Abstract
A purR::pGh9:ISS1 mutant of Lactococcus lactis was obtained following transposon mutagenesis of strain MG1363 and selection for purine auxotrophs. After determination of the nucleotide sequence and deduction of the purR reading frame, the PurR product was found to be highly similar to the purR-encoded repressor from Bacillus subtilis. The wild-type purR gene complemented the purine auxotrophy of a purR::ISS1 mutant, and it was shown that the purR::ISS1 mutation lowered the level of transcription from the purine-regulated L. lactis purD promoter. In a parallel study on the regulation of purC and purD expression in L. lactis (M. Kilstrup, S. G. Jessing, S. B. Wichmand-Jørgensen, M. Madsen, and D. Nilsson, J. Bacteriol. 180:3900–3906, 1998), we identified regions (PurBox sequences: AWWWCCGAACWWT) upstream of the promoters with a central G residue at exactly position −76 relative to the transcriptional start site. The PurBox sequences were found to be required for high-level promoter activity and purine regulation. We identified a PurBox sequence overlapping the −35 region of the L. lactis purR promoter and found, by studies of a purR-lacLM fusion plasmid, that purR is autoregulated. Because of the high degree of similarity of the PurR proteins from B. subtilis and L. lactis, we looked for PurBox sequences in the promoter regions of the PurR-regulated genes in B. subtilis and identified a perfectly matching PurBox sequence in the purA promoter region and slightly degenerate PurBox-like sequences in the promoter regions for the pur operon and the purR gene. Interestingly, the PurBox in the pur operon of B. subtilis is located almost identically, with respect to the promoter, to the PurBox sequences located in front of purC and purD in L. lactis. We present a hypothesis to explain how an ancestral PurR protein in B. subtilis could have evolved from an activator of the pur operon into a repressor which regulates transcription initiation from the same pur promoter by using the same PurR binding site and a similar response toward its effectors.
In nature, ATP and GTP are both derived from IMP, which is synthesized de novo from 5-phosphoribosyl-1-pyrophosphate (PRPP), glycine, glutamine, aspartate, and C1 units by the action of 10 enzymatic reactions (41). While the individual enzymatic steps in purine biosynthesis appear to be similar in all bacteria, the genetic organization and the regulation of the pur genes follow different rules in different bacteria.
In the gram-negative bacterium Escherichia coli, the purine biosynthetic genes are scattered around the chromosome. However, the transcription of all of these genes is repressed by a single regulatory protein, the purR-encoded purine repressor (11, 23, 29). Binding of the E. coli PurR repressor to its target DNA sequences (PurBox’s) is stimulated by the corepressors guanine and hypoxanthine (24, 30). In the gram-positive bacterium Bacillus subtilis, all genes involved in the synthesis of IMP are organized in a single transcriptional unit, the purine operon (6). The transcription of the operon is controlled by two independent mechanisms. Initiation is controlled by a repressor which is encoded by the purR gene and which, at low concentrations of PRPP, binds specifically to a DNA sequence in the promoter region (8, 39). The purR genes from B. subtilis and E. coli are unrelated, and the B. subtilis protein shows a high degree of similarity with purine phosphoribosyltransferases (39), while the E. coli protein is a classical lacI-type repressor (34). An attenuator model for the regulation of premature termination of transcription has been proposed (6). The formation of an antiterminator structure between the promoter and the structural genes has been suggested to be prevented by the binding of an unidentified RNA binding protein in the presence of a guanine nucleotide, thus resulting in the termination of transcription.
Recently, the purDEK operon (26) and the purC gene (18) were cloned from Lactococcus lactis CHCC285, and their nucleotide sequences were determined. A deletion analysis of the purD and purC promoters indicated that the regulatory element is a transcriptional activator and that the standard ς70 factor is involved (26).
In the present study, we have isolated ISS1 transposon mutants (21) with purine auxotrophic phenotypes. One of the mutated genes was identified as a homolog of the purR gene from B. subtilis. We report the characterization of the purR::ISS1 mutant, the nucleotide sequence of the wild-type purR gene, and the identification of the purR promoter. Moreover, we show that the purR gene encodes an activator required for the expression of the purine biosynthetic genes of L. lactis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in the present study are listed in Table 1. E. coli cells were grown in Luria-Bertani medium supplemented with ampicillin at 50 μg/ml and erythromycin at 150 μg/ml when required. Cultures of L. lactis were grown in SA medium (16) supplemented with 1% glucose (GSA medium) and erythromycin at 2 μg/ml when necessary.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Escherichia coli | ||
| JM83 | ara Δlac-pro rpsL thi φ80dlacZΔM15 | 37 |
| JM1000 | JM83/pJM1000 | This work |
| MC1000 | araD139 Δ(argF-leu)7679 galU galK Δ(lac)X74 rpsL thi-1 | 3 |
| MK163 | MT102/pMK1000 | This work |
| MT102 | MC1000 hsdR(r− m+) | Laboratory collection |
| Lactococcus lactis | ||
| JM1010 | MG1363 purR::pJM1010 | This work |
| MG1363 | Wild type | 9 |
| MK132 | MG1363 pur1::pGh9:ISS1 | This work |
| MK133 | MG1363 purD::pGh9:ISS1 | This work |
| MK134 | MG1363 pur3::pGh9:ISS1 | This work |
| MK135 | MG1363 purL::pGh9:ISS1 | This work |
| MK136 | MG1363 purR::pGh9:ISS1 | This work |
| MK137 | MG1363 pur6::pGh9:ISS1 | This work |
| MK138 | MG1363 purD::pGh9:ISS1 | This work |
| MK139 | MG1363 pur8::pGh9:ISS1 | This work |
| MK177 | MG1363 purR::ISS1 | This work |
| MK191 | MK177/pLN95 | This work |
| MK216 | MK177/pMK1030 | This work |
| MK218 | MG1363/pMK1000 | This work |
| MK219 | MK1363/pMK1033 | This work |
| MK221 | MK177/pMK1033 | This work |
| SH1 | MG1363/pLN95 | 18 |
Plasmids and primers.
The plasmids and oligonucleotide primers used in the present study are listed in Tables 2 and 3, respectively.
TABLE 2.
