Abstract
Schizosaccharomyces pombe cells divide by medial fission through contraction of an actomyosin ring and deposition of a multilayered division septum that must be cleaved to release the two daughter cells. Here we describe the identification of seven genes (adg1+, adg2+, adg3+, cfh4+, agn1+, eng1+, and mid2+) whose expression is induced by the transcription factor Ace2p. The expression of all of these genes varied during the cell cycle, maximum transcription being observed during septation. At least three of these proteins (Eng1p, Agn1p, and Cfh4p) localize to a ring-like structure that surrounds the septum region during cell separation. Deletion of the previously uncharacterized genes was not lethal to the cells, but produced defects or delays in cell separation to different extents. Electron microscopic observation of mutant cells indicated that the most severe defect is found in eng1Δ agn1Δ cells, lacking the Eng1p endo-β-1,3-glucanase and the Agn1p endo-α-glucanase. The phenotype of this mutant closely resembled that of ace2Δ mutants, forming branched chains of cells. This suggests that these two proteins are the main activities required for cell separation to be completed.
INTRODUCTION
Cytokinesis is the final stage of the cell cycle during which the daughter cells separate physically and become two independent entities. In a variety of organisms, the force necessary for cell cleavage is provided by an actomyosin-based contractile ring, coupled to the synthesis of new membrane, which is inserted at the division site (reviewed by Hales et al., 1999; Robinson and Spudich, 2000; Guertin et al., 2002). In yeast and fungi, the presence of a rigid cell wall that completely surrounds the cell gives rise to an additional step during cytokinesis, because a separation septum needs to be assembled to avoid cell lysis during cytokinesis.
Schizosaccharomyces pombe is an excellent organism to study cytokinesis because it divides by medial fission after assembly and contraction of an actomyosin ring, followed by cleavage of the septum (for a review, see Feierbach and Chang, 2001). Cytokinesis and cell division are brought about by the action of the actomyosin ring, whose constriction is perfectly coordinated with the synthesis of the primary septum. The isolation and characterization of cytokinesis mutants has allowed the identification of the many proteins involved in the different steps of this process. Establishment of the division site at the center of the cell begins early on the cell cycle— during the onset of mitosis—with the assembly of the contractile ring at the cell cortex adjacent to the nucleus (for a review, see Chang, 2001). The mid1+, plo1+, and pom1+ genes are required for the division plane to be established and for correct positioning of the actomyosin ring (Chang and Nurse, 1996; Sohrmann et al., 1996; Bähler and Pringle, 1998; Bähler et al., 1998a; Mulvihill et al., 1999; Bähler and Nurse, 2001). Coordination of ring contraction and the nuclear cycle requires a network of regulatory proteins that are collectively referred to as the Septation Initiation Network (SIN). These proteins also control the formation of the primary septum during constriction of the actomyosin ring. Genetic studies have indicated that activation of the SIN pathway might regulate Cps1p, a β-1,3-glucan synthase subunit essential for the assembly of the division septum (Le Goff et al., 1999; Liu et al., 2000). This septum has a three-layer structure (Johnson et al., 1982), with a central primary septum (mainly composed of linear β-1,3-glucan) surrounded on both sides by two secondary septa (composed of β-1,6-branched β-1,3-glucan, and β-1,6-glucan; Humbel et al., 2001).
Cell separation requires the dissolution of the primary septum for the daughter cells to become two independent entities. On completion of mitosis, the primary septum undergoes rapid degradation, accompanied by local erosion of the adjacent regions of the cell wall. During the last few years, the isolation of mutants affected to different extents in cell separation has provided some insight into the mechanistic details of this process. Mutants with a cell-cell separation phenotype include mutations in components of the exocyst complex (sec6+, sec8+, sec10+, and exo70+), an anillin homologue (mid2+), septins (spn3+ and spn4+), an endo-α-1,3-glucanase (agn1+), an endo-β-1,3-glucanase (eng1+), calcineurin (ppb1+), a MAPK (pmk1+), a MAPK phosphatase (pmp1+), two transcription factors (sep1+ and ace2+), and subunits of the mediator complex (sep10+, sep11+ and sep15+; Sipiczki et al., 1993; Yoshida et al., 1994; Toda et al., 1996; Ribár et al., 1997; Sugiura et al., 1998; Zilahi et al., 2000a; Szilagyi et al., 2002; Wang et al., 2002; Berlin et al., 2003; Martín-Cuadrado et al., 2003; Tasto et al., 2003; Dekker et al., 2004). The exocyst is an octameric protein complex present in many organisms and is involved in tethering vesicles to specific sites on the plasma membrane (Wang et al., 2002). Based on the fact that mutants in different subunits show a defect in cell separation, it was proposed that this complex might be involved in the delivery of the hydrolytic proteins that are important for cell cleavage to the septum. One such enzyme could be the product of the eng1+ gene, which codes for a protein with endo-β-1,3-glucanase activity, which has been shown to be the major enzymatic activity involved in the dissolution of the primary septum (Martín-Cuadrado et al., 2003). In addition, it has recently been reported that the endo-α-1,3-glucanase Agn1p is also required for cell separation (Dekker et al., 2004). The defect in cell separation produced by eng1+ deletion is not very severe but results in the formation of groups of four connected cells. A similar phenotype has also been reported for mutants lacking mid2+, an anillin homologue that is required for the assembly and stabilization of the septin ring during cytokinesis and for several septin mutants (Berlin et al., 2003; Tasto et al., 2003). It has been suggested that Mid2p and septins may be required for proper exocytosis of the enzymes involved in septum cleavage. Mutation in two transcription factors also interferes with cell separation. sep1+ encodes a transcription factor highly homologous to the HNF-3/forkhead family present in higher eukaryotic cells and also in other microorganisms (Ribár et al., 1997). ace2+, a homologue of the Saccharomyces cerevisiae Ace2p, codes for a transcription factor of the C2H2 zinc-finger family (Martín-Cuadrado et al., 2003). Deletion of each of these genes results in a severe cell separation defect, hyphal growth, and branching being observed. It has been shown that sep1+ controls the expression of mid2+, whereas the transcription of eng1+ is dependent on ace2+ (Martín-Cuadrado et al., 2003; Tasto et al., 2003). The fact that the phenotype of sep1Δ or ace2Δ mutants is more severe than that of cells lacking eng1+ or mid2+ suggests that these transcription factors also control the expression of other genes involved in cell-cell separation.
Here we have identified by microarray analysis a group of genes whose expression is dependent on the transcription factor Ace2p and hence is named AceII-dependent genes (adg). Northern analyses revealed that the expression of the adg genes is dependent on Ace2p and that ace2+ transcription requires Sep1p. Mutants lacking the identified genes showed modest cell separation defects, but a double mutant devoid of the Eng1p endo-β-1,3-glucanase and the Agn1p endo-1,3-α-glucanase showed a phenotype very similar to that of ace2Δ mutants. According to these observations, these two enzymes are likely to be the major enzymatic activities involved in the dissolution of the septum during cell separation.
MATERIALS AND METHODS
Strains, Growth Conditions, and Genetic Manipulations
The S. pombe strains used in this study are listed in Table 1. Yeast cells were grown on YES medium or minimal media (EMM) with appropriate supplements (Moreno et al., 1991). Yeast transformations were performed with the lithium acetate method (Ito et al., 1983). For overexpression experiments using the nmt1+ promoter, cells were grown in EMM containing 15 μM thiamine up to the logarithmic phase. Then, the cells were harvested, washed three times with EMM, and inoculated in fresh medium (without thiamine) at an OD595 = 0.05.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| A131 | sep1::ura4+ura4-D18 leu 1-32 h+ | M. Sipizcki |
| h20 | leu1-32 h- | S. Moreno |
| h123 | ura4-D18 h- | S. Moreno |
| KGY246 | ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY2061 | cdc7-24 ade6-M210 ura4-D18 leu1-32 h- | K. Gould |
| KGY4386 | cfh4-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4387 | eng1-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4390 | adg2-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4397 | adg1-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4398 | agn1-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4399 | adg3-GFP ade6-M210 ura4-D18 leu1-32 h- | This study |
| KGY4612 | adg1-GFP ace2::KanR ura4-D18 ade- leu 1-32 h+ | This study |
| KGY4614 | agn1-GFP ace2::KanR ura4-D18 ade- leu 1-32 h+ | This study |
| KGY4616 | adg2-GFP ace2::KanR ura4-D18 ade- leu 1-32 h- | This study |
| KGY4618 | eng1-GFP ace2::KanR ura4-D18 ade- leu 1-32 h+ | This study |
| KGY4620 | cfh4-GFP ace2::KanR ura4-D18 ade- leu 1-32 h- | This study |
| KGY4621 | adg3-GFP ace2::KanR ura4-D18 ade- leu 1-32 h+ | This study |
| LE25 | ace2::kanMX4 ura4-D18 h- | Lab Stock |
| PPG148 | ura4-D18 cdc25–22 h- | P. Pérez |
| YAB14 | ura4-D18 eng1::kanMX4 h- | Lab Stock |
| YAB53 | agn1::ura4+ura4-D18 h- | This study |
| YAB54 | eng1::kanMX4 agn1::ura4+ura4-D18 h–[ρ] | This study |
| YMAN2 | adg1::kanMX4 ura4-D18 h- | This study |
| YMAN4 | adg3::kanMX4 ura4-D18 h- | This study |
| YMAN5 | adg2::kanMX4 ura4-D18 h- | This study |
| YMAN7 | adg1::ura4+adg3::kanMX4 ura4-D18 h- | This study |
| YMAN9 | eng1::kanMX4 adg1::ura4+ura4-D18 h- | This study |
| YMAN11 | eng1::kanMX4 adg3::ura4+ura4-D18 h- | This study |
| YMAN19 | eng1::kanMX4 adg2::ura4+ura4-D18 h- | This study |
| YMAN25 | adg1::kanMX4 adg2::ura4+ura4-D18 h- | This study |
| YMAN26 | adg2::ura4+adg3::kanMX4 ura4-D18 h- | This study |
| YMAN28 | sep1::ura4+ura4-D18 leu 1-32 h+ (pREP3X-ace2) | This study |
| YMAN29 | leu1-32 h- (pREP3X-ace2) | This study |
| YMAN30 | ace2::kanMX4 ura4-D18 leu 1-32 h- | This study |
| YMAN31 | ace2::kanMX4 ura4-D18 leu 1-32h- (pREP3 × -ace2) | This study |
Synchronization of strains carrying the thermosensitive cdc25-22 mutation was achieved by growing the cells at the permissive temperature (25°C) to early log phase (OD595 = 0.5) and then shifting the cultures to 37°C for 4 h. Cells were released from arrest by transfer to 25°C, and samples were taken every 20 min. Cells were also synchronized by centrifugal elutriation in a Beckman JE 5.0 elutriator rotor (Fullerton, CA). Cells synchronized in early G2 were collected and inoculated into YE medium at 32°C. Synchrony was monitored by DAPI staining and estimation of the percentage of binucleate cells.
