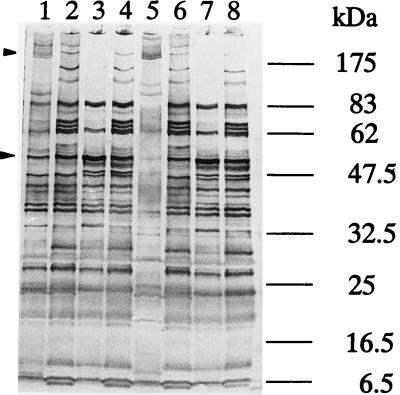
The results of SDS-PAGE analysis of the proteins expressed by the wild type and mutant K1 are shown in Fig. 4. In unheated samples, high-molecular-mass oligomeric proteins of 180 to 200 kDa were observed in the wild type (Fig. 4, lanes 1 and 5), and smaller amounts of low-molecular-mass proteins were also observed (lane 5). Pretreatment with PMSF, which prevents proteolysis in SDS-PAGE analysis (57), increased the intensity of the bands smaller than 100 kDa (Fig. 4, lane 1). On the other hand, the 180- to 200-kDa oligomeric proteins were less intense in the mutant with or without PMSF pretreatment (Fig. 4, lanes 2 and 6). The 78-kDa band was observed only in the mutant and not in the wild type. Boiling the samples resulted in alterations in the protein profiles for the wild-type and mutant cells. In the K1 mutant, multiple bands were combined around the position of Msp. The results of immunoblot analysis with antidentilisin antibody further indicated that antidentilisin serum reacted with a 72-kDa protein in boiled samples of the wild-type strain. However, this protein band was not observed in the prtP mutant extracts (Fig. 5).
FIG. 4.
SDS-PAGE analysis of sonicates of T. denticola ATCC 35405 and K1. Samples from lanes 1 to 4 were treated with PMSF. Lanes 1 and 5, T. denticola ATCC 35405 (without boiling); lanes 2 and 6, T. denticola K1 (without boiling); lanes 3 and 7, T. denticola ATCC 35405 (with boiling); lanes 4 and 8, T. denticola K1 (with boiling). After electrophoresis, the gel was stained with Coomassie brilliant blue R-250. Arrowheads indicate the high-molecular-mass oligomeric protein and the Msp band.
FIG. 5.
Immunoblot analysis of T. denticola ATCC 35405 (lane 1; boiled) and K1 (lane 2; boiled) with antidentilisin serum.
It was previously reported that a high-molecular-mass oligomeric protein consisted of polymers of Msp (11, 54). We analyzed the antigenic proteins with antiserum against T. denticola ATCC 35405 whole cells (Fig. 6). In the wild-type strain, the oligomeric protein band was observed from about 180 to 230 kDa with or without PMSF treatment (Fig. 6, lanes 1 and 5), but the reactivity of the antibody with the oligomeric protein from the mutant was decreased (lanes 2 and 6). Boiled samples of both strains contained the 53-kDa Msp and a similar band pattern below Msp. However, the 68-, 70-, 131-, 175-, and 200-kDa bands were observed only in unboiled samples of the mutant (Fig. 6, lanes 3 and 4). We also analyzed the size of Msp in unboiled and boiled samples with antiserum against T. denticola ATCC 35405 Msp. This serum showed reactivity with both T. denticola ATCC 35404 and T. denticola ATCC 35405 Msp (data not shown). As Fig. 7 indicates, anti-Msp serum reacted with 178- to 224-kDa oligomeric proteins in unboiled samples of wild-type cells. On the other hand, the corresponding bands appeared faint and a weak band was observed at 83 kDa in mutant cells. An Msp of 53 kDa was observed in boiled samples of both the wild type and the mutant. These results indicated that Msp was expressed in both the wild type and mutant K1 but that in the mutant the ability of the organization of the high-molecular-mass oligomeric protein decreased.
FIG. 6.
Immunoblot analysis of T. denticola ATCC 35405 and K1 with anti-T. denticola ATCC 35405 whole-cell serum. Samples from lanes 1 to 4 were treated with PMSF. Lanes 1 and 5, T. denticola ATCC 35405 (without boiling); lanes 2 and 6, T. denticola K1 (without boiling); lanes 3 and 7, T. denticola ATCC 35405 (with boiling); lanes 4 and 8, T. denticola K1 (with boiling). Arrowheads indicate the high-molecular-mass oligomeric protein and the Msp band.
