Abstract
Centrosomes are highly conserved organelles that act as the major microtubule-organizing center (MTOC) in animal somatic cells. Through their MTOC activity, centrosomes play various roles throughout the cell cycle, such as supporting cell migration in interphase and spindle organization and positioning in mitosis. Various approaches for removing centrosomes from somatic cells have been developed and applied over the past few decades to understand the precise roles of centrosomes. Centrinone, a reversible and selective PLK4 (polo-like kinase 4) inhibitor, has recently emerged as an efficient approach to eliminate centrosomes. In this review, we describe the latest findings on centrosome function that have been revealed using various centrosome-eliminating approaches. In addition, we discuss our recent findings on the mechanism of centrosome-independent spindle bipolarization, discovered through the use of centrinone.
Keywords: centrosome, centrinone, mitotic spindle, bipolarity, NuMA
Centrosome cycle and its function
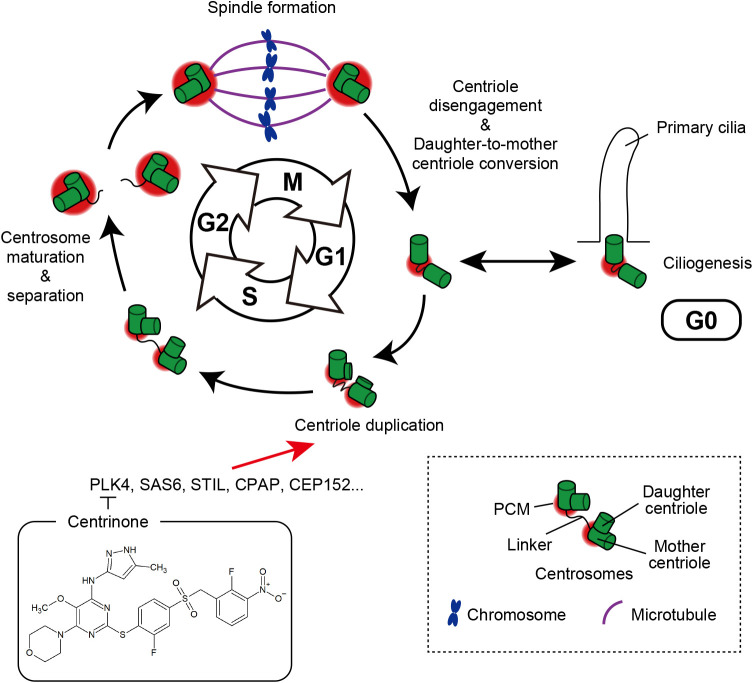
The centrosome is a highly conserved membrane-less organelle, which is composed of one or two centrioles surrounded by pericentriolar material (PCM) (Conduit et al., 2015; Nigg and Holland, 2018; Nigg and Raff, 2009). Centrosomes mainly function as the major microtubule-organizing center (MTOC) in somatic animal cells (Conduit et al., 2015), and undergo multiple events throughout the cell cycle (Fig. 1). Centrioles are duplicated once per cell cycle during S phase (Nigg and Holland, 2018). In this duplication process, only a single daughter centriole is formed next to each mother centriole (Ohta et al., 2014; Rattner and Phillips, 1973). Several factors critical for centriole duplication have been identified, such as PLK4, SAS6, STIL, CPAP, and CEP152 (Bettencourt-Dias et al., 2005; Cizmecioglu et al., 2010; Dammermann et al., 2004; Dzhindzhev et al., 2010; Habedanck et al., 2005; Hatch et al., 2010; Kleylein-Sohn et al., 2007; Leidel et al., 2005; Lin et al., 2013; Stevens et al., 2010; Tang et al., 2011). After centriole duplication, the process of centrosome maturation is initiated in G2 phase with an extreme expansion of PCM, allowing centrosomes to acquire robust MTOC activity (Conduit et al., 2014; Izquierdo et al., 2014; Paweletz et al., 1984). At the G2/M transition, the two centrosomes are separated from each other and move to the opposite sides of the mitotic cell (centrosome separation) (Raaijmakers et al., 2012; Rattner and Berns, 1976; Toso et al., 2009; Waters et al., 1993; Whitehead and Rattner, 1998). During mitosis, centrosomes localize at the mitotic spindle poles (Nigg and Holland, 2018). Following cell division, the connection between the mother and daughter centrioles is resolved (centriole disengagement) (Tsou et al., 2009; Tsou and Stearns, 2006; Vidwans et al., 1999), and the daughter centriole recruits PCM proteins and becomes a mother centriole (daughter-to-mother centriole conversion). After the conversion process, both centrioles obtain the ability to duplicate (Fu et al., 2016; Izquierdo et al., 2014; Tsou et al., 2009; Tsou and Stearns, 2006; Tsuchiya et al., 2016). Overall, centrosomes undergo many processes throughout the cell cycle and their number is strictly regulated in animal somatic cells.