Construction of Null Mutants and Plasmids
The entire coding sequences of adg1+ (SPAPJ760.03c), adg2+ (SPAC19G12.16c), and adg3+ (SPCC74.07c) were deleted by replacing the coding sequences with the kanMX4 cassette or the ura4+ gene. The deletion cassettes were constructed using the recombinant PCR approach described by Wach et al. (1996). For this purpose, DNA fragments of 300–500 base pairs corresponding to the 5′ and 3′ flanking regions of each gene were PCR-amplified using specific oligonucleotide pairs. The resulting fragments were then fused by recombinant PCR to the kanMX4 cassette (which confers resistance to the antibiotic G418) or to the ura4+ gene. The oligonucleotide sequences used are available upon request.
Plasmid pA16, containing the ace2+ ORF without promoter, was constructed by PCR amplification of the coding sequence with oligonucleotides that generated XhoI and SacI sites at the ends and then cloning the amplified fragment into the corresponding sites of vector pAU-KS. The wild-type ace2+ promoter was amplified with oligonucleotides that generated KpnI and XhoI sites at the ends and was cloned into the corresponding sites of plasmid pA16, yielding pB1. Deletion of the five copies of the TGTTTAC sequence was achieved by successive rounds of PCR, cloning the final product between the KpnI and XhoI sites of plasmid pA16, to generate plasmid pC4. The nmt1+ terminator was introduced at the KpnI site of the three plasmids.
Epitope Tagging
The eng1+, cfh4+, agn1+, adg1+, adg2+, and adg3+ genes were tagged at their chromosomal loci at their 3′ ends with sequences encoding the green fluorescent protein (GFP) by a PCR-mediated strategy as described previously (Bähler et al., 1998b). Proper integration of these epitope cassettes was confirmed by PCR and immunoblotting.
Microscopy Techniques
Microscopy was performed with a Leica DMRXA microscope (Deerfield, IL) equipped for Nomarski optics and epifluorescence and photographed with a Photometrics Sensys CCD camera (Tucson, AZ). For the imaging of the GFP-tagged strains, a Carl Zeiss MicroImaging, Axiovert II inverted microscope (Thornwood, NY) equipped with an UltraView LCI real-time scanning head confocal (PerkinElmer, Norwalk, CT) and a 488-nm argon ion laser (for GFP excitation) was used. Images were captured on an Orca-ER CCD camera (Hamamatsu, Bridgewater, NJ) using Ultra-View software (PerkinElmer) and were then processed using Volocity 2.0 software (Improvision, Lexington, MA). Visualization of nuclei was accomplished by staining cells fixed in 70% ethanol, washed two times in phosphate-buffered saline (PBS), and resuspended in PBS with DAPI (4′,6′-diamidino-2-phenylindole).
For transmission electron microscopy (TEM), the cells were stained with potassium permanganate according to the protocol described by Johnson et al. (1982). Electron photomicrographs were taken with a Jeol Jem-1010 electron microscope (Peabody, MA).
Protein Methods
Total cell extracts of S. pombe were prepared in SDS-lysis buffer (Gould et al., 1991) and immunoprecipitations were carried out using rabbit polyclonal anti-GFP serum. For immunoblotting, proteins were resolved by SDS-PAGE on 4–12% gradient gels. Protein transfer, blotting, and ECL detection were performed using standard procedures.
RNA Isolation, Northern Blot, and Microarray Analysis
Cells (1.3 × 109) were collected at different time intervals after release from the restrictive temperature (37°C) or from different mutant strains, and total RNA was prepared using the method described by Percival-Smith and Segall (1984). For Northern blot analysis, 12.5 μg of RNA was used. The DNA probes used to detect the different transcripts were DNA fragments obtained by PCR with specific oligonucleotides. To analyze gene expression in the cdc7 mutant, RNA was isolated by hot acid phenol extraction, as described previously (Burns et al., 1999). RNA, 20 μg, were transferred to a Duralon-UV membrane and probed with 32P-labeled probes corresponding to coding regions of eng1+, cdc22+, and his3+. The blots were exposed to Phosphor-Imager screens and visualized with ImageQuant 5.2 on an Amersham Biosciences Typhoon 9200 scanner (Piscataway, NJ).
For microarray analysis, mRNA was purified from wild-type or ace2Δ mutants during exponential growth. Hybridization to glass slides containing 4976 S. pombe ORFs was performed by Eurogentec (Serain, Belgium).
RESULTS
Identification of Genes Regulated by the Ace2p Transcription Factor
To identify the putative genes regulated by the Ace2p transcription factor in S. pombe, mRNA was purified from exponentially growing wild-type and ace2Δ mutants and used to hybridize a DNA microarray containing 4976 S. pombe ORFs (Eurogentec). The results, shown in Table 2, indicated the existence of seven genes whose expression was around two-fold lower in ace2Δ mutants. These included eng1+, which encodes an endo-β-1,3-glucanase required for dissolution of the primary septum and that has been previously shown to be regulated by this transcription factor (Martín-Cuadrado et al., 2003), agn1+, which has recently been identified as an endo-α-1,3-glucanase involved in cell separation (Dekker et al., 2004) and mid2+, an anillin homologue required for septin ring assembly (Berlin et al., 2003; Tasto et al., 2003). Further analysis of the role of Ace2p in mid2+ regulation will be described elsewhere (Petit and Gould, unpublished results). The other four genes have not been characterized previously, although one of them (SPBC3E7.12c, cfh4+) shows sequence similarity to the S. cerevisiae CHS4 gene (which encodes a regulatory subunit of the chitin synthase III). The other three genes (SPAPJ760.03c, SPAC19G12.16c, and SPCC74.07c) code for unknown proteins, although all three have the characteristic of secreted proteins, because they contain a putative signal sequence at the N-terminus, they are Ser-Thr rich proteins, and two of them (SPAPJ760.03c and SPAC19G12.16c) contain a predicted GPI-modification site (De Groot et al., 2003). For a complete description of the structure of the proteins and alignments to closely related proteins, see Supplementary Materials. Interestingly, analysis of the promoter region of these genes showed that they contain two or three copies of the hexanucleotide CCAGCC (separated by 100–200 nucleotides) in the promoter region. This sequence has been described as the binding site for S. cerevisiae Ace2p (Dohrmann et al., 1992; McBride et al., 1999), which suggests that they could also be targets for this transcription factor in S. pombe. Accordingly, the three unknown genes were named adg1+ (SPAPJ760.03c), adg2+ (SPAC19G12.16c), and adg3+ (SPCC74.07c), which stands for Ace2-dependent genes.
Table 2.
Genes down-regulated in ace2 mutants
| ORF | Gene name | Descriptiona |
|---|---|---|
| SPAPJ760.03c | adg1+ | Ser/Thr rich protein; predicted GPI anchored protein |
| SPCC74.07c | adg3+ | Serine-rich protein; SUN4 family; predicted β-glucosidase |
| SPBC3E7.12c | cfh4+ | Predicted chitin synthase regulatory factor; SEL1 repeat protein |
| SPAC14C4.09 | agn1+ | Putative α-glucanase |
| SPAC821.09 | eng1+ | Endo-β-1,3-glucanase required for dissolution of the primary septum |
| SPAC19G12.16c | adg2+ | Serine/threonine-rich protein; predicted GPI anchored protein |
| SPAPYUG7.03c | mid2+ | Pleckstrin homology domain; involved in cytokinesis and septation |
According to GeneDB (http://www.genedb.org/genedb/pombe)
Expression of the Putative ace2+ Targets Fluctuates during the Cell Cycle and Depends on Ace2p
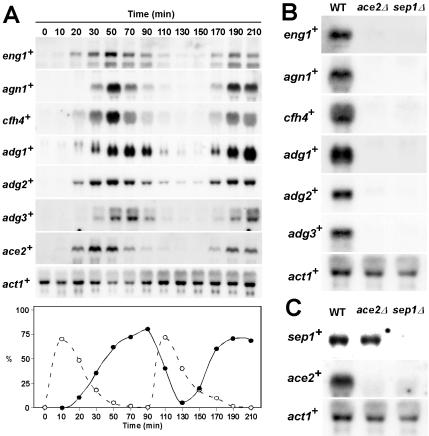
We have recently shown that expression of two of the identified genes, eng1+ and mid2+, fluctuates periodically during the cell cycle (Martín-Cuadrado et al., 2003; Tasto et al., 2003). To test whether the expression of the other putative targets showed a similar pattern, their expression was monitored by Northern blot analysis in cdc25-22 mutant cells that had been synchronized by arrest-release. When the level of the different mRNAs was examined, a periodic cell cycle variation was found for all of the genes tested (Figure 1A), with an expression pattern similar to that described for eng1+ and mid2+ (Martín-Cuadrado et al., 2003; Tasto et al., 2003). The maximum accumulation of mRNA was observed 20–40 min before the peak of septation, suggesting that their expression is coregulated during the cell cycle and that the products may exert their function during the last stages of the cell cycle, namely septum assembly or cell separation. The temporal pattern of transcription of ace2+ during the cell cycle was also analyzed in the same filters. We found that the expression of this gene also fluctuated periodically. As expected for a transcriptional activator controlling the expression of a group of genes, the peak of ace2+ transcription occurred slightly before that of the putative targets (30 min for ace2+ and 50–70 min for the other genes tested), suggesting that Ace2p might be required for correct expression of the putative targets.
