Abstract
hspA, a gene encoding a 16-kDa heat-induced protein from the thermophilic cyanobacterium Synechococcus vulcanus, has been cloned and sequenced. The deduced amino acid sequence of the gene product showed significant homology to sequences of the family of α-crystallin-related, small heat shock proteins. A monocistronic mRNA of hspA increased transiently in response to heat shock. The heat shock induction occurred at a vegetative promoter but without the CIRCE (controlling inverted repeat of chaperone expression) element.
Elevated temperatures as well as many other stresses such as high concentrations of ethanol, arsenite, and heavy metals elicit a physiological response resulting in the synthesis of a distinct set of proteins called heat shock proteins (HSPs) (16, 19). Among the most strongly induced HSP families are the α-crystallin-related, small HSPs, which are a group of proteins having molecular masses of 15 to 30 kDa (3, 11, 30). Unlike other groups of HSPs such as HSP70 and HSP60, small HSPs are not highly conserved (3, 11, 30). Sequence similarity is limited only to their carboxy-terminal regions. Various functions for small HSPs have been suggested, including protection against heat shock (3, 11, 30). Recent in vitro studies with recombinant small HSPs suggest that the small HSPs act in vivo as a type of molecular chaperone (3, 4, 11, 12, 30).
Prokaryotic homologs of small HSPs have been identified in Bacillus subtilis (10), Bradyrhizobium japonicum (17), Clostridium acetobutylicum (24), Escherichia coli (2), Mycobacterium leprae (18), Mycobacterium tuberculosis (29), Stigmatella aurantiaca (7, 8), and Streptomyces albus (25). Small HSPs from prokaryotes such as C. acetobutylicum (24), E. coli (2), Streptomyces albus (25), and Stigmatella aurantiaca (8) have been shown to be induced by elevated temperatures, although a homolog from Bacillus subtilis, CotM, was not induced by heat shock (10). Developmentally regulated expression of small HSPs from Bacillus subtilis (10), C. acetobutylicum (24), and Stigmatella aurantiaca (7, 8) have been demonstrated.
Like other organisms, cyanobacteria, which are oxygenic photosynthetic bacteria, synthesize a diverse range of HSPs upon exposure to high temperatures (31). However, the regulatory mechanism for the expression of cyanobacterial HSPs and their physiological functions remain poorly understood. Recently, an open reading frame (ORF), sll1514, which was designated hspA, was discovered during the genome sequencing project for Synechocystis sp. strain PCC 6803, a mesophilic cyanobacterium (13). The ORF has sequence homology with Stigmatella aurantiaca hspA, which encodes a small HSP homolog (8). This is the first indication of the presence of a small HSP in cyanobacteria. However, it is not known if the sll1514 ORF is actually expressed or not.
Synechococcus vulcanus, a thermophilic cyanobacterium, accumulates a 16-kDa protein along with GroEL and GroES as the major heat shock-induced proteins when the cells are shifted from 50 to 63°C (5, 21). Recently, we purified the 16-kDa protein to an apparent homogeneity (21). Size-exclusion chromatography and nondenaturing gel electrophoresis demonstrated that the 16-kDa protein formed large oligomeric structures of 280 kDa. The small HSPs from eukaryotes and a prokaryote (i.e., M. tuberculosis) are found in larger-molecular-mass complexes between 200 and 800 kDa that are most likely composed solely of small HSP subunits (3, 4, 11, 30).
In the present article, we report the isolation, sequence analysis, and in vivo transcription analysis of the gene encoding the 16-kDa protein from Synechococcus vulcanus.
Organisms and culture conditions.
The thermophilic unicellular cyanobacterium Synechococcus vulcanus (15) was grown at 50°C in a liquid medium used for another thermophilic cyanobacterium, Synechococcus sp. (14), under a light intensity of 50 μmol/m2/s. The culture was bubbled with air supplemented with 5% CO2. E. coli JM109 [recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15)] used in cloning experiments was grown in Luria-Bertani medium supplemented with ampicillin (50 μg/ml) when appropriate (22).
Cloning and sequencing of the gene encoding the 16-kDa protein from Synechococcus vulcanus.
