Abstract
A 2.3-kb BamHI-KpnI fragment was isolated from a partial genomic library and shown by nucleotide sequence analysis to contain the entire coding region of the gene encoding the β subunit of the Blastocladiella mitochondrial processing peptidase (β-MPP). The predicted β-MPP protein has 465 amino acids and a calculated molecular mass of 50.8 kDa. S1 nuclease protection assays revealed an intron, 209 bp in size, interrupting the coding region between the putative signal sequence and the mature protein. Northern blot analysis showed that β-MPP mRNA levels decrease significantly during B. emersonii sporulation, reaching basal levels in the zoospore stage. The amount of β-MPP protein, determined in Western blots, unlike its mRNA, does not vary significantly throughout the fungal life cycle.
The general mitochondrial processing peptidase (MPP) is a protein complex responsible for the processing of matrix-targeting signals from nucleus-encoded precursor proteins that are imported by the mitochondria. The MPP has been purified from the mitochondria of different organisms: yeast (30), Neurospora (9), rat liver (10, 12, 13, 21), potato tuber (2), and spinach leaves (5). In fungi and mammals, the MPP consists of two nonidentical but structurally related subunits, α-MPP and β-MPP, both of which are necessary for the processing of precursor proteins. MPP is a metalloendoprotease that requires divalent cations for activity and is inhibited by the metal chelators EDTA and o-phenanthroline (26). The two subunits share certain amino acid motifs, including a putative metal-binding sequence, HFLEH in the β subunit and HFLEK in the α subunit (12). MPP acts on hundreds of unrelated precursor proteins yet removes the presequence in a single specific cleavage reaction (23). Although there are no specific sequence motifs in mitochondrial import-targeting signals, these peptides have certain characteristic features. They are hydrophilic, rich in basic and hydroxylated amino acids, generally lacking acidic residues, and able to fold into an amphiphilic α-helix or β-sheet (24). They are usually between 20 and 35 residues long, and an arginine is found at position −2 or −3 relative to the cleavage site in most mitochondrial precursors from different species (20, 23, 24, 27, 28).
The aquatic fungus Blastocladiella emersonii is characterized by an interesting developmental cycle with well-defined stages: germination, vegetative growth, and sporulation (for a review, see reference 15). The cycle begins with the zoospore, a motile, uninucleated nongrowing cell which germinates rapidly and synchronously in the presence of nutrients. The germination process leads to the formation of the germling cell, which undergoes vegetative growth. During this stage, nuclear division is not accompanied by cell division, and so multinucleated cells, the sporangia, are generated. At any time during exponential growth, nutrient starvation induces another transitional stage, sporulation, which culminates in the intracellular formation of zoospores, which are then released to the medium. The zoospores contain a single giant mitochondrion, which is fragmented into several normal-sized mitochondria during germination in a process which is independent of protein synthesis (3). During sporulation, these multiple individual mitochondria fuse, giving rise to the huge single mitochondrion found in the zoospore (14).
The purpose of this work was to study the expression of the β-MPP gene throughout the B. emersonii life cycle in order to investigate possible variations during the drastic morphological changes experienced by the mitochondria in this organism.
Construction of the partial genomic library.
A partial cDNA, 0.6 kb in size, encoding the carboxy-terminal portion of the Blastocladiella β-MPP was fortuitously isolated from a λgt11 library, during the screening procedure used to clone the Blastocladiella hsp60 cDNA. To clone the entire β-MPP gene, B. emersonii genomic DNA was digested with BamHI and KpnI, size fractionated by agarose gel electrophoresis, transferred to nitrocellulose, and hybridized to the β-MPP cDNA labeled with 32P by random primed synthesis (6). A single band of hybridization in the region corresponding to 2.3 kb was observed. The DNA fragments from 2 to 4 kb were then excised from a similar agarose gel, electroeluted, and ligated to BamHI-KpnI-digested Bluescribe. After transformation into Escherichia coli, the resulting partial library was screened by colony hybridization with the β-MPP cDNA as probe.
DNA sequence analysis.
The genomic fragment containing the β-MPP gene was subjected to restriction endonuclease digestion, and the various fragments obtained were subcloned into M13mp18 and M13mp19 for nucleotide sequence analysis by the dideoxynucleotide chain termination method (25) with the Sequenase DNA sequencing kit (Amersham).