Plasmids
| Plasmid | Relevant featuresa | Source or reference |
|---|---|---|
| pAK80 | Reporter genes lacLM (L. lactis, E. coli) | 15 |
| pCI3340 | Shuttle vector (L. lactis, E. coli) | 10 |
| pGh9:ISS1 | Orits, used for ISS1 mutagenesis (L. lactis, E. coli) | 21 |
| pJM1000 | HindIII rescue plasmid from MK136 containing the purR promoter (E. coli) | This work |
| pJM1010 | Internal purR fragment in pRC1 (E. coli) | This work |
| pMK1000 | EcoRI rescue plasmid from MK136 containing the purR gene (E. coli) | This work |
| pMK1030 | purR gene and promoter inserted into the XhoI and HindIII sites of pCI3340 (L. lactis, E. coli) | This work |
| pMK1033 | purR promoter fused to the lacLM genes in pAK80 (L. lactis, E. coli) | This work |
| pRC1 | Ermr (E. coli) | 19 |
The plasmid can replicate in the genera shown in parentheses.
TABLE 3.
Oligonucleotide primers
| Primer | Sequencea | Comment or positionb |
|---|---|---|
| MKP55 | TCCTTTCAAAGTTACCC | pAK80 (downstream) |
| MKP56 | TTTCAACTGCCTGGC | pAK80 (upstream) |
| MKP72 | AAAACTGCAGCATTTGATTGGGATGATTAA | 161–141 |
| MKP90 | AAAAAGCTTGAATCTGGAAGCTGTCAG | 1–19 |
| MKP104 | CCCCCTCGAGCCAACCTTTACAGTCAAAGTCCC | 1122–1100 |
| PURR100 | GGCTCGAGAAACGAACGTTTAGTTGATT | 104–125 |
| PURR200 | GGGGATCCGATTGAAAGGCTCCG | 678–662 |
| ISSRS-3 | GAAGAAATGGAACGCTC | ISS1 |
Underlined sequences represent nonlactococcal DNA linkers carrying restriction endonuclease sites.
All numbers refer to the numbering of nucleotides in Fig. 1.
Transformation.
L. lactis was transformed by electroporation (13). E. coli cells were transformed as described before (31).
DNA isolation, manipulations, and sequencing.
Chromosomal lactococcal DNA was prepared as described by Johansen and Kibenich (17). The general in vitro DNA methods described by Sambrook et al. (31) were used. DNA sequences were determined from plasmid DNA by the dideoxy chain termination method (32) with a Thermosequenase radiolabeled terminator cycle sequencing kit (product US 79750; Amersham LifeScience) in accordance with the protocol of the manufacturer.
Southern blot analysis.
Southern blot analysis was performed with GeneScreen nylon membranes (New England Nuclear) and the digoxigenin (DIG) system (Boehringer Mannheim Biochemicals) for colorimetric detection of hybridized products in accordance with the protocols specified by the manufacturers.
PCR amplification of DNA.
L. lactis chromosomal DNA was amplified by PCR with 1 μg of template DNA in a final volume of 100 μl containing deoxyribonucleoside triphosphates (0.25 mM each), oligonucleotides (10 μM), and 2.5 U of AmpliTaq DNA polymerase (The Perkin-Elmer Corp.). Amplification was performed as follows: 30 cycles at 95°C for 1 min, 55°C for 1 min, and 1 min at 72°C.
Determination of specific activities of β-galactosidase.
Assays for β-galactosidase were performed as described by Miller (25). Specific activity is given as micromoles of ortho-nitrophenol (ONP) formed per minute per milligram of protein, with a molar extinction coefficient for ONP of 0.045 at 420 nm (25). Protein concentration was determined according to the method of Lowry et al. (20).
pGh9:ISS1 transposon mutagenesis and selection for purine auxotrophs.
A pool of pGh9:ISS1 (21) transposon mutants of L. lactis MG1363 was obtained as described by Maguin et al. (21), except that GSA medium containing hypoxanthine (10 μg/ml) and uracil (10 μg/ml) was used. After resuspension of the mutant library in GSA medium supplemented with erythromycin but without purine or pyrimidine addition, the cells were grown at 37°C for 30 min to stop the growth of purine or pyrimidine auxotrophs. Then, the culture was diluted 100-fold in the same medium, reaching an optical density at 450 nm of 0.08, and grown for an additional 1 h. Ampicillin counterselection for auxotrophs was performed overnight at 37°C after the addition of ampicillin (100 μg/ml) to the diluted culture. After washing and resuspension of the cells in 0.9% NaCl solution, 100-μl culture aliquots, both undiluted and 10-fold diluted, were plated on GSA medium containing hypoxanthine and uracil. After incubation at 37°C overnight, colonies were tested for purine auxotrophy on GSA medium with and without hypoxanthine.
Cloning and nucleotide sequencing of purR::pGh9:ISS1 sequences.
A standard plasmid rescue procedure was used to isolate genomic DNA adjacent to the transposon insertion point (40). Chromosomal DNA was extracted (1) from MK136 and digested with EcoRI. Following self-ligation of a diluted sample of the digested DNA and transformation of strain MT102, transformant MK163 was isolated. Plasmid pMK1000 isolated from MK163 contained a 1.1-kbp insert of Lactococcus DNA. Likewise, HindIII digestion of MK136 followed by ligation and transformation of JM83 resulted in plasmid pJM1000. The inserts of the plasmids were sequenced with the Thermosequenase kit as recommended by the manufacturer, and their lactococcal origin was verified by Southern blotting.
Isolation of temperature-resistant purR::ISS1 recombinants of purR::pGh9:ISS1 mutants.
Since pGh9:ISS1 mutagenesis creates a duplication of the ISS1 sequence, the total pGh9:ISS1 plasmid can be excised by homologous recombination. MK136 was purified to single-colony isolates on GM17 medium (36) and incubated at 28°C, the permissive temperature for plasmid replication, whereby all cells with integrated plasmids were counterselected. Single colonies were then repurified on GM17 medium and incubated at 37°C, the nonpermissive temperature for plasmid replication, so that without selection for erythromycin resistance, the plasmids would be lost. More than 80% of all colonies appearing were found to be erythromycin-sensitive, temperature-resistant recombinants.
Construction of an insertion mutant.
Competent L. lactis MG1363 cells were transformed with E. coli plasmid pJM1010, which is unable to replicate in L. lactis. The plasmid contains a selectable Emr marker and an internal fragment of the purR gene from pMK1000 (see Results). Transformants with plasmid pJM1010 integrated into the chromosome were selected and purified on plates containing 1 μg of erythromycin per ml. The correct insertion mutations in several transformants were verified by PCR analysis.
RNA extraction.