The following two degenerate oligonucleotide primers were used to amplify a part of the gene encoding the 16-kDa protein from Synechococcus vulcanus by PCR. One of the primers, 5′-GCIATICA(A/G)(C/A)GICA(A/G)ATGAA-3′ was based on the amino-terminal sequence of the 16-kDa protein, AIQRQMN, and the other primer, 5′-CCIGGIA(A/G)(C/T)TCIAC(C/T)TT-3′, was based on the internal amino acid sequence of the protein, KVELPG. These amino acid sequences (Fig. 1) were determined by Edman degradation by utilizing the purified 16-kDa protein from Synechococcus vulcanus (21). A single 140-bp product was amplified from the Synechococcus vulcanus genome (20 ng) after 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 2 min at 72°C. The PCR product was subcloned in pGEM-T vector (Promega, Madison, Wis.) to yield pGEMSH. Then, DNA sequencing of the PCR product was done by the dideoxy termination method (23) with an AutoRead sequencing kit (Pharmacia, Uppsala, Sweden) and a DNA sequencer (DSQ-1; Shimadzu, Kyoto, Japan). The deduced amino acid sequence of the PCR product contained the amino-terminal and internal amino acid sequences of the 16-kDa protein. The PCR fragment was recovered by digesting pGEMSH with PstI and SphI and radiolabeled with [α-32P]dCTP (ICN Biochemicals, Costa Mesa, Calif.) by the multiprime labeling method as directed by the manufacturer (Amersham International plc, Little Chalfont, England), purified through a NICK column (Pharmacia), and then used as a specific probe for genomic library screening and the Southern and Northern blot analyses described below.
FIG. 1.
Nucleotide sequence of the hspA gene from Synechococcus vulcanus. Putative −10 and −35 promoter sequences (underlined), the transcriptional start site (indicated by a downward arrow), a potential ribosome binding site (indicated by boldface type), and an inverted repeat thought to represent an hspA transcriptional terminator (indicated by horizontal arrows below the sequences) are marked. Only relevant restriction sites are depicted above the DNA sequence. The deduced amino acid sequence of the gene is shown below the DNA sequence (single-letter code). The translation stop signal is marked by an asterisk below the codon. The underlined amino acid sequences correspond to the sequences determined from the purified 16-kDa protein from Synechococcus vulcanus (21).
A Synechococcus vulcanus genomic library constructed in a bacteriophage λ-DASH vector, which was kindly provided by Yorinao Inoue of RIKEN, was screened through plaque hybridization (22) with the probe described above. Bacteriophage DNA from positive plaques was prepared by the liquid culture method, and further screening was performed by Southern blot analysis (22) after digestion of the DNA with EcoRI, PstI, and XbaI. EcoRI, PstI, and XbaI fragments of 1.1, 1.5, and 3.0 kbp, respectively, which were hybridized with the probe described above, were subcloned into pBluescript (pBS) II KS (+) (Stratagene, La Jolla, Calif.). A 3.0-kbp XbaI fragment was further digested with HincII, and the resulting fragments were subcloned into pBS II KS (+). The 1.1-kbp EcoRI, 1.5-kbp PstI, and 0.9- and 1.0-kbp HincII fragments and deletions obtained by exonuclease III digestion (Erase-a-Base system; Promega) of the 3-kbp XbaI fragment were used for sequencing both strands of the DNA containing the gene.
As shown in Fig. 1, an ORF of 438 bp encoding a putative polypeptide of 145 amino acids was found in the sequenced region of 1,302 bp. The amino-terminal and internal amino acid sequences determined chemically were present in the predicted amino acid sequence of the ORF (Fig. 1), thus confirming the ORF as the gene encoding the 16-kDa protein. The chemically determined amino-terminal sequence of the 16-kDa protein (Fig. 1) revealed that the amino-terminal methionine was removed in the mature protein. The molecular weight and pI value for the mature protein are predicted to be 16,519 and 5.26, respectively. The deduced molecular mass was in good agreement with an apparent molecular mass of 16 kDa that was estimated for the heat-induced protein on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
A putative ribosome-binding site highly homologous to the E. coli prototype (27) was identified 8 bp upstream from the initiation codon of the ORF (Fig. 1). Sequences homologous to E. coli ς70-dependent promoters (9) were found further upstream (Fig. 1). However, neither the heat shock promoter recognized by ς32 (32) nor CIRCE (controlling inverted repeat of chaperone expression) (33) was found. CIRCE has been detected around the transcriptional start site of groESL and dnaK operons of many bacterial species, including cyanobacteria which contain promoter sequences recognized by the vegetative sigma factor (6, 33). A 7-nucleotide palindromic sequence which has the potential to form a prokaryotic rho-independent transcriptional terminator (20) is located 50 bp downstream from the stop codon (Fig. 1).
