Abstract
In this work, the genes that encode the FliM and FliN proteins of Rhodobacter sphaeroides were characterized. These genes are part of a large flagellar gene cluster in which six additional open reading frames encoding products homologous to FliL, FliO, FliP, FliQ, FliR, and FlhB proteins from other bacteria were identified. The inactivation of the fliM gene gave a nonflagellate phenotype (Fla−), suggesting that FliM is required for flagellar assembly. Complementation analysis of this fliM mutant indicated that fliM and fliN transcription starts beyond the 5′ end of fliK and terminates after fliN.
Flagellar rotation alternates between the clockwise (CW) and counterclockwise (CCW) directions in some peritrichous species, like Escherichia coli. When flagella rotate in the CCW direction, the filaments coalesce in a bundle that acts as a propeller to push the bacterial cell body in a linear trajectory known as smooth swim. When the flagella reverse their direction of rotation, the bundle flies apart and the bacteria tumble; tumbling occurs every 2 to 5 s. In the presence of an attractant, the tumble frequency decreases and bacteria swim in the favorable direction. Ultimately, this behavior is controlled by the chemotactic protein CheY, which, in its phosphorylated form, binds to FliM, promoting the switch from CCW to CW rotation (for recent reviews, see references 13 and 24). Although the molecular details of switching are unknown, it is thought that the interaction between CheY-P and FliM is the first step in the control of this phenomenon (19, 30–32).
Interestingly, some bacteria rotate their flagellum in only one direction; consequently, reorientation at the end of a run is achieved by Brownian motion, which randomizes the direction of the next run. This kind of motility is observed in Rhodobacter sphaeroides (1). In the presence of an attractant, R. sphaeroides increases the duration of a run and, in contrast to enteric bacteria, also increases its swimming velocity. This phenomenon is known as chemokinesis (18).
Several chemotactic genes homologous to cheA, cheW, and cheR from other bacteria have been identified in R. sphaeroides (28, 29). Two cheY genes were found in the same operon. The deduced amino acid sequences of these genes showed that both putative CheY proteins carried residues D13, D57, and K109, which are essential for CheY function (4, 5, 28). This fact suggests that both putative CheY proteins may be functional.
Although in R. sphaeroides some flagellar genes have been characterized (2, 9, 21, 22), no information is available about the target of CheY in the cell. Our aim was to identify the fliM gene from this bacterium. We have previously reported the presence of a fliI homolog in R. sphaeroides. This gene is located in a 5.3-kb EcoRI fragment and is flanked by two open reading frames (ORFs) that show homology to fliH and fliJ from enteric bacteria (2). This gene arrangement encouraged us to search for the fliM gene downstream of fliJ. The sequence of the end of the 5.3-kb EcoRI fragment opposite to fliH revealed the 5′ end of fliM. A 4.6-kb SalI fragment carrying the complete fliM coding region was identified by Southern blot analysis (data not shown). This fragment was cloned in pTZ19R after digestion of WS8 chromosomal DNA with SalI. A positive clone was identified by colony hybridization and named pRS75. The sequence of this clone revealed the presence of fliM together with fliN, fliO, and part of fliP. To complete the sequence of fliP, we then cloned an EcoRI fragment of 4.4 kb in pTZ19R and performed colony hybridization with a fliP fragment as a probe. The new clone, pRS205, was then sequenced.
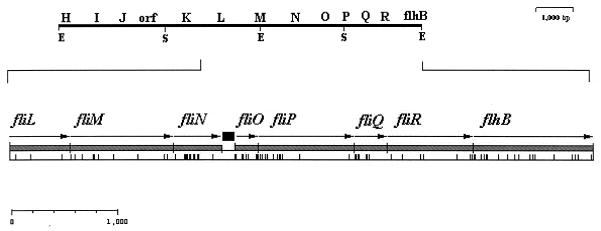
The analysis of pRS75 and pRS205 sequences was carried out with the Genetics Computer Group software package in conjunction with an R. sphaeroides codon usage table (7). Eight ORFs were identified (Fig. 1) and were then subjected to a BLAST search. As shown in Table 1, good homology between the flagellar proteins FliL, FliM, FliN, FliO, FliP, FliQ, FliR, and FlhB of different bacterial species was found.
FIG. 1.