FIG. 7.
Immunoblot analysis of T. denticola ATCC 35405 and K1 with anti-T. denticola ATCC 35404 Msp serum. Samples from lanes 1 to 4 were treated with PMSF. Lanes 1 and 5, T. denticola ATCC 35405 (without boiling); lanes 2 and 6, T. denticola K1 (without boiling); lanes 3 and 7, T. denticola ATCC 35405 (with boiling); lanes 4 and 8, T. denticola K1 (with boiling). Arrowheads indicate the high-molecular-mass oligomeric protein and the Msp band.
Hydrophobicity measurements.
Previous data suggested that the hydrophobicity of bacterial cells is sometimes correlated with colonization properties which may be relevant to the oral cavity (39). To determine the relationship between dentilisin and the cell surface architecture of T. denticola, the cell surface hydrophobicities of T. denticola wild-type and mutant K1 cells were compared. The cell surface hydrophobicity was dramatically decreased in the prtP mutant (23.0% ± 11.9%) compared with the wild type (60.0% ± 3.71%).
Coaggregation activity between T. denticola and other oral bacteria.
T. denticola was previously reported to coaggregate with P. gingivalis and F. nucleatum (17, 25, 44). To determine if the alteration of cell surface hydrophobicity following inactivation of the prtP gene influenced such interactions, the coaggregation reactions of the wild-type and mutant cells were evaluated (Table 2). The mutant exhibited autoaggregation activity in TYGVS medium but did not exhibit autoaggregation activity in coaggregation buffer. The coaggregation reaction between T. denticola K1 and F. nucleatum ATCC 25586 was enhanced relative to that of the wild-type strain. In fact, the interaction between wild-type strain ATCC 35405 and F. nucleatum ATCC 25586 was relatively weak. The coaggregation score for T. denticola K1 and P. gingivalis was not different from that of the wild-type strain, but a significant rapid coaggregation reaction was observed. Pretreatment of T. denticola cells with PMSF just prior to the reaction did not alter either coaggregation reaction.
TABLE 2.
Coaggregation activity between T. denticola and P. gingivalis or F. nucleatum
| T. denticola strain | Scorea for:
|
|
|---|---|---|
| P. gingivalis | F. nucleatum | |
| K1 | 3 | 4 |
| ATCC 35405 | 3 | 2 |
| K1 + 1 mM PMSF | 3 | 4 |
| ATCC 35405 + 1 mM PMSF | 3 | 2 |
0, no change in turbidity and no visible coaggregates; 1, finely dispersed coaggregation in a turbid background; 2, definite coaggregation without immediate settling; 3, formation of large settling coaggregates with a slightly turbid supernatant; 4, large coaggregates settling immediately, leaving a water-clear supernatant.
Virulence of the prtP mutant in the mouse abscess model system.
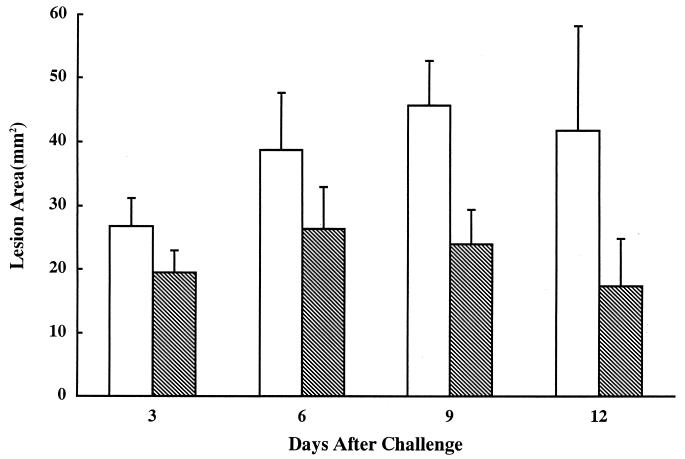
To evaluate the effects of the surface structure alterations displayed by mutant K1 on the virulence of the microorganism, the wild-type and mutant strains were injected s.c. into the posterior dorsolateral surface of two groups of mice (Fig. 8). The lesion areas of the group infected with the prtP mutant were smaller than those of the group injected with the wild type over a 3- to 14-day period after infection (days 3, 6, 9, 10, 11, 12, 13, and 14, P < 0.001; days 4, 5, and 8, P < 0.05). T. denticola ATCC 35404 and K1 were detected by microscopy following aspiration of samples of the abscesses at days 3 and 5. We also isolated on TYGVS plates viable T. denticola ATCC 35405 and K1 from the abscesses at days 3 and 5. No gross pathology of the animals was detected over this time period. However, the animals were not examined for any internal organ damage. The stability of the prtP mutation was not assessed, since the double-crossover recombination event used to construct mutant K1 should result in a stable mutation in the absence of antibiotic selection pressure.
