Abstract
The sorting nexin (SNX) family of proteins is characterized by sequence-related phox homology (PX) domains. A minority of PX domains bind with high affinity to phosphatidylinositol 3-phosphate [PI(3)P], whereas the majority of PX domains exhibit low affinity that is insufficient to target them to vesicles. SNX1 is located on endosomes, but its low affinity PX domain fails to localize in vivo. The NMR structure of the PX domain of SNX1 reveals an overall fold that is similar to high-affinity PX domains. However, the phosphatidylinositol (PI) binding pocket of the SNX1 PX domain is incomplete; regions of the pocket that are well defined in high-affinity PX domains are highly mobile in SNX1. Some of this mobility is lost upon binding PI(3)P. The C-terminal domain of SNX1 is a long helical dimer that localizes to vesicles but not to the early endosome antigen-1–containing vesicles where endogenous SNX1 resides. Thus, the obligate dimerization of SNX1 that is driven by the C-terminal domain creates a high-affinity PI binding species that properly targets the holo protein to endosomes.
INTRODUCTION
The sorting nexin (SNX) family of proteins, with >25 members in the human genome, is characterized by sequence-related phox homology (PX) domains that bind membrane-localized phosphatidylinositols (PIs) that are phosphorylated on the inositol ring (Sato et al., 2001; Teasdale et al., 2001; Worby and Dixon, 2002). These PX domains function in targeting SNX proteins to specific vesicular membranes. The descriptor “sorting nexin” was applied to the first discovered member, SNX1, based on the hypothesis that these proteins functioned in sorting vesicular cargo in the endosomal system (Kurten et al., 1996). SNX proteins also contain a variety of other functional domains, including coiled coils, regulator of G protein signaling, Src homology 3, and band four point one, ezrin, radixin, moesin homology domains (Worby and Dixon, 2002). Specific SNX family members are reported to bind various cargo proteins, including the tyrosine kinase receptors for epidermal growth factor, platelet-derived growth factor, and insulin (Kurten et al., 1996; Haft et al., 1998); the protease-activated G protein-coupled receptor (Wang et al., 2002); cargo receptors, including the low-density lipoprotein receptor and the transferrin receptor (Haft et al., 1998; Burden et al., 2004); Ser/Thr kinase receptors of the transforming growth factor-β family (Parks et al., 2001); leptin receptors (Haft et al., 1998); P-selectin (Florian et al., 2001); Down's syndrome cell adhesion molecule (Worby et al., 2001); enterophilin (Pons et al., 2003); and the Fanconi anemia complementation group A protein (Otsuki et al., 1999).
Evidence that these PX domain-containing proteins function as sorting nexins derives from genetic analysis in Saccharomyces cerevisiae where Vps5, the ortholog of mammalian SNX1 and SNX2, functions in a molecular complex, “the retromer,” that recycles Vps10p from the vacuole back to the trans-Golgi (Seaman et al., 1998). Grd19p, the ortholog of mammalian SNX3, functions to retrieve late-Golgi membrane proteins from the prevacuolar compartment (Voos and Stevens, 1998). Genetic analysis of mammalian SNX1 and SNX2, which are highly homologous and form homo- and heterodimers in vitro (Haft et al., 1998; Zhong et al., 2002), revealed that deletion of either gene in mice caused no discernible phenotype, but deletion of both SNX1 and SNX2 resulted in embryonic lethality with gross distortions of the endosomal system (Schwarz et al., 2002).
The major identified membrane target for PX domains is phosphatidylinositol 3-phosphate [PI(3)P]. Yu and Lemmon (2001) examined PI(3)P binding by all 15 S. cerevisiae PX domains and found that four bound with high-affinity (Kd of ∼2–3 μM), whereas the other 11 bound with only low affinity (Kd > 100 μM). Structures of the high-affinity PX domains of the p40 subunit of NADPH oxidase (Bravo et al., 2001), p47phox (Hiroaki et al., 2001; Karathanassis et al., 2002), Vam7p (Lu et al., 2002), Grd19p (Zhou et al., 2003), and CISK (Xing et al., 2004) indicate a common fold consisting of a three-stranded β-sheet followed by three or four helices, with a PI(3)P head group binding pocket containing conserved basic residues. Lemmon (2003) hypothesized that the low-affinity PX domain-containing proteins (and the FYVE domain-containing proteins) additionally require a membrane insertion loop or an oligomerization component for proper membrane targeting.
SNX1 and SNX2 are localized to endosomes (Kurten et al., 1996; Kurten et al., 2001; Zhong et al., 2002). Mutation of conserved residues of the PX domain of SNX1 involved in PI recognition abolished endosomal localization of holo SNX1, but the N terminus containing the PX domain did not itself localize (Zhong et al., 2002). Mutation of the C terminus of SNX1 that consists of a long helical domain with three coiled coil regions also abrogated endosomal localization. SNX1 thus seems to contain a low-affinity PX domain that is necessary but not sufficient for proper localization and function of SNX1; the C-terminal domain also is required.
In the present study, we first developed an in vivo assay to assess whether PI binding causes localization to vesicles by using microinjection of purified green fluorescent protein (GFP)-PX domains in the presence of competing PIs. The PX domains of SNX3 and CISK localize to endosomal membranes, and this localization is specifically blocked by PI(3)P and by inhibiting phosphoinositide 3-kinase (PI 3-kinase). The PX domains of SNX1 and SNX2 failed to localize and were diffusely distributed in the cytoplasm. Thus, similar to yeast PX domains, there are high- and low-affinity mammalian PX domains. To better understand the structural basis of the differential affinity of PX domains, we determined the solution structure of the SNX1 PX domain and compared this with the high-affinity PX domains of p40phox, Grd19p, and CISK. We also examined the effects of adding PI(3)P on the structure of the PX domain of SNX1.