Fig. 1.
The centrosome cycle. In mitosis, a bipolar spindle is formed and centrosomes function as the poles of the spindle. After cell division, the connection between the mother and daughter centrioles is resolved (centriole disengagement), and the daughter centriole recruits PCM proteins and becomes a mother centriole. When cells stop proliferating and enter G0 phase, centrosomes act as basal bodies for the growth of cilia (ciliogenesis). In S phase, the process of centriole duplication is promoted by several factors (e.g., PLK4, SAS6, STIL, CPAP, and CEP152). Starting at G2 phase, the two centrosomes experience extreme PCM expansions, which allow for stronger MTOC activity (centrosome maturation). At G2/M transition, the two centrosomes are separated from each other and move to the opposite sides of the mitotic cell (centrosome separation), followed by initiation of the next cell division process.
Centrosomes play various crucial roles in animal cells through their MTOC activity. During interphase, centrosomes regulate the microtubule network and govern cell motility (Werner et al., 2017). During mitosis, centrosomes function as the poles of the mitotic spindle to maintain spindle structure and spindle positioning for correct chromosome segregation (Conduit et al., 2015).
Centrosome-eliminating approaches developed over the past few decades
To investigate the necessity of centrosomes in animal somatic cells for proper mitotic spindle formation and chromosome segregation, several groups have eliminated centrosomes from these cells through different technical approaches. Traditionally, microsurgery or depletion of centriole duplication factors have been used to remove centrosomes (Table I). Khodjakov et al. succeeded in selectively removing centrosomes without damaging surrounding structures by laser microsurgery targeting GFP-tagged γ-tubulin (a centrosome marker) in mammalian cells (Khodjakov et al., 2000). They reported that bipolar mitotic spindles were formed without centrosomes. Basto et al. created DSas-4 (an ortholog of human CPAP) mutant flies, whose cells do not have centrosomes. They confirmed that centrosomes are not necessary for most aspects of Drosophila development (Basto et al., 2006). Hornick et al. performed microsurgery with a microneedle to obtain acentrosomal mammalian cells (Hornick et al., 2011). They found that the fidelity of bipolar mitotic spindle assembly is highly compromised in the absence of centrosomes. Sir et al. obtained chicken cells lacking centrosomes through knockout of CEP152 or STIL (Sir et al., 2013). These cells showed delays in bipolar spindle formation and high rates of chromosome segregation errors. In summary, the presence of centrosomes in animal somatic cells is important for proper mitotic progression to some extent, but, in many cases centrosomes are not essential. However, the detailed mechanisms of centrosome-independent bipolar spindle formation in somatic cells have not been extensively discussed.
Table I.
Examples of studies on acentrosomal somatic cells
| Species/cell | Approach | Main finding | Reference |
|---|---|---|---|
| Monkey/CVG-2 | Laser microsurgery | A bipolar mitotic spindle is formed in the absence of centrosomes. | Khodjakov et al., 2000 |
| Fly | Mutant of DSas-4 | Centrosomes are not necessary for the development of the fly. | Basto et al., 2006 |
| Monkey/BSC-1 | Needle microsurgery | The fidelity of the bipolar mitotic spindle is highly compromised in the absence of centrosomes. | Hornick et al., 2011 |
| Chicken/DT40 | Knockout of CEP152/STIL | Centrosome loss leads to delays in spindle formation and high rates of chromosomal instability. | Sir et al., 2013 |
Emergence of centrinone: a reversible and selective inhibitor of PLK4
All the above methods for the removal of centrosomes require special devices for microsurgery or efficient systems for gene knockdown or knockout. In contrast, centrinone, a reversible and selective PLK4 inhibitor developed in 2015 (Wong et al., 2015), has enabled the easy removal of centrosomes from living animal cells. Wong et al. selected VX-680, a pan-Aurora kinase inhibitor which also inhibits PLK4 (Harrington et al., 2004; Sloane et al., 2010), as a template to develop selective PLK4 inhibitors. Through the introduction of a methoxy substituent at the VX-680 C5 position and further optimization, they succeeded in synthesizing centrinone and centrinone-B. Both inhibitors exhibited >1,000-fold selectivity for PLK4 over Aurora kinases (Wong et al., 2015). After treatment with centrinone, they observed cell cycle arrest in G1 phase in normal human cells (RPE-1 cells) and confirmed that the arrest was not due to previously described stress responses to DNA damage, Hippo signaling, or chromosome segregation errors. On the other hand, they noted that cancer cell lines could proliferate in the absence of centrosomes, and the proliferation rates were not correlated with the basal frequency of centrosome amplification observed in those cell lines. These results suggest that numerous cancer cells have abolished the response system that arrests the cell cycle after centrosome loss.