Figure 1.
Transcription of Ace2-dependent genes. (A). Expression during the cell cycle. Synchrony was induced by arrest-release of a cdc25-22 mutant (PPG148), and samples were taken at the indicated time points (minutes) after the release for RNA extraction. RNA blots were probed with specific probes for eng1+, agn1+, cfh4+, adg1+, adg2+ adg3+, and ace2+, using act1+ as a loading control. The graph represents the anaphase index (○) or septation index (•) at each time point. In this experiment, the peak of septum formation occurred at 70–90 min. The anaphase index was determined by counting the percentage of anaphase cells (cells with two nuclei and without a septum) after DAPI staining. The septation index was determined by counting the percentage of cells with septum after calcofluor staining. (B) Expression of target genes in ace2Δ and sep1Δ strains. RNA was extracted from wild-type (h123), ace2Δ (LE25), or sep1Δ (A131) strains during exponential growth. After transfer to nitrocellulose membranes, RNA blots were sequentially hybridized with specific probes (prepared by PCR amplification of specific DNA fragments) for eng1+, agn1+, cfh4+, adg1+, adg2+, and adg3+, using act1+ as loading control. (C) Expression of ace2+ and sep1+ in ace2Δ and sep1Δ strains. RNA was extracted from wild-type (h123), ace2Δ (LE25), or sep1Δ (A131) strains during exponential growth. After transfer to nitrocellulose membranes, RNA blots were sequentially hybridized with specific probes (prepared by PCR amplification of specific DNA fragments) for sep1+ and ace2+, using act1+ as loading control.
The above observations suggested that the genes identified could be targets of Ace2p in S. pombe. We have previously shown that the expression of eng1+ is dependent on Ace2p (Martín-Cuadrado et al., 2003), but it has also been described that deletion of sep1+ strongly affects cell separation and results in a phenotype similar to that of cells lacking ace2+ (Sipiczki et al., 1993; Sipiczki and Bozsik, 2000; Martín-Cuadrado et al., 2003). To study the relationship between these two transcription factors and the putative target genes, Northern analyses were performed to compare their expression in wild-type, ace2Δ, and sep1Δ mutants. These analyses revealed that the expression of the seven genes identified in our microarray analysis was clearly reduced in both the ace2Δ and sep1Δ mutants (Figure 1B and unpublished data), suggesting that— directly or indirectly— both transcription factors are required for their expression.
A Transcriptional Cascade Controls the Last Steps of the Cell Cycle
In S. cerevisiae, a transcriptional cascade controls gene expression during the last stages of the cell cycle. Thus, the forkhead transcription factors Fkh1p and Fkh2p activate the expression of Ace2p, which in turn regulates the transcription of a group of genes that are specifically expressed in the daughter cell and are involved in cell separation (Zhu et al., 2000; Colman-Lerner et al., 2001; Simon et al., 2001; Baladrón et al., 2002). Because the expression of eng1+ and the other genes requires both the forkhead-like factor Sep1p and Ace2p, we decided to test whether a similar situation occurs in S. pombe cells. To this end, the expression of the two transcription factors was analyzed in wild-type and mutant cells lacking either of the two genes. Interestingly, sep1+ was transcribed at normal levels in ace2Δ mutants (Figure 1C), but ace2+ was not expressed in the sep1Δ deletion strain, suggesting that in fission yeast there is also a transcriptional cascade controlling gene expression during the last stages of the cell cycle, ace2+ transcription being dependent on Sep1p.
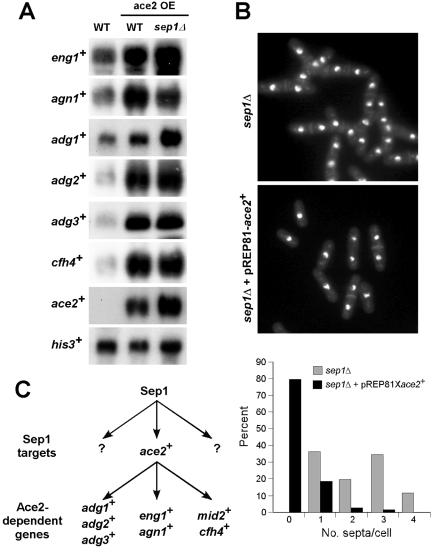
This result suggests that Ace2p is the most direct activator of gene expression for these seven genes and that the defect in expression observed in sep1Δ mutants could be indirect and due to a failure to activate the transcription of ace2+. One prediction of this hypothesis is that expression of ace2+ from an heterologous promoter should restore the expression of these genes in sep1Δ cells, and it should also complement the cell separation defect of this mutant. To test this idea, the expression of the target genes was analyzed in wild-type and sep1Δ cells carrying the ace2+ gene under the control of the nmt1+ promoter. When cells were grown in medium lacking thiamine to induce Ace2p production, an increase in transcript levels was detected in all the target genes in comparison with those found in wild-type cells and sep1Δ mutants (Figure 2A), suggesting that Ace2p is the most direct transcriptional activator of this group of genes. In addition, we also tested whether overexpression of ace2+ complements the separation defect of sep1Δ cells. Microscopic observation of sep1Δ cells in which ace2+ was expressed from a heterologous promoter revealed that the cell separation defect was almost completely suppressed in comparison with the hyphal growth seen for sep1Δ mutants (Figure 2B). In comparison with the sep1Δ mutant, in which most of the cell contain three or four septa, mild ace2+ overexpression suppressed this separation defect, most of the cells containing no septum (around 80%) or only a single septum (15%). Thus, these results are in good agreement with the model of a transcriptional cascade in the last steps of the S. pombe cell cycle.
Figure 2.
Relationship between Ace2p and Sep1. (A) Overexpression of ace2+ restores transcription in sep1Δ cells. RNA was extracted from wild-type (h123) or sep1Δ (A131) strains carrying pREP3X-ace2+ grown for 22 h in the absence of thiamine (ace2 OE). The wild-type strain carrying empty vector grown under the same conditions (first lane) was used to determine the endogenous transcript levels. After transfer to nitrocellulose membranes, RNA blots were sequentially hybridized with specific probes for eng1+, agn1+, adg1+, adg2+, adg3+, cfh4+, and ace2+, using his3+ as loading control. (B) Moderate ace2+ overexpression complements the cell separation defect of sep1Δ cells. Representative microscopic images of the sep1Δ mutant (top) or the sep1Δ mutant carrying pREP81X-ace2+ (middle) are shown. Quantitation of the percentage of septa in both cultures is shown in the bottom panel. Cells were stained with aniline blue and DAPI before counting. For each sample, 200 cells of were counted. (C) Model depicting transcriptional regulation during the last stages of the S. pombe cell cycle.
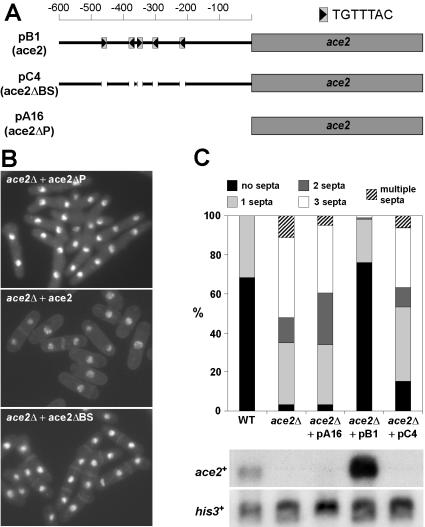
The Fkh1p and Fkh2p proteins recognize and bind the consensus sequence TGTTTAC in S. cerevisiae (Pic et al., 2000; Hollenhorst et al., 2001). Interestingly, analysis of the ace2+ promoter region revealed the presence of five copies of this sequence in a 300-base pairs region (Figure 3A), suggesting that they could be the putative binding sites for the forkhead factor Sep1p. To obtain additional evidence that Sep1p controls the expression of ace2+ in fission yeast, the effects of deleting the five repeats of the TGTTTAC sequence were tested. To this end, we constructed a version of the ace2+ gene in which the five copies were precisely deleted by PCR (ace2ΔBS, plasmid pC4). As controls for the experiment, we used another two constructs: one that contained the wild-type promoter (ace2, plasmid pB1) and another that contained the ace2+ ORF without promoter (ace2ΔP, plasmid pA16). The three constructs were introduced into an ace2Δ mutant and their ability to complement the cell separation defect was tested. Microscopic observation of cells revealed that cells carrying the wild-type ace2 were able to separate, whereas those containing ace2ΔBS or ace2ΔP constructs were not (Figure 3B). This result was further confirmed when the number of septa in the cells was quantified (Figure 3C). Cells carrying the wild-type ace2 on plasmid pB1 were almost identical to wild-type cells, whereas the percentage of cells with two or more septa in cells containing the ace2ΔBS or ace2ΔP constructs closely resembled those found in the deletion strain. Northern analysis confirmed that no ace2+ expression was seen in strains carrying the deletion of the five putative Sep1p binding sites (plasmid pC4), similar to those cells carrying the deletion of the whole promoter (Figure 3C, bottom panel). Thus, these results suggest that the five copies of the TGTTTAC sequence are important for ace2+ expression.