Holo SNX1, in contrast to its isolated PX domain, binds to PI(3)P with high affinity in vitro (∼1 μM) and localizes to endosomes in vivo (Cozier et al., 2002; Zhong et al., 2002). However, holo SNX1 and holo SNX2 seem to function as obligate dimers, and therefore we investigated the localization of the C-terminal domain, which, by homology with the Bin/Amphiphysin/Rvs (BAR) domain of amphiphysin, should itself be an obligate dimer (Peter et al., 2004). The C terminus localizes to membranes but not specifically to early endosome antigen (EEA)1-positive endosomes, where SNX1 is localized. These results support a process in which SNX1 localizes to specific endosomal membranes as a dimer, where protein–protein interactions and the binding of the C-terminal domain to curved vesicle membrane surfaces contribute to a PI(3)P affinity enhancement of the PX domain.
MATERIALS AND METHODS
Preparation of Constructs Based on PET14b
The cDNAs coding for PX domain proteins were amplified using polymerase chain reaction (PCR) and inserted into the pET14b vector (Novagen, La Jolla, CA) by using NdeI and BamHI restriction sites. The resulting constructs encode His-tagged fusion proteins. The PCR fragment of His-glutathione S-transferase (GST) or His-green fluorescent protein (GFP) was used to replace the His-tagged region of pET14b between the NcoI and NdeI restriction sites to yield His-GST or His-GFP fusion PX proteins, respectively. The amino acid boundaries of the PX domains are SNX1, 139–269; SNX2, 118–266; SNX3, full length; and CISK, 1–121.
Expression of His-GFP-PX Proteins in Escherichia coli
Frozen stocks of transformed E. coli cells were prepared according to standard procedures, and 1 liter of LB culture was inoculated with approximately 100 μl of frozen stock solution and cultured at 37°C. When the optical density (OD) (600 nm) reached 0.6–0.8, isopropyl β-d-thiogalactoside (IPTG) was added to the medium to a final concentration of 0.5–1.0 mM. The culture was allowed to grow at 15°C overnight. The E. coli cells are collected by centrifugation at 15,000 rpm for 15 min, and the cell pellet was resuspended in buffer A (20 mM Tris-HC1, pH 8.0, and 0.5 M NaC1).
Creation of a GFP-PX-Leucine Zipper Dimer and Mutants
The cDNA of the leucine zipper from GCN4 was amplified in a PCR reaction and appended to the 3′ end of the GFP fusion construct of SNX1 PX. The resulting DNA was ligated into the pEYFP-c1 vector. The leucine zipper fragment of GCN4 covers amino acid residues 241–281. The double mutations on the fusion protein of SNX1 PX and leucine-zipper were made on the PX domain at positions 44 and 45, changing from ArgArg to SerGly, and at positions 69 and 70, changing from ProPro to AlaAla. The Quikchange kit (Stratagene, La Jolla, CA) was used to generate the mutants. The primers used were 5′tttgcagtaaaaagcggatttagtgacttt3′ and 5′ggcttcattgtcgctgcacccccggagaag3′.
Purification of His-GFP-PX
The His-tag fusion proteins were purified following the manufacturer's suggested procedures. The eluted protein from a nitrilotriacetic acid column was concentrated using Centriprep (molecular weight cut-off [MWCO] 3000) (Millipore, Billerica, MA). Protein samples at approximate 10 mg/ml were dialyzed against 1 liter of 10 mM phosphate buffer, pH 7.5, with 0.1 M KCl at 4°C overnight. A concentration step was performed if necessary after dialysis using a Centricon (MWCO 3000) (Millipore). The final sample had a protein concentration between 10 and 20 mg/ml.
Microinjection Experiments
HeLa cells were maintained on coverslips. The protein samples were injected into the cytoplasm of cells at 40% confluence by using a microinjector (Bio-Rad, Hercules, CA). Approximately 200 cells on each coverslip were injected for each protein. After injection, cells were allowed to recover at 37°C for 1 h and then fixed using 2% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in phosphate-buffered saline (PBS) at room temperature for 60 min. The coverslips were mounted on the glass slides and visualized with a 63×/1.4 numerical aperture (NA) oil immersion objective as described below. PI(3)P and phosphatidylinositol 3, 5 diphosphate [PI(3,5)P2] were dioctanoylglyceryl derivatives dissolved as 1 mM solutions in 1 mM NaOH. The PI 3-kinase inhibitor LY294002 was added at 50 μM 1 h before microinjection.
NMR Sample Preparation
The cDNA coding for the PX domain of SNX1(aa 139–269) was amplified using PCR and ligated into pTYB11 (New England Biolabs, Beverly, MA) between the restriction sites SapI and XhoI. Constructs were transformed into BL21(DE3) CodonplusRIL E. coli competent cells (Invitrogen, Carlsbad, CA) to express the protein. A frozen stock was prepared by collecting cells from 100-ml cultures when OD = 600 nm was between 0.2 and 0.3 and resuspending cells in LB broth with 15% glycerol and antibiotics. To prepare the protein, 1 liter of LB was inoculated with 100 μl of frozen stock and cultured at 37°C until OD 600 nm was between 0.8 and 1.0. IPTG was then added to the medium to a final concentration of 1 mM, and the temperature was changed to 15°C. The culture was incubated at 15°C with shaking overnight. The E. coli cells were collected by centrifugation at 15,000 rpm for 15 min on Ananti J-20 (Beckman Coulter, Fullerton, CA), and the cell pellet was resuspended in buffer A. After French Press breakage and high-speed centrifugation at 20,000 rpm for 30 min by using a J-20 rotor (Beckman Coulter), the supernatant was obtained and a proteinase inhibitor, 4-(2-aminoethyl)benzenesulfonyl fluoride (Calbiochem, San Diego, CA), was added to the supernatant to a final concentration of 0.2 mg/ml. The protein was purified using a Chitin affinity column (New England Biolabs). The pure protein was eluted in buffer A with 50 mM dithiothreitol. The eluent was dialyzed against 20 mM Tris-HC1, pH 8.0, with 0.2 M NaCl at 4°C overnight and concentrated to 1 ml by using Centriprep (MWCO 3000) (Millipore). The concentrated protein sample was applied to a Superdex S75 column (Amersham Biosciences, Piscataway, NJ) equilibrated with 0.2 M Tris-HC1, pH 8.0, and 0.2 M NaC1. The peak fractions were pooled and concentrated. If desirable, a dialysis step was used to change buffers. For isotopically labeled protein, 15N and 13C were introduced uniformly using Martek M9 media (Spectra Stable Isotopes, Columbia, MD).