Strains MK219 (MK177 harboring plasmid pMK1033) and MK221 (MG1363 harboring plasmid pMK1033) were grown exponentially in GSA medium supplemented with erythromycin and guanosine alone (GSA + GR) in one experiment and with the subsequent addition of adenine plus hypoxanthine (GSA + GR + A + Hx) in another. Erythromycin at 2 μg/ml, guanosine at 30 μg/ml, hypoxanthine at 15 μg/ml, and adenine at 15 μg/ml were added. At an optical density at 450 nm of 0.6, RNA was extracted from 5 ml of culture as described previously (18).
Primer extension mapping.
About 10 pmol of oligonucleotide primer (MKP55) was used in a primer extension experiment as described previously (18); 10 μg of RNA was used in each reaction.
RESULTS
Isolation of purine auxotrophic mutants after ISS1 transposon mutagenesis of MG1363.
From a library of approximately 4,500 pGh9:ISS1 transposon mutants, we obtained 8 purine auxotrophic mutants by ampicillin counterselection. Using plasmid rescue, we cloned the DNA regions flanking pGh9:ISS1 from these mutants and determined the nucleotide sequence of the Lactococcus chromosomal DNA. Putative open reading frames identified in MK134, MK138, and MK133 showed extensive similarity to the last part of purK, the middle of purD, and the last part of purD from L. lactis (26) and B. subtilis (7) (data not shown). Interestingly, the product of an open reading frame from MK136 showed extensive similarity to the B. subtilis PurR repressor (see below).
Nucleotide sequence of the wild-type purR gene and purR::pGh9:ISS1 fusion junction from L. lactis.
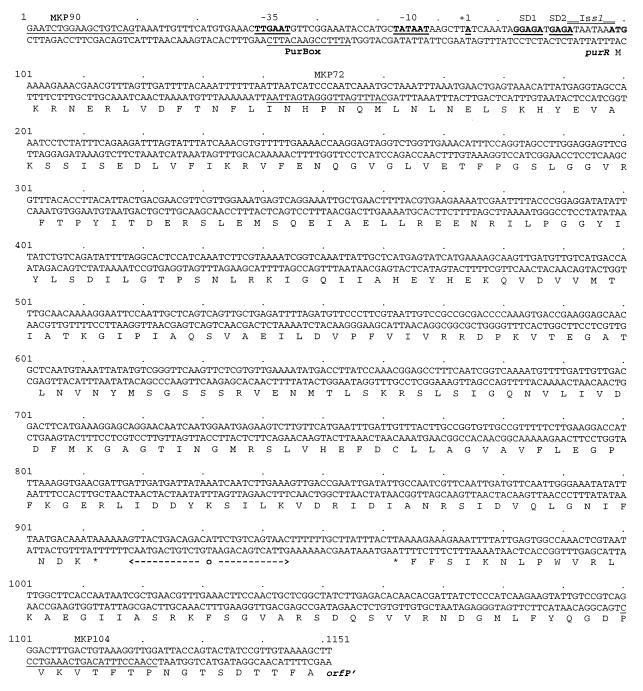
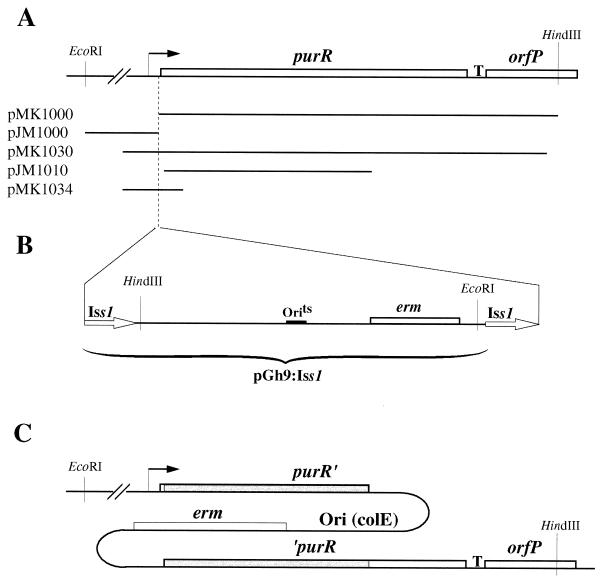
The nucleotide sequence of the purR region from L. lactis MG1363 is shown in Fig. 1. The sequence was determined from the rescued plasmids pMK1000 and pJM1000, which contain DNA from either site of the pGh9:ISS1 fusion (see Materials and Methods for details and Fig. 2 for an overview), and the wild-type sequence of the junction was confirmed by use of PCR fragments amplified from the purR gene on the MG1363 chromosome with primers MKP72 and MKP90 (Fig. 2). The DNA region includes an open reading frame of 271 codons marked as purR in Fig. 1, with a poor putative ribosome binding site (SD2: TGAGA) at the right distance from the start codon. An alternative ribosome binding site (SD1: GGAGA), which is closer to the consensus Shine-Dalgarno sequence, is situated upstream of SD2, but this may be too far away from the start codon. The reading frame encodes a 30.4-kDa protein with an isoelectric point of 5.3. Downstream of the purR gene is a region of dyad symmetry (Fig. 1) which could serve as a terminator of purR transcription as well as of transcription in the opposite direction. The downstream region includes an open reading frame (marked orfP in Fig. 1) reading toward purR. This reading frame appears to be homologous to those for signal peptidases from various organisms (data not shown), including the spsB gene from Staphylococcus aureus (5). The site of insertion of the pGh9:ISS1 element was found to be located between the ribosome binding site and the translational start codon, with the ISS1 element and the chromosomal nucleotides 92-ATAATAAA-98 duplicated so that they are present on each side of the pGh9 plasmid. This arrangement creates the sequence CAGAACCATAATAAATG in front of the purR reading frame, with the start codon underlined and the ISS1 sequence in boldface (Fig. 1).
FIG. 1.
Nucleotide sequence of the purR region from L. lactis MG1363. The nucleotide sequence of a 1,151-bp fragment is shown with the translated sequence of two open reading frames (purR and orfP) aligned with the coding sequence. Putative ribosome binding sites are underlined and marked by SD (Shine-Dalgarno). The transcriptional start site is underlined and marked with +1, and the presence of putative −10 and −35 regions is likewise indicated by underlining. The location of a PurBox sequence overlapping the −35 region is indicated by underlining. An inverted repeat which might function as a ρ-independent terminator structure for both purR and orfP transcription is shown by arrows under the nucleotide sequence. The location of the ISS1 insertion point in MK177 is shown by double overlining of the duplicated purR sequence (ATAATAAA). Primers MKP90, MKP72, and MKP104 are also shown by underlining.
FIG. 2.