Figure 3.
The consensus sequence TGTTTAC is important for ace2+ function. (A) Schematic representation of the position of the five copies of the TGTTTAC sequence in the ace2+ promoter and the constructs used. Plasmid pB1 contains the wild-type ace2+ promoter upstream of the ace2+ ORF (ace2), plasmid pC4 contains the ace2+ promoter in which the five copies of the TGTTTAC sequence were deleted (ace2ΔBS) and pA16 carries the ace2+ ORF without promoter (ace2ΔP). The arrow indicates the orientation of the TGTTTAC sequence. (B) Representative microscopic images of the ace2Δ mutants carrying ace2ΔP (top), ace2 (middle), or ace2ΔBS (bottom) are shown. (C) Quantification of the percentage of septa in wild-type, ace2Δ mutants and ace2Δ mutants containing ace2ΔP (pA16), ace2 (pB1) or ace2ΔBS (pC4). Cells were stained with aniline blue and DAPI before counting. For each sample, 200 cells of were counted. The bottom panel shows the expression of the ace2+ gene in wild-type (WT), ace2Δ mutants (ace2Δ), or ace2Δ mutants containing ace2ΔP (pA16), ace2 (pB1), or ace2ΔBS (pC4). The his3+ gene was used as loading control.
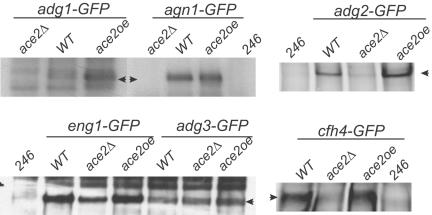
We also analyzed the effect of altering ace2+ expression levels on the abundance of the target proteins. To this end, adg1+, adg2+, adg3+, cfh4+, agn1+, and eng1+ were tagged with GFP at their C-termini and the protein levels were monitored in strains lacking or overexpressing ace2+. Changes in ace2+ levels affected the abundance of all the protein products, except Adg3p, which remained more or less constant in all three strains tested (Figure 4). The protein levels of four gene products (Adg1p, Adg2p, Agn1p, and Cfh4p) were almost undetectable in ace2Δ mutants, whereas Eng1p was clearly reduced (although there was still some protein present in the ace2 deletion strain). When Ace2p was overproduced using the nmt1+ promoter, Adg1p and Adg2p were markedly up-regulated, whereas Cfh4p protein levels also increased. These results support the idea that adg1+, adg2+, adg3+, cfh4+, agn1+, eng1+, and mid2+ are direct targets of the Ace2p transcription factor in S. pombe.
Figure 4.
Effects of ace2 levels on protein abundance of microarray targets. Protein lysates were prepared from wild-type untagged control strain KGY 246, adg1-GFP (KGY 4397), agn1-GFP (KGY 4398), adg2-GFP (KGY 4390), eng1-GFP (KGY 4387), adg3-GFP (KGY 4399), cfh4-GFP (KGY 4386) cells carrying a WT copy of ace2+ (WT); cells overexpressing ace2+ from the nmt1 promoter of pREP3X (ace2oe) or lacking ace2+ (ace2Δ): adg1-GFP ace2Δ (KGY 4612), agn1-GFP ace2Δ (KGY 4614), adg2-GFP ace2Δ (KGY 4616), eng1-GFP ace2Δ (KGY 4618), adg3-GFPace2Δ (KGY 4621) cfh4-GFPace2Δ (KGY 4620). Immunoprecipitates of adg2-GFP (top right panel) and cfh4-GFP (bottom right panel) were resolved on a 3–8% Tris-acetate gel. Immunoblotting was performed using the Odyssey Infrared Imaging System (Cincinnati, OH) and protocols (LI-COR). Primary anti-GFP serum was used at 1:1000 concentration. Goat anti-rabbit Alexa680 (LI-COR) was used at 1:5000 dilution. Image files were exported from the Odyssey software in TIFF format. The rest of the immunoprecipitates were resolved on a 4–12% Bis-Tris gel, immunoblotted with anti-GFP serum, and visualized by ECL.
Taken together, all the above results strongly support the idea that a transcriptional cascade governs the last stages of the cell cycle in S. pombe, which is depicted in Figure 2C. Thus, Sep1p controls the expression of ace2+ (and other genes), and Ace2p directly activates the transcription of at least seven genes (adg1+, adg2+, adg3+, cfh4+, agn1+, eng1+, and mid2+), whose products are important for cell separation to be completed.
Relationship between Ace2p and SIN
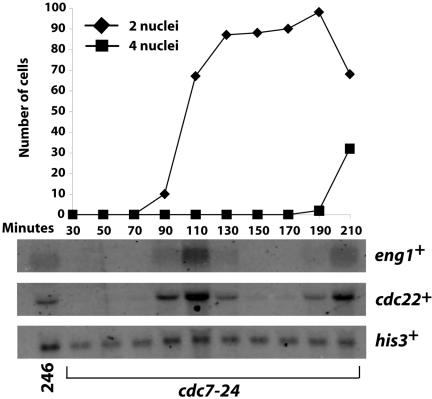
Because Ace2p-directed gene expression occurred at late stages in the cell cycle, we considered the possibility that Ace2p activity might depend on the Septation Initiation Network (SIN). The SIN triggers actomyosin ring constriction and septation and is also important for coordinating nuclear and cell division (Krapp et al., 2004). One of the components of the SIN is the Cdc7p protein kinase, and hence we tested whether the expression of Ace2p target genes occurred in a synchronized population of the cdc7-24 temperature-sensitive strain. Although Cdc7p function was clearly impaired, as monitored by failed cytokinesis, the expression of eng1+ (Figure 5) and other Ace2p targets (Petit and Gould, unpublished results) was unperturbed. Thus, the SIN is not required for Ace2p function.
Figure 5.
Relationship between Ace2p and the SIN network. cdc7–24 (KGY2061) cells, grown at 25°C, were synchronized in early G2 phase by centrifugal elutriation. Cells were then filtered and placed in medium prewarmed to 36°C. Samples were collected every 20 min, beginning 30 min after the shift. RNA was extracted from cell pellets, and Northern analysis was used to determine the level of eng1+ mRNA. The levels of cdc22+ mRNA were used to demonstrate cell cycle progress, and his3+ mRNA was used as a loading control. A small sample of cells from each time point was also fixed in ethanol and then stained with DAPI to determine the number of nuclei. The same RNA samples were utilized to determine the levels of other mRNAs in a separate study (Petit et al., unpublished results).
Localization of ace2+ Targets
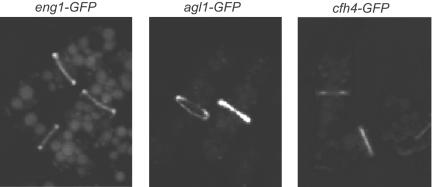
To better understand the role of these proteins during the last stages of the cell cycle, their localization was examined. Carboxy-terminal GFP fusions were constructed by homologous recombination at the endogenous locus of eng1+, adg1+, adg2+, adg3+, cfh4+, and agn1+ in order to maintain the physiological expression level and the temporal pattern of transcription. We found that Eng1p-GFP localizes as a ring to the septum region (Figure 6), as previously described (Martín-Cuadrado et al., 2003). The other glucanase, Agn1p-GFP, showed a similar pattern of fluorescence, i.e., a ring surrounding the septum, whereas Cfh4p-GFP was also seen in the septum region, suggesting that these proteins might also exert their function during cell separation. When the localization of the products of the three adg genes was analyzed, different patterns of intracellular fluorescence were observed. Adg1p and Adg2p mainly appeared as small speckles and/or vesicles, consistent with the remnants of GPI anchoring (unpublished data; Sazer and Sherwood, 1990). Indeed, Adg1p and Adg2p are predicted to be GPI-anchored proteins (De Groot et al., 2003), and hence the localization of the modified full-length proteins probably cannot be determined by N- or C-terminal tagging although it was possible to ascertain their levels of production (see Figure 4). Adg3p-GFP was visualized in a pattern similar to Adg1p, which would be consistent with an ER localization or processing of the C-terminal tag in the ER (unpublished data). In light of these observations, we cannot rule out that at least some portion of Adg1p, Adg2p, and Adg3p might be present and might function in the septal region together with Eng1p, Agn1p, and Cfh4p.
Figure 6.
Localization of Ace2p targets. Confocal images of the indicated strains, eng1-GFP (KGY4387), agn1-GFP (KGY4398), and cfh4-GFP (KGY4386). Cells were grown to midlog phase, and live cell images were obtained.