NMR Analysis
Samples for NMR structural determinations were concentrated to 1 mM in 20 mM phosphate buffer containing 0.1 M NaCl, 10 mM NaN3, and 8% D2O, at pH 6.5. All experiments were recorded at 25°C on Bruker DRX-500, DRX-600, and DRX 800 spectrometers. Data processing and analysis were performed using Felix 2000 (Accelrys, San Diego, CA), supplemented with in-house macros and UNIX scripts. The backbone chemical shifts were obtained using 15N-HSQC (acquisition times: H 265 ms, N 120 ms), HNCA (H 141 ms, N 32 ms, C 10 ms), HNCO (H 141 ms, N 30 ms, C 25 ms), HN(CA)CO (H 141 ms, N 24 ms, C 24 ms), CBCA(CO)NH (H 141 ms, N 38 ms, C 5 ms), and HNCACB (H 141 ms, N 24 ms, C 35 ms) spectra, as described previously (Thaw et al., 2001). Sequential resonance assignment was performed using the asstool package (Reed et al., 2003). The side chain resonance assignment was performed using CCH-TOCSY (H 163 ms, C 17 ms, Ctoc 11 ms; mixing time 14 ms) and HCCH-TOCSY (H 163 ms, C 14 ms, Htoc 29 ms; mixing time 14 ms) spectra in conjunction with aromatic (H 228 ms, C 38 ms) and aliphatic (H 163 ms, C 52 ms) versions of the 13C-ctHSQC, as described previously (Thaw et al., 2001). Nuclear Overhauser effects (NOEs) were assigned in a 15N- and 13C-edited nuclear Overhauser effect spectroscopy spectrum (H 163 ms, N 34 ms, C 12 ms, Hnoe 26 ms; mixing time 100 ms). NOE restraints were calibrated on known distances in the helices and strands and divided into two categories: <3.5 and <5 Å. Dihedral angle restraints were introduced using TALOS (Corlinescu et al., 1999) with upper and lower bounds set at twice the TALOS standard deviations. Structures were calculated using the simulated annealing protocol in CNS 1.1 (Brunger et al., 1998) and analyzed using PRO-CHECK-NMR (Laskowski et al., 1996). Changes in NMR parameters of the SNX1-PX domain on phosphoinositide binding were investigated by the addition of dioctanoyl-PI(3)P in 0.1 equivalent aliquots to a 400 μM solution of SNX1-PX in the buffer used for structure determination.
Preparation of Human Embryonic Kidney (HEK)293 Cells Expressing Inducible GFP-SNX1 and GFP-SNX2
The cDNAs containing SNX1 and SNX2 were subcloned into plasmid pEGFP-c1 (Stratagene), and the resulting plasmids were digested with the restriction enzymes NcoI and DraI (New England Biolabs) at 37°C overnight. The DNA fragment containing GFP-SNX1 was isolated by agarose gel electrophoresis and purified using QIAquick gel extraction kit (QIAGEN, Valencia, CA). The plasmid SNX2/pEGFP-c1 was transformed into E. coli strain SCS110 (Stratagene) to obtain unmethylated DNA. A similar digestion procedure was followed using the restriction enzymes NcoI and BclI (New England Biolabs) at 37°C overnight, and the DNA fragment was isolated from agarose gels and purified. The GFP-SNX1 and GFP-SNX2 fragments were ligated to the pcDNA5/FRT/TO-TOPO vector (Invitrogen) following the manufacturer's procedure. The resulting GFP-SNX1/pcDNA5/FRT/TO-TOPO and GFP-SNX2/pcDNA5/FRT/TO-TOPO plasmids were transfected into Flp-in T-REx-293 cells (Invitrogen) by using Transfectene (QIAGEN). Cells were cultured in DMEM with 10% fetal bovine serum (FBS) and antibiotics, including zeocin and balsticidin (Invitrogen). Stable cell lines were selected using hygromycin. Protein expression was induced by the addition of tetracycline.
Immunofluorescence Microscopy
The cDNAs coding for PX domain proteins were ligated into pEGFP-c1 expression vectors (Strategene) by using either EcoR I and BglII or BglII and BamHI restriction sites. Transfections are carried out using Effectene (QIA-GEN) on COS 7cells grown to 40–50% confluence. Cells were fixed and examined 16 h after transfection. Cells were fixed with 2% paraformaldehyde in PBS, blocked with 2.5% FBS, and permeabilized with 0.1% Triton X-100. Cells were stained with goat anti-SNX1 (Santa Cruz Biotechnology, Santa Cruz, CA) (1:250); goat anti-SNX2 (Santa Cruz Biotechnology) (1:250), or mouse monoclonal anti-EEA1 (1:100). Coverslips were washed and incubated with donkey anti-goat IgG H + L chains conjugated to Alexa Fluor 594 (Molecular Probes, Eugene, OR) or goat anti-mouse IgG H + L chains conjugated to Alexa Fluor 488 or Alexa Fluor 350. Omission of primary antibodies was used as negative controls. Coverslips were viewed using a 63×/1.4 NA Zeiss oil immersion objective on a Zeiss Axioskop fluorescence microscope equipped with a 640 × 480 pixel COHU interline transfer charge-coupled device camera (Cohu, San Diego, CA). Deconvolution images were captured with a Delta Vision deconvolution microscope (Applied Precision, Issaguah, WA). Approximately 20 optical sections spaced by 0.2 μm were taken using a 100×/NA lens. Data sets were deconvoluted and analyzed using SOFT-WORK software (Applied Precision) on a Silicon Graphics (Mountain View, CA) Octane workstation. Colocalization of proteins on vesicles was determined by blinded counting to determine the fraction of vesicles expressing both GFP proteins and endogenous SNX1 (n = 250).