Physical maps of the purR regions in wild-type and mutant strains. (A) Physical map of the purR region in L. lactis MG1363. Boxes represent structural genes, and the transcriptional start site is marked with an arrow. The putative terminator of purR and orfP transcription is indicated by T. The purR-derived inserts in a number of plasmids are shown below the physical map. (B) Integration point and structure of the pGh9:ISS1 integrative plasmid after replicative insertion into the chromosome. During the insertion event, the ISS1 element is duplicated. (C) Physical map of the purR region in strain JM1010. The shaded regions indicate duplicated DNA.
The purR gene from L. lactis is homologous to the purR gene from B. subtilis.
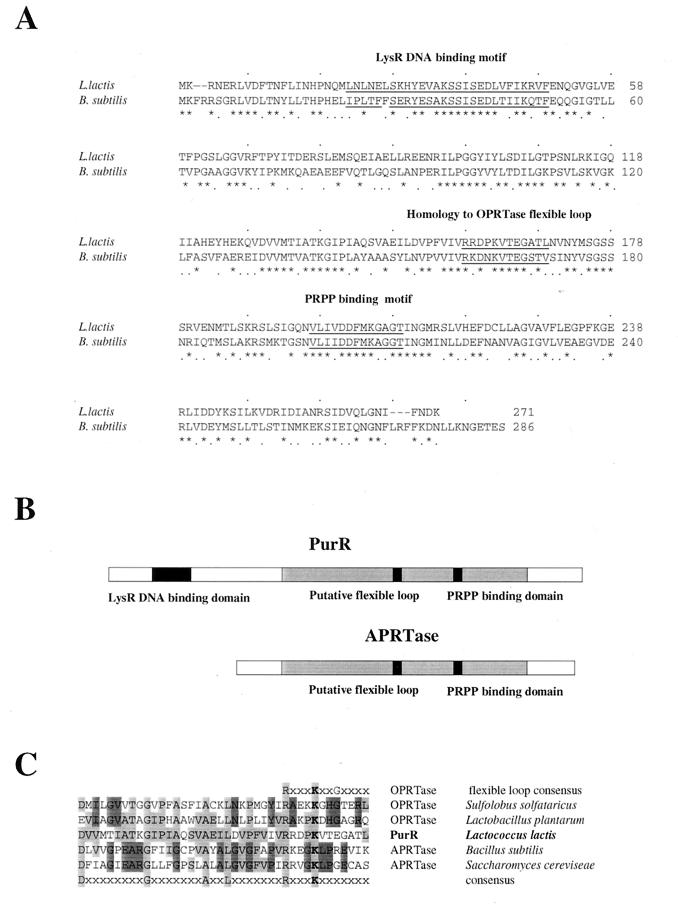
Figure 3A shows a comparison between the purR gene products from L. lactis and B. subtilis. The homology of the two genes appears to span the entire open reading frames, including a PRPP binding motif previously identified for B. subtilis PurR (39). The two proteins show 80% overall similarity, with 51% identical amino acids. When the nucleotide sequence of the purR gene from B. subtilis was reported, it was noted that the encoded protein showed homology in certain regions with purine phosphoribosyltransferases (39), and later Christiansen et al. (4) noted a high level of similarity between PurR and the xanthine phosphoribosyltransferase from B. subtilis. Figure 3B shows an outline of the PurR protein in which the areas with homology to phosphoribosyltransferases (exemplified by the adenine phosphoribosyltransferase from B. subtilis [38]) are shaded. By homology to the helix-turn-helix domains of LysR family transcriptional activators (33) a possible DNA binding region was suggested during computer-aided similarity searches (data not shown). The position of this motif is also shown in Fig. 3A and B. It should be noted that the homology to the LysR family is restricted to the helix-turn-helix DNA binding domain.
FIG. 3.
Comparison of PurR from L. lactis with homologous proteins. (A) Alignment of PurR from L. lactis with PurR from B. subtilis. A PRPP binding motif is underlined. Also, a region fulfilling the consensus requirements for a LysR family DNA binding motif (see text) is underlined. A region with similarity to a flexible loop found in the three-dimensional structure of the orotate phosphoribosyltransferase (OPRTase) of Salmonella typhimurium is also underlined. Asterisks and dots indicate identical amino acids and semiconservative substitutions, respectively. (B) Comparison of PurR with phosphoribosyltransferases exemplified by the adenine phosphoribosyltransferase (APRTase) from B. subtilis. The shaded regions show the extent of similarity between the two proteins. The locations of the putative LysR family DNA binding motif and the PRPP binding motif are indicated by black boxes. Also shown by a black box is a region with similarity to a flexible loop found in the three-dimensional structure of the OPRTase from S. typhimurium. The loop was inferred to make contact with PRPP. (C) Comparison of the PurR region from amino acids D130 to L169 with the homologous regions of the OPRTases from Sulfolobus solfataricus and Lactobacillus plantarum (accession no. g2065443 and e199390, respectively) and the APRTases from B. subtilis and Saccharomyces cerevisiae (accession no. U86377 and z46659, respectively). The flexible loop consensus sequence (also shown in panel B) was reported previously (28). A consensus amino acid sequence for this region is shown in the bottom row.
The purR gene encodes a transcriptional activator of purine gene expression.
It came as a surprise to discover that a transposon insertion in a gene homologous to a repressor gene would lead to a purine auxotrophic phenotype. The trivial explanation, that the insertion prevents transcription of a downstream purine gene, could be rejected, since no sequences similar to known pur genes were found downstream of purR. The only open reading frame found downstream of the purR gene is transcribed in the opposite direction and shows high similarity to the SpsB peptidase gene from S. aureus. We were therefore left with two different explanations. Either PurR is a transcriptional activator and the pGh9:ISS1 insertion abolishes the translation of the gene or PurR is a repressor and the insertion of the transposon leads to elevated levels of the repressor protein, with transcription and the ribosome binding site originating in the ISS1 element. The elevated levels of the PurR protein could then shift the binding equilibrium, resulting in constitutive repression of the purine biosynthetic genes. To test these possibilities, we transformed parental strain MG1363 with the multiple-copy rescue plasmid pMK1000, which contained the same genetic organization as MK136 but at a higher gene dose. If MK136 were auxotrophic due to elevated repressor levels, then MG1363 transformed with pMK1000 should also exhibit purine auxotrophy, since it would contain even higher levels of the repressor. If, however, PurR is a transcriptional activator, then the presence of an inactivated gene on the plasmid should not lead to auxotrophy. The presence of pMK1000 in MK218 was found not to result in a purine requirement when samples were plated on solid GSA medium, in accordance with the first explanation.