Deletion of adg Genes Affects Cell Separation to Different Extents
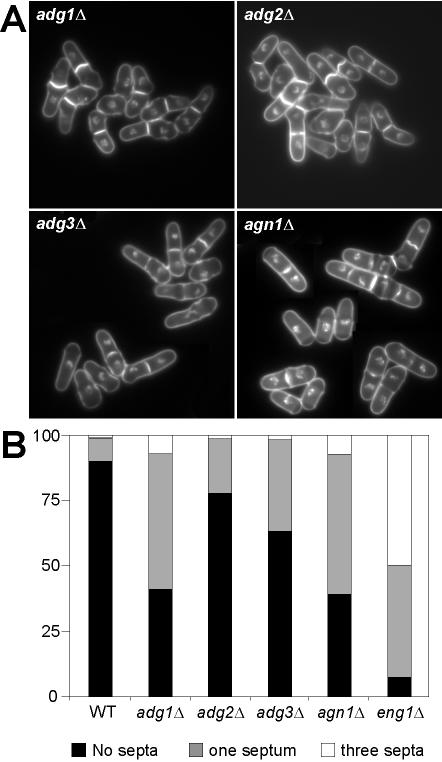
The results of the localization experiments suggested that at least some of these proteins would be required for cell separation. To test this prediction, deletion strains were generated using a PCR-based homologous recombination gene targeting system (Bähler et al., 1998b). All the mutants generated grew at wild-type rates, both on plates and in liquid medium, and at different temperatures (25–37°C). Microscopic examination of the mutant cells revealed that they had slight defects in cell separation (Figure 7A). When grown in rich medium, 10% of asynchronous wild-type cells showed a septum, whereas this percentage was increased in all of the mutants, ranging from 21% (adg2Δ) to more than 50% (adg1Δ and agn1Δ; Figure 7B). A small percentage of cells (∼1% in wild-type cells) had two or three septa and formed short chains of cells. This percentage was fairly similar to the wild-type in two of the mutants (adg2Δ and adg3Δ) but increased in adg1Δ (>6%) and agn1Δ (8%) cells. In no case were cells with more than three septa seen. Actually, cells lacking cfh4+ do not have any defect related to cell separation (H. Valdivieso, personal communication). As controls, we used mutants lacking the eng1+ endo-β-1,3-glucanase, whose product is involved in dissolution of the primary septum, in which a large percentage of the cells (>50%) were found in short chains of four connected cells, as described previously (Martín-Cuadrado et al., 2003). Analysis of the ultrastructure of the septum of the mutant cells did not reveal any significant difference between the adg1Δ, adg2Δ, or adg3Δ mutants and the wild-type (unpublished data). The normal growth rate of all of the mutants suggested that they were not undergoing a significant delay in cell cycle progression and that the accumulation of pairs of cells could be due to a specific delay in digestion of the septum during cell separation.
Figure 7.
Phenotype of mutants lacking Ace2p target genes. (A) Microscopic appearance of adg1Δ (YMAN2), adg2Δ (YMAN5), adg3Δ (YMAN4), and agn1Δ (YAB53) deletion mutants. Cells were grown in rich medium (YES), washed, and stained with aniline blue and DAPI before photographs were taken. (B) Number of septa in wild-type, adg1Δ (YMAN2), adg2Δ (YMAN5), adg3Δ (YMAN4), agn1Δ (YAB53), and eng1Δ (YAB14) mutants. More than 300 cells were counted for each strain.
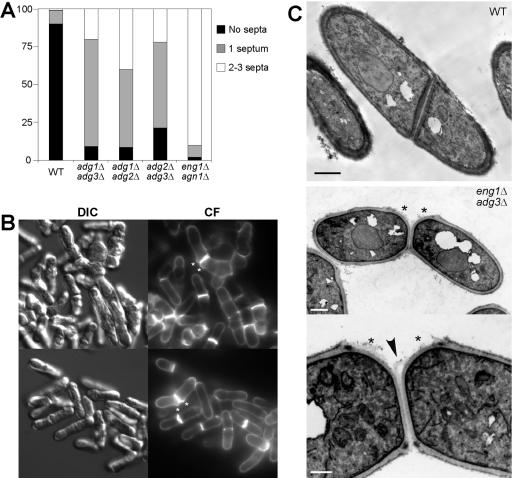
Because the phenotype of the single mutants analyzed was much less severe than that of ace2Δ cells, we decided to examine whether deletion of two of these genes would result in a more severe phenotype. We first constructed the three possible combinations between adg1+, adg2+, and adg3+. The three mutants grew at wild-types rates in solid and liquid medium, and at different temperatures (from 25 to 37°C). Microscopic examination of the double mutant cells revealed that in general they had more severe cell separation defects than the single mutants (Figure 8A). adg1Δ adg3Δ cells showed an increased percentage of cells with one septum (>70%), or two or more septa (20%). The adg1Δ adg2Δ and adg2Δ adg3Δ mutants also presented higher percentages of cells with one septum (52 and 57%, respectively) or with two or more septa (40 and 22%). These results indicate that the products of the three adg genes are also required for efficient cell separation. However, groups of more than four cells were rarely seen in any of the three mutants, suggesting that their roles would be minor.
Figure 8.
Phenotype of different double mutants. (A) Number of septa in wild-type (h123), adg1Δ adg3Δ (YMAN7), adg1Δ adg2Δ (YMAN25), adg2Δ adg3Δ (YMAN26), and eng1Δ agn1Δ (YAB54) mutants. More than 300 cells were counted for each strain. (B) Microscopic appearance of eng1Δ adg3Δ (YMAN11) mutant cells. Cells were grown in rich medium (YES), washed, and stained with Calcofluor. Photographs of differential interference contrast microscopy (DIC) or Calcofluor-stained cells (CF) are shown. (C) Electron microscopy ultrastructure of wild-type (top panel) and eng1Δ adg3Δ (YMAN7) mutant cells (middle and bottom panels). The arrowhead marks the uncleaved primary septum, and asterisks indicate points of lateral growth. Scale bars, (top and middle) 1 μm; (bottom) 0.5 μm.
Because eng1+ codes for the only enzymatic activity required for degradation of the primary septum that has been described, double mutants were constructed by combining the deletion of this gene with the deletion of adg1+, adg2+, or adg3+. The phenotype of the eng1Δ adg1Δ and eng1Δ adg2Δ mutants was almost identical to that of the single eng1Δ mutant; that is, most of the cells were present in groups of four connected cells (55–60% of the cells). However, the appearance of the eng1Δ adg3Δ mutant was slightly different. This mutant grew forming clumps of cells, with a marked defect in cell separation (Figure 8B). In addition, in many cells it was possible to observe a lateral activation of growth at the new pole, even though the septum was not degraded and the cells had not separated, resulting in a bending of the cells (Figure 8B, asterisks). This defect was never observed in eng1Δ mutants, although it is clearly reminiscent of the morphology observed in ace2Δ cells, but less severe. The ultrastructure of the cells was also analyzed using electron microscopy. In contrast to wild-type cells, in some of the eng1Δ adg3Δ cells the activation of a new growth pole next to the septum was observed (Figure 8C, asterisks). In addition, in some cells we also observed the phenotype described for eng1Δ; i.e., the primary septum was still present between the two sister cells (Figure 8C, arrowhead). Thus, these results indicate that the defects observed in ace2Δ cells could arise from a combination of the phenotypes observed in several of the mutants analyzed.
eng1Δ agn1Δ Mutants Are Completely Defective in Cell Separation
One of the genes identified in our screening, agn1+, codes for a protein with sequence similarity to Trichoderma harzianum and Penicillium purpurogenum mutanases (Fuglsang et al., 2000). Because the major structural components of the S. pombe cell wall are β-1,3-glucan (50–54% of total polysaccharides) and α-1,3-glucan (28–32%; Kopecka et al., 1995; Manners and Meyer, 1977; Humbel et al., 2001; Sugawara et al., 2003), it is possible that complete cell separation at the end of mitosis might be achieved through the concerted action of enzymes capable of degrading these two types of polymers. To test this hypothesis, the double eng1Δ agn1Δ deletion mutant was constructed and the morphology of the mutant cells was analyzed. Microscopic observation of the mutant cells revealed the presence of large clumps of cells that were largely defective in cell separation, a phenotype very similar to that observed for ace2Δ cells (Figure 9, A and B). In both mutants, groups of more than four connected cells in which lateral branching was observed were frequently seen (Figure 9, D and E) in comparison with wild-type cells (Figure 9C). In spite of this similarity, we also noticed that the morphology of the ace2Δ mutant cells was more elongated than that of the eng1Δ agn1Δ mutant. These results are in good agreement with the recent report that Agn1p codes for an endo-α-1,3-glucanase involved in cell separation (Dekker et al., 2004) and suggest that Eng1p and Agn1p would be the main activities required for cell separation.
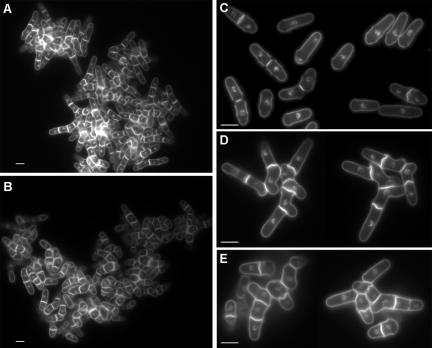
Figure 9.
eng1Δ agn1Δ cells are defective in cell separation. Morphology of ace2Δ (LE25, A) and eng1Δ agn1Δ (YAB54, B) cells. (C–E) Magnifications of representative groups of ace2Δ (D) and eng1Δ agn1Δ (E) cells, with the wild-type (C) as control. Cells were grown in rich medium to early-log phase, fixed with ethanol, and stained with aniline blue and DAPI. eng1Δ agn1Δ mutants show a mycelial and branched phenotype, similar to ace2Δ mutants, although the cells are less elongated. Scale bar, 10 μm.