RESULTS
In Vivo Analysis of PX Domain Binding Affinity
In vitro analyses of the lipid-binding specificities of the PX domains of SNX1 and CISK have given different results by using lipid overlay and liposome binding assays (Virbasius et al., 2001; Xu et al., 2001a; Cozier et al., 2002; Zhong et al., 2002). To clarify PI target specificities and to assess relative binding affinities in vivo, we prepared GFP-PX domains and microinjected these into HeLa cells in the presence or absence of competitor lipids. The PX domains of SNX3 and CISK localized to vesicular structures, whereas the PX domains of SNX1 and SNX2 were diffusely distributed and did not localize to vesicles (Figure 1, A, D, G, and H). These results are consistent with the PX domains of SNX3 and CISK exhibiting high-affinity binding, whereas the PX domains of SNX1 and SNX2 bind with low affinity to PIs. Vesicular localizations of the PX domains of SNX3 and CISK were abolished by PI(3)P but not by PI(3,5)P2 (Figure 1, B, C, E, and F), confirming that these high-affinity PX domains preferentially bind to PI(3)P-containing vesicles in vivo (Virbasius et al., 2001; Xu et al., 2001b; Gillooly et al., 2003). The specificity of CISK for PI(3)P was verified by treating cells with the PI 3-kinase inhibitor LY294002. As shown in Figure 1I, this abolished localization to vesicles. Overall, these results indicate that the failure of the isolated PX domains of SNX1 and SNX2 to localize to vesicles in vivo is due to their low lipid binding affinity.
Figure 1.
Distribution and binding specificities of PX domains. GFP-PX domains were expressed in E. coli, purified, and microinjected into HeLa cells without or with PI(3)P or PI(3,5)P2. (A) SNX3. (B) SNX3 + PI(3)P. (C) SNX3 + PI(3,5)P2. (D) PX domain of CISK. (E) PX domain of CISK + PI(3)P. (F) PX domain of CISK + PI(3,5)P2. (G) PX domain of SNX1. (H) PX domain of SNX2. (I) Cells were treated with the PI 3-kinase inhibitor LY294002 (50 μM) for 1 h before microinjection of GFP-CISK.
Structural Analysis of PX Domain Binding Affinity
According to sequence alignments of PX domains of known structure (Worby and Dixon, 2002), it is not clear why SNX1 and SNX2 are low-affinity binding domains, whereas CISK and SNX3 are high-affinity binding domains. Despite only modest levels of sequence identity between, for example, SNX1 and SNX3 (28%), the residues identified to be key in the binding of PI(3)P in structures of PX domain-phosphoinositol complexes, all have counterparts in the SNX1 sequence. To investigate the sources of differential affinity, we determined the solution structure of the PX domain of SNX1 by using heteronuclear NMR methods.
1H, 13C, and 15N resonances from 94% of residues were assigned. Using a restraints list of 952 unambiguous NOE distance restraints and 146 dihedral angle restraints, 100 structures were calculated, and an ensemble of the 10 lowest energy structures is shown in Figure 2A. Statistics for the structure calculations are given in Table 1. The overall fold of the SNX1 PX domain (Figure 2B) is similar to previously determined PX domain structures and consists of a three-stranded β-sheet packed against a helical subdomain consisting of three α-helices and a proline-rich loop. A representative conformer of the SNX1 PX ensemble structure overlays well with the structures of the p40phox and Grd19p PX domains (1.8 and 1.7 Å Cα r.m.s.d. [root mean square deviation], respectively, for the secondary structure elements β1-β3, α1, and α3).
Figure 2.
Solution structure of the SNX1 PX domain. (A) Superposition of the backbone atoms of 10 refined solution structures for the SNX1-PX domain, highlighting in the lower part of the representation, the relatively low definition of the proline-rich loop connecting helices 1 and 2. (B) A ribbon representation of the lowest energy member of the ensemble shown at top, illustrating the topology within the tertiary fold of the SNX1-PX domain, the nomenclature of the secondary structure elements, and the location of residues predicted to be involved in phosphoinositide binding. The β-strands are yellow, and the α helices are green and blue. The proline-rich loop (labeled Pro-rich) is red except for the region of high mobility (orange). The β1-β2 loop is also highly mobile and orange.
Table 1.
Structural statistics
| Experimental restraints | |
| NOE distances | 952 |
| Intraresidue | 449 |
| Sequential | 235 |
| Medium range | 105 |
| Long range (Δ >4) | 164 |
| Dihedral restraints | 146 |
| Deviations of experimental restraintsa | |
| NOE distance violations (>0.2 Å) | 0 |
| Dihedral restraint violations (>2.5°) | 5 |
| RMS deviation from the mean structureb (Å) | |
| Backbone | 0.72 |
| Heavy atom | 1.42 |
| Ramachandran analysisc | |
| Most favored region (%) | 85.5 |
| Additionally allowed region (%) | 11.5 |
| Generously allowed region (%) | 0.4 |
| Disallowed regions (%) | 2.5 |
Ensemble of the 10 lowest energy structures out of 100 calculated. The r.m.s. deviations of bond lengths and bond or improper angles from the ideal CNS values were 0.0022 ± 0.00013 Å, 0.3927 ± 0.0129°, and 0.2254 ± 0.0136°, respectively.
r.m.s. NOE violation 0.0161 ± 0.0008 Å; r.m.s. dihedral violation 0.477 ±0.0955°
For residues in the well ordered region
From PROCHECK-NMR (Laskowski et al., 1996) analysis of the well ordered region in the ensemble of structures
The key difference between the SNX1 PX domain and the high-affinity PX domains is in the proline-rich loop, which is between helices α1 and α2. This loop contains residues identified by homology with other PX domain structures (Figure 3) to be important in the phosphoinositide binding site (residues 80, 82, and 85 in SNX1). In the SNX1 PX domain, much of this loop is not well defined owing to high-frequency mobility in solution for the region that should make direct contact with phosphoinositides. 15N resonances from residues in the region 78–90 have increased transverse relaxation times corresponding to mobility considerably in excess of the overall rotational correlation time of the domain, i.e., there is extensive conformational inter-conversion on a subnanosecond time scale. Similar high-frequency mobility is observed for residues in the loop between strands β1 and β2 (residues 13–21), which packs adjacently to the residue 78–90 region in other PX domains (Figure 2B). Hence, although residues capable of binding phosphoinositide are present within the appropriate area of the primary sequence of the SNX1 PX domain, the binding site is not preformed.