The amino acid sequence deduced from the gene encoding the 16-kDa protein was analyzed with the National Center for Biotechnology Information BLAST network server to search for a homologous sequence. The 16-kDa protein exhibited sequence similarity to proteins belonging to the family of small HSPs (data not shown). These proteins include prokaryotic homologs from several species of Mycobacterium, Stigmatella aurantiaca, Streptomyces albus, and the plant class I and class II small HSPs, chloroplast small HSPs, and plant class IV HSPs. The 16-kDa protein from Synechococcus vulcanus had the closest sequence homology to the protein encoded by hspA from Synechocystis sp. strain PCC 6803 (52% overall identity). Thus, we designated the gene encoding the 16-kDa protein from Synechococcus vulcanus hspA.
Southern blot analysis.
We determined the number of copies of hspA in the Synechococcus vulcanus genome by Southern blot analysis. Synechococcus vulcanus genomic DNA digested with several different restriction endonucleases was hybridized with the radiolabeled 140-bp PCR-generated fragment (data not shown). The probe hybridized to only one DNA fragment from each restriction endonuclease digest. Similar experiments were repeated with another probe prepared with a 0.6-kbp EcoRI fragment containing a 3′ portion of the hspA coding region (Fig. 1) that includes sequence encoding the conserved consensus region I (30). Results with this probe corresponding to a higher conserved region together with the 140-bp PCR-generated fragment demonstrated that there is no other gene in the genome that is homologous to hspA.
Northern blot analysis.
A 500-ml culture of exponentially growing Synechococcus vulcanus cells at 50°C whose optical density at 730 nm was approximately 0.5 was divided into three portions. Under the same aeration and light conditions, one portion was kept at 50°C and the others were shifted to a 63°C bath and incubated for 15 or 60 min. After heat treatment, cells harvested by centrifugation at 4°C were immediately frozen in liquid nitrogen. Total RNA was isolated by the hot-phenol extraction method described previously (1). Total RNAs (5 μg) were electrophoresed on a denaturing 1.5% (wt/vol) agarose gel containing 6.6% (wt/vol) formaldehyde, transferred to a BA-S 83 nitrocellulose membrane in 20× SSC (1× SSC is 0.15 M NaCl containing 0.015 M sodium citrate) by a capillary transfer method (22), and then cross-linked to the membrane by UV illumination (FUNA-UV-Linker; Funakoshi, Tokyo, Japan). Prehybridization with the membrane was performed in a solution containing 6× SSC–5× Denhardt’s solution (22)–0.1% SDS for 1 h at 65°C, and then hybridization was done in a solution of the same constituents containing the denatured radiolabeled 140-bp PCR fragment and 100 μg of denatured salmon sperm DNA per ml at 65°C overnight. After hybridization, the membrane was washed once in 6× SSC at room temperature for 5 min and, thereafter, twice in 6× SSC at 65°C for 30 min. The size of each mRNA was determined by using an RNA ladder (Gibco-BRL, Gaithersburg, Md.). The hybridization signals were detected and quantified with a BAS1000 Mac bio-imaging analyzer (Fuji Film, Tokyo, Japan).