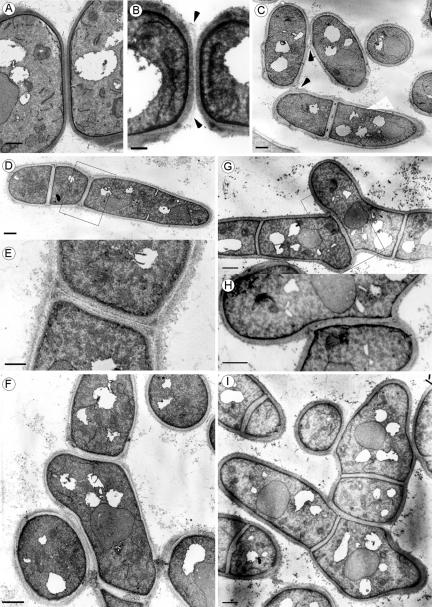
To further assess the nature of the separation defect of the eng1Δ agn1Δ mutants, transmission electron microscopy was used to compare the morphology of the septum region between the wild-type and the mutant strains. In wild-type cells, the three-layer structure of the septum was apparent, with a clear primary septum surrounded by two darker layers corresponding to the secondary septum (Figure 10A). In wild-type cells, it was observed that the primary septum was being degraded centripetally, from the cortex to the midpoint of the septum, and no remnants of this structure were seen in the region from which the two cells had already detached themselves. The septum of the eng1Δ mutants showed the presence of an uncleaved primary septum (Figure 10B), as previously reported (Martín-Cuadrado et al., 2003). In mutants lacking agn1+, the primary septum was always degraded, but some cells remained attached by the remnants of the cell wall on one side (Figure 10C), suggesting that this protein might be required for dissolution of the cell wall that surrounds the septum. In ace2Δ mutants, the cells had a branched morphology as a consequence of the complete inability to degrade the septum (Figure 10, G–I), a phenotype that is almost identical to that reported for sep1 mutants (Sipiczki et al., 1993; Sipiczki and Bozsik, 2000). Inspection of the morphology of the eng1Δ agn1Δ mutants (Figure 10, D–F) revealed the presence of branched cells resembling those of the ace2Δ mutants. In these cells, the typical three-layer structure that forms the septum was present, indicating that this structure had been normally assembled. However, in this case it was also evident that cell separation had not progressed, because the septum and the cylinder of cell wall that surrounds it had not been cleaved at all. These observations therefore suggest that cell separation in S. pombe is mainly achieved through the concerted action of the endo-β-1,3-glucanase Eng1p and the α-1,3-glucanase Agn1p.
Figure 10.
Electron microscopy ultrastructure of wild-type and mutant cells. Wild-type (A), eng1Δ (B), agn1Δ (C), eng1Δ agn1Δ (D–F), and ace2Δ (G–I) cells were grown to midlog phase before preparation for electron microscopy. The septum region of eng1Δ mutants (B) shows the presence of uncleaved primary septum (arrowheads). In agn1Δ mutants, the primary septum is degraded, but some cells remain attached by the cell wall at one of the ends (C, arrowheads). Double eng1Δ agn1Δ mutants grew as branched filaments, in which the septum was not cleaved (D–F). ace2Δ shows similar defects in cell separation, forming branching filaments of cells (G–I). The rectangles in D and G indicate the region magnified in E and H, respectively. The seemingly abnormal shape of cells (F and I) is the consequence of their different spatial positions in the branched filaments. Scale bars, 0.5 μm (A, B, E, and H) or 1 μm (C, D, F, G, and I).
DISCUSSION
In this work we describe the identification and characterization of a group of genes whose expression is regulated by the Ace2p transcription factor. Based on its sequence similarity to the S. cerevisiae Ace2p, this protein was identified as the transcription factor required for the expression of eng1+ (Martín-Cuadrado et al., 2003). eng1+ codes for an endo-β-1,3-glucanase, belonging to family 81 glycosyl hydrolases, whose activity is required for dissolution of the primary septum. The deletion of both eng1+ or ace2+ resulted in a cell separation defect, but to a different extent. Although the phenotype of ace2Δ mutants is severe and results in the formation of filaments of cells similar to what has been described for sep1Δ mutants (Sipiczki et al., 1993; Sipiczki and Bozsik, 2000), mutants lacking the endo-β-1,3-glucanase Eng1p form groups of four connected cells (Martín-Cuadrado et al., 2003). This difference suggested that Ace2p might regulate the expression of other genes whose products are also required for cell separation, including additional hydrolytic enzymes required for the dissolution of other components of the septum.
In fission yeast, the septum is a three-layered structure that is synthesized in two steps (Johnson et al., 1982). The first part to be laid down in a centripetal manner is the primary septum, which is rich in linear β-1,3-glucan (Horisberger and Rouver-Vauthey, 1985; Humbel et al., 2001), and its deposition is dependent on constriction of the actomyosin ring (Liu et al., 1999, 2000). After this, each daughter contributes cell wall material to its own side of the primary septum, building a secondary septum that is mainly composed of β-1,6-branched β-1,3-glucan and β-1,6-glucan (Horisberger and Rouver-Vauthey, 1985; Humbel et al., 2001). Separation of the sister cells once the septum has been assembled requires two degradative processes: erosion of the surrounding cylinder of the cell wall at its junction with the septum and dissolution of the primary septum (Johnson et al., 1982), in which the participation of hydrolytic enzymes is required. The first enzymatic activity described is that of the endo-β-1,3-glucanase encoded by eng1+, which is required for dissolution of the β-1,3-glucan-rich primary septum (Martín-Cuadrado et al., 2003). Whereas this article was under revision, Dekker et al. (2004) reported the characterization of Agn1p as en endo-α-1,3-glucanase and showed that it is involved in the hydrolysis of the cylinder of cell wall that surrounds the septum, a region that was named the septum edging. These findings are in complete agreement with our own.
The absence of information about other proteins required for cell separation when this work was begun prompted us to attempt to identify the genes regulated by Ace2p using S. pombe DNA microarrays. Of all the genes whose expression was lower in the ace2Δ mutants than in the wild-type cells identified, we focused on seven of them showing the characteristics of extracellular proteins, and these were designated adg genes (for Ace2-dependent genes). As expected, one of the genes was eng1+, which had previously been shown to be dependent on Ace2p. In addition, another six genes were identified, one of which (mid2+) was already known and which encodes an anillin homologue required for assembly of the septin ring. Deletion of mid2+ results in a phenotype similar to eng1Δ mutants (Berlin et al., 2003; Tasto et al., 2003). The other five genes encode hitherto uncharacterized proteins, either of unknown function (adg1+, adg2+, or adg3+) or displaying sequence similarity to other proteins in the databases (agn1+ or cfh4+). By Northern analysis we confirmed that the expression of these genes, including mid2+ (Petit and Gould, unpublished results), was dependent on Ace2p, because their expression was clearly reduced in ace2Δ mutants and both the mRNA and the protein levels were induced when ace2+ was overexpressed using the nmt1+ promoter. In addition, expression of the target genes is cell cycle-regulated, with maximum accumulation coinciding with the septation process. Interestingly, the expression of ace2+ was also cell cycle-regulated and the peak of maximum expression occurred before that of the other genes, as expected for a transcriptional regulator. Our results are in good agreement with the recent findings by Rustici et al. (2004), who have described the whole transcriptional program of the fission yeast cell cycle. The seven genes described here were grouped in Cluster 2, their expression being dependent on Ace2p and Sep1, and their expression occurred slightly after the peak of ace2+ transcription. Analysis of the promoter region of the seven target genes indicated the existence of two or three copies of the hexanucleotide CCAGCC separated by 100–200 nucleotides in the promoter of each gene. This sequence has been described as the consensus binding site for S. cerevisiae Ace2p and is necessary for correct transcriptional regulation of the target genes (Dohrmann et al., 1992, 1996). Based on the sequence similarity of the S. cerevisiae and S. pombe Ace2p factors, especially in the zinc-finger region (43.2% identity in 148 aa region), it is very likely that these regions would also be important for the induction of expression mediated by Ace2p in fission yeast. The importance of these sites in transcriptional regulation is currently under investigation.
Deletion of a second transcription factor, sep1+, affords a phenotype very similar to ace2Δ mutants and also results in branched cells (Sipiczki et al., 1993; Sipiczki and Bozsik, 2000; Martín-Cuadrado et al., 2003). We observed that none of the Ace2p-target genes was expressed in sep1Δ mutants, suggesting that the cell separation defect of the sep1Δ mutants could be indirect and could be due to their inability to activate ace2+ expression, which in turn controls the expression of the genes required for cell separation. Several lines of evidence support this model. First, ace2+ was not expressed in sep1Δ mutants, whereas sep1+ was normally transcribed in ace2Δ cells, indicating that ace2+ could be one of the genes regulated by Sep1p. Second, the expression of ace2+ from an heterologous promoter restored the expression of the target genes in a sep1Δ strain and suppressed the cell separation defect of this mutant, indicating that the genes involved in cell separation were more directly regulated by Ace2p. However, suppression of the separation defect was not complete, indicating that Ace2p might be regulated by other mechanisms. Although we have ruled out the contribution of the SIN in controlling Ace2p function, it is possible that, by analogy with S. cerevisiae Ace2p, Cdk1p, or Orb6p (similar to S. cerevisiae Cbk1p) phosphorylation might be involved in its regulation (O'Conallain et al., 1999; Weiss et al., 2002; Nelson et al., 2003; Ufano et al., 2004). It is also possible that Sep1p not only regulates ace2+ transcription, but also the activity or localization of Ace2p. In fact, many interactions between Sep1p and cell cycle regulators, such as wee+, cdc25+, or cdc2+, have been described (Sipiczki et al., 2000; Zilahi et al., 2000b). Finally, the promoter of ace2+ contained five copies of the sequence TGTTTAC in a 300-nt region. In S. cerevisiae, this sequence is the binding site for the forkhead transcription factors Fkh1p and Fkh2p (Pic et al., 2000; Hollenhorst et al., 2001), which suggested that they could also be the binding sites for the forkhead-like factor Sep1p. We have shown that deletion of the five repeats eliminated ace2+ transcription and function, indicating that they are important for ace2+ expression. Interestingly, two of these sites (at -375 base pairs and -220 base pairs) are located 8 and 15 nt downstream of the GCAACG/A sequence (the PCB element), which is the binding site for the MADS box protein Mbx1p (Buck et al., 2004). These authors have proposed that Mbx1p might form a complex with Sep1p and a second forkhead-like factor, Fkh2p, to control periodic gene transcription in M phase. Surprisingly, they found that deletion of sep1+ or fkh2+ abolished periodic expression of ace2+ and other genes, but deletion of mbx1+ only reduced the amplitude of the expression, suggesting that Sep1p and Fkh2p may control periodic gene transcription in the absence of the associated MADS box protein. These results, together with our data, indicate that correct temporal regulation of ace2+ expression requires both the TGTTTAC sequences, possibly as binding sites for Sep1p and/or Fkh2p, and the PCB element (for Mbx1p binding). The transcriptional cascade in the M/G1 transition is not a unique feature of S. pombe cells, because a similar situation has been described for S. cerevisiae, in which the fork-head transcription factors Fkh1p and Fkh2p regulate the expression of ACE2 and SWI5 (Hollenhorst et al., 2000, 2001; Hollenhorst et al., 2001). In fact, the use of microarrays to analyze the transcriptional pattern of gene expression during the fission yeast cell cycle has led to a model similar to that proposed here (Rustici et al., 2004). The sequential activation of different groups of genes could be a mechanism for the delivery of different protein products to the same region of the cell (in this case, the septum region) to perform their function in the correct order and using the same secretory system. This will ensure that activation of the genes involved in cell separation does not occur until septum synthesis has been completed.
