Abstract
The chemical structure of the lipid A of the lipopolysaccharide component isolated from Flavobacterium meningosepticum IFO 12535 was elucidated. Methylation and nuclear magnetic resonance analyses showed that two kinds of hydrophilic backbone exist in the free lipid A: a β (1→6)-linked 2-amino-2-deoxy-d-glucose, which is usually present in enterobacterial lipid A’s, and a 2-amino-6-O-(2,3-diamino-2,3-dideoxy-β-d-glucopyranosyl)-2-deoxy-d-glucose, in a molar ratio of 1.00:0.35. Both backbones were α-glycosidically phosphorylated in position 1, and the hydroxyl groups at positions 4, 4′, and 6′ were unsubstituted. Liquid secondary ion-mass spectrometry revealed a pseudomolecular ion at m/z 1673 [M-H]− as a major monophosphoryl lipid A component carrying five acyl groups. Fatty acid analysis showed that the lipid A contained 1 mol each of amide-linked (R)-3-OH iC17:0, ester-linked (R)-3-OH iC15:0, amide-linked (R)-3-O-(iC15:0)-iC17:0, and both amide- and ester-linked (R)-3-OH C16:0. Fatty acid distribution analyses using several mass spectrometry determinations demonstrated that the former two constituents were distributed on positions 2 and 3 of the reducing terminal unit of the backbones and that the latter two were attached to the 2′ and 3′ positions in the nonreducing terminal residue.
Lipopolysaccharide (LPS) is known to act as an endotoxin that mediates pathophysiological changes such as fever and shock which occur in the course of severe gram-negative bacterial infection (5, 18, 27). The pathophysiological activity of LPS depends on the chemical structure of the hydrophobic portion called lipid A, the biologically active center of LPS (12, 14), which generally consists of a β(1→6)-linked 2-amino-2-deoxy-d-glucose (GlcN) disaccharide carrying phosphate and fatty acid residues; many fine structural variations are observed in different bacterial families (38).
Since many of the LPSs from various gram-negative bacteria cause similar endotoxic effects despite differences in chemical composition and positions of substitution, the chemical structure required for the activity does not seem to be very strict. It has been reported, however, that several lipid A forms, isolated from the LPSs of Porphyromonas gingivalis (16), Rhodobacter sphaeroides (22, 26) and Rhodobacter capsulatus (20), as well as chemically synthesized lipid A analogs (6, 12), which are structurally similar to the active-type lipid A, exhibit dramatically low endotoxicity. Biologically active lipid A has been found to be changed to completely nontoxic derivatives by simple chemical modifications (28, 29). These findings indicate that the biological activity of lipid A is controlled by the fine structural variations. The nontoxic or low-toxicity lipid A preparations are very important for the determination of the relationship between the chemical structure and biological activity of lipid A, and also for the systematic development of LPS antagonists. However, the essential structural requirements for the complete activity or nontoxicity of lipid A are still uncertain. It is, therefore, meaningful to study the chemical and biological properties of naturally occurring lipid A’s which possess a unique structure.
Flavobacterium meningosepticum is an aerobic gram-negative rod which is known to cause meningitis and septicemia in newborn infants (4, 21). Interestingly, the bacterium does not induce Limulus gelation activity when tested with whole cells (32), strongly suggesting that the LPS is of low toxicity or nontoxic and that the lipid A must have a unique structure relative to other enterobacterial lipid A’s.
In the present study, the chemical structure of lipid A isolated from F. meningosepticum LPS was characterized by compositional study, methylation analysis, mass spectrometry (MS), and nuclear magnetic resonance (NMR) spectroscopy.
MATERIALS AND METHODS
Bacteria and preparation of LPS.
F. meningosepticum IFO 12535 strain was obtained from the Institute for Fermentation Osaka. The bacterium was cultivated in a fermentor at 30°C for 16 h in a medium consisting of 1% (wt/vol) polypeptone, 0.2% (wt/vol) beef extract, and 0.1% (wt/vol) MgSO4 · 7H2O, at pH 7.0. The heat-killed cells were harvested by centrifugation and washed with distilled water, acetone, ethanol, and diethyl ether, then acetone dried (yield, 0.5 g of cells/liter).
LPS (7.1 mg/g [dry weight] of cells) was extracted from the acetone-dried cells with a mixture of phenol-chloroform-petroleum ether (2:5:8 [vol/vol/vol]) according to the method described by Galanos et al. (7) and was purified by RNase and DNase (Sigma) treatments (36) and repeated ultracentrifugation (105,000 × g, 3 h, six times).
Isolation of lipid A.
Purified LPS (1.23 g) was hydrolyzed with 1% (vol/vol) aqueous acetic acid at 100°C for 2 h, followed by centrifugation (14,000 × g, 10 min). The sediment was washed with distilled water and crude lipid A was obtained after lyophilization. The crude lipid A (708.6 mg) was purified by Sephadex LH-20 column chromatography according to the method described in reference 16 to yield 420 mg of purified lipid A.
Chemical modification of LPS and lipid A.
Lipid A backbone was prepared according to the method of Hase and Rietschel (11). Briefly, LPS (20 mg) was treated with 0.17 M NaOH (100°C, 1 h) and 0.1 M HCl (100°C, 30 min), followed by reduction with NaBH4 (37°C, 16 h), complete hydrazinolysis (103°C, 40 h), and N acetylation to obtain the reduced and N-acetylated lipid A backbone. Escherichia coli F654 R3 LPS was treated by the same method as reference material.
De-O-acylation of LPS and lipid A was performed by treatment with anhydrous hydrazine at 60°C for 30 min, and the de-O-acylated preparations were recovered by a method reported previously, (13).
De-O-acylated LPS was hydrolyzed in 0.1 M HCl at 100°C for 30 min. The centrifugal sediment recovered was treated with acetic anhydride-pyridine (1:1 [vol/vol]) containing small amounts of 4-dimethylaminopyridine at room temperature for 16 h, followed by purification using silica gel column chromatography (column, 1 by 60 cm) with toluene-ethyl acetate (20:1 [vol/vol]) as the eluent at a flow rate of 1 ml/min. Each 1-ml volume was fractionated, and fractions 32 to 36 were collected to yield the peracetylated derivative of hybrid backbone carrying three N-acyl and no phosphate groups.
Monophosphoryl-methylated lipid A was prepared by treating lipid A with diazomethane according to the method of Qureshi et al. (23).
GLC conditions.