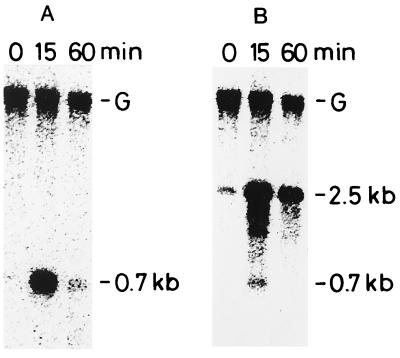
A single signal, corresponding to a 700-nucleotide RNA, was detected after heat shock (Fig. 2A). This corresponds to the predicted size of a monocistronic transcript which starts from the transcription start point determined in this study (see Fig. 4) and ends at an inverted repeat located immediately downstream of hspA (Fig. 1). The accumulation of the hspA transcript was enhanced more than 10-fold when cells were exposed to temperatures of 63°C for 15 min (Fig. 2A). This high level of transcript accumulation was transient; even during continued heat shock, it declined to almost preshock levels within 60 min (Fig. 2A). We detected a correspondingly rapid accumulation of the 16-kDa protein by SDS-polyacrylamide gel electrophoresis analysis. It could be detected 15 min after heat shock (21), and after 30 min, accumulation of the protein reached a plateau. Therefore, expression of hspA appeared to be regulated by heat shock at the level of transcription.
FIG. 2.
Northern blot analysis of the hspA (A) and groEL1 (B) transcripts from Synechococcus vulcanus. Total RNA was isolated from cells grown at 50°C (0 min) or incubated for different time intervals (15 and 60 min) after cultures were shifted from 50 to 63°C. Five micrograms of RNA from each sample was electrophoresed on a 1.5% (wt/vol) agarose gel containing 6.6% (wt/vol) formaldehyde. The 140-bp PCR product complementary to a part of the hspA coding region (Fig. 1) was used as a specific probe of the hspA transcripts. After the analysis, the same membrane was used for reprobing with a radiolabeled 540-bp PCR product complementary to a part of the Synechococcus vulcanus groEL1 coding region (28) (B). The 0.7-kb signal originates from the previous hybridization with the hspA probe since the same membrane was used. The uppermost band (labeled G) is due to contaminating genomic DNA as judged from its size. It disappeared when the sample was treated with DNase I before being loaded onto the agarose gel (data not shown).
FIG. 4.
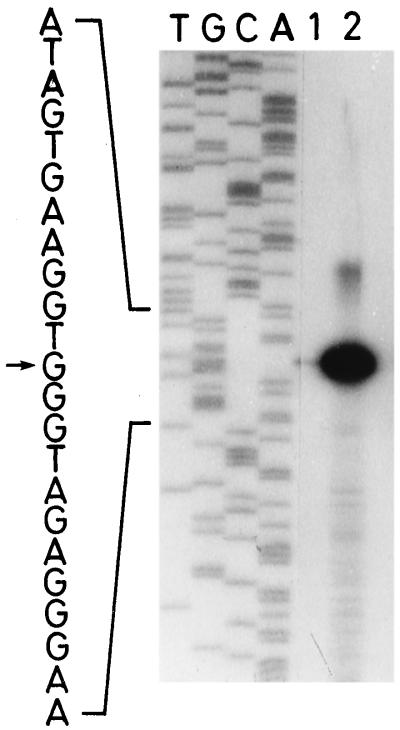
Mapping of the 5′ end of hspA mRNA by primer extension analysis. Primer extension reactions were performed with the radiolabeled oligonucleotide primer complementary to the 5′-end coding region of hspA, and total RNA (20 μg) which was isolated from Synechococcus vulcanus cells before (lane 1) and after (lane 2) shifting from 50 to 63°C for 15 min. T, G, C, and A indicate the dideoxy sequencing ladder obtained with the same primer and the pBS II KS (+) derivative described in the text as a template. The potential transcription start site is indicated by an arrow.
The expression of hspA was compared with that of groESL1. CIRCE has been detected upstream of all the cyanobacterial groESL genes, including the gene from Synechococcus vulcanus (28). The Northern blot was reprobed with a 32P-labeled 540-bp DNA fragment containing a 5′ portion of the groEL1 coding region from Synechococcus vulcanus (28). The probe hybridized to a 2.5-kb transcript (Fig. 2B) carried by the groESL1 operon. Previously, we showed that groES and groEL1 from Synechococcus vulcanus are organized in an operon and cotranscribed (28). The level of the groESL1 transcript expression was also transiently induced during heat shock and declined to low levels within 60 min (Fig. 2B).
Effect of rifampin on the stability of hspA mRNA.