Organization of the R. sphaeroides flagellar cluster. At the top is shown the general gene arrangement for this region (2, 9). The flagellar genes reported in this work are shown below; each ORF is represented as a shaded box. The arrows indicate the direction of transcription. A black square in the middle of fliN and fliO indicates a noncoding region containing a putative ς54 promoter sequence. Vertical bars below the schematic ORFs indicate the presence of rare codons. This analysis was done with CODONPREFERENCE from the Genetics Computer Group Wisconsin software package with a R. sphaeroides codon usage table (7).
TABLE 1.
Comparison of encoded proteins of R. sphaeroides flagellar genes with those of other bacterial species
| Gene | Function | Size of product (amino acids)a | % of unusual codons | % Homology (identity/similarity) to the gene from:
|
|||
|---|---|---|---|---|---|---|---|
| S. typhimurium | B. subtilis | C. crescentus | B. burgdorferi | ||||
| fliL | Unknown | 190 | 1.5 | 28/48 | 23/61 | 18/43 | 16/45 |
| fliM | Motor switch | 323 | 3.4 | 26/48 | 21/45 | 21/44 | 23/48 |
| fliN | Motor switch/export | 152 | 9.0 | 38/64 | — | 31/61 | 44/72 |
| fliO | Unknown | 85 | 11.7 | 21/51 | — | 34/58 | — |
| fliP | Unknownc | 301 | 5.3 | 53/74 | 48/71 | 46/68 | 38/66 |
| fliQ | Unknownc | 88 | 5.6 | 44/70 | 33/57 | 31/56 | 32/66 |
| fliR | Unknownc | 269 | 2.6 | 29/55 | 29/58 | 23/52 | 23/56 |
| flhB | Unknownc | >376 | — | 43/64 | 30/56 | — | 28/58 |
Number of amino acids deduced from the conceptual translation.
—, not determined.
Suspected to be involved in flagellum assembly; the product shows homology to virulence proteins (13).
The highest degree of identity was obtained when these ORFs were compared to those obtained from Salmonella typhimurium, with the exception of FliN and FliO, which showed a higher identity to their homologs from Borrelia burgdorferi and Caulobacter crescentus, respectively.
Figure 1 and Table 1 show that the frequency of codon usage for all these ORFs is in good agreement with that observed for R. sphaeroides genes with the exception of fliN and fliO; in the fliO ORF, 11.7% of the codons are unusual ones like ATT, TTT, CTT, and ACA.
The start codons for the fliL, fliM, and fliN ORFs are located only a few bases downstream of, or overlapping, the stop codon of the preceding ORF, suggesting that these genes belong to the same transcriptional unit. We also found that the putative ribosome binding site for fliL overlaps the stop codon of the recently reported fliK gene (9); this result suggests that fliK is part of this putative operon.
Also shown in Fig. 1 is the arrangement of fliO, fliP, fliQ, fliR, and flhB. These genes seem to form a single operon, given that the start codon overlaps the preceding stop codon in an ATGA or TGATG arrangement, which are the first and the second most frequent stop-start overlaps in E. coli (3). The initiation codons that could be used to translate the fliO mRNA are TTG or GTG, which predict a protein of 85 or 60 amino acids, respectively. Alignment of the NH2 end of FliO sequences from E. coli, S. typhimurium, C. crescentus, and R. sphaeroides shows a number of conserved residues in all these species (Fig. 2); however, if the R. sphaeroides FliO started at the GTG codon, some of these conserved residues would be missing. This fact allows us to conclude that the TTG codon is the most probable initiation site. Furthermore, a good ribosome binding site is located 7 bp upstream of this codon. The use of TTG as start codon, together with the high content of unusual codons in fliO, suggests a particular translational control of fliO that could maintain its product at a low molar concentration compared to the other genes of the operon.
FIG. 2.
Alignment of the NH2-end sequences of FliO from E. coli (GenBank accession no. L22182 and L21994) (FliOEc) S. typhimurium (L49021) (FliOSt), and C. crescentus (U20387) (FliOCc) with the deduced sequence of FliO from R. sphaeroides (FliORs) starting from the TTG codon. The second possible start codon encodes V25. Identical and similar residues present in all the species are shaded. The difference in length between E. coli and S. typhimurium corresponds to the controversy between the actual translation start codon for FliO (for details, see references 14 and 17).
Detailed sequence analysis did not reveal a good match with the consensus promoter sequence known for sigma 70 (ς70). However, a putative ς54 promoter sequence was located 83 bp downstream of the stop codon for fliN. This sequence (GGCACN5TTGC) matches 9 of 10 bp of the ς54 consensus promoter sequence (10). No inverted repeated sequences were found 100 bp upstream of this putative ς54 promoter. Although this may argue against its functionality, it is known that transcriptional activation of Eς54 still occurs if the activator binding sites are moved 2,000 bp upstream or downstream the ς54 promoter (16). Therefore, the possibility that remote activation sites exist to promote initiation of this putative promoter still remains.