Gas-liquid chromatography (GLC) analysis was performed on a model GC-14A (Shimadzu) with a HiCap-CBM5 fused silica capillary column (25 m by 0.25 mm; GL Science) and temperature programs A (120°C for 3 min increasing to 250°C at 3°C/min), B (150°C for 3 min, increasing to 300°C at 5°C/min), C (140°C for 3 min, increasing to 250°C at 3°C/min), and D (180°C for 3 min, increasing to 300°C at 5°C/min). A chemically bonded DB-5 fused silica capillary column (DB5MS, 30 m by 0.32 mm; J & W Scientific) was used for the determination of the absolute configurations of amino sugars with temperature program E (180°C for 3 min, increasing to 250°C at 3°C/min). Nitrogen was used as the carrier gas.
Analytical methods.
Each temperature program is described under “GLC conditions” above. Total fatty acids were determined by means of GLC and GLC-MS using temperature program A, described under “GLC conditions” above as the methyl ester according to the method of Haeffner et al. (8). 2-Hydroxydecanoic acid and nonadecanoic acid (GL Science) were used as internal standards. Ester- and amide-linked fatty acids were also assigned by GLC and GLC-MS using temperature programs A and B, respectively, according to methods described elsewhere (13, 16, 24, 37).
Neutral and amino sugars were analyzed by GLC and GLC-MS (program C) as the alditol acetate derivatives (9). The absolute configurations of amino sugar constituents were determined by GLC analysis (program E) of the peracetylated S-2-butylglycoside derivatives according to the method described in reference 13. Amino sugars present in the lipid A possessed a d-configuration.
Colorimetric determination of GlcN was performed by using the Morgan-Elson reaction (25). Total phosphorus was measured by the method of Lowry et al. (17).
Methylation of the lipid A backbone was performed by the method of Hakomori (10, 13), and the methylated products were analyzed by GLC-MS using temperature program D.
Protein contents were determined with an L-8500 amino acid analyzer (Hitachi) after hydrolysis at 110°C for 24 h in 6 M HCl containing 1% (wt/vol) phenol.
Determination of the R,S configuration of 3-hydroxy fatty acid.
Total fatty acids obtained from lipid A (10 mg) were carboxymethylated with diazomethane at 0°C for 1 min. To determine the R,S configuration, the product (1 mg) was directly analyzed by 1H NMR spectroscopy under the usual conditions and in the presence of a shift reagent (1 mg), Tris-[3-(heptafluoroprolyl-hydroxymethylene)-(+)-camphorate] europium(III) derivative (2, 13). Racemic 3-hydroxyhexadecanoic acid methyl ester was used as a reference compound.
MS.
Liquid secondary ion (LSI)-mass spectra were measured on a ZAB-2SEQ instrument (VG Analytical) in the positive- and negative-ion modes under the conditions described previously (13). A mixture of m-nitrobenzyl alcohol for the positive-ion mode and m-nitrobenzyl alcohol–triethanolamine (1:1 [vol/vol]) for the negative-ion mode, each containing a small amount of Kryptofix 222 (Aldrich), was used as the matrix.
GLC-MS and fast atom bombardment-tandem MS (FAB-MS/MS) were carried out by the methods previously reported (16).
NMR spectroscopy.
1H NMR spectra of 3-hydroxy fatty acid methyl ester were recorded at 400 MHz (Varian UNITY plus-400) in CDCl3 at room temperature. Tetramethylsilane (TMS) was used as an internal standard (0.00 ppm) of chemical shifts.
One- and two-dimensional NMR spectra of de-O-acylated and peracetylated lipid A and monophosphoryl-methylated lipid-A derivatives were measured with a JEOL α-600 instrument in CDCl3 and C6D6–(CD3)2SO (9:1 [vol/vol]) at 25°C. TMS (0.00 ppm) was used as an internal standard for 1H and 13C NMR experiments, and 31P NMR spectra were referenced to triphenyl phosphate (−18.0 ppm) as an external standard. Assignments were made by a field gradient 1H,1H-homonuclear correlation spectroscopy (COSY) experiment and by 13C,1H- and 31P,1H-heteronuclear multiple quantum coherence (HMQC) analyses.
RESULTS
Chemical analysis of free lipid A.
The chemical composition of lipid A isolated from F. meningosepticum LPS is shown in Table 1. Fatty acid analysis of the lipid A revealed the presence of 13-methyltetradecanoic (iC15:0), 3-hydroxy-13-methyltetradecanoic (3-OH iC15:0), 3-hydroxyhexadecanoic (3-OH C16:0), and 3-hydroxy-15-methylhexadecanoic (3-OH iC17:0) acids in a ratio of approximately 1:1:1:2. 1H NMR analysis of the fatty acid fraction obtained from the lipid A indicated that these 3-hydroxy fatty acids possessed the R configuration, because the carboxymethyl proton of 3-hydroxy fatty acid methyl esters, resonating at 3.79 ppm under usual conditions, was dose-dependently shifted to a lower field by addition of europium(III) complex shift reagent, and the shift behavior was identical to that of authentic (R)3-hydroxyhexadecanoic acid methyl ester.
TABLE 1.
Chemical composition of lipid A isolated from F. meningosepticum LPS
| Component | Amt (nmol/mg) |
|---|---|
| d-Glucosamine | 611.0 |
| 2,3-Diamino-hexosea | 86.5 |
| Total phosphate | 467.0 |
| Fatty acids | |
| iC15:0 | 344.1 |
| (R)-3-OH iC15:0 | 442.1 |
| (R)-3-OH C16:0 | 391.2 |
| (R)-3-OH iC17:0 | 957.7 |
2,3-Diamino-hexose was identified as 2,3-dideoxy-2,3-di-amino-glucose by NMR analyses.
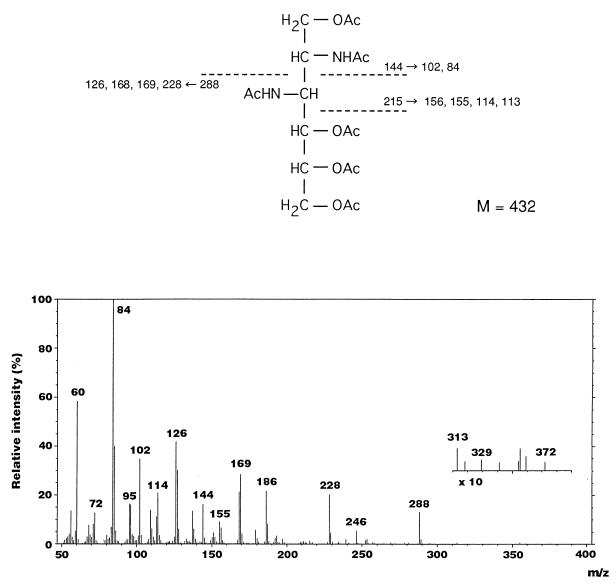
The lipid A also contained 467.0 nmol of total phosphate/mg, 416.7 nmol of GlcN/mg, and 86.5 nmol of 2,3-diamino-2,3-dideoxy-d-glucose (GlcN3N)/mg the latter being identified by GLC-MS and NMR analyses. In the electron impact (EI)-mass spectrum of the alditol acetate derivative of GlcN3N, characteristic fragment ions that originated from the cleavage between C-2 and C-3 were detected at m/z 144 (C-1 to C-2 [C1-C2] fragment) and m/z 288 (C3-C6 fragment), as shown in Fig. 1. Several daughter ions were observed at m/z 228, 169, 168, 126, 102, and 84, caused by elimination of acetic acid (−60) or an N-acetyl group (−42) from C3-C6 and C1-C2 fragment ions. Ions at m/z 156, 155, 114, and 113 were assigned as daughter ions that originated from the C1-C3 fragment.