FIG. 8.
Mean areas of lesions at infection sites after challenge with T. denticola ATCC 35405 and prtP mutant K1. Mice were injected with live T. denticola ATCC 35405 (open bars) or mutant K1 (hatched bars), and lesion areas were determined at the indicated times following infection.
DISCUSSION
Several reports on the proteases of T. denticola have appeared in the literature (1, 23, 31, 32–35, 38, 43, 45, 53). One of these proteases, dentilisin, is present in the outer sheath of the organism (23). Dentilisin has been proposed to participate in the adhesion of the microorganism to epithelial cells (27), to interfere with the host immune response (53) and infiltration of the tissues (18), and to have cytotoxic effects on human epithelial cells (52). Therefore, this protease may be involved in the etiology of periodontal diseases. However, it is not yet clear what role this enzyme plays in the normal physiology of T. denticola. It has been suggested that proteases play a role in the formation of the surface layer of some bacteria. For example, the activity of the protease Arg-gingipain is required for the maturation and expression of the fimbriae of P. gingivalis (42, 50). It has been demonstrated that there is an outer sheath surrounding T. denticola and other spirochete cells (20) and that the sheath contains high-molecular-mass oligomeric proteins (6, 55, 58). Masuda and Kawata (36) reported that a major protein component was observed in the outer sheath. Weinberg and Holt (58) reported that oligomeric proteins were observed in the outer sheath of T. denticola and were not dissociated with sarcosine extraction. Furthermore, Fenno et al. (11) determined the DNA sequence of the gene expressing Msp, the major protein of the outer sheath, and stated that Msp in its native form is an oligomeric protein of 150 to 200 kDa. Nevertheless, in the case of recombinant Msp, an oligomeric form could not be detected. Therefore, it is possible that the proteases in the outer sheath play a role in organizing high-molecular-mass oligomeric proteins. As a result, in the present study, by use of a defective strain of T. denticola that did not express the dentilisin following site-specific mutagenesis, it was possible to assess the role of dentilisin in the pathogenicity of this microorganism and the conversion of Msp into its normal oligomeric form in the outer sheath.
When the dentilisin-deficient mutant, K1 was compared with the wild-type strain by means of SDS-PAGE, it was found that a high-molecular-mass oligomeric protein was present in the wild-type strain in unboiled samples but that in the K1 mutant there was a marked decrease in the size of this complex. This finding suggests that dentilisin plays a role in the formation of the oligomeric complex. Dentilisin may serve as a component of the oligomeric complex or may contribute to the formation of the oligomeric complex via processing of another protein. On the basis of the results of zymography, which revealed no protease activity associated with the high-molecular-mass oligomeric protein complex, it is unlikely that dentilisin is a component of this complex with Msp. Immunoblotting with a polyclonal antibody against strain ATCC 35405 whole cells showed that there was less high-molecular-mass oligomeric protein in the unboiled samples of the mutant strain than in those of the wild-type strain. However, in the boiled samples, the size and amount of Msp were the same in both strains. This result suggests that Msp does not require processing by dentilisin. The 83-kDa protein was weakly observed in the PMSF-treated wild-type strain. It is possible that in the dentilisin-deficient mutant, Msp associated with an 83-kDa complex but not with higher-molecular-mass complexes. Fenno et al. (12) suggested that the amino acid sequence of Msp resembled those of bacterial porins and that the protein exhibited properties consistent with this proposal (37). It is possible that in the K1 mutant, part of Msp associated with the 83-kDa complex because of the formation of porins (high-molecular-weight oligomers) or because of autodegradation. This suggestion may indicate that dentilisin assists in the formation of high-molecular-mass oligomeric proteins. Sela et al. (47) analyzed the lipoproteins of T. denticola and reported that Msp is also a lipoprotein. Cox et al. (7) proposed a model in which the N-terminal portions of the mature polypeptides of Treponema pallidum are inserted into lipid sites of the cytoplasmic membrane. We propose that dentilisin assists in the formation of high-molecular-mass oligomeric proteins by a mechanism which has not been determined.