To demonstrate that the purine auxotrophy of the purR::ISS1 mutation was due to the absence of the PurR protein, a mutant (JM1010) which synthesizes a truncated PurR protein was constructed. This mutant was constructed by integration of a plasmid (pJM1010) which carries an internal purR fragment covering nucleotides 104 to 678 (Fig. 2A) by a single crossover event, as described in Materials and Methods. Integrant JM1010 produces a truncated PurR protein lacking the 74 carboxy-terminal amino acids created by the disruption of the purR gene at position 678 (Fig. 2C). The strain was found to be purine auxotrophic, demonstrating that an intact PurR protein is required for growth in the absence of exogenous purines.
As a second test of the positive role of the purR gene in purine gene expression, we tested the ability of the wild-type purR gene to complement the purR::ISS1 mutation in MK136. To do this, we created an erythromycin-sensitive, temperature-resistant purR::ISS1 derivative of MK136 in the following way. The ISS1 element is duplicated during transposition (Fig. 2A). Therefore, if homologous recombination occurs between the ISS1 elements, the plasmid will be deleted from the chromosome, leaving a single ISS1 element in the purR gene. The resulting strain, MK177, was found to retain the purine auxotrophy of MK136, as judged by its requirement for purines when plated on solid GSA medium.
Subsequently, the intact purR gene was amplified from the chromosome of MG1363 with primers MKP90 and MKP104 (Fig. 1). The primers were constructed so that a HindIII site was attached adjacent to position 1 and an XhoI site was attached adjacent to position 1122 on the sequence map shown in Fig. 1 (Table 3). The PCR-amplified fragment was digested with XhoI and HindIII restriction enzymes and inserted between the XhoI and HindIII sites in the E. coli-L. lactis shuttle vector pCI3340 (10). Plasmid pMK1030 was found to complement the mutation in MK177, since MK216 (MK177/pMK1930) could grow in the absence of purines while MK177 required a purine source, such as hypoxanthine, for growth.
To see how the purR::ISS1 mutation affects the regulation of purine genes, we transformed MK177 with a purD-lacLM fusion plasmid, pLN95 (26), and measured the production of β-galactosidase. Table 4 shows the levels of β-galactosidase produced from the plasmid in the wild-type background (SH1) and in the purR mutant (MK191). The strains were grown in GSA + GR and GSA + GR + A + Hx. The purR mutation resulted in a low level of expression of the purD gene (Table 4), in accordance with the purine auxotrophic phenotype of MK177. Guanosine was added to support the growth of MK177 and had been found to deactivate PurR-regulated promoters to a lesser extent than other purine sources (data not shown).
TABLE 4.
β-Galactosidase production from fusion plasmids
| Strain | Promoter fusion | β-Galactosidase production (μmol of ONP formed/min/mg of protein in medium with the following additiona:
|
|
|---|---|---|---|
| None | Adenine plus hypoxanthine | ||
| SH1 (wild type) | purD-lacLM | 210 | 36 |
| MK191 (purR) | purD-lacLM | 9 | 2 |
| MK219 (wild type) | purR-lacLM | 78 | 78 |
| MK221 (purR) | purR-lacLM | 360 | 180 |
The basal growth medium was GSA medium containing erythromycin at 2 μg/ml and supplemented with guanosine at 30 μg/ml.
Autoregulation of purR transcription and mapping of the transcriptional start site.
Since plasmid pMK1030, which contains the purR sequence from position 1 to position 1122 (Fig. 1), was found to complement the purR::ISS1 mutation, the purR gene must be expressed from the plasmid. This suggestion strongly indicates the presence of a promoter in front of purR, between nucleotides 1 and 90 (Fig. 1). We constructed a fusion between the purR promoter and a promoterless copy of the lacLM genes from Leuconostoc mesenteroides in plasmid pAK80 (15). The source of the purR promoter was a PCR-amplified fragment from MG1363 incorporating a PstI site downstream and a HindIII site upstream of the promoter. In Table 4 we present data on the production of β-galactosidase from the resulting plasmid, pMK1033, when present in MG1363 or in MK177. The transformants were grown in GSA + GR (derepressing conditions; see above) or in GSA + GR + A + Hx (repressing conditions; see above). The fact that transcription from the promoter was elevated in the purR background (compare MK219 to wild-type MK221) suggests that PurR does repress its own synthesis. It is interesting that both repressing and derepressing conditions resulted in the same level of transcription from the purR promoter in the MG1363 background (MK221). This finding could indicate that purR is autoregulated independently of the effector molecule. In other words, PurR appears to bind to the purR promoter under all growth conditions. For unknown reasons, the derepressed level of transcription in the purR background (MK219) was reduced by about 50% by the addition of adenine and hypoxanthine.
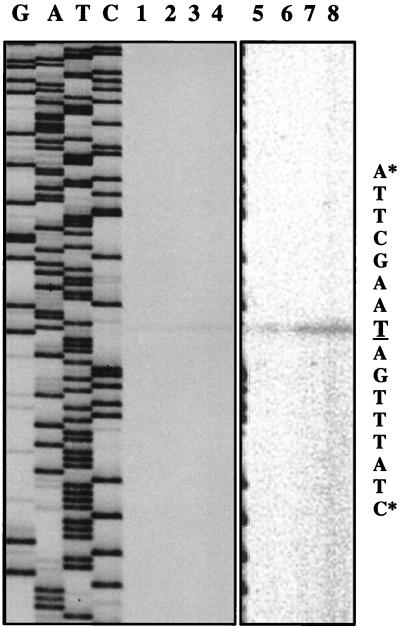
In order to map the precise location of the promoter, we determined the 5′ end of the purR transcript by primer extension analysis of RNA isolated from MK219 and MK221 using a primer which hybridized to the beginning of the lacL gene (MKP55). The results are presented in Fig. 4. This experiment showed that the first nucleotide to be transcribed (+1) is the adenine nucleotide at position 75 in Fig. 1. Just upstream of the start site a consensus −10 sequence (TATAAT) can be identified with a spacing of 6 nucleotides. Furthermore, spaced by 17 nucleotides a consensus-like −35 sequence is present (TTGAAT). In addition, a TGN sequence, which is often found in front of the −10 element in lactococcal promoters, is seen at the correct position. The amount of extension product was higher in the purR background (Fig. 4, lane 7) than in the wild-type background (lane 5) but was independent of purine additions to the growth medium (compare lanes 5 and 6 and lanes 7 and 8). Although we did not see a reduction in purR transcript levels in the purR background upon purine additions, the primer extension experiments confirmed the data concerning β-galactosidase production from fusion plasmid pMK1033.
FIG. 4.