FIG. 1.
EI-mass spectrum and fragmentation pattern of the peracetylated derivative of GlcN3N found in F. meningosepticum lipid A.
The protein content of the lipid A determined by amino acid analysis was 0.36% (wt/wt).
Determination of linkage type of fatty acid.
Ester-linked fatty acids were investigated by trans-esterification of the lipid A with 0.25 M sodium methylate treatment. One mole each of iC15:0 and (R)-3-OH iC15:0, and a part of (R)-3-OH C16:0 methyl esters, were detected as O-acyl residues, and no free fatty acid was detected in the methanolysates by GLC analysis, indicating that iC15:0, (R)-3-OH iC15:0, and a part of (R)-3-OH C16:0 exist as ester-linked residues in F. meningosepticum and that the hydroxyl groups of 3-hydroxy fatty acids bound directly to the hydroxyl groups of lipid A backbone are not esterified by the second acyl residue. Two moles of (R)-3-OH iC17:0 and a significant amount of (R)-3-OH C16:0 were quantitatively recovered as amide-linked fatty acids from the residual de-O-acylated lipid A.
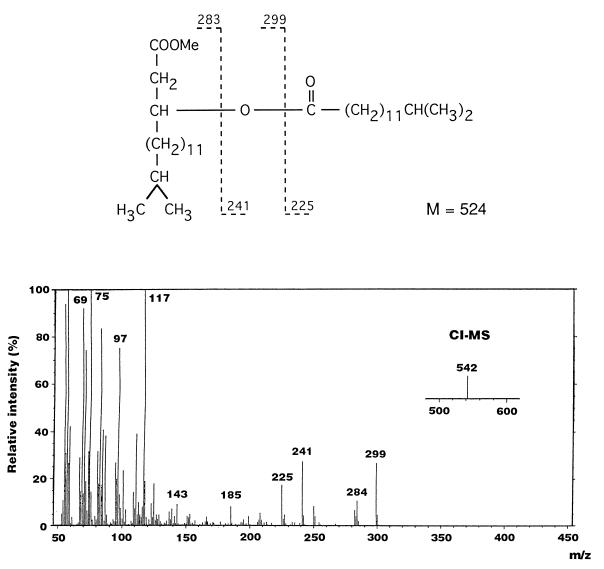
In the analysis of amide-linked 3-acyloxyacyl residues, an acyloxyacyl group was detected on GLC (retention time, 29 min) and was identified as (R)-3-O-(iC15:0)-iC17:0 methyl ester by GLC-MS. In the EI-mass spectrum (Fig. 2), characteristic fragment ions generated from the cleavage of the second acyl residue of the methyl ester were observed at m/z 225, 241, and 299. The molecular mass of the derivative was confirmed to be 524 Da (m/z 542 [M+NH4]+) in chemical ionization-MS. These results indicate that ester-linked iC15:0 exists as a second acyl residue of 1 mol of amide-linked (R)-3-OH iC17:0 in the lipid A.
FIG. 2.
EI-mass spectrum and fragmentation pattern of methyl ester derivatives of (R)-3-O-(13-methyltetradecanoyl)-15-methylhexadecanoic acid found in F. meningosepticum lipid A.
Analysis of lipid A backbone.
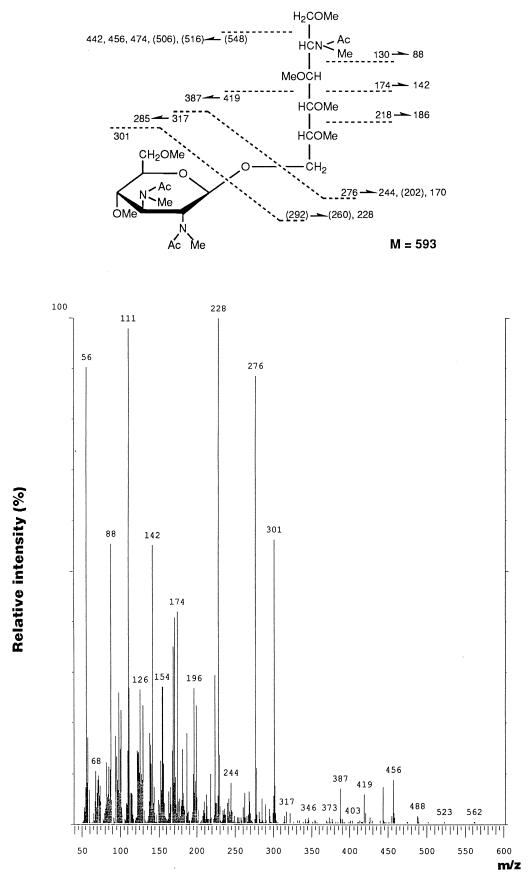
The chemical structure of the lipid A backbone was determined by methylation analysis. Two major peaks were observed at 17.48 and 22.1 min on GLC at an intensity ratio of 1.00:0.35. The former was identified as 6-O- [2-deoxy-3,4,6-tri-O-methyl-2-(N-methylacetamido)-β-d- glucopyranosyl)-2-deoxy-1,3,4,5-tetra-O-methyl-2-(N-methyl- acetamido)-d-glucitol, which originated from the usual GlcN disaccharide backbone, because the EI-mass spectrum and retention time were identical to those of the same derivative prepared from E. coli R3 LPS possessing a β(1→6)-linked GlcN disaccharide lipid A backbone. On the other hand, the latter peak was identified as 6-O-[2,3-dideoxy-4,6-di-O-methyl- 2,3-di-(N-methylacetamido)-d-glucopyranosyl]-2-deoxy- 1,3,4,5-tetra-O-methyl-2-(N-methylacetamido)-d-glucitol. In the EI-mass spectrum (Fig. 3), fragment ions representing cleavage of the glycosidic linkage of the hybrid disaccharide were recognized at m/z 276 and 301. A significant fragment ion at m/z 218, corresponding to cleavage of the C-4–C-5 bond, which was characteristic of the derivative of β(1→6)-linked GlcN disaccharide (3), was also detected in the spectrum. As shown in Fig. 3, other characteristic fragment ions were obtained at m/z 130 (C1-C2 fragment) and m/z 174 (C1-C3 fragment), as well as several daughter ions based on the loss of methanol (−32), acetic acid (−60), or an N-acetyl group (−42). This EI-mass spectrum was almost identical to that of the same derivative of β(1→6)-linked GlcN3N-GlcN disaccharide reported by Moran et al. (19). These results indicate that two kinds of lipid A backbones exist in the F. meningosepticum lipid A: one β(1→6)-linked GlcN disaccharide backbone that is normally present in enterobacterial lipid A’s and a (1→6)-linked GlcN3N-GlcN disaccharide, the GlcN3N residue of which was found to possess a β configuration by NMR analysis as described below.