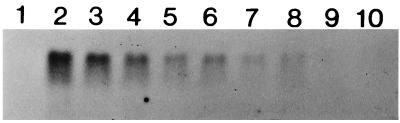
To examine whether increased transcript stability could contribute to heat shock induction of Synechococcus vulcanus hspA, the decay of hspA mRNA which accumulated during 15 min of heat shock was monitored, at 50 and 63°C, after inhibition of transcription by the addition of rifampin (Fig. 3). It appeared that Synechococcus vulcanus hspA mRNA was less stable at 50 than at 63°C (Fig. 3, compare lanes 3 and 4 with lanes 5 and 6). The half-life of Streptomyces albus hsp18 mRNA at 30°C was also shorter than that at 41°C (26). Interestingly, the stability of the mRNA was significantly increased by the addition of rifampin (Fig. 3, compare lanes 3 and 4 with lanes 7 and 8). It suggests that expression of an unknown gene(s) affects the stability of hspA mRNA.
FIG. 3.
Effect of rifampin on the stability of hspA mRNA in Synechococcus vulcanus. A 500-ml culture of exponentially growing cells at 50°C was shifted to a 63°C bath after cells were harvested (0 time) (lane 1) and incubated for 15 min to induce hspA expression. Immediately after cells were harvested (lane 2), the culture was divided into four 80-ml portions. An 0.8-ml volume of rifampin (30 mg dissolved in 1 ml of 100% methanol) was added to each of two cultures, and 0.8 ml of 100% methanol was added to each of the other ones. Then, a pair of cultures with (lanes 3, 4, 5, and 6) or without (lanes 7, 8, 9, and 10) rifampin was left at 63°C (lanes 3, 4, 7, and 8), and the other pair of cultures was shifted back to 50°C (lanes 5, 6, 9, and 10). Cells were harvested at 5 (lanes 3, 5, 7, and 9) or 10 (lanes 4, 6, 8, and 10) min after rifampin or methanol addition. Harvested cells were immediately frozen in liquid nitrogen, and total RNA was isolated by the hot-phenol extraction method as described in the text. A 7.5-μg amount of RNA from each sample was electrophoresed on a 1.5% (wt/vol) agarose gel containing 6.6% (wt/vol) formaldehyde. The 0.6-kbp EcoRI fragment containing the 3′ region of hspA (see Fig. 1) was used as a specific probe of the hspA transcripts. Hybridization and detection of the hybridization signals were performed by the nonradioactive method (AlkPhos Direct) as directed by the manufacturer (Amersham International plc.).
The transcriptional initiation site of hspA in Synechococcus vulcanus.
To determine the 5′ end of the hspA transcript, an oligonucleotide primer, 5′-GGTTCCCAACGAACGAGTGC-3′, complementary to the 5′ terminus of the hspA coding strand sequence (Fig. 1, nucleotides 433 to 452) was radiolabeled with [γ-32P]dATP (ICN Biochemicals) by using T4 polynucleotide kinase. Ten microliters (20 μg) of total RNA isolated from heat-shocked or non-heat-shocked cells, 4 μl of fivefold-concentrated buffer, which was supplied with the reverse transcriptase described below, and 2 μl (4 pmol) of the labeled primer were mixed together, heated at 65°C for 10 min, and then incubated at room temperature for 1 to 2 h. Two microliters of 0.1 M dithiothreitol and 1 μl of 20 mM deoxynucleoside triphosphates were added to the annealed primer-template mixture, and the mixture was incubated at 42°C for 2 min. Primer extension was carried out by the addition of 1 μl (200 U) of SuperScript II RNase H− reverse transcriptase (Gibco-BRL) and subsequent incubation at 42°C for 1 h. The extended products were precipitated with ethanol and resuspended in 4 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–6 μl of formamide loading buffer (80% formamide, 10 mM EDTA [pH 8.0], 1 mg of xylenecyanol per ml, and 1 mg of bromophenol blue per ml). After heat denaturation, a 5-μl sample was loaded onto a 6% polyacrylamide–7 M urea sequencing gel for electrophoresis (Fig. 4). Products of dideoxynucleotide sequencing reactions performed with the same primer and the cloned 3.0-kbp XbaI fragment as a template were run in parallel to allow determination of the end points of the primer extension products.