The ability of bacteria to adhere to cell surfaces is a fundamental aspect of their pathogenicity. Surface hydrophobicity has been reported to contribute to bacterial adhesion (15). In the present study, we demonstrated that there was a distinct decrease in cell surface hydrophobicity in the dentilisin-deficient mutant. The result of immunoblotting with an antiserum against T. denticola ATCC 35405 whole cells indicated that the oligomeric protein band was observed from 180 to 230 kDa. This change may have been caused by the loss of dentilisin activity. The change in hydrophobicity was paralleled by the observed decreased concentration of the high-molecular-mass oligomeric protein band in unboiled samples in mutant K1 and the increased concentration of additional bands. These results suggest that the hydrophobicity of T. denticola is influenced by the high-molecular-mass proteins of the outer sheath.
Some microorganisms in the oral cavity undergo coaggregation reactions and may colonize the oral cavity following interaction with early-colonizing bacteria. Some of these intraoral bacteria possess cryptic receptors (cryptitopes) (16) whose function is related to their colonization. T. denticola has been reported to coaggregate with F. nucleatum and P. gingivalis (17, 25, 44). The strong coaggregation of P. gingivalis with T. denticola was given a score of 3, in contrast to the data reported earlier by Grenier (17). This difference may result from the use of higher cell numbers in the present study than in the earlier study. Kolenbrander et al. (25) reported visible coaggregation between F. nucleatum and T. denticola ATCC 35405. The reaction varied depending upon the strain of F. nucleatum examined. An increase in aggregation may reflect differences in the expression of surface receptors between the strains. It is possible that dentilisin digests the surface proteins of the microorganisms and exposes cryptitopes. However, our observations indicated that coaggregation was strengthened in a dentilisin-deficient strain of T. denticola. Grenier (17), on analyzing the coaggregation mechanisms of T. denticola and P. gingivalis, demonstrated that chymotrypsin and trypsin treatments reduced the degree of coaggregation and suggested that this result was due to the degradation of receptor protein. In our study, coaggregation activity was not decreased by PMSF treatment, indicating that dentilisin did not affect coaggregation activity. Coaggregation activity did not decrease in the dentilisin-deficient mutant, although the hydrophobicity of the cells in the mutant decreased. This result indicated that hydrophobicity did not play a role in coaggregation between T. denticola and F. nucleatum or P. gingivalis. The decrease in the hydrophobicity of the K1 mutant may reflect the change in surface structure. This change may increase the exposure of the receptor protein for its coaggregation reaction with P. gingivalis and F. nucleatum.
It has been suggested that surface structures such as capsules, the S-layer, and the outer sheath contribute to the pathogenicity of microorganisms (10, 11, 14, 56). In this regard, Msp, the major protein of the outer sheath of T. denticola, exhibits fibronectin adhesiveness (11), cytopathogenicity for host cells (37), and pore formation in artificial membrane systems (9). On the other hand, dentilisin exhibited a cytopathic effect for epithelial cells. We demonstrated that when mice were inoculated s.c., strain ATCC 35405 produced larger lesions than did dentilisin-deficient mutant K1. No significant differences could be detected in the structures of the outer sheaths by electron microscopy (data not shown), while both the cellular protein profile and the cell surface hydrophobicity of the mutant were altered. The ultrastructure of the outer sheath of mutant K1 was not visibly different from that of the wild type when both were viewed by transmission electron microscopy. The results from SDS-PAGE analysis revealed that the concentration of the high-molecular-mass oligomeric protein complex decreased in the mutant but was still detectable. Therefore, the decreased level of the complex may still have been sufficient to maintain the structure of the cell surface in mutant K1. In addition, immunoblot analysis indicated that Msp was not degraded by dentilisin.