FIG. 3.
EI-mass spectrum and fragmentation pattern of the permethylated 2-acetamido-6-O-(2,3-diacetamido-2,3-dideoxy-d-glucopyranosyl)-2-deoxy-d-glucitol derivative originating from the hybrid backbone present in F. meningosepticum lipid A.
Distribution of fatty acid and phosphate residues.
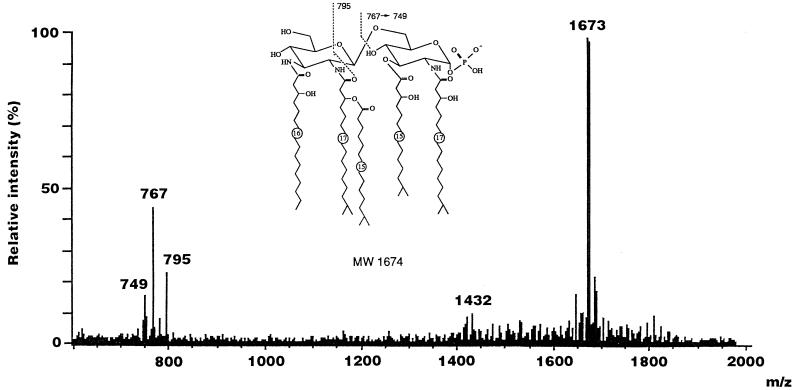
The distribution pattern of fatty acid and phosphate residues on the F. meningosepticum lipid A backbone was determined by LSI-MS and FAB-MS/MS. The LSI-mass spectrum of F. meningosepticum lipid A in the negative-ion mode is shown in Fig. 4. A predominant ion observed at m/z 1673 [M-H]− corresponds to monophosphoryl lipid A species carrying 1 mol each of (R)-3-OH iC15:0, (R)-3-OH C16:0, (R)-3-OH iC17:0, and (R)-3-O-(iC15:0)-iC17:0 on the lipid A backbone. Characteristic ions originating from the reducing terminal unit of the lipid A were also detected at m/z 767 and 795, which arise from the cleavage of the glycosidic linkage and C-1′–C-2′—C-1′–O bond, respectively (Fig. 4). The fragment ion at m/z 749 corresponds to the daughter ion caused by elimination of H2O from the ion at m/z 767. These fragment ions indicate that the reducing terminal unit of F. meningosepticum lipid A consists of monophosphoryl GlcN replaced by 1 mol each of (R)-iC15:0 and (R)-3-OH iC17:0 and that a nonreducing terminal residue contains 1 mol each of GlcN (or GlcN3N), (R)-3-OH C16:0, and (R)-3-O-(iC15:0)-iC17:0, respectively. These results also showed that the hydroxyl groups at positions 4 and 6 in the nonreducing terminal unit of the lipid A species having a GlcN3N-GlcN hybrid backbone exist in the free form, because the amide-linked (R)-3-OH C16:0 and (R)-3-O-(iC15:0)-iC17:0 are attached to positions 3 and 2 of the GlcN3N residue, respectively. Several species based on the loss of acyl and phosphate residues were usually observed in LSI-mass spectra in most of the lipid A preparations, but F. meningosepticum lipid A appears to be extremely homogeneous.
FIG. 4.
LSI-mass spectrum of F. meningosepticum lipid A in the negative-ion mode. A mixture of triethanolamine and 3-nitrobenzyl alcohol (1:1 [vol/vol]) containing a small amount of Kryptofix 222 was used as the matrix.
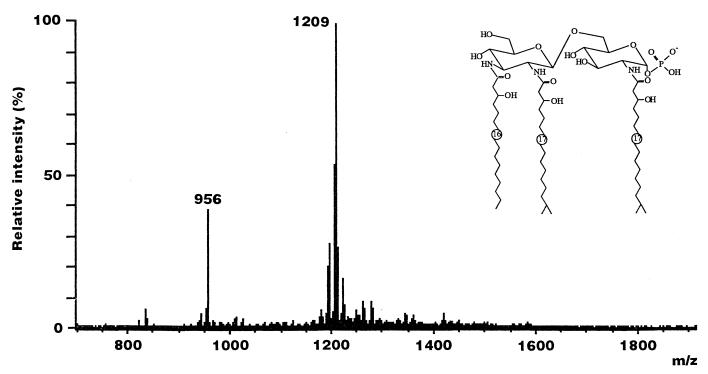
Since the difference in molecular mass between a GlcN disaccharide backbone and an identically acylated hybrid backbone is only 1 Da, which is within the uncertainty of mass scale calibration, it was necessary to degrade the material further to differentiate between the two backbone disaccharides. As shown in Fig. 5, two peaks were predominantly detected in the LSI-mass spectrum of the de-O-acylated lipid A of F. meningosepticum. A molecular ion was observed at m/z 1209 [M-H]−; it corresponds to the monophosphoryl lipid A species carrying three N-acyl residues, 1 mol of (R)-3-OH C16:0, and 2 mol of (R)-3-OH iC17:0 on the β(1→6)-linked GlcN3N-GlcN hybrid backbone. Another ion at m/z 956 [M-H]− was identified as a monophosphoryl GlcN disaccharide replaced by 2 mol of amide-linked (R)-3-OH iC17:0. Thus, the presence of a β(1→6)-linked GlcN3N-GlcN hybrid backbone in addition to a GlcN disaccharide backbone was again recognized in the LSI-mass spectrum of the de-O-acylated lipid A.
FIG. 5.
LSI-mass spectrum of F. meningosepticum de-O-acylated lipid A in the negative-ion mode. A mixture of triethanolamine and 3-nitrobenzyl alcohol (1:1 [vol/vol]) containing a small amount of Kryptofix 222 was used as the matrix.