A unique 5′ end of hspA transcripts was identified 121 nucleotides upstream from the hspA translational initiation codon (Fig. 4). The 5′ end was preceded by −35 and −10 regions which display features similar to E. coli ς70-dependent promoters. These results suggest that the 5′ end is the transcriptional start point of the Synechococcus vulcanus hspA gene. Despite being transcribed from an apparent ς70-dependent promoter sequence, mRNA was barely detected at 50°C under non-heat-shock conditions. Instead, the transcript was clearly heat inducible (Fig. 2A and 4).
Concluding remarks.
In the present study, we have cloned and sequenced hspA, a gene encoding the 16-kDa protein. The deduced amino acid sequence of the gene product showed significant homology to the sequence of other small HSPs from prokaryotes and eukaryotes. Thus, the sequence comparison as well as the structural analysis (21) confirmed that the 16-kDa protein belongs to the family of small HSPs.
In Bradyrhizobium japonicum, there are three different genes for small HSP homologs, hspA, hspB, and hspC. hspA and rpoH1, and hspB and hspC, form respective operons which are transcribed as bicistronic mRNAs (17). ibpA and ibpB, encoding two small HSPs in E. coli, also appear to form an operon (2). Contrary to the case with Bradyrhizobium japonicum and E. coli, only one copy of the hspA gene was found in the Synechococcus vulcanus genome; this gene was transcribed as a monocistronic mRNA (Fig. 2A). In C. acetobutylicum, hsp18 was shown to be transcribed as a monocistronic mRNA (24). However, the gene copy number in this organism is not known.
Although genes for small HSPs have been isolated from several prokaryotes, expression of those genes induced by heat stress has not been analyzed in detail. In C. acetobutylicum and Bradyrhizobium japonicum, Northern blot analysis and/or primer extension analysis detected no small HSP mRNA under non-heat-shock conditions but showed increased mRNA levels in heat-shocked cells (17, 24). However, the data are from only one time point after heat shock. The kinetics of mRNA accumulation in Streptomyces albus was analyzed previously (25). Expression was induced upon heat shock, a peak was reached after 20 min of heat shock, and thereafter the amount of hsp18 mRNA began to decrease slowly. Two hours later, the mRNA reached a plateau of still-high-level expression (25). The continuously enhanced levels of hsp18 mRNA accumulation in Streptomyces albus are in marked contrast to the results with Synechococcus vulcanus, in which the level of the hspA transcripts is only transiently induced during heat shock and declines to almost preshock levels within 60 min (Fig. 2A).
In E. coli, ς32 was shown to be involved in the regulation of ibpA and ibpB (2). In other prokaryotes such as Bradyrhizobium japonicum (17), C. acetobutylicum (24), and Streptomyces albus (25), vegetative promotors were shown to be involved in the expression of the small HSP genes. Primer extension analysis with mRNA of heat-shocked cells revealed that the potential transcription initiation site of the Synechococcus vulcanus hspA gene is also preceded by vegetative −10 and −35 sequences. Recently, Servant and Mazodier (26) showed that orfY, located upstream of Streptomyces albus hsp18, is involved in the transcriptional regulation of the hsp18 gene. orfY is present in the opposite orientation to the hsp18 gene, and the start codons of the two genes are separated by only 150 bp. The sequenced region upstream from Synechococcus vulcanus hspA does not contain (part of) an ORF in the opposite orientation to the hspA (Fig. 1). Furthermore, we found no ORF that shows significant homology to orfY in the entire genome of Synechocystis sp. strain PCC 6803. In total, our results suggest that a novel regulatory mechanism suppresses the expression of hspA in cyanobacteria under non-heat-shock conditions.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB002666.
Acknowledgments
This work was supported in part by a grant for the improvement of education from Saitama University.
We are grateful to Tetsuo Hiyama for his encouragement throughout this study, Naoki Tanaka for his help in initial screening and sequencing experiments, Garrett J. Lee and Elizabeth Vierling for reading this manuscript, and Eiji Suzuki for his advice in performing primer extension analysis.