Characterization of the β-MPP gene from B. emersonii.
A 2.3-kb BamHI-KpnI genomic fragment, containing the entire coding region of the β-MPP gene plus sequences upstream and downstream of it, was isolated from a partial genomic library. Nucleotide sequence determination showed a single open reading frame, interrupted by a small intron of 209 bp (Fig. 1). The predicted amino acid sequence of the Blastocladiella β-MPP, encompassing 465 amino acids, has 65% identity and 79% similarity to the sequence of the β-MPP from Neurospora crassa (9) and 60% identity to the corresponding proteins from yeast (29) and from rat liver (11, 22). The putative metal-binding sequence (HXXEH), essential for the catalytic activity of the MPP (12), is conserved in the Blastocladiella protein.
FIG. 1.
Nucleotide sequence of the B. emersonii β-MPP gene and deduced amino acid sequence. Capital letters indicate deoxynucleotides in exons or sequences upstream and downstream of the coding region of the gene; lowercase letters show the deoxynucleotides in the intron. The deduced protein sequence is shown below the nucleotide sequence. Endonuclease restriction sites are shown in boldface type. Nucleotide +1 denotes the A of the ATG of the initiator methionine. Residues preceding it are indicated by negative numbers. Transcription initiation sites predicted by primer extension analysis (see Fig. 3) are indicated (•). The putative helix-loop-helix transcription factor-binding motifs (CANNTG) are boxed. The underlined sequences are complementary to the oligonucleotides used for primer extension and S1 protection experiments. The arrows indicate the putative signal sequence cleavage sites. The putative metal-binding sequence (HFLEH) is shown in boldface type. The putative polyadenylation signal is doubly underlined.
S1 mapping of the 5′ and 3′ ends of the intron interrupting the coding region of the β-MPP gene.
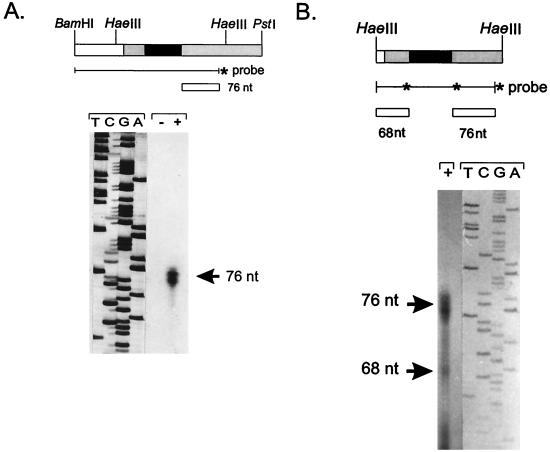
The open reading frame of the B. emersonii β-MPP gene is interrupted by an in-frame stop codon located at nucleotides +249 to +251 relative to the initiator methionine codon. This fact was indicative of the existence of an intron in this region. The presence of an intron, of 209 bp, was confirmed by S1 nuclease protection assays. The 3′ end of the intron was determined with a 5′-end-labeled probe, which was prepared by labeling an 18-residue synthetic oligonucleotide (C-4) complementary to nucleotides (nt) +310 to +327 of the β-MPP gene with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). The labeled 18-mer was then annealed to a single-stranded DNA from M13mp19 containing a 1.2-kb BamHI-PstI fragment (see Fig. 2) from the genomic clone (coding strand) and extended with the Klenow polymerase (Boehringer Mannheim). The probe (3 × 106 cpm) was ethanol precipitated with 50 μg of total RNA isolated from B. emersonii zoospores, and the nucleic acid pellet was resuspended in 28 μl of formamide plus 7 μl 40 mM PIPES buffer (pH 6.4) containing 400 mM NaCl and 1 mM EDTA. The annealing reaction was carried out for 3 h at 52°C; then the samples were diluted with 350 μl of 30 mM sodium acetate (pH 4.6) containing 250 mM NaCl, 1 mM ZnSO4 and 20 μg of salmon testis DNA per ml and digested at 37°C for 30 min with 50 U of S1 nuclease (Amersham). After digestion, the nucleic acids were analyzed by electrophoresis in 7 M urea–7.5% polyacrylamide gels followed by autoradiography. Sizing of the protected fragments was carried out by comparison with a sequencing ladder generated by using the 18-mer C-4 as primer and M13mp19 containing the BamHI-PstI fragment (coding strand). Figure 2A shows the protected fragment obtained, of 76 nt, which localizes the 3′ end of the intron to position +251. A splice acceptor site (gtagC) can be found at this position, which conforms to the consensus for B. emersonii introns (4).