Since the advent of centrinone, other various studies have used this agent (Table II). Meitinger et al. performed a genome-wide CRISPR/Cas9 screen in RPE-1 cells and identified a 53BP1-USP28 module which induces G1-phase cell cycle arrest after centrosome loss (Meitinger et al., 2016). Two other groups identified the same pathway by using another method to remove centrosomes from human somatic cells (Fong et al., 2016; Lambrus et al., 2016). Gheiratmand et al. performed BioID experiments targeting seven centriolar satellite components while treating cells with centrinone. BioID is a method to screen for protein interactions in living cells using biotin ligase. The ligase fused to the protein of interest biotinylates proximal proteins and enables their isolation and identification (Roux et al., 2012). Using this technique, they revealed that the interactions among centriolar satellite proteins are not affected by centrosome removal (Gheiratmand et al., 2019). Another group performed similar experiments using the knockout of STIL or CEP152 to remove centrosomes, and reached the same conclusion (Quarantotti et al., 2019). Collectively, these results suggest that treatment with centrinone is as effective as other existing methods. The Golgi apparatus also exhibits MTOC activity and localizes around centrosomes (Chabin-Brion et al., 2001; Efimov et al., 2007; Rivero et al., 2009; Ríos et al., 2004). Centrinone has been used in several studies to investigate the relationship and dependency between the two organelles (Guizzunti and Seemann, 2016; Wu et al., 2016). Moreover, Dudka et al. produced human cells which have only one centrosome during mitosis through treatment with centrinone (Dudka et al., 2019). They used these cells as a tool for studying k-fiber dynamics in cells with asymmetric bipolar spindles and found that the centrosome regulated k-fiber plus-ends in a HURP-dependent manner. Overall, owing to its ease of use, centrinone has made a huge impact on the latest developments in cell biology. However, a detailed study on the mechanisms of acentrosomal spindle formation using centrinone has not been performed.
Table II.
Examples of studies using centrinone and human cells
| Cell | Approach | Main finding | Reference |
|---|---|---|---|
| BT549, Calu-6, HCT116, HeLa, MDA-MB-231, RPE-1, U2OS | Centrinone | Centrosome loss leads to G1-phase cell cycle arrest in normal cells, but not in cancer cells. | Wong et al., 2015 |
| SV589 HeLa |
Centrinone | The link between the Golgi apparatus and centrosomes must be dissolved to reach metaphase. | Guizzunti and Seemann, 2016 |
| RPE-1 | Centrinone | A single Golgi can be maintained in the absence of centrosomes. | Wu et al., 2016 |
| RPE-1 | Centrinone CRISPR/Cas9 screen | The 53BP1-USP28 module induces G1-phase cell cycle arrest after centrosome loss. | Meitinger et al., 2016 |
| HEK293T | Centrinone BioID |
The interactions of centriolar satellite proteins are not affected by centrosome loss. | Gheiratmand et al., 2019 |
| RPE-1 | Centrinone | Centrosomes indirectly regulate k-fiber plus-ends via spindle length-dependent accumulation of HURP. | Dudka et al., 2019 |
| A549, DU145, GI-1, HCT116, HeLa, MCF-7, PANC-1, SKOV-3, U2OS | Centrinone | NuMA-mediated pathways promote spindle bipolarity independently of centrosomes. | Chinen et al., 2020 |
A centrosome-independent mechanism of spindle bipolarity establishment revealed using centrinone
Based on the aforementioned evidence, we used centrinone to investigate the machinery of bipolar spindle formation in acentrosomal human cells. The establishment of a bipolar spindle is crucial for equational chromosome segregation (Prosser and Pelletier, 2017). In somatic cells, spindle bipolarity is achieved through centrosome separation, which occurs ahead of or following nuclear envelope breakdown (NEBD) (Beaudouin et al., 2002; Kaseda et al., 2012; Raaijmakers et al., 2012; Rattner and Berns, 1976; Toso et al., 2009; Waters et al., 1993; Whitehead and Rattner, 1998). During centrosome separation, two centrosomes linked by antiparallel microtubules are pushed away from each other through microtubule crosslinking and the sliding activity of kinesin Eg5 (Bertran et al., 2011; Hata et al., 2019; Kapitein et al., 2005; Kaseda et al., 2012; Smith et al., 2011). During mitosis, these separated centrosomes function as the core of spindle poles and organize microtubules into a bipolar spindle.