Primer extension analysis of the purR transcriptional start site. An autoradiogram shows primer extension experiments performed with 10 μg of RNA extracted from MK219 (MG1363 purR-lacLM, lanes 1 and 2) and MK221 (MK177 purR-lacLM, lanes 3 and 4). RNA was extracted from cells growing exponentially in GSA medium (lanes 1 and 3) or in the same medium supplemented with purines (lanes 2 and 4). Lanes G, A, T, and C, sequencing reactions. Asterisks indicate the limits of the nucleotide sequence, shown on the right, with the transcriptional start site underlined. The picture was scanned at 400 dpi with a Scan Jet 4c/T (Hewlett-Packard Co.) and DeskScan II version 2.3 software. The TIF file was imported into Top Draw version 3.1 for the addition of text. Lanes 5, 6, 7, and 8 are identical to lanes 1, 2, 3, and 4, respectively, except that the image was acquired with a Packard Instant Imager. The Instant Imager measures the radioactivity over the surface of the gel and is more sensitive than autoradiography.
DISCUSSION
In the present work, we identified a number of purine auxotrophic mutants after pGh9:ISS1 mutagenesis of strain MG1363. Most of the mutants had the transposon element inserted in genes involved in purine de novo synthesis, but in one mutant the transposon impaired the expression of a gene for a putative regulatory protein which showed a high degree of similarity to the purR gene from B. subtilis. By complementation of the purR:ISS1 mutant MK177 from L. lactis with plasmid pMK1030 containing the wild-type L. lactis purR gene, it was verified that the purine auxotrophic phenotype was the result of the purR mutation.
Does PurR activate purC and purD transcription through binding to PurBox’s?
In the accompanying report on the regulation of the purC and purD operons in L. lactis (18), a 13-nucleotide-long DNA sequence (PurBox; Table 5) which is present in front of both promoters was found to be important for high-level expression and purine regulation from the promoters. It was concluded that the PurBox sequences are binding sites for a transcriptional activator. In the present study, we showed that PurR most likely is the transcriptional activator for purC and purD, since the absence of PurR resulted in decreased expression of β-galactosidase synthesis from the purD promoter on plasmid pLM95 (26), compared to the expression in the wild-type parental strain MG1363. The purine regulation of the residual promoter activity was not expected and may indicate that the mutation present in MK177 does not totally abolish PurR production. Transcription from the purR promoter was found to be repressed by PurR, a result which strengthens the inferred bond between PurR and PurBox’s, since the −35 region of the purR promoter overlaps a PurBox (Fig. 1).
TABLE 5.
Comparison of PurBox sequences from L. lactis and B. subtilis
| Operator sitea | Sequenceb | Locationc | Organism | Verification |
|---|---|---|---|---|
| PurBox C1 | ATTTCCGAACATT | −93 | L. lactis | Deletion |
| PurBox C2 | AAAACCGAACAAT | −76 | L. lactis | Deletion, mutation |
| PurBox C3 | AAAACCGAACAAT | +263 | L. lactis | None |
| PurBox D1 | AATACCGAACAAT | −76 | L. lactis | Deletion, mutation |
| PurBox D2 | ATTTCCGAACTAT | +289 | L. lactis | None |
| PurBox R1 | ATTTCCGAACATT | −26R | L. lactis | None |
| Consensus sequence | AWWWCCGAACWWT | |||
| Nucleotide position | 1.....7.....13 | |||
| PurBox O | AAACACGAACATT | −75 | B. subtilis | Binding studiesd |
| PurBox A | AAATCCGAACATT | −46R | B. subtilis | Binding studiese |
| PurBox R | AAATCCGAATATT | −20R | B. subtilis | Binding studiesf |
PurR operator sites are labeled as follows: C1 to C3, purC; D1 and D2, purD; R1, purR; O, purine operon; A, purA; R, purR.
Given from the 5′ end and numbered from nucleotides 1 to 13.
The location of the PurBox is given for the G7 residue relative to the transcriptional start site. The notation −26R indicates that the PurBox sequence is found at position −26 on the opposite strand.
PurBox O has a putative core PurR binding site in front of the purine operon (8).
The PurBox sequence located at position −76 relative to the transcriptional start site (18) in both the purC and the purD promoter regions was shown to be required for the activation of transcription. The apparent constraint on the location of the PurBox suggests that PurR and the initiating RNA polymerase have to form a specific complex involving protein-protein interactions between the PurR protein and the RNA polymerase. The autorepression of purR was found to be identical whether the cells were grown in the presence or in the absence of purine bases (Table 4), and only the absence of PurR led to elevated activity. These data suggest that PurR remains bound to the PurBox under all conditions. If this is the case for all PurBox sequences, which is a reasonable assumption, then the activation of transcription at the purC and purD promoters may involve a conformational change of PurR from a nonactivating to an activating conformation which recognizes the RNA polymerase. Such an activation mechanism is known for members of the LysR family of transcriptional activators (33).
It is interesting to note that two regions in phosphoribosyltransferases show a high degree of similarity to PurR (Fig. 3B and C). One of these is the PRPP binding region previously noted by Weng et al. (39). Another region, which was found in a homology search of GenBank (Fig. 3C), was previously shown for orotate phosphoribosyltransferases to constitute a flexible loop. This loop forms a contact with a PRPP molecule, situated in the PRPP binding site, through a conserved lysine residue (12, 27, 28), thereby promoting a conformational change. In PurR, a lysine residue in a similar context was detected at amino acid position 162 (nucleotide position 582 in Fig. 1; shown in boldface in Fig. 3C); this finding could suggest that the movement of a flexible loop is involved in the conformational change leading to the activation of transcription by binding to PRPP.
Do PurR from L. lactis and PurR from B. subtilis share common mechanisms of action?
We face an interesting dilemma when we consider that the PurR proteins from L. lactis and B. subtilis show high overall similarity (Fig. 3A). The PurR protein from B. subtilis was reported to function as a repressor (7, 35, 39) which represses the pur operon only when purines are present in the growth medium. The activation of transcription by the PurR protein from L. lactis, on the other hand, is seen only when purines are absent from the medium. It has been reported that the DNA binding ability of the PurR protein from B. subtilis is inhibited by PRPP (39), since the ability of PurR to bind DNA to the promoter region from the purine operon was detected only at lower PRPP concentrations. It is reasonable to suppose that the activity of the PurR protein from L. lactis is also regulated by the level of PRPP, since it shows 80% overall similarity with the PurR protein from B. subtilis and since both protein sequences contain a consensus PRPP binding domain (14, 39). The addition of adenine and hypoxanthine to the growth medium of both bacteria resulted in reduced expression of the purine genes, and in both bacteria this regulation was dependent upon a PurR homolog. It must be kept in mind, however, that when one is interpreting levels of enzymes encoded by the intact purine operon in B. subtilis, the regulation of the pur operon involves an additional guanine- and hypoxanthine-responsive attenuation system (6).