To test the biological role of the identified genes, strains carrying single and double deletions in different combinations were constructed. We found that the deletion of most of the genes regulated by Ace2p had an effect on cell separation, although to different extents. This phenotype was subtle in cells lacking adg1+, adg2+, adg3+, or agn1+ and mainly produced a delay in cell separation that was reflected in an increase in the percentage of cells containing a septum in the population (ranging from 22 to 52%, vs. 10% in the wild-type). As previously described, mid2Δ and eng1Δ mutants had a more severe phenotype, with the formation of groups of four connected cells. When double mutants were analyzed, in most cases the severity of the cell separation defect was increased. The most interesting phenotypes were seen in eng1Δ adg3Δ and in eng1Δ agn1Δ mutants. In eng1Δ adg3Δ cells, growth activation at the new end before separation had been completed led to the formation of “bent” cells, a phenotype that was not normally seen in any of the single mutants. This phenotype may have arisen from some inability to cleave the septum, which would prevent tip growth at the time of new-end take off (NETO). Often, many cell pairs showed septum cleavage only on one side (where the cells were starting to grow), which could reflect a physical tearing of the cell wall as a consequence of cell growth rather than enzymatic dissolution, because in many cells the remnants of the primary septum were still present. This phenotype is fairly similar to that previously described for sep1Δ mutants (Ribár et al., 1999) and for the ace2Δ mutants reported here.
Cell separation in yeast and fungi requires controlled hydrolysis of the septum, a process that is mediated by different glycosyl hydrolases. Sequence analysis of all of the proteins identified indicated that Eng1p and Agn1p were the only ones containing motifs from glycosyl hydrolase families: Eng1p is a member of family 81, whereas Agn1p is grouped in family 71. Interestingly, the double mutant eng1Δ agn1Δ showed the most severe phenotype of all the combinations tested. eng1Δ agn1Δ cells were largely deficient in cell separation and showed the same growth pattern as that observed in ace2Δ mutants (and also in sep1Δ mutants), suggesting that Eng1p and Agn1p are the two main enzymatic activities that are required for cell separation to be completed. Agn1p showed strong sequence similarity to fungal α-glucanases and has recently been shown to possess endo-α-1,3-glucanase activity in S. pombe cells (Dekker et al., 2004). Because α-glucan is one of the major components of S. pombe cells (28–32% of total polysaccharides, Manners and Meyer, 1977; Kopecka et al., 1995), it was not surprising to find that an α-glucanase also participated in cell separation. Although agn1Δ mutants showed only a very weak phenotype (50% of the cells with septum and 7% with two or three septa), the observation that the cells often remained attached by the cell wall encourages the view that this enzyme might play a role in the dissolution of the cell wall that surrounds the septum. Indeed, similar results recently reported by Dekker et al. (2004) suggest that Agn1p might be required for the degradation of the cylinder of cell wall that surrounds the septum: the so called septum edging. The finding that eng1Δ agn1Δ mutants show a similar separation defect to that seen in ace2Δ or sep1Δ cells suggests that Eng1p and Agn1p might be the two main enzymatic activities required for cell separation in fission yeast. Thus, the concerted action of two enzymes with different substrate specificity (α- and β-glucans) but similar mechanism of action (endo-glucanases) is required to hydrolyze the main components of the septum and septum edging, that is, the α- and β-glucans. Site-specific dissolution of the mother cell wall at its junction with the primary septum could be the most important event in cell separation. Once the cell wall has been broken, either enzymatically dissolved or through physically rupture by mechanical forces, cell separation can be achieved. Consistent with this interpretation, it has been reported that the calcofluor-stainable material rapidly disappears from the cell ends when the septum is physically broken in sep1Δ cells (Sipiczki and Bozsik, 2000). In sum, the phenotype of ace2Δ mutants (and that of sep1Δ mutants) can largely be explained as a defect in the transcription of eng1+ and agn1+ and, perhaps, a minor contribution of adg3+.
As expected for proteins with a role in cell separation, the Eng1p, Agn1p, and Cfh4p proteins localized to the septum region, and Agn1p showed a ring-like structure very similar to that described for Eng1p (Martín-Cuadrado et al., 2003). C-terminal tagging of the products of the other three genes described (Adg1p, Adg2p, and Adg3p) revealed different cellular localizations, although the predictions from sequence analysis indicated that they have the features of extracellular proteins and the results of deletion analysis indicate that they are involved in the cell separation process. Two of them, Adg1p and Adg2p, may contain a glycosylphosphatidyl inositol (GPI) attachment site at the C-terminus and thus the localization detected may be misleading, representing the site of protein processing rather than function. We also cannot rule out the possibility that the addition of the GFP sequence at this position might interfere with the correct destination and function of Adg1p and Adg2p, although the strains carrying the fusions did not show the phenotypes of the deletion. Interestingly, Adg3p contains a domain of the SUN family of proteins close to the N-terminus of the protein, followed by a large region that is very rich in Ser/Thr. The SUN family of proteins, present in yeasts and fungi, is involved in a diverse set of functions, including DNA replication, aging, mitochondrial biogenesis, and cell septation. One of the members of this family, Candida wickerhamii bglA, has been characterized as a β-glucosidase (Skory and Freer, 1995), although this activity has not been reported for other members of the family. S. cerevisiae sun4Δ mutants show a delay in cell septation, which results in the formation of clumps of cells (Mouassite et al., 2000). Sun4p (also known as Scw3p) has a dual localization: it has been identified as a cell wall protein, although it is also present in mitochondria (Cappellaro et al., 1998; Velours et al., 2002). Further experiments will be required to test whether Adg3p also has β-glucosidase activity or whether it has a dual cell localization.