FIG. 2.
S1 protection mapping of the 5′ and 3′ ends of the intron in the B. emersonii β-MPP gene. (A) Determination of the 3′ ends of the intron. The 5′-end-labeled probe depicted in the figure was annealed to 50 μg of total RNA isolated from zoospores (lane +) or 50 μg of yeast tRNA (lane −) and digested with nuclease S1, as described in the text. (B) Determination of the 5′ end of the intron. The uniformly labeled probe depicted in the figure was annealed to 50 μg of total RNA from zoospores (lane +) and treated as described in the text. The arrows indicate the protected fragments. A scheme of part of the genomic clone is shown on the top of the figure, where black rectangles depict the intron and gray rectangles depict the coding region.
S1 nuclease protection assays were also carried out to determine the 5′ end of the intron. In this case, a uniformly labeled probe was obtained by 5′-end labeling of the 18-mer C-4, which was then annealed to a single-stranded DNA from M13mp19 containing a 0.4-kb HaeIII fragment from the genomic clone (coding strand [Fig. 2]) and extended with Klenow polymerase. The probe was then digested with PstI at the restriction site present in the M13mp19 polylinker and located upstream of the SmaI site where the blunt-ended HaeIII fragment was originally cloned. The labeled probe, isolated after denaturing polyacrylamide gel electrophoresis and electroelution, was ethanol precipitated with 50 μg of total RNA isolated from B. emersonii zoospores and processed as described above for the determination of the 3′ end of the intron. As shown in Fig. 2B, two protected fragments were obtained. The 68-nt fragment indicated the position of the 5′ end of the intron, which corresponds to position +43 where there is a splice donor site (Ggtacg), which also follows the consensus for B. emersonii introns (4). The 76-nt fragment also obtained confirms the 3′ end of the intron. The 14-amino-acid sequence preceding the intron has all the characteristic features of a typical mitochondrial signal sequence (24). It is rich in basic (two Arg and one Lys) and hydroxylated (one Ser and one Thr) amino acid residues and has no negatively charged amino acids, and its amino-terminal segment (between Ala-5 and Arg-15) has the potential to form an amphiphilic α-helical structure in hydrophobic environments, as determined with the NNPREDICT program (18).
Primer extension mapping of the transcription start sites.
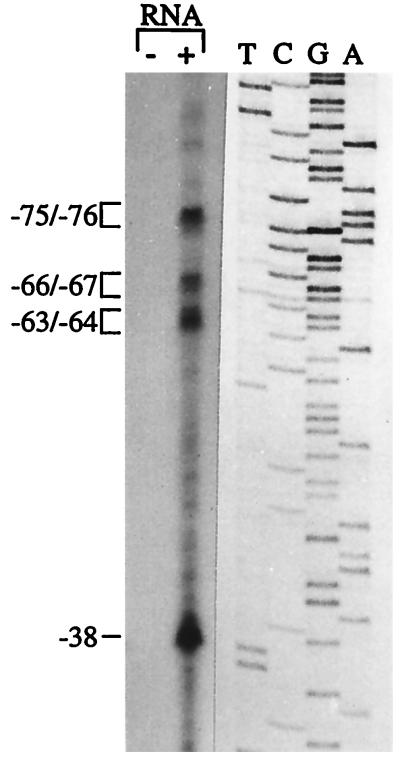
To determine the transcription start sites of the Blastocladiella β-MPP gene, primer extension experiments were performed. An 18-nt primer (C-5), complementary to positions +10 to +27 of the β-MPP gene coding region, was 5′-end labeled with [γ-32P]ATP and hybridized with either total RNA from Blastocladiella cells at 2 h of sporulation or yeast tRNA as a control. The hybrids were then extended with reverse transcriptase, as previously described (1). Multiple bands were detected in Blastocladiella RNA, whereas no bands were detected in the control (Fig. 3). The extension products were distributed in four groups (positions −75/−76, −66/−67, −63/−64, and −38) relative to the adenine of the initiator methionine codon. Consistent with the presence of multiple transcription start sites, the 5′ noncoding region of Blastocladiella β-MPP gene revealed no TATA box or CCAAT box sequences (Fig. 1). No other characteristic features could be observed in the 5′ regulatory region, except for two putative core sequences (CANNTG; positions −291 to −286 and −39 to −34) for binding of helix-loop-helix transcription factors (19).