How the PRPP concentration in B. subtilis was influenced by the addition of adenine to the growth medium was suggested for B. subtilis to be the consequence of two separate events (39). Conversion of adenine to AMP by the adenine phosphoribosyltransferase requires PRPP, so a massive conversion of adenine to AMP could lead to PRPP depletion. At the same time, the high AMP pool levels could lead to elevated levels of ADP, which is known to be a potent inhibitor of the PRPP synthetase from B. subtilis (2). Therefore, the model for PurR regulation in B. subtilis suggests that a high adenine concentration leads to a low PRPP concentration, which allows PurR binding. Accordingly, a low extracellular concentration of adenine would lead to a high PRPP concentration, which would inhibit DNA binding of PurR. We believe that DNA binding of PurR from L. lactis is not inhibited by PRPP, since the high PRPP level which is present when no adenine is added results in PurR activation and thus DNA binding. Moreover, PurR appears to bind to PurBox’s both in the presence and in the absence of exogenous purine bases (Table 4 and Fig. 4, lanes 5 and 6).
In B. subtilis, PurR has been shown to protect a region of more than 60 bp from DNase I attack upon binding to a poorly defined operator site in front of the pur operon (8, 35). In the core of the protected region (8) we have detected at position −75 relative to the transcriptional start site a degenerate PurBox (Table 5) which could serve as the primary binding site. We have also detected DNA sequences resembling PurBox’s in front of two additional PurR-regulated genes from B. subtilis, purA (22) and purR (39), at about position −40 relative to the suggested transcriptional start sites, as for the purR gene in L. lactis (this work). The purA-specific PurBox is a perfect match for the L. lactis consensus sequence, while the PurBox in the purR promoter region has a C10-to-T mismatch (Table 5). If the PurBox consensus sequence also applies to the B. subtilis PurR repressor, then the degenerate PurBox in the purR gene would lead to weaker autorepression and elevated levels of PurR in B. subtilis relative to L. lactis, in which autorepression involves a perfect PurBox (Table 5). Actually, Shin et al. (35) have presented data suggesting that PurR from B. subtilis does recognize the PurBox sequence. A consensus sequence for PurR binding (GAAC-N24–25-GTTC) was proposed based on its presence in the pur, purA, and purR promoter regions. By site-directed mutagenesis and studies of binding of the region from the pur operon, Shin et al. (35) obtained data showing that the GAAC sequence was required for efficient PurR binding, whereas the GTTC sequence was not. Interestingly, the GAAC sequence required for PurR binding in B. subtilis is situated in the core of the PurBox sequence proposed above (from G7 to C10). The authors showed that mutating the GAAC sequence to GAAT, AAAT, or ATCG resulted in an increase in the apparent Kd from 7 nM to 16, 52, or 78 nM, respectively (35). Additionally, they found that methylation of the G7 on one strand and the G residue complementary to the C10 on the other strand resulted in a loss of PurR binding (35).
A unifying evolutionary model for PurR action in L. lactis and B. subtilis.
If the PurR proteins from L. lactis and B. subtilis are homologous, then both proteins have evolved from a common PurR ancestor. Whether this nearest ancestor was more similar to one or the other is at present impossible to deduce, since we have only two PurR homologs to compare, but the question can be addressed when more PurR proteins are analyzed. It is reasonable to assume that the ancestral PurR was an activator which could activate transcription when bound to a PurBox located at about 76 bp upstream of the transcriptional start site, since both the L. lactis purC and purD promoters and the B. subtilis pur operon promoter share this organization. There is no reason to doubt that this activation would have been inhibited by PRPP, as it is for the L. lactis protein. Whether the ancestral PurR protein, upon PRPP binding, would have been able to repress the promoter from the same binding site cannot be answered with our limited data, but this type of PurR repression is known for the pur operon in B. subtilis.
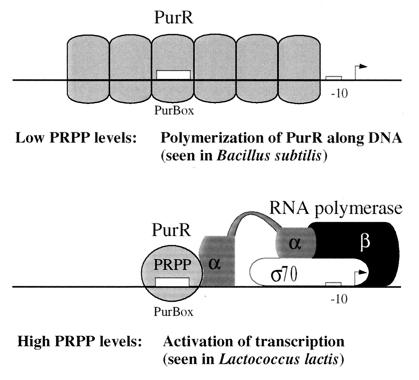
Somewhere in the evolutionary transition from an activator to a repressor, there must have been a bifunctional PurR protein, which could both activate and repress transcription from the same site. A hypothetical model for this transit PurR protein is shown in Fig. 5. The protection of more than 60 bp by PurR binding in B. subtilis was suggested to be the result of the DNA being wrapped around the PurR repressor (35). Alternatively, it could involve more than one PurR unit, so that massive protection could be the result of PurR multimerization along the DNA, leading to sequestering of the −35 region in B. subtilis. This possibility, which is in accord with the PurR binding stoichiometry of six PurR homodimers per 196 bp of the pur operon in the absence of PRPP (35), is indicated in Fig. 5. Such a polymerization would be dependent upon a high PurR concentration, since no additional PurBox’s can be found in the promoter regions. That multimerization and transcription activation in the transit PurR must have been mutually exclusive phenomena is almost self-evident, since a PurR multimer blocks access to the −10 region. Thus, multimerization could be favored at low PRPP concentrations, leading to repression, while RNA polymerase binding could be favored at high PRPP concentrations, leading to enhanced transcription of the purine biosynthetic genes (Fig. 5).
FIG. 5.
Hypothetical bifunctional regulator as an intermediate in the evolution of PurR. The hypothetical bifunctional regulator is proposed to bind to a PurBox under all conditions, but it can change between two conformations, depending on the presence of PRPP. The conformational changes in the transit PurR upon PRPP binding might favor polymerization at low PRPP concentrations and RNA polymerase binding at high PRPP concentrations. The model incorporates data for PurR binding in both B. subtilis and L. lactis. In B. subtilis, PurR binding to PurBox DNA is only detected at low PRPP concentrations and only with an extended footprint from multiple PurR homodimers bound to the DNA. In L. lactis, PurR binds to DNA with both high and low PRPP concentrations, but activation is detected only with high PRPP concentrations. The bifunctional transit PurR protein could be the intermediary state in the evolution of an activator-regulated system into a repressor-regulated system with the same regulatory protein, promoter, and regulator binding site.