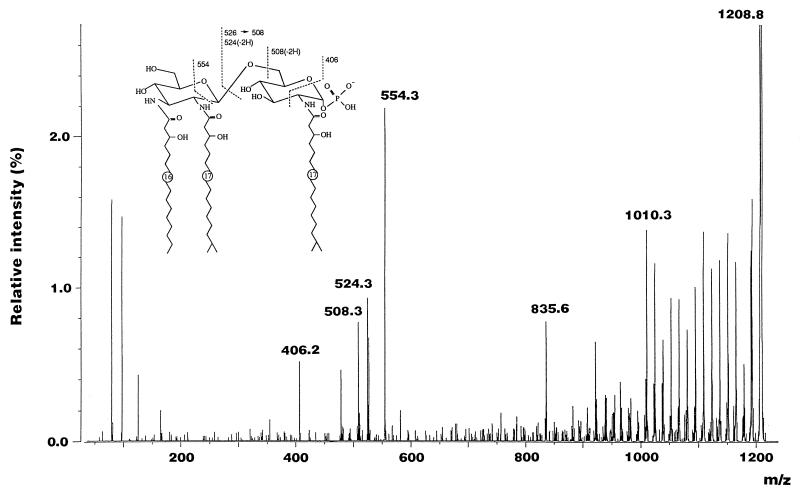
The pattern of distribution of acyl residues was also confirmed by FAB-MS/MS of the molecular ions at m/z 1209 [M-H]− and 956 [M-H]−, detected in the LSI-mass spectrum of the de-O-acylated lipid A (Fig. 5). The tandem spectrum of the ion at m/z 1209 [M-H]− is shown in Fig. 6. A fragment ion at m/z 406.2 caused by the cleavage of the C-1–O—C-2–C-3 bond of the reducing terminal unit of the backbone revealed that a phosphate residue was linked to position 1 of the backbone and that an amino group at position 2 was N acylated with (R)-3-OH iC17:0 (Fig. 6). Other characteristic fragment ions were observed at m/z 508.3 and 524.3 and m526.3z and 554.3; they were generated by the cleavage of the glycosidic linkage and C-1′–C-2′—C-5′-O bond, respectively (Fig. 6). The tandem spectrum of the ion at m/z 956 [M-H]− was almost identical to that of the ion at m/z 1209 [M-H]− (data not shown).
FIG. 6.
Negative-ion mode FAB-MS/MS of the molecular ion at m/z 1209 [M-H]− detected in the LSI-mass spectrum of F. meningosepticum de-O-acylated lipid A.
Structure of the peracetylated derivative of de-O-acylated lipid A having a hybrid backbone.
LSI-MS and NMR analyses of the peracetylated derivative of the hybrid backbone carrying three N-acyl and no phosphate residues were performed. In positive-ion mode LSI-MS, a molecular ion was detected at m/z 1467 [M+H]+, and a fragment ion originating from the nonreducing terminal unit was also observed at m/z 853, indicating that 1 mol each of (R)-3-OH C16:0 and (R)-3-OH iC17:0 was distributed on the nonreducing terminal unit and the remaining 1 mol of (R)-3-OH iC17:0 was present in the reducing terminal residue. The 1H and 13C NMR data are shown in Tables 2 and 3. In the 1H NMR analysis (Table 2), the J2,3 (10.4 Hz), J3,4 (9.9 Hz), and J4,5 (9.9 Hz) values of the nonreducing terminal unit indicated that this sugar constituent possesses the gluco-conformation, and the J1,2 value (8.4 Hz) revealed the β-configuration. Two amide protons were assigned at 6.48 ppm and 6.37 ppm, and they were cross-coupled with H-2 (3.96 ppm) and H-3 (4.09 ppm), respectively, indicating that this sugar unit possesses two amino groups at positions 2 and 3. On the other hand, the α- and β-anomers of the reducing terminal unit, which were produced during acetylation, were assigned. In both anomers, the J2,3, J3,4, and J4,5 values indicated the gluco-configuration, and an amino group exists at position 2, because the amide proton cross-coupled with H-2 was detected at 6.04 ppm for the α-anomer and 6.34 ppm for the β-anomer. H-6a and H-6b were slightly shifted to a higher field, because the nonreducing terminal unit was linked to position 6. These findings were also supported by the 13C NMR data shown in Table 3. These results clearly showed that the hybrid backbone present in F. meningosepticum lipid A consists of a β(1→6)-linked GlcN3N-GlcN hybrid disaccharide.
TABLE 2.
1H NMR data for the peracetylated derivative of F. meningosepticum de-O-acylated lipid A-HCl containing a hybrid backbonea
| Unit | Value for chemical shift (coupling constant)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3 | H-4 | H-5 | H-6a | H-6b | 2-NH | 3-NH | |
| Reducing terminal unit | |||||||||
| α-d-Glucosamine | 6.10 (3.3) | 4.41 (10.8) | 5.25 (9.4) | 5.15 (9.9) | 3.91 (2.2) | 3.41 (11.7) | 4.00 (4.8) | 6.04 (8.8) | |
| β-d-Glucosamine | 5.68 (8.8) | 4.29 (10.8) | 5.20 (9.2) | 4.91 (9.7) | 3.79 (2.3) | 3.68 (12.5) | 3.80 (NDc) | 6.34 (9.2) | |
| Nonreducing terminal unit, 2,3-di-amino-glucose | 4.38 (8.4) | 3.96 (10.4) | 4.09 (9.9) | 4.83 (9.9) | 3.70 (2.2) | 4.05 (12.6) | 4.24 (3.1) | 6.48 (7.7) | 6.37 (8.4) |
Spectra were recorded at 600 MHz in CDCl3 relative to TMS (0.00 ppm). Assignments were made by 1H,1H-COSY NMR. The α-, β-, γ-, methylene, and methyl protons of fatty acid residues were detected at 2.15 to 2.60, 4.94 to 5.24, 1.50 to 1.70, 1.15 to 1.35, and 0.85 to 0.90 ppm, respectively, and methyl protons of acetyl groups were detected at 2.00 to 2.25 ppm.
Chemical shift is expressed in parts per million; coupling constant (J) is expressed in hertz.
ND, not defined.
TABLE 3.
13C NMR data for the peracetylated derivative of F. meningosepticum de-O-acylated lipid A-HCl containing a hybrid backbonea
| Unit | Chemical shift (ppm)
|
|||||
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| Reducing terminal unit | ||||||
| α-d-Glucosamine | 91.4 | 51.6 | 71.0 | 69.2 | 71.4 | 67.7 |
| β-d-Glucosamine | 94.3 | 62.8 | 72.3 | 68.8 | 68.2 | 67.7 |
| Nonreducing terminal unit, 2,3-di-amino-glucose | 102.9 | 54.2 | 62.8 | 70.0 | 73.7 | 63.2 |
Spectra were recorded at 150.8 MHz in CDCl3 relative to TMS (0.00 ppm). Assignments were made by HMQC experiment. The β-, methylene, and methyl carbons of fatty acid residues were detected at 71.7 to 72.1, 27.4 to 39.1, and 20.3 to 26.5 ppm, respectively, and the methyl and carbonyl carbons of acetyl groups were detected at 19.2 to 25.0 and 168.5 to 172.4 ppm, respectively.