Construction and characterization of an R. sphaeroides mutant with a fliM mutation.
To test the functionality of these genes, we decided to isolate a fliM mutant strain. For this purpose, a Spcr cassette, ′uidA-aad (uidA encodes β-glucuronidase, and aad encodes the aminoglycoside-3:adenyltransferase and confers Spcr) (15), was cloned in the middle of fliM in plasmid pRS75. The resulting fragment, carrying fliM::uidA-aad, was subcloned into the SalI site of pJQ200mp18 (Gmr) (20), which is unable to replicate in R. sphaeroides. This new plasmid was then transferred to R. sphaeroides by mating (8), and Spcr Gms transconjugants were selected. One colony was purified and named NG1.
That a successful double recombination event had occurred to yield NG1 was confirmed by a Southern blot experiment (data not shown). To test if the fliM::uidA-aad allele affected cell motility, NG1 cells were plated on tryptone motility plates and incubated at 30°C in a humid chamber. After 48 h, these cells were unable to swarm, in contrast to the wild-type cells (Fig. 3). It should be noted that these strains showed similar growth rates; therefore, the difference in swarming behavior cannot be explained by growth differences. To rule out the possibility that this mutant could still swim in liquid medium, an aliquot from a NG1 culture grown under aerobic and anaerobic conditions was observed directly under the microscope; swimming was not detected after growth under either condition. To determine if the inability to swim or swarm could be ascribed to a Fla− or paralyzed (Mot−) phenotype, an aliquot of NG1 cells was negatively stained and observed under the electron microscope. Since no flagellar structure was visible in more than 90% of cells in the sample, Fla− was the phenotype assigned to NG1. A few cells showed a small filament-like structure clearly different from the wild-type filament. These cells may represent secondary suppressors of the fliM::uidA-aad allele.
FIG. 3.
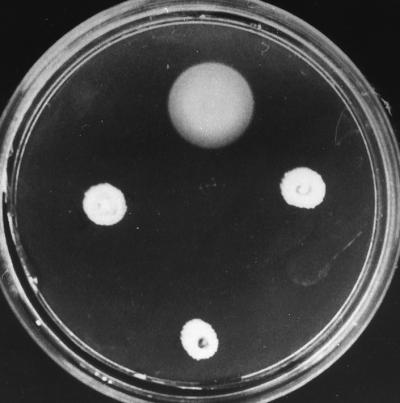
Motility plates showing the phenotype of NG1 cells. WS8 was included as a control (top). The negative control (bottom) corresponds to strain PG2 (fliK::TnphoA) (9). NG1 cells were spotted twice (middle row).
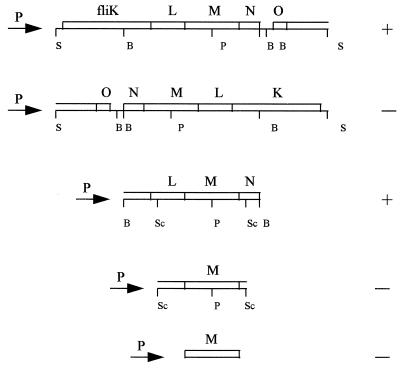
Full motility was recovered when pRK415 (11) carrying the 4.6-kb SalI fragment was introduced into the mutant strain NG1. Both swimming and swarming were indistinguishable from wild-type behavior (Fig. 4). However, when this DNA fragment was cloned in the opposite orientation, complementation of NG1 cells was not observed (Fig. 4). In this last construction, the fragment containing fliLMNOP′ was in the opposite orientation to the direction of transcription from the lac and tet promoters (lacp and tetp) of pRK415. The fact that this SalI fragment was able to complement NG1 in just one orientation suggests that fliM does not have a promoter in this fragment and that its transcription depends on the lac and tet promoters of pRK415. This result strongly suggests that fliM expression is dependent on a promoter located upstream of fliK.
FIG. 4.
Complementation analysis of NG1 cells with the 4.6-kb SalI fragment and subclones. Some of the relevant restriction sites are indicated: S, SalI; B, BamHI; P, PstI; Sc, SacII. Restoration of swimming behavior is recorded by +; no complementation is indicated by −. The arrows indicate the direction of transcription starting from the lac and tet promoters located in the pRK415 vector.