If the Bacillus PurR regulator has gone through such a transition, one could envision that the pur operon was originally under activation control but that the evolving bifunctional transit PurR allowed the introduction of a −35 region, which created an activator-independent, repressor-controlled promoter. We are currently testing whether the PurR protein in B. subtilis can activate transcription from the lactococcal purC promoter, thus resembling the postulated transit PurR protein. We are also testing whether the PurR activator from L. lactis forms extended footprints at high protein concentrations and whether it can repress the pur operon promoter when supplied at elevated levels.
ACKNOWLEDGMENTS
We sincerely appreciate the expert technical assistance of Kristina Brandborg Jensen and Susan Outzen Jørgensen. We thank Hans Henrik Saxild for help on the plasmid rescues and many stimulating discussions. We also thank Martin Willemoës for many fruitful discussions and for directing our attention to the orotate phosphoribosyltransferase flexible loop.
REFERENCES
- 1.Arnau J, Sørensen K I, Appel K F, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. doi: 10.1099/13500872-142-7-1685. [DOI] [PubMed] [Google Scholar]
- 2.Arnvig K, Hove-Jensen B, Switzer R L. Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem. 1990;192:195–200. doi: 10.1111/j.1432-1033.1990.tb19214.x. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen L C, Schou S, Nygaard P, Saxild H H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cregg K M, Wilding I, Black M T. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J Bacteriol. 1998;178:5712–5718. doi: 10.1128/jb.178.19.5712-5718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebbole D J, Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987;262:8274–8287. [PubMed] [Google Scholar]
- 7.Ebbole D J, Zalkin H. Characterization of the Bacillus subtilis pur operon: new insights into gene regulation. In: Hoch J A, editor. Genetics and biotechnology of bacilli. Vol. 2. New York, N.Y: Academic Press, Inc.; 1988. pp. 51–55. [Google Scholar]
- 8.Ebbole D J, Zalkin H. Interaction of a putative repressor protein with an extended control region of the Bacillus subtilis pur operon. J Biol Chem. 1989;264:3553–3561. [PubMed] [Google Scholar]
- 9.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes F, Daly C, Fitzgerald G F. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl Environ Microbiol. 1990;56:202–219. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He B, Shiau A, Choi K Y, Zalkin H, Smith J M. Genes of the Escherichia coli pur regulon are negatively controlled by a repressor-operator interaction. J Bacteriol. 1990;172:4555–4562. doi: 10.1128/jb.172.8.4555-4562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriksen A, Aghajari N, Jensen K F, Gajhede M. A flexible loop at the dimer interface is part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase. Biochemistry. 1996;35:3803–3809. doi: 10.1021/bi952226y. [DOI] [PubMed] [Google Scholar]
- 13.Holo H, Nes I F. Transformation of Lactococcus by electroporation. Methods Mol Biol. 1995;47:195–199. doi: 10.1385/0-89603-310-4:195. [DOI] [PubMed] [Google Scholar]
- 14.Hove-Jensen B, Harlow K W, King C J, Switzer R L. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- 15.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen E, Kibenich A. Isolation and characterization of IS1165, an insertion sequence of Leuconostoc mesenteroides subsp. cremoris and other lactic acid bacteria. Plasmid. 1992;27:200–206. doi: 10.1016/0147-619x(92)90022-3. [DOI] [PubMed] [Google Scholar]
- 18.Kilstrup M, Jessing S G, Wichmand-Jørgensen S B, Madsen M, Nilsson D. Activation control of pur gene expression in Lactococcus lactis: proposal for a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions. J Bacteriol. 1998;180:3900–3906. doi: 10.1128/jb.180.15.3900-3906.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 20.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Maguin E, Prévost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mäntsälä P, Zalkin H. Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP. J Bacteriol. 1992;174:1883–1890. doi: 10.1128/jb.174.6.1883-1890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng L M, Kilstrup M, Nygaard P. Autoregulation of PurR repressor synthesis and involvement of purR in the regulation of purB, purC, purL, purMN and guaBA expression in Escherichia coli. Eur J Biochem. 1990;187:373–379. doi: 10.1111/j.1432-1033.1990.tb15314.x. [DOI] [PubMed] [Google Scholar]
- 24.Meng L M, Nygaard P. Identification of hypoxanthine and guanine as the corepressors for the purine regulon genes of Escherichia coli. Mol Microbiol. 1990;4:2187–2192. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Assay of β-galactosidase. In: Miller J H, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 26.Nilsson, D., and M. Kilstrup. Cloning and expression of the Lactococcus lactis purDEK genes, required for growth on milk. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 27.Ozturk D H, Dorfman R H, Scapin G, Sacchettini J C, Grubmeyer C. Locations and functional roles of conserved lysine residues in Salmonella typhimurium orotate phosphoribosyltransferases. Biochemistry. 1998;34:10755–10763. doi: 10.1021/bi00034a007. [DOI] [PubMed] [Google Scholar]
- 28.Ozturk D H, Dorfman R H, Scapin G, Sacchettini J C, Grubmeyer C. Structure and function of Salmonella typhimurium orotate phosphoribosyltransferase: protein complementation reveals shared active sites. Biochemistry. 1998;34:10764–10770. doi: 10.1021/bi00034a008. [DOI] [PubMed] [Google Scholar]
- 29.Rolfes R J, Zalkin H. Escherichia coli gene purR encoding a repressor protein for purine nucleotide synthesis. J Biol Chem. 1988;263:19653–19661. [PubMed] [Google Scholar]
- 30.Rolfes R J, Zalkin H. Purification of the Escherichia coli purine regulon repressor and identification of corepressors. J Bacteriol. 1990;172:5637–5642. doi: 10.1128/jb.172.10.5637-5642.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schell M A. Molecular biology of the LysR family of transcriptional activators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 34.Schumacher M A, Choi K Y, Zalkin H, Brennan R G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 35.Shin B S, Stein A, Zalkin H. Interaction of Bacillus subtilis purine repressor with DNA. J Bacteriol. 1997;179:7394–7402. doi: 10.1128/jb.179.23.7394-7402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 38.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- 39.Weng M, Nagy P L, Zalkin H. Identification of the Bacillus subtilis pur operon repressor. Proc Natl Acad Sci USA. 1995;92:7455–7459. doi: 10.1073/pnas.92.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youngman P, Perkins J B, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- 41.Zalkin H, Dixon J E. De novo purine nucleotide biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1992. pp. 258–287. [DOI] [PubMed] [Google Scholar]