NMR analysis of monophosphoryl-methylated lipid A.
In order to determine the positions of free-hydroxyl groups and the attachment site of phosphate residue, one- and two-dimensional 1H NMR analyses of the monophosphoryl-methylated derivative of F. meningosepticum lipid A were performed (Table 4). The H-1 signal (6.01 ppm; J1,2 = 3.12 Hz) of the reducing terminal unit of the lipid A was shifted at 0.55 ppm to a lower field in comparison to the unsubstituted H-1 signal (5.46 ppm) (23), indicating that the phosphate residue was α-glycosidically linked to position 1. Since direct J coupling of the hydroxyl proton with H-4 (3.98 ppm) was detected at 5.86 ppm, the hydroxyl group at position 4 of the reducing terminal unit was identified as being free form. Furthermore, an amide proton was assigned at 8.12 ppm (2-NH; JNH,2 = 9.34 Hz), and it was cross-coupled with the proton at position 2 (4.68 ppm). The other signals that originated from the unit were detected at 4.10 ppm (H-6b), 4.19 ppm (H-5), 4.39 ppm (H-6a), and 5.64 ppm (H-3). An H-1′ proton was detected at 5.05 ppm (J1′,2′ = 7.51 Hz) as a signal originating from the nonreducing terminal unit of the lipid A. The resonation was shifted to a higher field (0.41 ppm) by glycosidical substitution, and the J1′,2′ value (7.51 Hz) revealed a β-configuration. However, no other signals were able to be clearly assigned because of the heterogeneity.
TABLE 4.
1H NMR data for F. meningosepticum phosphoryl-methylated lipid Aa
| Proton | Value for chemical shift (ppm)
|
|
|---|---|---|
| GlcN I | Fatty acid | |
| H-1 | 6.01b | |
| H-2 | 4.68 | |
| H-3 | 5.64 | |
| H-4 | 3.98 | |
| H-5 | 4.19 | |
| H-6a | 4.39 | |
| H-6b | 4.10 | |
| 2-NH | 8.12 | |
| 4-OH | 5.86 | |
| Hα1 | 2.53 | |
| Hα2 | 2.58 | |
| Hα3 | 2.69 | |
| Hα4 | 2.79 | |
| Hβ1 | 4.19 | |
| Hβ2 | 4.27 | |
| Hβ3 | 4.28 | |
| Hβ4 | 5.64 | |
| Hβ1-OH | 4.82 | |
| Hβ2-OH | 4.92 | |
| Hβ3-OH | 4.96 | |
Spectra were recorded at 600 MHz in C6D6–(CD3)2 SO (9:1) relative to TMS (0.00 ppm). Assignments were made by field gradient 1H,1H-COSY NMR. Chemical shifts for the nonreducing terminal unit were not clearly assigned because of heterogeneity.
J = 3.12 Hz.
The location of phosphate groups was directly determined by 31P NMR analysis. One signal was predominantly observed at −0.669 ppm in the 31P NMR spectrum. This signal was cross-coupled with the H-1 proton in 31P,1H-HMQC analysis, proving that the phosphate residues are attached to position 1 of the lipid A backbone.
DISCUSSION
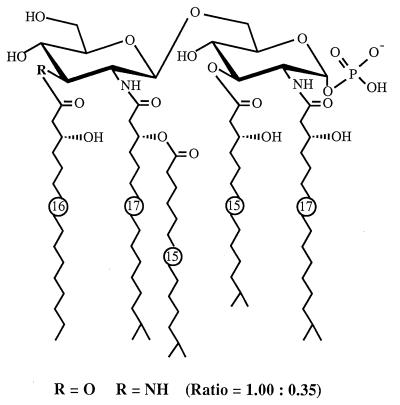
The proposed chemical structure of F. meningosepticum lipid A, determined by chemical and physicochemical analysis in the present study, is shown in Fig. 7. It was noted that the hydrophilic backbone consisted of an unusual hybrid backbone identified as a β(1→6)-linked GlcN3N-GlcN disaccharide in addition to a β(1→6)-linked GlcN disaccharide backbone, which is widely distributed in many LPS molecules (38). Both backbones are 1-O-α-glycosidically phosphorylated, and (R)-3-OH iC15:0 and (R)-3-OH iC17:0 attached to the reducing terminal unit are linked to positions 3 and 2, respectively. The hydroxyl group at position 4 of the unit exists in the free form in the lipid A molecule, and position 6 is the site to which the nonreducing terminal unit is linked. Although the exact attachment sites of (R)-3-OH C16:0 and (R)-3-O-(iC15:0)-iC17:0 on the nonreducing terminal unit were not determined, they are assumed to link to positions 3′ and 2′ of the unit based on the following results: (i) all (R)-3-O-(iC15:0)-iC17:0 existed as an amide-linked acyl residue, while (R)-3-OH C16:0 was detected as both amide- and ester-linked residues; (ii) monophosphoryl GlcN disaccharide carrying 2 mol of amide-linked (R)-3-OH iC17:0 at positions 2 and 2′ was determined by LSI-MS to be a single component of de-O-acylated lipid A; and (iii) the hydroxyl groups at positions 4′ and 6′ of lipid GlcA species having N3N-GlcN hybrid backbones were identified as free form by LSI-MS.
FIG. 7.
Proposed structure of F. meningosepticum lipid A.