On the other hand, meiotic spindles are formed without centrosomes in the oocytes of many species, including flies, frogs, mice, and humans (Calarco-Gillam et al., 1983; Heald et al., 1996; Holubcová et al., 2015; Matthies et al., 1996). In addition, the somatic cells of flies and vertebrates can organize functional bipolar spindles following the removal of centrosomes by lasers, microneedles, or mutations of centriole duplication factors (Basto et al., 2006; Bonaccorsi et al., 2000; Hornick et al., 2011; Khodjakov et al., 2000; Sir et al., 2013). Therefore, it appears that the establishment of spindle bipolarity is not completely dependent on centrosome separation, and an acentrosomal pathway may exist as a compensation mechanism. The in vitro reconstitution of meiotic spindles utilizing Xenopus egg extracts has been a useful model system for the study of acentrosomal spindle formation (Cross and Powers, 2009). In this system, microtubules are nucleated via chromatin- and Ran-based pathways (Carazo-Salas et al., 1999; Heald et al., 1996; Kalab et al., 1999; Karsenti et al., 1984; Wilde, 1999; Zhang et al., 1999) and are presumably rearranged into a bipolar state in a microtubule- and motor protein-dependent manner (Groen et al., 2008; Heald et al., 1996; Loughlin et al., 2010; Petry et al., 2011). In addition, in mouse oocytes, multiple MTOCs that contain PCM are formed de novo, and these MTOC foci are eventually clustered into a bipolar state in an Eg5-dependent manner (Schuh and Ellenberg, 2007). However, other factors involved in acentrosomal spindle bipolarization, besides microtubules, motor proteins, and PCM components have not been extensively investigated thus far.
NuMA (nuclear mitotic apparatus) protein, one of the spindle pole components, directly interacts with microtubules (Du et al., 2002; Haren and Merdes, 2002) and presumptively organizes centrosome-independent microtubule asters (Du et al., 2002; Gaglio et al., 1995; Haren and Merdes, 2002). This protein is necessary for focusing microtubules at spindle poles (spindle pole organization) (Merdes et al., 2000, 1996). In addition, cortical NuMA creates spindle-pulling forces in coordination with the dynein-dynactin complex, placing the spindle at an appropriate position within the cell (Du and Macara, 2004; Okumura et al., 2018). However, the role of NuMA in spindle bipolarization, rather than spindle pole organization and spindle positioning, is not completely understood.
We treated human cells with centrinone to create artificial acentrosomal cells (Chinen et al., 2020). We confirmed that these cells displayed prolonged mitotic duration and increased frequency of chromosome segregation errors, consistent with previous studies using centrinone (Meitinger et al., 2016; Wong et al., 2015). These results suggest that mitotic fidelity is compromised in the absence of centrosomes in human cells as well as other vertebrates (Sir et al., 2013). Subsequently, we investigated the role of NuMA in spindle bipolarization in acentrosomal cells. We found that in acentrosomal cells, shortly after NEBD, NuMA formed several aggregates that organized microtubule asters. Subsequently, these aggregates assembled into two NuMA structures (initial bipolarity establishment), in a manner dependent on microtubules, dynein, and the clustering activity of NuMA. These two structures, located close to each other, organized a radial array of microtubules around them. This radial array of microtubules incorporated Eg5. Eventually, these two NuMA structures were separated to form a bipolar spindle. The separation of the two NuMA structures is dependent on Eg5 motor activity and kinetochore-microtubule attachment. Disruption of Eg5 motor activity or kinetochore-microtubule attachment resulted in fusion of the two NuMA structures after the initial bipolarity was established, leaving cells in a monopolar-like state. Following the depletion of NuMA in acentrosomal cells by auxin-inducible degradation (Natsume et al., 2016; Nishimura et al., 2009; Okumura et al., 2018), cells failed to establish bipolar spindles.