Figure 3.
Phosphoinositide binding region. Superimposition of the PX domains of SNX1 (blue backbone, cyan side chains) and p40phox (red backbone, orange side chains), focusing on the phosphoinositide binding region. The phosphoinositide bound to p40phox is shown in green. The PX domain of p40phox is representative of the high-affinity class of PX domains, all of which overlay closely. Residues proposed to be important for phosphoinositide binding in p40phox, and their counterparts in SNX1, are labeled (where joint labeling occurs, the label for SNX1 is first).
On addition of PI(3)P, the increased transverse relaxation times are lost in the region 79–85, indicating that the high-frequency mobility is removed on phosphoinositide binding, when a lysine side chain from this region becomes involved in coordination of PI(3)P, as observed in the high-affinity PX domains. Large changes in mobility reported by the resonances of V81-V83 suggest that it is the side chain of K82 that fulfills the role equivalent to that of K92 in the phosphoinositide-p40phox complex (Figure 3). The changes in NMR chemical shifts observed on addition of PI(3)P are consistent with other PX domain-phosphoinositide complexes (Bravo et al., 2001; Zhou et al., 2003).
In Vivo Localization of Holo SNX1 and SNX2
Although the PX domains of SNX1 and SNX2 do not localize to vesicles, the endogenous holo proteins are localized to vesicles, and both colocalize with the FYVE domain-containing endosomal protein EEA1 (Figure 4). Reports of the extent of colocalization of transfected SNX1 and SNX2 with EEA1 vary (Kurten et al., 2001; Cozier et al., 2002; Wang et al., 2002; Zhong et al., 2002; Gullapalli et al., 2004). In the present study, in which antibodies were used to examine the distribution of endogenous SNX1 and SNX2 with endogenous EEA1, extensive colocalization was observed. At the light microscopy level, we failed to observe endogenous SNX1 on the tubular vesicles that were seen with immunoelectron microscopy of overexpressor cells (Zhong et al., 2002).
Figure 4.
Colocalization of SNX1 with EEA1-positive endosomes. (A and B) COS 7 cells were fixed, permeabilized, and stained with goat anti-SNX1 or anti-SNX2 and mouse monoclonal anti-EEA1 antibodies. Detection was via donkey anti-goat and goat anti-mouse secondary antibodies conjugated to Alexa Fluor 594 and488. (C and D) Merged images of A and B. Inset is a close-up of the merged image of the boxed area (D).
To verify the immunological localization of endogenous SNX1 and SNX2 with EEA1, we introduced a single copy of an inducible GFP-SNX1 and GFP-SNX2 into HEK293 cells by using homologous recombination. Permanent cell lines were selected, and the expression of GFP-SNX1 and GFP-SNX2 was induced by tetracycline under conditions that limit the amount of protein expressed and avoid formation of large vesicles where mixing of compartment markers is seen with overexpression of transfected proteins (Barr et al., 2000). As shown in Figure 5, both GFP-SNX1 and GFP-SNX2 are expressed in small vesicles that contain EEA1. Using deconvolution microscopy, both SNX proteins seem to localize to microdomains of endosomes that are distinct from microdomains containing EEA1 (Figure 5, C and F). These results resemble the localization of Rab proteins and of PI(3)P to microdomains of endosomes (Zerial and McBride, 2001; Pfeffer, 2003; Gillooly et al., 2003).
Figure 5.
Endosomal colocalization of GFP-SNX1 and GFP-SNX2 with EEA1. HEK293 cells containing a chromosomally integrated copy of GFP-SNX1 or GFP-SNX2 were treated with 1 μg/ml tetracycline for 12 h to induce protein expression. Cells were fixed, permeabilized, and EEA1 was detected using mouse monoclonal anti-EEA1 and visualized using goat anti-mouse IgG H + L chains conjugated to Alexa Fluor 350. Cells were examined by deconvolution microscopy. Insets in C and F are close-ups of the boxed areas. Arrows show localization of GFP-SNX1 or GFP-SNX2 to microdomains of endosomes that are distinct from EEA1.
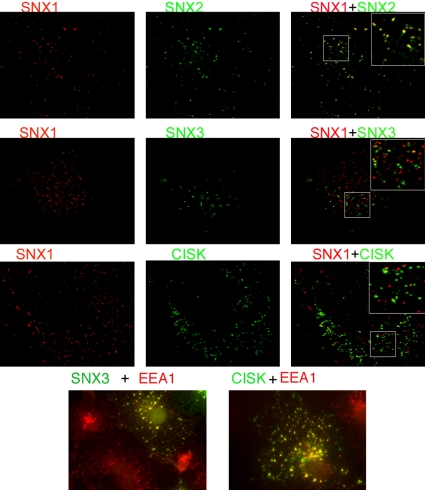
Comparison of the Subcellular Localization of SNX1 with SNX2, SNX3, and CISK
We next compared the subcellular localization of SNX1 with SNX2, SNX3, and CISK. As shown in Figure 6, top, deconvolution microscopy indicates that transfected SNX2 is largely colocalized with endogenous SNX1, confirming previous data using two transfected proteins (Zhong et al., 2002). There is an ∼80% concordance of SNX1 and SNX2 on endosomal vesicles. Similar colocalization was observed using transfected SNX1 with detection of endogenous SNX2 (our unpublished data). In contrast, transfected SNX3- and CISK-containing vesicles are largely distinct from vesicles that contain endogenous SNX1 (Figure 6, two middle panels). Approximately 90% of SNX3- and ∼80% of CISK-positive vesicles are distinct from endogenous SNX1-positive vesicles. These results indicate that SNX1 and SNX2 colocalize and are located on EEA1-containing endosomes; SNX3 and CISK are localized to a different subset of endosomes than SNX1. The SNX3 and CISK endosomes also contain EEA1 (Figure 6, bottom). These results suggest heterogeneity in the composition of PX domain containing proteins in EEA1-positive endosomes.