It has been previously shown that the uidA-aad fragment is highly polar in E. coli as well as in other bacteria (15). Therefore, we decided to determine if the uidA-aad cassette inserted in fliM exerted a polar effect on the transcription of downstream genes. For this, a 1.3-kb SacII fragment and a PCR product, both containing fliM, were each cloned in pRK415 in the proper orientation to allow its expression from lacp and tetp. The resulting plasmids, pRS14 and pRS10, respectively, did not complement NG1 cells, indicating that the Fla− phenotype observed in the NG1 strain was caused by inactivation of the fliM gene, which in turn caused lack of transcription of downstream genes.
Since the polar effect of the uidA-aad cassette will affect only genes that belong to the same transcriptional unit, we introduced into NG1 cells a 2.3-kb BamHI fragment in pRK415. This fragment includes the fliLMN genes under control of the pRK415 promoters. We observed that this construction restored the swimming and swarming abilities of NG1 cells. This result suggests that fliN must be the last gene of a single transcriptional unit. In consequence, fliOPQR and flhB may form an independent operon. Alternatively, it is possible that fliOPQR and flhB are not functional genes; however, we consider this explanation less probable given that we have isolated a mutant with an fliR mutation that showed the nonmotile phenotype (6).
Finally, to determine the phenotype of a strain carrying a lesion in fliM, we introduced into NG1 cells a 980-bp PstI-BamHI fragment cloned in pRK415. This fragment contains the 3′ end of fliM and the complete fliN gene. The phenotype of NG1 carrying this plasmid should correspond to a fliM mutant, avoiding any polar effect on fliN. Phenotypic characterization of NG1 cells carrying this construction shows that they have the Fla− phenotype (data not shown). This result suggests that FliM plays some role in the export of flagellar components. It was recently shown that FliM and FliN proteins require each other for their integration into the switch structure (12). In this regard, Vogler et al. (27) showed that a temperature-sensitive fliN mutant cannot export flagellar proteins at the restrictive temperature. These data suggest that the switch complex may contribute to the stability of the export apparatus. Nevertheless, the role of the switch complex in the export process has yet to be solved.
Is a single copy of fliM present in R. sphaeroides?
The presence of two cheY genes in the chromosome of R. sphaeroides, as well as the high degree of conservation of some residues involved in CheY functionality such as D13, D57, and K109 (28) in both cheY products, raises the question whether more than one copy of the fliM gene would be necessary to form a switch complex carrying two different species of FliM protein. Using the complete fliM gene as a probe, we carried out a Southern blot experiment under low hybridization stringency and gentle washing conditions. We did not find evidence for a second fliM gene in the chromosomal DNA of R. sphaeroides, suggesting that CheY-mediated chemotaxis control may be achieved by competition between the two different CheY proteins for FliM or for CheA binding, as has been suggested for Rhizobium meliloti (23). Alternatively, it is possible that a gene encoding a protein carrying just the CheY-P binding site could not be detected by Southern blotting.
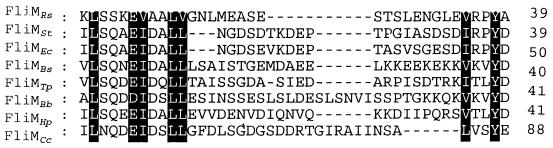
On the other hand, it has been shown that CheY binds to the first 40 to 50 amino acids of FliM (25, 26). Alignment of our deduced FliM sequence with seven other FliM sequences shows some highly conserved residues among the first 50 amino acids, i.e., a tyrosine approximately 40 residues from the N terminus, and the presence of an LN3(E/D)N3L(L/V) motif, which predicts an α-helix (Fig. 5). The importance of these conserved amino acids in the function of FliM is currently under investigation in our laboratory.
FIG. 5.
Alignment of the first 39 amino acids of FliM from R. sphaeroides with FliM sequences of other bacterial species. Identical and similar residues conserved in all the sequences are shaded. (The GenBank accession numbers are as follows: U46011 and U00096, FliMEc [E. coli]; M24465, FliMSt, [S. typhimurium]; L75945, FliMBb [B. burgdorferi]; U28219, FliMTp [T. pallidum]; M37691, FliMBs [B. subtilis]; AE000611 FliMHp [H. pylori]; M85232, FliMCc [C. crescentus]).