Importantly, we found that NuMA promoted the initial steps of spindle bipolarization in early mitosis even in cells with centrosomes (Chinen et al., 2020). We observed that the time from NEBD to bipolarity establishment prolonged upon depletion of NuMA. The results of our study suggest that the canonical centrosomal pathway and the NuMA-mediated acentrosomal pathway complementally regulate bipolar spindle assembly in somatic cells, and that the latter becomes predominant in the absence of centrosomes (Fig. 2). Understanding the multiple pathways that ensure robust bipolar spindle formation will assist in the design of anticancer drugs that target spindle assembly (Henriques et al., 2019; Tischer and Gergely, 2019).
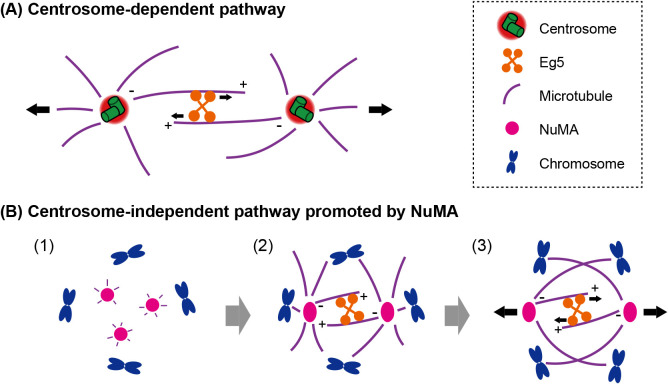
Fig. 2.
Models for two pathways that promote spindle bipolarization in human cells. (A) The canonical centrosomal pathway. At G2/M transition, the two centrosomes are pushed apart by the plus-end-directed motor activity of kinesin Eg5. (B) The NuMA-mediated pathway, which occurs independently of centrosomes. (1) At the onset of mitosis, NuMA aggregates and organizes microtubule asters. (2) Dynein activity and the clustering activity of NuMA assembles the NuMA aggregates into two poles; subsequently, Eg5 is loaded onto the antiparallel microtubules. (3) Spindle poles are separated through Eg5 motor activity and kinetochore-microtubule attachments.
Concluding remarks
Since its development in 2015, centrinone has allowed for great advances in our understanding of centrosome function and the consequences of centrosome loss. Its ease of use has led to the success of numerous large-scale screens that shed light on the interplay between centrosomes and other organelles or cellular systems. Moreover, centrinone has enabled a detailed analysis of the cell division machinery in acentrosomal cells. This analysis assists us in understanding the complemental mechanisms through which the mitotic spindle is regulated by both centrosome-dependent and centrosome-independent pathways.
References
- Basto, R., Lau, J., Vinogradova, T., Gardiol, A., Woods, C.G., Khodjakov, A., and Raff, J.W.. 2006. Flies without Centrioles. Cell, 125: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Beaudouin, J., Gerlich, D., Daigle, N., Eils, R., and Ellenberg, J.. 2002. Nuclear Envelope Breakdown Proceeds by Microtubule-Induced Tearing of the Lamina. Cell, 108: 83–96. [DOI] [PubMed] [Google Scholar]
- Bertran, M.T., Sdelci, S., Regué, L., Avruch, J., Caelles, C., and Roig, J.. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J., doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Rodrigues-Martins, A., Carpenter, L., Riparbelli, M., Lehmann, L., Gatt, M.K., Carmo, N., Balloux, F., Callaini, G., and Glover, D.M.. 2005. SAK/PLK4 Is Required for Centriole Duplication and Flagella Development. Curr. Biol., 15: 2199–2207. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S., Giansanti, M.G., and Gatti, M.. 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol., 2: 54–56. [DOI] [PubMed] [Google Scholar]
- Calarco-Gillam, P.D., Siebert, M.C., Hubble, R., Mitchison, T., and Kirschner, M.. 1983. Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell, 35: 621–629. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Guarguaglini, G., Gruss, O.J., Segref, A., Karsenti, E., and Mattaj, I.W.. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature, 400: 178–181. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion, K., Marceiller, J., Perez, F., Settegrana, C., Drechou, A., Durand, G., and Poüs, C.. 2001. The Golgi Complex Is a Microtubule-organizing Organelle. Mol. Biol. Cell, 12: 2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen, T., Yamamoto, S., Takeda, Y., Watanabe, K., Kuroki, K., Hashimoto, K., Takao, D., and Kitagawa, D.. 2020. NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J., 39: e102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu, O., Arnold, M., Bahtz, R., Settele, F., Ehret, L., Haselmann-Weiß, U., Antony, C., and Hoffmann, I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol., 191: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Richens, J.H., Wainman, A., Holder, J., Vicente, C.C., Pratt, M.B., Dix, C.I., Novak, Z.A., Dobbie, I.M., Schermelleh, L., and Raff, J.W.. 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife, 3: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman, A., and Raff, J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol., 16: 611–624. [DOI] [PubMed] [Google Scholar]