The major components of the outer sheath appear to be lipid material and Msp (58). Therefore, these latter components may be sufficient to maintain the outer surface of spirochetes, at least as far as can be detected by such analysis. The decreased virulence displayed by mutant K1 may directly result from a decrease in dentilisin activity, from alterations in the expression of the high-molecular-mass oligomeric complex containing Msp in the outer sheath of T. denticola, or from a decreased growth rate in vivo. As the results indicated that the growth rate of the mutant was the same as that of the wild type, protease activity and Msp are major factors in the change in pathogenicity. The multiple defects of the dentilisin-deficient mutant make it difficult to clearly explain the molecular bases for virulence attenuation. However, the isolation of a dentilisin-deficient mutant now makes it possible to genetically analyze the role of proteases in oral spirochete pathogenicity.
ACKNOWLEDGMENTS
We thank Y. Nakano for providing pMCL191. We thank M. Kitamura for assistance in the statistical analysis. We thank T. Umemoto for supplying anti-Msp serum.
This study was partially supported by Oral Health Science Center grant 961A02 from Tokyo Dental College and grant 06671833 from the Ministry of Education, Science, Culture and Sport of Japan.
REFERENCES
- 1.Arakawa S, Kuramitsu H K. Cloning and sequence analysis of a chymotrypsinlike protease from Treponema denticola. Infect Immun. 1994;62:3424–3433. doi: 10.1128/iai.62.8.3424-3433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 3.Chan E C S, Soiboo R, Keng T, Psarra N, Hurley R, Cheng S L, Iugovaz I. Treponema denticola (ex Brumpt 1925) sp. nov. nom. rev. and identification of new spirochete isolates from periodontal pockets. Int J Syst Bacteriol. 1993;43:196–203. doi: 10.1099/00207713-43-2-196. [DOI] [PubMed] [Google Scholar]
- 4.Choi B-K, Nattermann H, Grund S, Haider W, Gröbel U B. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int J Syst Bacteriol. 1997;47:175–181. doi: 10.1099/00207713-47-1-175. [DOI] [PubMed] [Google Scholar]
- 5.Cisar J O, Kolenbrander P E, McIntire F C. Specificity of coaggregation reaction between human oral streptococcus and strains of Actinomyces viscosus and Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cockayne A, Sanger R, Ivic A, Strugnell R A, MacDougall J H, Russell R R, Penn C W. Antigenic and structural analysis of Treponema denticola. J Gen Microbiol. 1989;135:3209–3218. doi: 10.1099/00221287-135-12-3209. [DOI] [PubMed] [Google Scholar]
- 7.Cox D L, Chang P, McDowall A W, Radolf J D. The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect Immun. 1992;60:1076–1083. doi: 10.1128/iai.60.3.1076-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson J R, Ellen R P. Tip-oriented adherence of Treponema denticola to fibronectin. Infect Immun. 1990;58:3924–3928. doi: 10.1128/iai.58.12.3924-3928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egli C, Leung W K, Mäller K-H, Hancock R E W, McBride B C. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect Immun. 1993;61:1694–1699. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evenberg D, Lugtenberg B. Cell surface of the fish pathogenic bacterium Aeromonas salmonicida. II. Purification and characterization of a major cell envelope protein related to autoagglutination, adhesion, and virulence. Biochim Biophys Acta. 1982;684:249–254. doi: 10.1016/0005-2736(82)90013-x. [DOI] [PubMed] [Google Scholar]
- 11.Fenno J C, Muller K H, McBride B C. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–2497. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenno J C, Wong G W, Hannam P M, Muller K H, Leung W K, McBride B C. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol. 1997;179:1082–1089. doi: 10.1128/jb.179.4.1082-1089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto S, Takade A, Amako K, Blaser M J. Correlation between molecular size of the surface array protein and morphology and antigenicity of the Campylobacter fetus S-layer. Infect Immun. 1991;59:2017–2022. doi: 10.1128/iai.59.6.2017-2022.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons R J, Etherden I, Skobe S. Association of fimbriae with the hydrophobicity of Streptococcus sanguis FC-1 and adherence to salivary pellicles. Infect Immun. 1983;41:414–417. doi: 10.1128/iai.41.1.414-417.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbons R J, Hay D I, Childs W C, Davis G. Role of cryptic receptors (cryptitopes) in bacterial adhesion to oral surfaces. Arch Oral Biol. 1990;35:107S–224S. doi: 10.1016/0003-9969(90)90139-2. [DOI] [PubMed] [Google Scholar]
- 17.Grenier D. Demonstration of a bimodal coaggregation reaction between Porphyromonas gingivalis and Treponema denticola. Oral Microbiol Immunol. 1992;7:280–284. doi: 10.1111/j.1399-302x.1992.tb00589.x. [DOI] [PubMed] [Google Scholar]