It will be interesting to understand the interactions between FliM and both CheY proteins as well as FliG in order to understand how chemokinesis and the stop-start processes are controlled in this organism.
Nucleotide sequence accession numbers.
The DNA sequences of the R. sphaeroides fliL, fliM, fliN, fliO, fliP, fliQ, fliR, and flhB genes have been deposited in the GenBank data base under accession no. AF044254 and AF044580.
Acknowledgments
We are indebted to Luis Servín and Laura Velázquez for helpful discussions and critical review of the manuscript. We thank T. Ballado for technical assistance. We also thank M. West and E. Silva-Herzog for their comments and helpful discussions and J. Sepúlveda for technical support with electron microscopy. We also thank the Unidad de Biologia Molecular (IFC) for the synthesis of some oligonucleotides used in this work.
This work was supported in part by CONACyT grants 4739-N9406 to L.C. and 3290P-N9607 to G.D.
ADDENDUM
In the course of this work, the sequences of the R. sphaeroides fliM and fliN genes appeared in the GenBank database (9a). A comparison of these sequences with ours showed an inversion of GC to CG in the fliM sequence, producing a change of a cysteine (C152) for a valine (V152) in our sequence.
REFERENCES
- 1.Armitage J P, Macnab R M. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol. 1987;169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballado T, Campos A, Camarena L, Dreyfus G. Flagellar genes from Rhodobacter sphaeroides are homologous to genes of the fliF operon of Salmonella typhimurium and to the type-III secretion system. Gene. 1996;170:69–72. doi: 10.1016/0378-1119(95)00855-1. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 6.Camarena, L., S. Poggio, and G. Dreyfus. Unpublished data.
- 7.Campos, A., L. Camarena, and G. Dreyfus. Codon usage table of R. sphaeroides of more than one hundred genes. Unpublished data.
- 8.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a puf mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Pedrajo B, Ballado T, Campos A, Sockett R E, Camarena L, Dreyfus G. Structural and genetic analysis of a mutant of Rhodobacter sphaeroides WS8 deficient in hook length control. J Bacteriol. 1997;179:6581–6588. doi: 10.1128/jb.179.21.6581-6588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Goodfellow, I. G., et al. GenBank accession no. Y14335.
- 10.Helmann J D, Chamberline M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 11.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 12.Kubori T, Yamaguchi S, Aizawa S-I. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macnab R M. Flagella and motility. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 14.Malakooti J, Ely B, Matsumura P. Molecular characterization, nucleotide sequence, and expression of the fliO, fliP, fliQ, and fliR genes of Escherichia coli. J Bacteriol. 1994;176:189–197. doi: 10.1128/jb.176.1.189-197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metcalf W W, Wanner B L. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 16.Ninfa A J, Reitzer L J, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi K, Fan F, Schoenhals G J, Kihara M, Macnab R M. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J Bacteriol. 1997;179:6092–6099. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Packer H L, Armitage J P. The chemokinetic and chemotactic behavior of Rhodobacter sphaeroides: two independent responses. J Bacteriol. 1994;176:206–212. doi: 10.1128/jb.176.1.206-212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkinson J S, Parker S R, Talbert P B, Houts S E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983;155:265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 21.Shah D S H, Sockett R E. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol Microbiol. 1995;17:961–969. doi: 10.1111/j.1365-2958.1995.mmi_17050961.x. [DOI] [PubMed] [Google Scholar]
- 22.Shah D S H, Armitage J P, Sockett R E. Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J Bacteriol. 1995;177:2929–2932. doi: 10.1128/jb.177.10.2929-2932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sourjik V, Schmitt R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol. 1996;22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 24.Stock J, Surette M G. Chemotaxis. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 25.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 27.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward M J, Bell A W, Hamblin P A, Packer H L, Armitage J P. Identification of a chemotaxis operon with two cheY genes in Rhodobacter sphaeroides. Mol Microbiol. 1995;17:357–366. doi: 10.1111/j.1365-2958.1995.mmi_17020357.x. [DOI] [PubMed] [Google Scholar]
- 29.Ward M J, Harrison D M, Ebner M J, Armitage J P. Identification of a methyl-accepting chemotaxis proteins in R. sphaeroides. Mol Microbiol. 1995;18:115–121. doi: 10.1111/j.1365-2958.1995.mmi_18010115.x. [DOI] [PubMed] [Google Scholar]
- 30.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, Aizawa S-I, Kihara M, Jones C J, Macnab R M. Genetic evidence for a switch an energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]