- Cross, M.K. and Powers, M.A.. 2009. Learning about cancer from frogs: analysis of mitotic spindles in Xenopus egg extracts. Dis. Model. Mech., 2: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., Müller-Reichert, T., Pelletier, L., Habermann, B., Desai, A., and Oegema, K.. 2004. Centriole Assembly Requires Both Centriolar and Pericentriolar Material Proteins. Dev. Cell, 7: 815–829. [DOI] [PubMed] [Google Scholar]
- Du, Q., Taylor, L., Compton, D.A., and Macara, I.G.. 2002. LGN Blocks the Ability of NuMA to Bind and Stabilize Microtubules. Curr. Biol., 12: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Du, Q. and Macara, I.G.. 2004. Mammalian Pins Is a Conformational Switch that Links NuMA to Heterotrimeric G Proteins. Cell, 119: 503–516. [DOI] [PubMed] [Google Scholar]
- Dudka, D., Castrogiovanni, C., Liaudet, N., Vassal, H., and Meraldi, P.. 2019. Spindle-Length-Dependent HURP Localization Allows Centrosomes to Control Kinetochore-Fiber Plus-End Dynamics. Curr. Biol., 29: 3563–3578.e6. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., Yu, Q.D., Weiskopf, K., Tzolovsky, G., Cunha-Ferreira, I., Riparbelli, M., Rodrigues-Martins, A., Bettencourt-Dias, M., Callaini, G., and Glover, D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature, 467: 714–718. [DOI] [PubMed] [Google Scholar]
- Efimov, A., Kharitonov, A., Efimova, N., Loncarek, J., Miller, P.M., Andreyeva, N., Gleeson, P., Galjart, N., Maia, A.R.R., McLeod, I.X., Yates, J.R., Maiato, H., Khodjakov, A., Akhmanova, A., and Kaverina, I.. 2007. Asymmetric CLASP-Dependent Nucleation of Noncentrosomal Microtubules at the trans-Golgi Network. Dev. Cell, 12: 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, C.S., Mazo, G., Das, T., Goodman, J., Kim, M., O’Rourke, B.P., Izquierdo, D., and Tsou, M.-F.B.. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife, 5: e16270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., Lipinszki, Z., Rangone, H., Min, M., Mykura, C., Chao-Chu, J., Schneider, S., Dzhindzhev, N.S., Gottardo, M., Riparbelli, M.G., Callaini, G., and Glover, D.M.. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol., 18: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., Saredi, A., and Compton, D.A.. 1995. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol., 131: 693–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheiratmand, L., Coyaud, E., Gupta, G.D., Laurent, E.M., Hasegan, M., Prosser, S.L., Gonçalves, J., Raught, B., and Pelletier, L.. 2019. Spatial and proteomic profiling reveals centrosome‐independent features of centriolar satellites. EMBO J., 38: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen, A.C., Needleman, D., Brangwynne, C., Gradinaru, C., Fowler, B., Mazitschek, R., and Mitchison, T.J.. 2008. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J. Cell Sci., 121: 2293–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizzunti, G. and Seemann, J.. 2016. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc. Natl. Acad. Sci., 113: E6590–E6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck, R., Stierhof, Y.-D., Wilkinson, C.J., and Nigg, E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol., 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Haren, L. and Merdes, A.. 2002. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci., 115: 1815–24. [DOI] [PubMed] [Google Scholar]
- Harrington, E.A., Bebbington, D., Moore, J., Rasmussen, R.K., Ajose-Adeogun, A.O., Nakayama, T., Graham, J.A., Demur, C., Hercend, T., Diu-Hercend, A., Su, M., Golec, J.M.C., and Miller, K.M.. 2004. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med., 10: 262–267. [DOI] [PubMed] [Google Scholar]
- Hata, S., Pastor Peidro, A., Panic, M., Liu, P., Atorino, E., Funaya, C., Jäkle, U., Pereira, G., and Schiebel, E.. 2019. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat. Cell Biol., 21: 1138–1151. [DOI] [PubMed] [Google Scholar]
- Hatch, E.M., Kulukian, A., Holland, A.J., Cleveland, D.W., and Stearns, T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol., 191: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., and Karsenti, E.. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature, 382: 420–425. [DOI] [PubMed] [Google Scholar]