- 18.Grenier D, Uitto V J, McBride B C. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect Immun. 1990;58:347–351. doi: 10.1128/iai.58.2.347-351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haapasalo M, Singh U, McBride B C, Uitto V J. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect Immun. 1991;59:4230–4237. doi: 10.1128/iai.59.11.4230-4237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishihara K, Takazoe I, Okuda K. Immunomodulating activity of oral Treponema strains on proliferation of mouse lymphocyte. Bull Tokyo Dent Coll. 1992;33:45–50. [Google Scholar]
- 22.Ishihara K, Kuramitsu H K. Cloning and expression of a neutral phosphatase gene from Treponema denticola. Infect Immun. 1995;63:1147–1152. doi: 10.1128/iai.63.4.1147-1152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishihara K, Miura T, Kuramitsu H K, Okuda K. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin) Infect Immun. 1996;64:5178–5186. doi: 10.1128/iai.64.12.5178-5186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kesavalu L, Walker S G, Holt S C, Crawley R R, Ebersole J L. Virulence characteristics of oral treponemes in a murine model. Infect Immun. 1997;65:5096–5102. doi: 10.1128/iai.65.12.5096-5102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolenbrander P E, Parrish K D, Andersen R N, Greenberg E P. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intergeneric coaggregation among Fusobacterium spp. Infect Immun. 1995;63:4584–4588. doi: 10.1128/iai.63.12.4584-4588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Leung W K, Haapasalo M, Uitto V-J, Hannam P M, McBride B C. The surface proteinase of Treponema denticola may mediate attachment of the bacteria to epithelial cells. Anaerobe. 1996;2:39–46. [Google Scholar]
- 28.Li H, Ruby J, Charon N, Kuramitsu H K. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loesche W J. Periodontal disease and the treponemes. In: Johnson R C, editor. The biology of parasitic spirochetes. New York, N.Y: Academic Press, Inc.; 1976. pp. 261–275. [Google Scholar]
- 30.Loesche W J, Syed S A, Schmidt E, Morrison E C. Bacterial profiles of subgingival plaques in periodontitis. J Periodontol. 1985;56:447–456. doi: 10.1902/jop.1985.56.8.447. [DOI] [PubMed] [Google Scholar]
- 31.MacDougall J H, Beighton D, Russell R R B. Cloning and expression of protease genes from Treponema denticola in Escherichia coli. Oral Microbiol Immunol. 1991;6:270–274. doi: 10.1111/j.1399-302x.1991.tb00490.x. [DOI] [PubMed] [Google Scholar]
- 32.Mäkinen K K, Chen C-Y, Mäkinen P-L. Proline iminopeptidase from the outer cell envelope of the human oral spirochete Treponema denticola ATCC 35405. Infect Immun. 1996;64:702–708. doi: 10.1128/iai.64.3.702-708.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mäkinen K K, Mäkinen P-L, Syed S A. Purification and substrate specificity of an endopeptidase from human oral spirochete Treponema denticola ATCC 35405, active on furylacryloyl-Leu-Gly-Pro-Ala and bradykinin. J Biol Chem. 1992;267:14285–14293. [PubMed] [Google Scholar]
- 34.Mäkinen P-L, Mäkinen K K, Syed S. Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect Immun. 1995;63:3567–3575. doi: 10.1128/iai.63.9.3567-3575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mäkinen P-L, Mäkinen K K, Syed S A. An endo-acting proline-specific oligopeptidase from Treponema denticola ATCC 35405: evidence of hydrolysis of human bioactive peptides. Infect Immun. 1994;62:4938–4947. doi: 10.1128/iai.62.11.4938-4947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda K, Kawata T. Isolation, properties, and reassembly of outer sheath carrying a polygonal array from an oral treponeme. J Bacteriol. 1982;150:1405–1413. doi: 10.1128/jb.150.3.1405-1413.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathers D A, Leung W K, Fenno J C, Hong Y, McBride B C. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun. 1996;64:2904–2910. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikx F H, Jacobs F, Satumalay C. Cell-bound peptidase activities of Treponema denticola ATCC 33520 in continuous culture. J Gen Microbiol. 1992;138:1837–1842. doi: 10.1099/00221287-138-9-1837. [DOI] [PubMed] [Google Scholar]
- 39.Naito Y, Tohda H, Okuda K, Takazoe I. Adherence and hydrophobicity of invasive and noninvasive strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1993;8:195–202. doi: 10.1111/j.1399-302x.1993.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 40.Nakano J Y, Kuramitsu H K. Mechanism of Streptococcus mutans glucosyltransferases: hybrid enzyme analysis. J Bacteriol. 1992;174:5639–5646. doi: 10.1128/jb.174.17.5639-5646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano, Y. Personal communication.