FIG. 3.
Primer extension mapping of the transcription start sites of B. emersonii β-MPP gene. An 18-nt primer (C-5) complementary to positions +10 to +27 of the β-MPP coding region was 5′-end-labeled and hybridized to 50 μg of total RNA from B. emersonii cells at 2 h of sporulation (lane +) or 50 μg of yeast tRNA (lane −). The hybrids were then extended with reverse transcriptase, and the extension products were resolved by denaturing gel electrophoresis and autoradiography. The sequencing ladder was generated with the same 18-mer as a primer and M13mp19 containing the 5′ end of the β-MPP gene (coding strand).
Expression of the β-MPP gene at the RNA and protein level.
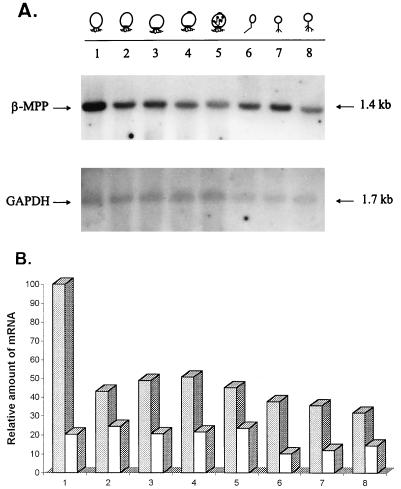
The relative amount of β-MPP mRNA was determined in cells at different stages of Blastocladiella development. Total RNA was isolated, as described by Maniatis et al. (16), from synchronized cells at different times during the fungal life cycle. The RNA was subjected to electrophoresis under denaturing conditions and then transferred to a Hybond N+ membrane (Amersham), as previously described (1). Northern blot analysis, with the β-MPP cDNA as probe, showed a single hybridization band, of 1.4 kb, which was present throughout the life cycle of the fungus. However, its amount decreased significantly during sporulation, reaching basal levels in the zoospore stage, as determined by densitometer scanning of the autoradiogram (Fig. 4). As a control, the same blot was hybridized to a 32P-labeled cDNA clone encoding rat glyceraldehyde-3-phosphate dehydrogenase (7), previously shown to hybridize to a constitutively expressed B. emersonii gene (17).
FIG. 4.
β-MPP mRNA levels during B. emersonii development. Total RNA (15 μg/lane) isolated from cells at different stages of B. emersonii life cycle were subjected to electrophoresis in a formaldehyde-agarose gel and transferred to an Hybond-N+ membrane. Lanes: 1 to 5, sporulating cells 0, 1, 1.5, 2, and 3 h after starvation, respectively; 6, zoospores; 7 and 8, germinating cells 45 and 90 min after the beginning of germination, respectively. The Northern blot was probed with β-MPP cDNA, and as a control, the same blot was hybridized to rat GAPDH cDNA (7). (A) Autoradiograms of the blot. (B) Relative levels of β-MPP (gray rectangles) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (white rectangles) mRNAs as determined by scanning of the autoradiograms.