REFERENCES
- 1.Aiba H, Adhya S, De Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Allen S P, Polazzi J O, Gierse J K, Easton A M. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo A-P, Landry J. Expression and function of the low-molecular-weight heat shock proteins. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. pp. 335–373. [Google Scholar]
- 4.Chang Z, Primm T P, Jakana J, Lee I H, Serysheva I, Chiu W, Gilbert H F, Quiocho F A. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- 5.Furuki M, Tanaka N, Hiyama T, Nakamoto H. Cloning, characterization and functional analysis of groEL-like gene from thermophilic cyanobacterium Synechococcus vulcanus, which does not form an operon with groES. Biochim Biophys Acta. 1996;1294:106–110. doi: 10.1016/0167-4838(96)00037-4. [DOI] [PubMed] [Google Scholar]
- 6.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 7.Heidelbach M, Skladny H, Schairer H U. Purification and characterization of SP21, a development-specific protein of the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1993;175:905–908. doi: 10.1128/jb.175.3.905-908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidelbach M, Skladny H, Schairer H U. Heat shock and development induce synthesis of a low-molecular-weight stress-responsive protein in the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1993;175:7479–7482. doi: 10.1128/jb.175.22.7479-7482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 10.Henriques A O, Beall B W, Moran C P., Jr CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–1897. doi: 10.1128/jb.179.6.1887-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakob U, Buchner J. Assisting spontaneity: the role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 12.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 14.Katoh S. Photosystem I and photosystem II preparations from thermophilic Synechococcus. Methods Enzymol. 1988;167:263–269. [Google Scholar]
- 15.Koike H, Inoue Y. Preparation of oxygen-evolving photosystem II particles from a thermophilic blue-green alga. In: Inoue Y, Crofts A R, Govindjee, Murata N, Renger G, Satoh K, editors. The oxygen evolving system of photosynthesis. Tokyo, Japan: Academic Press; 1983. pp. 257–263. [Google Scholar]
- 16.Morimoto R I, Tissières A, Georgopoulos C. Progress and perspectives on the biology of heat shock proteins and molecular chaperones. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The Biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. pp. 1–30. [Google Scholar]
- 17.Narberhaus F, Weiglhofer W, Fischer H-M, Hennecke H. The Bradyrhizobium japonicum rpoH1 gene encoding a ς32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J Bacteriol. 1996;178:5337–5346. doi: 10.1128/jb.178.18.5337-5346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nerland A H, Mustafa A S, Sweetser D, Godal T, Young R A. A protein antigen of Mycobacterium leprae is related to a family of small heat shock proteins. J Bacteriol. 1988;170:5919–5921. doi: 10.1128/jb.170.12.5919-5921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parcell D A, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 20.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 21.Roy, S. K., T. Hiyama, and H. Nakamoto. Unpublished data.
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer U, Dürre P. Sequence and molecular characterization of a DNA region encoding a small heat shock protein of Clostridium acetobutylicum. J Bacteriol. 1993;175:3394–3400. doi: 10.1128/jb.175.11.3394-3400.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Servant P, Mazodier P. Characterization of Streptomyces albus 18-kilodalton heat shock-responsive protein. J Bacteriol. 1995;177:2998–3003. doi: 10.1128/jb.177.11.2998-3003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servant P, Mazodier P. Heat induction of hsp18 gene expression in Streptomyces albus G: transcriptional and posttranscriptional regulation. J Bacteriol. 1996;178:7031–7036. doi: 10.1128/jb.178.24.7031-7036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka N, Hiyama T, Nakamoto H. Cloning, characterization and functional analysis of groESL operon from thermophilic cyanobacterium Synechococcus vulcanus. Biochim Biophys Acta. 1997;1343:335–348. doi: 10.1016/s0167-4838(97)00159-3. [DOI] [PubMed] [Google Scholar]
- 29.Verbon A, Hartskeerl R A, Schuitema A, Kolk A H J, Young D B, Lathigra R. The 14,000-molecular-weight antigen of Mycobacterium tuberculosis is related to the alpha-crystallin family of low-molecular-weight heat shock proteins. J Bacteriol. 1992;174:1352–1359. doi: 10.1128/jb.174.4.1352-1359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters E R, Lee G J, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- 31.Webb R, Sherman L A. The cyanobacterial heat-shock response and the molecular chaperones. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 751–767. [Google Scholar]
- 32.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 33.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]