- 42.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine protease (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohta K, Mäkinen K K, Loesche W J. Purification and characterization of an enzyme from Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun. 1986;53:213–220. doi: 10.1128/iai.53.1.213-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onagawa M, Ishihara I, Okuda K. Coaggregation between Porphyromonas gingivalis and Treponema denticola. Bull Tokyo Dent Coll. 1994;35:171–181. [PubMed] [Google Scholar]
- 45.Rosen G, Nator R, Kutner S, Sela M N. Characterization of fibrinolytic activities of Treponema denticola. Infect Immun. 1994;62:1749–1754. doi: 10.1128/iai.62.5.1749-1754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg M, Gutnik D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. [Google Scholar]
- 47.Sela M N, Bolotin A, Noar R, Weinberg A, Rosen G. Lipoproteins of Treponema denticola: their effect on human polymorphonuclear neutrophils. J Periodontal Res. 1997;32:455–466. doi: 10.1111/j.1600-0765.1997.tb00558.x. [DOI] [PubMed] [Google Scholar]
- 48.Shenker B J, Listgarten M A, Taichman N S. Suppression of human lymphocyte responses by oral spirochetes: a monocyte-dependent phenomenon. J Immunol. 1984;132:2039–2045. [PubMed] [Google Scholar]
- 49.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 50.Tokuda M, Duncan M, Cho M-I, Kuramitsu H K. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uitto V-J, Pan Y-M, Leung W K, Larjava H, Ellen R P, Finlay B B, McBride B C. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect Immun. 1995;63:3401–3410. doi: 10.1128/iai.63.9.3401-3410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uitto V J, Grenier D, Chan E C, McBride B C. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect Immun. 1988;56:2717–2722. doi: 10.1128/iai.56.10.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umemoto T, Namikawa I, Asai S. A major antigen on the outer envelope of a human oral spirochete, Treponema denticola. Infect Immun. 1989;57:2470–2474. doi: 10.1128/iai.57.8.2470-2474.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker S G, Ebersole J L, Holt S C. Identification, isolation, and characterization of the 42-kilodalton major outer membrane protein (MompA) from Treponema pectinovorum ATCC 33768. J Bacteriol. 1997;179:6441–6447. doi: 10.1128/jb.179.20.6441-6447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang E, Garcia M M, Blake M S, Pei Z, Blaser M J. Shift in S-layer protein expression responsible for antigenic variation in Campylobacter fetus. J Bacteriol. 1993;175:4979–4984. doi: 10.1128/jb.175.16.4979-4984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weidner M-F, Grenier D, Mayrand D. Proteolytic artifact in SDS-PAGE analysis of selected periodontal pathogens. Oral Microbiol Immunol. 1996;11:103–108. doi: 10.1111/j.1399-302x.1996.tb00343.x. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg A, Holt S C. Chemical and biological activities of a 64-kilodalton outer sheath protein from Treponema denticola strains. J Bacteriol. 1991;173:6935–6947. doi: 10.1128/jb.173.21.6935-6947.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]