F. meningosepticum lipid A has a number of chemically unique characteristics compared to other enterobacterial lipid A’s (12, 14, 38). F. meningosepticum lipid A mainly contains relatively longer-chain and isoform fatty acids, in contrast to the enterobacterial lipid A’s, which contain (R)-3-hydroxytetradecanoic acid as the main constituent of acyl residues. Regarding the location of fatty acids, F. meningosepticum lipid A contained only 1 mol of an acyloxyacyl group at position 2′, while two or sometimes three acyloxyacyl residues are present at positions 2′ and 3′, or additionally at position 2 (in the case of Salmonella), of the nonreducing terminal in enterobacterial lipid A’s. A pattern of distribution of phosphate groups different from that of enterobacterial lipid A’s was also recognized in F. meningosepticum lipid A, which completely lacked an ester-linked phosphate residue attached to position 4′ of the lipid-A backbone. Interestingly, a quite similar structure has been found in the lipid A isolated from oral anaerobic gram-negative bacteria such as Bacteroides fragilis (34) and P. gingivalis (16). These lipid A’s have the same fatty acid composition and phosphate group distribution pattern as F. meningosepticum lipid A, although a small amount of ester-linked phosphate group was recognized in P. gingivalis lipid A. However, F. meningosepticum lipid A could be obviously distinguished chemically from these oral bacterial lipid A’s based on its hybrid backbone consisting of β(1→6)-linked GlcN3N-GlcN disaccharide in addition to the usual β(1→6)-linked GlcN disaccharide. Such an unusual GlcN3N has been found to be a component of the lipid A backbone in Brevundimonas (Pseudomonas) diminuta, Brevundimonas vesicularis, Rhodopseudomonas viridis, Rhodopseudomonas sulfoviridis, Rhodopseudomonas palustris, Legionella pneumophila, and Campylobacter jejuni (1, 15, 19, 33, 35). The backbones of R. diminuta and B. vesicularis consist of the disaccharide of the diamino sugar, in which position 1 may be replaced by d-glucuronic acid. R. viridis, R. sulfoviridis, and R. palustris lipid A’s have unique backbones consisting of the diamino monosaccharide only. The backbone of C. jejuni lipid A is similar to that of F. meningosepticum, which contains a β(1→6)-linked GlcN3N-GlcN disaccharide forming the lipid A backbone in addition to a β(1→6)-linked GlcN disaccharide and a GlcN3N disaccharide. With the exception of these structural similarities, F. meningosepticum lipid A has a unique fatty acid composition, phosphate distribution, and hybrid backbone. Moreover, the lipid A preparation seems to contain unknown compounds in the backbone structure, because the total amount of GlcN3N recovered by the compositional analysis does not match the theoretical quantity obtained from the other structural analysis, which indicates the existence of a hybrid backbone in the lipid A. Recovery was not increased by any hydrolysis or degradation procedures tested. The reason for this is not clear, but it may be based on the tight linkage of the unknown compounds to the GlcN3N residue, which may make the detection of the amino sugar impossible.
We have recently proposed the complete lipid A structure of P. gingivalis (16). Using the lipid A, we demonstrated that the lipid A moiety of P. gingivalis LPS, which exhibited relatively lower activity in LPS-responsive mice than lipid A moieties from enteric bacteria, specifically mediates the activation of LPS-unresponsive C3H/HeJ mice (30, 31). Since the chemical structure of F. meningosepticum has similarities to that of P. gingivalis, as found in the present study, the biological properties of this lipid A are of especially great interest, and studies are currently in progress in our laboratory.
REFERENCES
- 1.Arata S, Kasai N, Klein T W, Friedman H. Legionella pneumophila growth restriction and cytokine production by murine macrophages activated by a novel Pseudomonas lipid A. Infect Immun. 1994;62:729–732. doi: 10.1128/iai.62.2.729-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bednarski M, Danishefsky S. Interactivity of chiral catalysts and chiral auxiliaries in the cycloaddition of activated dienes with aldehydes: a synthesis of l-glucose. J Am Chem Soc. 1986;108:7060–7067. [Google Scholar]
- 3.Brade H, Moll H, Rietschel E T. Structural investigations on the inner core region of lipopolysaccharides from Salmonella minnesota rough mutants. Biomed Mass Spectrom. 1985;12:602–609. doi: 10.1002/bms.1200121007. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan R E, Gibbons N E, editors. Bergey’s manual of determinative bacteriology. 8th ed. Baltimore, Md: The Williams & Wilkins Co.; 1974. pp. 357–364. [Google Scholar]
- 5.Fong Y, Marano M A, Moldawer L L, Wei H, Calvano S E, Kenney J S, Allison A C, Cerami A, Shires G T, Lowry S F. The acute splanchnic and peripheral tissue metabolic response to endotoxin in humans. J Clin Invest. 1990;85:1896–1904. doi: 10.1172/JCI114651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galanos C, Lüderitz O, Freudenberg M A, Rietschel E T, Westphal O, Brade H, Brade L, Shade U, Imoto M, Yoshimura H, Kusumoto S, Shiba T. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur J Biochem. 1985;148:1–5. doi: 10.1111/j.1432-1033.1985.tb08798.x. [DOI] [PubMed] [Google Scholar]
- 7.Galanos C, Lüderitz O, Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. 1969;9:245–249. doi: 10.1111/j.1432-1033.1969.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 8.Haeffner N, Chaby R, Szabo L. Identification of 2-methyl-3-hydroxydecanoic and 2-methyl-3-hydroxytetradecanoic acids in the ‘lipid X’ fraction of the Bordetella pertussis endotoxin. Eur J Biochem. 1977;77:535–544. doi: 10.1111/j.1432-1033.1977.tb11696.x. [DOI] [PubMed] [Google Scholar]
- 9.Haishima Y, Holst O, Brade H. Structural investigation on the lipopolysaccharide of Escherichia coli rough mutant F653 representing the R3 core type. Eur J Biochem. 1992;203:127–134. doi: 10.1111/j.1432-1033.1992.tb19837.x. [DOI] [PubMed] [Google Scholar]
- 10.Hakomori S. A rapid permethylation of glycolipid and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J Biochem. 1964;51:205–208. [PubMed] [Google Scholar]
- 11.Hase S, Rietschel E T. Isolation and analysis of the lipid A backbone. Lipid A structure of lipopolysaccharides from various bacterial groups. Eur J Biochem. 1976;63:101–107. doi: 10.1111/j.1432-1033.1976.tb10212.x. [DOI] [PubMed] [Google Scholar]