- Henriques, A.C., Ribeiro, D., Pedrosa, J., Sarmento, B., Silva, P.M.A., and Bousbaa, H.. 2019. Mitosis inhibitors in anticancer therapy: When blocking the exit becomes a solution. Cancer Lett., 440–441: 64–81. [DOI] [PubMed] [Google Scholar]
- Holubcová, Z., Blayney, M., Elder, K., and Schuh, M.. 2015. Error-Prone Chromosome-Mediated Spindle Assembly Favors Chromosome Segregation Defects in Human Oocytes. Science, 348: 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick, J.E., Mader, C.C., Tribble, E.K., Bagne, C.C., Vaughan, K.T., Shaw, S.L., and Hinchcliffe, E.H.. 2011. Amphiastral Mitotic Spindle Assembly in Vertebrate Cells Lacking Centrosomes. Curr. Biol., 21: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo, D., Wang, W.-J., Uryu, K., and Tsou, M.-F.B.. 2014. Stabilization of Cartwheel-less Centrioles for Duplication Requires CEP295-Mediated Centriole-to-Centrosome Conversion. Cell Rep., 8: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., Pu, R.T., and Dasso, M.. 1999. The Ran GTPase regulates mitotic spindle assembly. Curr. Biol., 9: 481–484. [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman, E.J.G., Kwok, B.H., Kim, J.H., Kapoor, T.M., and Schmidt, C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature, 435: 114–118. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., Newport, J., Hubble, R., and Kirschner, M.. 1984. Interconversion of metaphase and interphase microtubule arrays, as studied by the injection of centrosomes and nuclei into Xenopus eggs. J. Cell Biol., 98: 1730–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda, K., McAinsh, A.D., and Cross, R.A.. 2012. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open, 1: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., Cole, R.W., Oakley, B.R., and Rieder, C.L.. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol., 10: 59–67. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., Westendorf, J., Le Clech, M., Habedanck, R., Stierhof, Y.-D., and Nigg, E.A.. 2007. Plk4-Induced Centriole Biogenesis in Human Cells. Dev. Cell, 13: 190–202. [DOI] [PubMed] [Google Scholar]
- Lambrus, B.G., Daggubati, V., Uetake, Y., Scott, P.M., Clutario, K.M., Sluder, G., and Holland, A.J.. 2016. A USP28–53BP1–p53–p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol., 214: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel, S., Delattre, M., Cerutti, L., Baumer, K., and Gönczy, P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol., 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Lin, Y.-C., Chang, C.-W., Hsu, W.-B., Tang, C.-J.C., Lin, Y.-N., Chou, E.-J., Wu, C.-T., and Tang, T.K.. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J., 32: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin, R., Heald, R., and Nédélec, F.. 2010. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol., 191: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, H.J.G., McDonald, H.B., Goldstein, L.S.B., and Theurkauf, W.E.. 1996. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol., 134: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F., Anzola, J.V., Kaulich, M., Richardson, A., Stender, J.D., Benner, C., Glass, C.K., Dowdy, S.F., Desai, A., Shiau, A.K., and Oegema, K.. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol., 214: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J.D., and Cleveland, D.W.. 1996. A Complex of NuMA and Cytoplasmic Dynein Is Essential for Mitotic Spindle Assembly. Cell, 87: 447–458. [DOI] [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W.C., and Cleveland, D.W.. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol., 149: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume, T., Kiyomitsu, T., Saga, Y., and Kanemaki, M.T.. 2016. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep., 15: 210–218. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. and Raff, J.W.. 2009. Centrioles, Centrosomes, and Cilia in Health and Disease. Cell, 139: 663–678. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. and Holland, A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol., 19: 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., and Kanemaki, M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods, 6: 917–922. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Ashikawa, T., Nozaki, Y., Kozuka-Hata, H., Goto, H., Inagaki, M., Oyama, M., and Kitagawa, D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun., 5: 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, M., Natsume, T., Kanemaki, M.T., and Kiyomitsu, T.. 2018. Dynein–Dynactin–NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. Elife, 7: e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweletz, N., Mazia, D., and Finze, E.-M.. 1984. The centrosome cycle in the mitotic cycle of sea urchin eggs. Exp. Cell Res., 152: 47–65. [DOI] [PubMed] [Google Scholar]