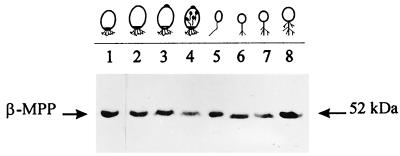
To investigate if the β-MPP protein presented the same pattern of accumulation as its corresponding mRNA, antiserum against the B. emersonii protein was used in Western blots of total extracts of synchronized cells, isolated at different times during the life cycle of the fungus. The anti-β-MPP antiserum was obtained from a rabbit immunized with a fusion protein overexpressed in E. coli, corresponding to the maltose-binding protein encoded by the pMAL-C vector (New England Biolabs) and about 15 kDa of the carboxy-terminal portion of the B. emersonii β-MPP. A single 52-kDa band, whose levels did not change significantly during B. emersonii development, was recognized by the anti-β-MPP antiserum (Fig. 5). The same 52-kDa polypeptide band was recognized in Western blots with antiserum against the N. crassa β-MPP (data not shown).
FIG. 5.
Western blot of total protein extracts from cells at different stages of the Blastocladiella life cycle. Protein blots from cells at the indicated stages of the fungal life cycle (15 μg/lane) were probed with anti-β-MPP antiserum and an enhanced chemiluminescence kit. Lanes: 1 to 4, sporulating cells 0, 1, 2, and 3 h after starvation, respectively; 5, zoospores; 6, 45-min germling cells; 7 and 8, growing cells 1.5 and 3.5 h after the beginning of germination, respectively.
Conclusions.
The B. emersonii β-MPP gene encodes a hydrophilic polypeptide with a calculated molecular mass of 50.8 kDa. A single intron which interrupts the coding region between the putative signal sequence and the mature protein was identified. From an evolutionary point of view, an intron positioned between the signal sequence and the mature protein could be a very good example of a protein encoded in the mitochondrial genome whose gene was transferred to the nucleus sometime after the endosymbiotic event (8). There are two possible sites for the processing of the signal sequence, if we consider the need for an arginine residue at position −2 relative to the cleavage site (20, 23, 24, 27, 28). The first putative cleavage site, which is located in the region preceding the intron, between Asn-13 and Val-14, results in a signal sequence of 13 amino acids, whereas the second site, located immediately after the 3′ splicing sequence, between Ser-16 and Leu-17, results in a signal sequence with 16 amino acids. This second cleavage site would produce a mature protein with the amino-terminal sequence Leu-Ala-Thr, which is identical to that determined for the mature N. crassa β-MPP (9).
The drastic morphological changes occurring in the mitochondria during the B. emersonii life cycle, which include fragmentation during the germination of a giant single mitochondrion present in the zoospores into several normal-sized mitochondria and the fusion of these multiple mitochondria during sporulation, giving rise to the huge single mitochondrion of the zoospore (3), led us to investigate possible changes in β-MPP mRNA and protein levels, throughout the B. emersonii life cycle. However, even though some change in the amount of β-MPP mRNA was observed during sporulation, no significant variation was detected in the level of β-MPP protein during the fungal life cycle. These results indicate that despite the profound alterations in morphology, mitochondria are still capable of importing proteins during all stages of B. emersonii development.
Nucleotide sequence accession number.
The nucleotide sequence of the B. emersonii β-MPP gene has been submitted to the GenBank/EMBL Data Bank and assigned accession no. U41300.
Acknowledgments
We thank W. Neupert for the kind gift of N. crassa β-MPP antiserum, M. V. Marques for critical reading of the manuscript, and Elisety de Andrade Silva for manuscript preparation.
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-PADCT). C. R. C. Rocha was a predoctoral fellow of CNPq, and S. L. G. was partially supported by CNPq.