Figure 6.
Comparison of the localization of SNX1 with SNX2, SNX3, and CISK. The vesicular localization of endogenous SNX1 was compared with the vesicular localization of transfected GFP-SNX2, GFP-SNX3, and GFP-CISK by using immunofluorescent deconvolution microscopy. Endogenous SNX1 plus GFP-SNX2. Endogenous SNX1 plus GFP-SNX3. Endogenous SNX1 plus GFP-CISK. Endogenous EEA1 plus GFP-SNX3 and GFP-CISK. Panels on the right are merged images, and insets are higher magnification of the boxed areas.
Role of the C-Terminal Domain of SNX1
Although the PX domain of SNX1 fails to bind to PI(3)P-containing endosomes, holo SNX1 localizes precisely indicating other structural features of SNX1 are essential.
SNX1 and SNX2 form both homo- and heterodimers with the C-terminal domain of these proteins contributing strongly to formation of dimers (Haft et al., 1998; Wang et al., 2002; Zhong et al., 2002). The C-terminal domain of SNX2, which is highly homologous to that of SNX1, is proposed to function like the obligate dimeric BAR domains of amphiphysin that binds to curved membrane surfaces (Peter et al., 2004). The C-terminal domain is located on vesicle membranes (Teasdale et al., 2001) but, as shown in Figure 7A, these vesicles do not correspond to EEA1-containing endosomes where holo SNX1 resides (Figure 7B). The C-terminal domain of SNX1 thus targets to vesicles, but the PX domain is necessary for proper endosomal localization.
Figure 7.
Vesicular localization of dimeric SNX1 PX domains. (A) GFP-C terminal domains of SNX1 (aa 272–522) was transfected into COS 7 cells, and its localization was compared with EEA1 by using immunofluorescence microscopy. (B) Comparison of the localization of holo SNX1 with EEA1.
Because holo SNX1 is an obligate dimer, we hypothesized that dimerization of the PX domain is necessary to increase its affinity for PIs and properly localize to endosomal membranes. To test this hypothesis, we dimerized the SNX1 PX domain by adding the 40-residue leucine zipper of GCN4 (Oas et al., 1990) to its C terminus. Dimerization via this short leucine zipper minimally increased targeting of the SNX1 PX domain to vesicles (<10% of expressor cells; our unpublished data). We also fused the SNX1-PX domain to the self-dimerizing domain of the lambda repressor with similar results. The long C-terminal domain dimers of SNX1, which have affinity for membranes but not specificity, seem necessary for proper targeting of the PX domain-containing holo SNX1 protein.
DISCUSSION
The sorting nexin family of proteins are essential components of vesicle membrane external coats that, in the best studied examples, function in movement of cargo through the endosomal system. Proper membrane localization depends on the PX domains of SNX proteins that bind to PI(3)P, although involvement of phosphorylation of other sites on the inositol ring has not been rigorously excluded. Although a minority of PX domains bind with high-affinity to PI(3)P in vitro and localize to membranes in vivo, most exhibit low-affinity binding in vitro and do not localize in vivo (Yu and Lemmon, 2001). The PX domains of SNX1 and SNX2 fall into this latter class. Although SNX1 localizes to EEA1-containing endosomes, its PX domain fails to localize.
Interestingly, holo SNX1 may localize to a different microdomain of endosomal vesicles than EEA1. Analysis of various intracellular membrane compartments indicates these contain distinct subdomains (Pfeffer 2003). In endosomes, Rab proteins localize to different microdomains that likely represent sites of vesicle entry and exit (Zerial and McBride, 2001). PI(3)P also localizes to microdomains of vesicles, and EEA1 and CISK colocalize with PI(3)P in these microdomains (Gillooly et al., 2003). SNX1 seems to localize to a different population of endosomes than SNX3 and CISK, but it also colocalizes with EEA1. It is possible that SNX1 and SNX2 colocalize due to their proclivity to form heterodimers, whereas the lack of colocalization of SNX1 with SNX3 and CISK may result from competition for PI(3)P. Because EEA1 also binds to PI(3)P and all four PX domain-containing proteins colocalize with EEA1, endosomal heterogeneity seems more likely.
Despite its low-affinity binding to PI(3)P, the SNX1 PX domain retains the fold of high-affinity PX domains. In the high-affinity domains, phosphoinositide binding occurs in a conserved pocket between the β1-β2 loop, the β3-α1 loop, the proline-rich α2 loop, and the N-terminal part of α2 (Figure 3), through the interaction of the phosphates and inositol hydroxyls with a number of basic residues, and the stacking of the inositol ring against a conserved aromatic residue (Bravo et al., 2001). In p40phox, for example, the 3-phosphate of the phosphoinositide head-group interacts with the side chain of R58 and the backbone NH groups of Y59 and R60. The 4- and 5-hydroxyls form hydrogen bonds with R105, and the 1-phosphate is bound by the side chain of K92. The inositol ring forms a hydrophobic stacking interaction against Y59. R58, Y59, K92, and R105 are conserved or have an equivalent in all of the high-affinity PX domains and are also retained in the low-affinity SNX1-PX domain corresponding to R45, F46, K82 (or K80 or K85), and R97. However, in the SNX1-PX domain, parts of this region of the protein (for example that containing K82) are not fully folded, whereas in the high-affinity PX domains in the absence of phosphoinositide, the binding pocket is largely preformed (Bravo et al., 2001; Lu et al., 2002; Zhou et al., 2003). This points to a likely source of the attenuation in the affinity of the SNX1 PX domain for phosphoinositide, particularly as the region containing K82 becomes immobilized on phosphoinositide binding. Thermodynamically, the cost in free energy of making the complete binding pocket of the SNX1-PX domain reduces the binding affinity otherwise achievable. The packing of the β1-β2 loop against the polyproline-α2 loop and steric hindrance around the conserved arginine that binds the 4- and 5-positions of the inositol (R97 in SNX1) have been proposed to be factors that influence ligand specificity in PX domains (Bravo et al., 2001; Karathanassis et al., 2002; Xing et al., 2004). The mobility of this region in the isolated SNX1 PX domain also may contribute to the relaxed specificity of SNX1-PX in in vitro assays (Zhong et al., 2002; Cozier et al., 2002).