Supplementary Material
Acknowledgments
We thank Encarnación Dueñas for help in microarray analysis, Rachel Roberts and Courtney Lovejoy for experiment assistance, Shelley Sazer for advice, and Nick Skinner for language revision of the manuscript. This research was supported by the HHMI of which K.L.G. is an investigator and grants from the Comisión Interministerial deCiencia y Tecnología (BIO2000-1573 and BIO2001-2041), from the European Community (QKL3-2000-01537) and fromthe Hungarian grant agency OTKA (T 042694). M.L.A.-N. is recipient of a fellowship from the Ministerio deEducación y Ciencia (Spain).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0442) on February 2, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bähler, J., and Nurse, P. (2001). Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. EMBO J. 20, 1064-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., and Pringle, J. R. (1998). Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12, 1356-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., Steever, A. B., Wheatley, S., Wang, Y., Pringle, J. R., Gould, K. L., and McCollum, D. (1998a). Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143, 1603-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998b). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Baladrón, V., Ufano, S., Dueñas, E., Martín-Cuadrado, A. B., Rey, F.D., and Vázquez de Aldana, C. R. (2002). Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1, 774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin, A., Paoletti, A., and Chang, F. (2003). Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 160, 1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, V., Ng, S., Ruiz-Garcia, A. B., Papadopoulou, K., Bhatti, S., Samuel, J. M., Anderson, M., Millar, J. B., and McInerny, C. J. (2004). Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 117, 5623-5632. [DOI] [PubMed] [Google Scholar]
- Burns, C. G., Ohi, R., Krainer, A. R., and Gould, K. L. (1999). Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96, 13789-13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro, C., Mrsa, V., and Tanner, W. (1998). New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180, 5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F. (2001). Studies in fission yeast on mechanisms of cell division site placement. Cell Struct. Funct. 26, 539-544. [DOI] [PubMed] [Google Scholar]
- Chang, F., and Nurse, P. (1996). How fission yeast fission in the middle. Cell 84, 191-194. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner, A., Chin, T. E., and Brent, R. (2001). Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107, 739-750. [DOI] [PubMed] [Google Scholar]
- De Groot, P. W., Hellingwerf, K. J., and Klis, F. M. (2003). Genome-wide identification of fungal GPI proteins. Yeast 20, 781-796. [DOI] [PubMed] [Google Scholar]
- Dekker, N., Speijer, D., Grün, C. H., van den Berg, M., de Haan, A., and Hochstenbach, F. (2004). Role of the α-glucanase Agn1p in fission-yeast cell separation. Mol. Biol. Cell 15, 3903-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann, P. R., Butler, G., Tamai, K., Dorland, S., Greene, J. R., Thiele, D. J., and Stillman, D. J. (1992). Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6, 93-104. [DOI] [PubMed] [Google Scholar]
- Dohrmann, P. R., Voth, W. P., and Stillman, D. J. (1996). Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol. Cell. Biol. 16, 1746-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach, B., and Chang, F. (2001). Cytokinesis and the contractile ring in fission yeast. Curr. Opin. Microbiol. 4, 713-719. [DOI] [PubMed] [Google Scholar]
- Fuglsang, C. C., Berka, R. M., Wahleithner, J. A., Kauppinen, S., Shuster, J. R., Rasmussen, G., Halkier, T., Dalboge, H., and Henrissat, B. (2000). Biochemical analysis of recombinant fungal mutanases. A new family of α1,3-glucanases with novel carbohydrate-binding domains. J. Biol. Chem. 275, 2009-2018. [DOI] [PubMed] [Google Scholar]
- Gould, K. L., Moreno, S., Owen, D. J., Sazer, S., and Nurse, P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10, 3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin, D.A., Trautmann, S., and McCollum, D. (2002). Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66, 155-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, K. G., Bi, E., Wu, J.-Q., Adam, J. C., Yu, I.-C., and Pringle, J. R. (1999). Cytokinesis: an emerging unified theory for eukaryotes? Curr. Opin. Cell Biol. 11, 717-725. [DOI] [PubMed] [Google Scholar]
- Hollenhorst, P. C., Bose, M. E., Mielke, M. R., Muller, U., and Fox, C. A. (2000). Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154, 1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst, P. C., Pietz, G., and Fox, C. A. (2001). Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 15, 2445-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger, M., and Rouver-Vauthey, M. (1985). Cell wall architecture of the fission yeast Schizosaccharomyces pombe. Experientia 41, 748-750. [Google Scholar]
- Humbel, B. M., Konomi, M., Takagi, T., Kamasawa, N., Ishijima, S. A., and Osumi, M. (2001). In situ localization of β-glucans in the cell wall of Schizosaccharomyces pombe. Yeast 18, 433-444. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, K., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cation. J. Bacteriol. 153, 163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B. F., Calleja, G. B., Zuker, M., and McDonald, T. J. (1982). Cell division: key to cellular morphogenesis in the fission yeast Schizosaccharomyces pombe. Int. Rev. Cytol. 75, 167-208. [Google Scholar]
- Kopecka, M., Fleet, G. H., and Phaff, H. J. (1995). Ultrastructure of the cell wall of Schizosaccharomyces pombe following treatment with various glucanases. J. Struct. Biol. 114, 140-152. [DOI] [PubMed] [Google Scholar]
- Krapp, A., Gulli, M. P., and Simanis, V. (2004). SIN and the art of splitting the fission yeast cell. Curr. Biol. 14, R722-R730. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., Woollard, A., and Simanis, V. (1999). Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262, 163-172. [DOI] [PubMed] [Google Scholar]
- Liu, J., Wang, H., and Balasubramanian, M. K. (2000). A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 113, 1223-1230. [DOI] [PubMed] [Google Scholar]
- Liu, J., Wang, H., McCollum, D., and Balasubramanian, M. K. (1999). Drc1p/Cps1p, a 1,3-β-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics 153, 1193-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners, D. J., and Meyer, M. T. (1977). The molecular structures of some glucans from the cell wall of Schizosaccharomyces pombe. Carbohydr. Res. 57, 189-203. [Google Scholar]
- Martín-Cuadrado, A. B., Dueñas, E., Sipiczki, M., Vázquez de Aldana, C. R., and Rey, F. d. (2003). The endo-β-1,3-glucanase Eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 116, 1689-1698. [DOI] [PubMed] [Google Scholar]
- McBride, H. J., Yu, Y., and Stillman, D. J. (1999). Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J. Biol. Chem. 274, 21029-21036. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetics analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Mouassite, M., Camougrand, N., Schwob, E., Demaison, G., Laclau, M., and Guerin, M. (2000). The'SUN′ family: yeast SUN4/SCW3 is involved in cell septation. Yeast 16, 905-919. [DOI] [PubMed] [Google Scholar]
- Mulvihill, D. P., Petersen, J., Ohkura, H., Glover, D. M., and Hagan, I. M. (1999). Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell 10, 2771-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B. et al. (2003). RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14, 3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conallain, C., Doolin, M. T., Taggart, C., Thornton, F., and Butler, G. (1999). Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262, 275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith, A., and Segall, J. (1984). Isolation of DNA sequences preferentially expressed during sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 4, 142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic, A., Lim, F. L., Ross, S. J., Veal, E. A., Johnson, A. L., Sultan, M. R., West, A. G., Johnston, L. H., Sharrocks, A. D., and Morgan, B. A. (2000). The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19, 3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribár, B., Bánrévi, A., and Sipiczki, M. (1997). sep1+ encodes a transcription-factor homologue of the HNF-3/forkhead DNA-binding-domain family in Schizosaccharomyces pombe. Gene 202, 1-5. [DOI] [PubMed] [Google Scholar]
- Ribár, B., Grallert, A., Oláh, E., and Szállási, Z. (1999). Deletion of the sep1+ forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 263, 465-474. [DOI] [PubMed] [Google Scholar]
- Robinson, D. N., and Spudich, J. A. (2000). Towards a molecular understanding of cytokinesis. Trends Cell. Biol. 10, 228-237. [DOI] [PubMed] [Google Scholar]
- Rustici, G., Mata, J., Kivinen, K., Lió, P., Penkett, C. J., Burns, G., Hayles, J., Brazma, A., Nurse, P., and Bähler, J. (2004). Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36, 809-817. [DOI] [PubMed] [Google Scholar]
- Sazer, S., and Sherwood, S. W. (1990). Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97, 509-516. [DOI] [PubMed] [Google Scholar]
- Simon, I. et al. (2001). Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697-708. [DOI] [PubMed] [Google Scholar]
- Sipiczki, M., and Bozsik, A. (2000). The use of morphomutants to investigate septum formation and cell separation in Schizosaccharomyces pombe. Arch. Microbiol. 174, 386-392. [DOI] [PubMed] [Google Scholar]
- Sipiczki, M., Grallert, B., and Miklos, I. (1993). Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 104, 485-493. [DOI] [PubMed] [Google Scholar]
- Sipiczki, M., Yamaguchi, M., Grallert, A., Takeo, K., Zilahi, E., Bozsik, A., and Miklos, I. (2000). Role of cell shape in determination of the division plane in Schizosaccharomyces pombe: random orientation of septa in spherical cells. J. Bacteriol. 182, 1693-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skory, C. D., and Freer, S. N. (1995). Cloning and characterization of a gene encoding a cell-bound, extracellular β-glucosidase in the yeast Candida wickerhamii. Appl. Environ. Microbiol. 61, 518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann, M., Fankhauser, C., Brodbeck, C., and Simanis, V. (1996). The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 10, 2707-2719. [DOI] [PubMed] [Google Scholar]
- Sugawara, T., Sato, M., Takagi, T., Kamasaki, T., Ohno, N., and Osumi, M. (2003). In situ localization of cell wall α-1,3-glucan in the fission yeast Schizosaccharomyces pombe. J. Electron. Microsc. 52, 237-242. [DOI] [PubMed] [Google Scholar]
- Sugiura, R., Toda, T., Shuntoh, H., Yanagida, M., and Kuno, T. (1998). pmp1+, a suppressor of calcineurin deficiency, encodes a novel MAP kinase phosphatase in fission yeast. EMBO J. 17, 140-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilagyi, Z., Grallert, A., Nemeth, N., and Sipiczki, M. (2002). The Schizosaccharomyces pombe genes sep10 and sep11 encode putative general transcriptional regulators involved in multiple cellular processes. Mol. Genet. Genom. 268, 553-562. [DOI] [PubMed] [Google Scholar]
- Tasto, J. J., Morrell, J. L., and Gould, K. L. (2003). An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Dhut, S., Superti-Furga, G., Gotoh, Y., Nishida, E., Sugiura, R., and Kuno, T. (1996). The fission yeast pmk1+ gene encodes a novel mitogen activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16, 6752-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufano, S., Pablo, M. E., Calzada, A., Rey, F. d., and Vázquez de Aldana, C. R. (2004). The Swm1p subunit of the APC/Cyclosome is required for activation of the daughter-specific gene expression program mediated by Ace2p during growth at high temperature in Saccharomyces cerevisiae. J. Cell Sci. 117, 545-557. [DOI] [PubMed] [Google Scholar]
- Velours, G., Boucheron, C., Manon, S., and Camougrand, N. (2002). Dual cell wall/mitochondria localization of the'SUN′ family proteins. FEMS Microbiol. Lett. 207, 165-172. [DOI] [PubMed] [Google Scholar]
- Wach, A. (1996). PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12, 259-265. [DOI] [PubMed] [Google Scholar]
- Wang, H., Tang, X., Liu, J., Trautmann, S., Balasundaram, D., McCollum, D., and Balasubramanian, M. K. (2002). The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 13, 515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E. L., Kurischko, C., Zhang, C., Shokat, K., Drubin, D. G., and Luca, F. C. (2002). The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158, 885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T., Toda, T., and Yanagida, M. (1994). A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci. 107, 1725-1735. [DOI] [PubMed] [Google Scholar]
- Zhu, G., Spellman, P. T., Volpe, T., Brown, P. O., Botstein, D., Davis, T. N., and Futcher, B. (2000). Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406, 90-94. [DOI] [PubMed] [Google Scholar]
- Zilahi, E., Miklos, I., and Sipiczki, M. (2000a). The Schizosaccharomyces pombe sep15+ gene encodes a protein homologous to the Med8 subunit of the Saccharomyces cerevisiae transcriptional mediator complex. Curr. Genet. 38, 227-232. [DOI] [PubMed] [Google Scholar]
- Zilahi, E., Salimova, E., Simanis, V., and Sipiczki, M. (2000b). The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett. 481, 105-108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.