REFERENCES
- 1.Avedissian M, Gomes S L. Expression of the groESL operon is cell-cycle controlled in Caulobacter crescentus. Mol Microbiol. 1996;19:79–89. doi: 10.1046/j.1365-2958.1996.347879.x. [DOI] [PubMed] [Google Scholar]
- 2.Braun H P, Emmermann M, Kruft V, Schmitz U. The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J. 1992;11:3219–3227. doi: 10.1002/j.1460-2075.1992.tb05399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromberg R. Mitochondrial fragmentation during germination in Blastocladiella emersonii. Dev Biol. 1974;36:187–194. doi: 10.1016/0012-1606(74)90201-2. [DOI] [PubMed] [Google Scholar]
- 4.De Oliveira J C F, Borges A C C, Marques M V, Gomes S L. Cloning and characterization of the gene for the catalytic subunit of cAMP-dependent protein kinase in the aquatic fungus Blastocladiella emersonii. Eur J Biochem. 1994;219:555–562. doi: 10.1111/j.1432-1033.1994.tb19971.x. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson A C, Glasser E. Mitochondrial processing proteinase: A general processing proteinase of spinach leaf mitochondria is a membrane-bound enzyme. Biochem Biophys Acta. 1992;1140:208–214. [Google Scholar]
- 6.Feinberg A P, Volgelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 7.Fort P, Marty L, Piechiczyk M, El Sabrout S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray M W. The evolutionary origins of organelles. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 9.Hawlitscheck G, Schneider H, Schmidt B, Tropschug M, Hartl F U, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- 10.Isaya G, Kalousek F, Fenton W A, Rosenberg L E. Cleavage of precursors by the mitochondrial processing peptidase requires a compatible mature protein or an intermediate octapeptide. J Cell Biol. 1991;113:65–76. doi: 10.1083/jcb.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitada S, Niidome T, Nagano T, Ogishima T, Ito A. Molecular cloning of the smaller subunit (P52) of rat liver mitochondrial processing peptidase. Biochem Biophys Res Commun. 1993;190:289–293. doi: 10.1006/bbrc.1993.1044. [DOI] [PubMed] [Google Scholar]
- 12.Kitada S, Shimokafa K, Niidome T, Ogishima T, Ito A. A putative metal-binding site in the β subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. J Biochem. 1995;117:1148–1150. doi: 10.1093/oxfordjournals.jbchem.a124836. [DOI] [PubMed] [Google Scholar]
- 13.Kleiber J, Kalousek F, Swaroop M, Rosenberg L E. The general mitochondrial matrix processing protease from rat liver: structural characterization of the catalytic subunit. Proc Natl Acad Sci USA. 1990;87:7978–7982. doi: 10.1073/pnas.87.20.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessie P E, Lovett J S. Ultrastructural changes during sporangium formation and zoospore differentiation in Blastocladiella emersonii. Am J Bot. 1968;55:220–236. [PubMed] [Google Scholar]
- 15.Lovett J S. Growth and differentiation of the water mold Blastocladiella emersonii: cytodifferentiation and the role of ribonucleic acid and protein synthesis. Bacteriol Rev. 1975;39:345–404. doi: 10.1128/br.39.4.345-404.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Marques M V, Borges A C C, de Oliveira J C F, Gomes S L. Coordinate pretranslational control of cAMP-dependent protein kinase subunit expression during development in Blastocladiella emersonii. Dev Biol. 1992;149:432–439. doi: 10.1016/0012-1606(92)90297-t. [DOI] [PubMed] [Google Scholar]
- 18.McClelland J L, Rumelhart D E. Explorations in parallel distributed processing. Vol. 3. Cambridge, Mass: MIT Press; 1988. pp. 318–362. [Google Scholar]
- 19.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, Myo D and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 20.Niidome T, Kitada S, Shimokata K, Ogishima T, Ito A. Arginine residues in the extension peptide are required for cleavage of a precursor by mitochondrial processing peptidase. J Biol Chem. 1994;269:24719–24722. [PubMed] [Google Scholar]
- 21.Ou W-J, Ito A, Okasaki H, Omura T. Purification and characterization of a processing protease from rat liver mitochondria. EMBO J. 1989;8:2605–2612. doi: 10.1002/j.1460-2075.1989.tb08400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paces V, Rosenberg L E, Fenton W A, Kalousek F. The β subunit of the mitochondrial processing peptidase from rat liver: cloning and sequencing of a cDNA and comparison with a proposed family of metallopeptidases. Proc Natl Acad Sci USA. 1993;90:5355–5358. doi: 10.1073/pnas.90.11.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfanner N, Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- 24.Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt S, Wachter E, Sebald W, Neupert W. Processing peptidase of Neurospora mitochondria—two-step cleavage of imported ATPase subunit 9. Eur J Biochem. 1984;144:581–588. doi: 10.1111/j.1432-1033.1984.tb08505.x. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Heijne G, Steppuhn J, Hermann R G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 29.Witte C, Robert E J, Yaffe M P, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M, Jensen R E, Yaffe M P, Opplinger W, Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988;7:3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]