Because holo SNX1 is dimeric, we tested whether creating a dimeric PX domain would increase binding affinity, as assessed by in vivo localization to endosomal membranes whose principal phosphatidylinositol is phosphorylated at the 3-position (Lemmon, 2003). Dimerization via the leucine zipper of GCN4 or the dimerization domain of the lambda repressor had little to no effect. In holo SNX1, dimerization is driven by the C-terminal domain, which has homology to the BAR domain of amphiphysin, a long obligate helical dimer that binds to curved membrane surfaces (Peter et al., 2004). However, the C-terminal domain of SNX1, although it localizes to vesicle membranes, lacks the targeting specificity of holo SNX1. Endosomal specificity is provided by the PX domain. There are additional proteins that form the “retromer” complex in both yeast and mammalian cells (Seaman et al., 1998; Haft et al., 2000). These proteins also may stabilize dimers of holo SNX1 and SNX2 to favor PI(3)P binding. Although two tandem copies of the PH domain of phospholipase C-δ1 increased plasma membrane localization (Levine and Munro, 2002), this chimeric construct likely acts differently from the PX domains of SNX1 where the dimeric C-terminal domain may act as a more dominant anchor to the membrane and may alter the confirmation of the PX domain. We cannot distinguish whether the affinity enhancement of holo SNX1 versus the isolated PX domain is a result of a conformational change in the PI binding site or whether it is a chelating effect from the anchoring of the C-terminal domain to the membrane, or a combination of both.
Together, these results support a model in which the C-terminal domain of SNX1 (and of SNX2) binds to vesicle membranes as an obligate dimer; dimerization enhances PI binding affinity to provide proper endosomal targeting specificity. Future structural studies will be necessary to determine how dimerization affects the binding pocket of the PX domain and the lipid binding specificity of such dimers.
Acknowledgments
We thank Carolan Buckmaster for expert help in microinjection studies, Steve McMullen and Dr. James Feramisco for expert help with deconvolution microscopy, and Drs. Jeremy Craven and Lee Higgins for specialized NMR assistance. We thank Dr. Kevin Struhl for the GCN4 plasmid and Accelrys for the provision of Felix. These studies were supported by National Institutes of Health grant PO1CA58689 (to G.N.G.) and Biotechnology and Biological Sciences Research Council grant C18024 (to J.P.W.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0504) on January 26, 2005.
References
- Barr, V. A., Phillips, S. A., Taylor, S. I., and Haft, C. R. (2000). Overexpression of a novel sorting Nexin, SNX15, affects endosome morphology and protein trafficking. Traffic 1, 904-916. [DOI] [PubMed] [Google Scholar]
- Bravo, J., et al. (2001). The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol. Cell 8, 829-839. [DOI] [PubMed] [Google Scholar]
- Brunger, A. T., et al. (1998). Crystallography and NMR System. Acta Crystallogr. D54, 905-921. [DOI] [PubMed] [Google Scholar]
- Burden, J. J., Sun, X.-M., Garcia, A. B. G., and Soutar, A. K. (2004). Sorting motifs in the intracellular domain of the low density lipoprotein receptor interact with a novel domain of sorting Nexin 17. J. Biol. Chem. 279, 16237-16245. [DOI] [PubMed] [Google Scholar]
- Corlinescu, G., Delaglio, F., and Bax, A. (1999). Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- Cozier, G. E., Carlton, J., McGregor, A. H., Gleeson, P. A., Teasdale, R. D., Mellor, H., and Cullen, P. J. (2002). The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting Nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 48730-48736. [DOI] [PubMed] [Google Scholar]
- Florian, V., Schluter, T., and Bohnensack, R. (2001). A new member of the sorting Nexin family interacts with the C-terminus of P-selectin. Biochem. Biophys. Res. Commun. 281, 1045-1050. [DOI] [PubMed] [Google Scholar]
- Gillooly, D. J., Raiborg, C., and Stenmark, H. (2003). Phosphatidyl Inositol 3-phosphate found in microdomains of early endosomes. Histochem. Cell Biol. 120, 445-453. [DOI] [PubMed] [Google Scholar]
- Gullapalli, A., Garrett, T. A., Paing, M. M., Griffin, C. T., Yang, Y., and Trejo, J. A. (2004). A role for sorting Nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting Nexin 1 and 2 in protein trafficking. Mol. Biol. Cell 15, 2143-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft, C. R., de la Luz Sierra, M., Barr, V. A., Haft, D. H., and Taylor, S. I. (1998). Identification of a family of sorting Nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 18, 7278-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft, C. R., de la Luz Sierra, M., Bafford, R., Lesniak, M. A., Barr, V. A., and Taylor, S. I. (2000). Human orthologs of yeast vacuolar protein sorting proteins Vps 26, 29 and 35: assembly into multimeric complexes. Mol. Biol. Cell 11, 4105-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroaki, H., Ago, T., Ito, T., Sumimoto, H., and Kohda, D. (2001). Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 8, 526-530. [DOI] [PubMed] [Google Scholar]
- Karathanassis, D., Stahelin, R. V., Bravo, J., Perisic, O., Pacold, C. M., Cho, W., and Williams, R. L. (2002). Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21, 5057-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurten, R. C., Cadena, D. L., and Gill, G. N. (1996). Enhanced degradation of EGF receptors by a sorting Nexin, SNX. Science 272, 1008-1010. [DOI] [PubMed] [Google Scholar]