- 12.Homma J Y, Matsuura M, Kanegasaki S, Kawakubo Y, Kojima Y, Shibukawa N, Kumazawa Y, Yamamoto A, Tanamoto K, Yasuda T, Imoto M, Yoshimura H, Kusumoto S, Shiba T. Structural requirements of lipid A responsible for the functions: a study with chemically synthesized lipid A and its analogues. J Biochem. 1985;98:395–406. doi: 10.1093/oxfordjournals.jbchem.a135294. [DOI] [PubMed] [Google Scholar]
- 13.Iida T, Haishima Y, Tanaka A, Nishiyama K, Saito S, Tanamoto K. Chemical structure of lipid A isolated from Comamonas testosteroni lipopolysaccharide. Eur J Biochem. 1996;237:468–475. doi: 10.1111/j.1432-1033.1996.0468k.x. [DOI] [PubMed] [Google Scholar]
- 14.Kanegasaki S, Tanamoto K, Yasuda T, Homma J Y, Matsuura M, Nakatsuka M, Kumazawa Y, Yamamoto A, Shiba T, Kusumoto S, Imoto M, Yoshimura H, Shimamoto Y. Structure-activity relationship of lipid A: comparison of biological activities of natural and synthetic lipid A’s with different fatty acid compositions. J Biochem. 1986;99:1203–1210. doi: 10.1093/oxfordjournals.jbchem.a135583. [DOI] [PubMed] [Google Scholar]
- 15.Kasai N, Arata S, Mashimo J, Akiyama Y, Tanaka C, Egawa K, Tanaka S. Pseudomonas diminuta LPS with a new endotoxic lipid A structure. Biochem Biophys Res Commun. 1987;142:972–978. doi: 10.1016/0006-291x(87)91509-9. [DOI] [PubMed] [Google Scholar]
- 16.Kumada H, Haishima Y, Umemoto T, Tanamoto K. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J Bacteriol. 1995;177:2098–2106. doi: 10.1128/jb.177.8.2098-2106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry O H, Roberts N R, Leiner K Y, Wu M L, Farr A L. The quantitative histochemistry of brain. I. Chemical methods. J Biol Chem. 1954;207:1–17. [PubMed] [Google Scholar]
- 18.Miche H R, Manogue K R, Spriggs D R, Revhaug A, O’Dwye S, Dinarello C A, Cerami A, Wolff S M, Wilmore D W. Detection of circulating tumor necrosis factor after endotoxin administration. N Engl J Med. 1988;318:1481–1486. doi: 10.1056/NEJM198806093182301. [DOI] [PubMed] [Google Scholar]
- 19.Moran A P, Zäringer U, Seydel U, Scholz D, Stütz P, Rietschel E T. Structural analysis of the lipid A component of Campylobacter jejuni CCUG 10936 (serotype 0:2) lipopolysaccharide. Eur J Biochem. 1991;198:459–469. doi: 10.1111/j.1432-1033.1991.tb16036.x. [DOI] [PubMed] [Google Scholar]
- 20.Omar A S, Flammann H T, Borowiak D, Weckesser J. Lipopolysaccharide of two strains of the phototrophic bacterium Rhodopseudomonas capsulata. Arch Microbiol. 1983;134:212–216. doi: 10.1007/BF00407760. [DOI] [PubMed] [Google Scholar]
- 21.Pokrywka M, Viazanko K, Medvick J, Knabe S, McCool S, Pasculle A W, Dowling J N. A Flavobacterium meningosepticum outbreak among intensive care patients. Am J Infect Control. 1993;21:139–145. doi: 10.1016/0196-6553(93)90005-o. [DOI] [PubMed] [Google Scholar]
- 22.Qureshi N, Takayama K, Kurtz R. Diphosphoryl lipid A obtained from the nontoxic lipopolysaccharide of Rhodopseudomonas sphaeroides is an endotoxin antagonist in mice. Infect Immun. 1991;59:441–444. doi: 10.1128/iai.59.1.441-444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi N, Mascagni P, Ribi E, Takayama K. Mono-phosphoryl lipid A obtained from lipopolysaccharides of Salmonella minnesota R595. Purification of the dimethyl derivative by high performance liquid chromatography and complete structural determination. J Biol Chem. 1985;260:5271–5278. [PubMed] [Google Scholar]
- 24.Rietschel E T, Gottert H, Lüderitz O, Westphal O. Nature and linkages of fatty acids present in the lipid A component of Salmonella lipopolysaccharides. Eur J Biochem. 1972;28:166–173. doi: 10.1111/j.1432-1033.1972.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 25.Rondle C J M, Morgan W T J. The determination of glucosamine and galactosamine. Biochem J. 1955;61:586–589. doi: 10.1042/bj0610586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strittmatter W, Weckesser J, Salimath P V, Galanos C. Nontoxic lipopolysaccharide from Rhodopseudomonas sphaeroides ATCC 17023. J Bacteriol. 1983;155:153–158. doi: 10.1128/jb.155.1.153-158.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suffredini A F, Fromm R E, Parker M M, Brenner M, Kovacs J A, Wesley R A, Parillo J E. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 28.Tanamoto K. Predominant role of the substituents on the hydroxyl groups of 3-hydroxy fatty acids of non-reducing glucosamine in lipid A for the endotoxic and antagonistic activity. FEBS Lett. 1994;351:325–329. doi: 10.1016/0014-5793(94)00857-4. [DOI] [PubMed] [Google Scholar]
- 29.Tanamoto K. Free hydroxyl groups are not required for endotoxic activity of lipid A. Infect Immun. 1994;62:1705–1709. doi: 10.1128/iai.62.5.1705-1709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanamoto K, Azumi S, Haishima Y, Kumada H, Umemoto T. The lipid A moiety of Porphyromonas gingivalis LPS specifically mediates the activation of C3H/HeJ mice. J Immunol. 1997;158:4430–4436. [PubMed] [Google Scholar]
- 31.Tanamoto K, Azumi S, Haishima Y, Kumada H, Umemoto T. Endotoxic properties of free lipid A from Porphyromonas gingivalis. Microbiology. 1997;143:63–71. doi: 10.1099/00221287-143-1-63. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchiya, M., F. Kobayashi, N. Asahi, A. Yokota, and S. Matsuura. 1994. Reactivities of gram-negative bacteria and gram-positive bacteria with Limulus amoebocyte lysate and silkworm larvae plasma. J. Endotoxin Res. 1(Suppl. 1):70.
- 33.Weckesser J, Drews G, Roppel J, Mayer H, Fromme I. The lipopolysaccharides (O-antigens) of Rhodopseudomonas viridis. Arch Microbiol. 1974;101:233–245. doi: 10.1007/BF00455941. [DOI] [PubMed] [Google Scholar]
- 34.Weintraub A, Zäringer U, Wollenweber H W, Seydel U, Rietschel E T. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur J Biochem. 1989;183:425–431. doi: 10.1111/j.1432-1033.1989.tb14945.x. [DOI] [PubMed] [Google Scholar]
- 35.Weisshaar R, Lingens F. The lipopolysaccharide of a chloridazon-degrading bacterium. Eur J Biochem. 1983;137:155–161. doi: 10.1111/j.1432-1033.1983.tb07809.x. [DOI] [PubMed] [Google Scholar]
- 36.Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 37.Wollenweber H W, Broady K W, Lüderitz O, Rietschel E T. The chemical structure of lipid A. Demonstration of amide-linked 3-acyloxyacyl residues in Salmonella minnesota Re lipopolysaccharide. Eur J Biochem. 1982;124:191–198. doi: 10.1111/j.1432-1033.1982.tb05924.x. [DOI] [PubMed] [Google Scholar]
- 38.Zäringer U, Lindner B, Rietschel E T. Molecular structure of lipid A, the endotoxic center of bacterial lipopolysaccharides. Adv Carbohydr Chem Biochem. 1994;50:211–276. [PubMed] [Google Scholar]