- Kurten, R. C., Eddington, A. D., Chowdhury, P., Smith, R. D., Davidson, A. D., and Shank, B. B. (2001). Self assembly and binding of a sorting Nexin to sorting endosomes. J. Cell Sci. 114, 1743-1756. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., Rullman, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477-486. [DOI] [PubMed] [Google Scholar]
- Lemmon, M. A. (2003). Phosphoinositide recognition domains. Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (2002). Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase dependent and independent components. Curr. Biol. 12, 695-704. [DOI] [PubMed] [Google Scholar]
- Lu, L., Garcia, J., Dulubova, I., Sudhof, T. C., and Rizo, J. (2002). Solution structure of the Vam7p PX domain. Biochemistry 41, 5956-5962. [DOI] [PubMed] [Google Scholar]
- Oas, T. G., McIntosh, L. P., O'Shea, E. K., Dahlquist, F. W., and Kim, P. S. (1990). Secondary structure of a leucine zipper determined by nuclear magnetic resonance spectroscopy. Biochemistry 29, 2891-2894. [DOI] [PubMed] [Google Scholar]
- Otsuki, T., Kajigaya, S., Ozawa, K., and Liu, J. M. (1999). SNX5, a new member of the sorting Nexin family, binds to the fanconi anemia complementation group A protein. Biochem. Biophys. Res. Commun. 265, 630-635. [DOI] [PubMed] [Google Scholar]
- Parks, W. T., et al. (2001). Sorting Nexin 6, a novel SNX, interacts with the transforming growth factor-β family of serine-threonine kinases. J. Biol. Chem. 276, 19332-19339. [DOI] [PubMed] [Google Scholar]
- Peter, B. J., Kent, H. M., Mills, I. G., Vallis, Y., Butler, P. J. G., Evans, P. R., and McMahon, H. T. (2004). BAR domains and sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495-499. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. (2003). Membrane domains in secretary and endocytic pathways. Cell 112, 507-517. [DOI] [PubMed] [Google Scholar]
- Pons, V., Hullin-Matsuda, F., Nauze, M., Barbaras, R., Peres, C., Collet, X., Perret, B., Chap, H., and Gassama-Diagne, A. (2003). Enterophilin-1, a new partner of sorting Nexin 1, decreases cell surface epidermal growth factor receptor. J. Biol. Chem. 278, 21155-21161. [DOI] [PubMed] [Google Scholar]
- Reed, M. A. C., et al. (2003). Effects of domain dissection on the folding and stability of the 43 kDa protein PGK probed by NMR. J. Mol. Biol. 330, 1189-1201. [DOI] [PubMed] [Google Scholar]
- Sato, T. K., Overduin, M., and Emr, S. D. (2001). Location, location, location: membrane targeting by PX domains. Science 294, 1881-1885. [DOI] [PubMed] [Google Scholar]
- Schwarz, D. G., Griffin, C. T., Schneider, E. A., Yee, D., and Magnuson, T. (2002). Genetic analysis of sorting nexins 1 and 2 reveals a redundant and essential function in mice. Mol. Biol. Cell 13, 3588-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M. N. J., McCaffery, J. M., and Emr, S. D. (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, R. D., Loci, D., Houghton, F., Karlsson, L., and Gleeson, P. A. (2001). Large family of endosome-localized proteins related to sorting Nexin 1. Biochem. J. 358, 7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaw, P., Baxter, N. J., Hounslow, A. M., Price, C., Waltho, J. P., and Craven, C. J. (2001). Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat. Struct. Biol. 8, 701-704. [DOI] [PubMed] [Google Scholar]
- Virbasius, J. V., Song, X., Pomerleau, D. P., Zhan, Y., Zhou, G. W., and Czech, M. P. (2001). Activation of the Akt-related cytokine-independent survival kinase requires interaction of its phox domain with endosomal phosphatidyl inositol 3-phosphate. Proc. Natl. Acad. Sci. USA 98, 12908-12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos, W., and Stevens, T. H. (1998). Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd 19p. J. Cell Biol. 140, 577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhou, Y. Szabo, K., Haft, C. R., and Trejo, J. A. (2002). Down regulation of protease-activated receptor-1 is regulated by sorting Nexin 1. Mol. Biol. Cell 13, 1965-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby, C. A., and Dixon, J. E. (2002). Sorting out the cellular function of sorting Nexins. Nat. Rev. Mol. Cell. Biol. 3, 919-931. [DOI] [PubMed] [Google Scholar]
- Worby, C. A., Simonson-Laff, N., Clemens, J. C., Kruger, R. P., Muda, M., and Dixon, J. E. (2001). The sorting nexin DSH3PX1, connects the axonal guidance receptor, DSCAM, to the actin cytoskeleton. J. Biol. Chem. 276, 41782-41789. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Liu, D., Zhang, R., Joachimiak, A., Songyang, Z., and Xu, W. (2004). Structural basis of membrane targeting by the phox homology domain of cytokine-independent survival kinase (CISK-PX). J. Biol. Chem. 279, 30662-30669. [DOI] [PubMed] [Google Scholar]
- Xu, J., Liu, D., Gill, G. N., and Songyang, Z. (2001a). Regulation of protein kinase cytokine-independent survival kinase (CISK) by the phox homology domain and phosphoinositides. J. Cell Biol. 154, 699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Hortsman, H., Seet, L., Wong, S. H., and Hong, W. (2001b). SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns (3)P. Nat. Cell Biol. 3, 658-666. [DOI] [PubMed] [Google Scholar]
- Yu, J. W., and Lemmon, M. A. (2001). All phox homology (PX) domains from Saccharomyces cervisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179-44184. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107-119. [DOI] [PubMed] [Google Scholar]
- Zhong, Q., Lazar, C. S., Tronchere, H., Sato, T., Meerloo, T., Yeo, M., Songyang, Z., Emr, S. D., and Gill, G. N. (2002). Endosomal localization and function of sorting Nexin 1. Proc. Nat. Acad. Sci USA 99, 6767-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C.-Z., et al. (2003). Crystal structure of the yeast phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278, 50371-50376. [DOI] [PubMed] [Google Scholar]