- Petry, S., Pugieux, C., Nedelec, F.J., and Vale, R.D.. 2011. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci., 108: 14473–14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S.L. and Pelletier, L.. 2017. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol., 18: 187–201. [DOI] [PubMed] [Google Scholar]
- Quarantotti, V., Chen, J., Tischer, J., Gonzalez Tejedo, C., Papachristou, E.K., D’Santos, C.S., Kilmartin, J.V, Miller, M.L., and Gergely, F.. 2019. Centriolar satellites are acentriolar assemblies of centrosomal proteins. EMBO J., 38: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, J.A., Van Heesbeen, R.G.H.P., Meaders, J.L., Geers, E.F., Fernandez-Garcia, B., Medema, R.H., and Tanenbaum, M.E.. 2012. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J., 31: 4179–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner, J.B. and Phillips, S.G.. 1973. INDEPENDENCE OF CENTRIOLE FORMATION AND DNA SYNTHESIS. J. Cell Biol., 57: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner, J.B. and Berns, M.W.. 1976. Centriole behavior in early mitosis of rat kangaroo cells (PTK2). Chromosoma, 54: 387–395. [DOI] [PubMed] [Google Scholar]
- Rivero, S., Cardenas, J., Bornens, M., and Rios, R.M.. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J., 28: 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos, R.M., Sanchís, A., Tassin, A.M., Fedriani, C., and Bornens, M.. 2004. GMAP-210 Recruits γ-Tubulin Complexes to cis-Golgi Membranes and Is Required for Golgi Ribbon Formation. Cell, 118: 323–335. [DOI] [PubMed] [Google Scholar]
- Roux, K.J., Kim, D.I., Raida, M., and Burke, B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol., 196: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh, M. and Ellenberg, J.. 2007. Self-Organization of MTOCs Replaces Centrosome Function during Acentrosomal Spindle Assembly in Live Mouse Oocytes. Cell, 130: 484–498. [DOI] [PubMed] [Google Scholar]
- Sir, J.-H., Pütz, M., Daly, O., Morrison, C.G., Dunning, M., Kilmartin, J.V., and Gergely, F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol., 203: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane, D.A., Trikic, M.Z., Chu, M.L.H., Lamers, M.B.A.C., Mason, C.S., Mueller, I., Savory, W.J., Williams, D.H., and Eyers, P.A.. 2010. Drug-Resistant Aurora A Mutants for Cellular Target Validation of the Small Molecule Kinase Inhibitors MLN8054 and MLN8237. ACS Chem. Biol., 5: 563–576. [DOI] [PubMed] [Google Scholar]
- Smith, E., Hégarat, N., Vesely, C., Roseboom, I., Larch, C., Streicher, H., Straatman, K., Flynn, H., Skehel, M., Hirota, T., Kuriyama, R., and Hochegger, H.. 2011. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J., 30: 2233–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, N.R., Dobbelaere, J., Brunk, K., Franz, A., and Raff, J.W.. 2010. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol., 188: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, C.J.C., Lin, S.Y., Hsu, W. Bin, Lin, Y.N., Wu, C.T., Lin, Y.C., Chang, C.W., Wu, K.S., and Tang, T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J., 30: 4790–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer, J. and Gergely, F.. 2019. Anti-mitotic therapies in cancer. J. Cell Biol., 218: 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso, A., Winter, J.R., Garrod, A.J., Amaro, A.C., Meraldi, P., and McAinsh, A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol., 184: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, M.-F.B. and Stearns, T.. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature, 442: 947–951. [DOI] [PubMed] [Google Scholar]
- Tsou, M.-F.B., Wang, W.-J., George, K.A., Uryu, K., Stearns, T., and Jallepalli, P.V.. 2009. Polo Kinase and Separase Regulate the Mitotic Licensing of Centriole Duplication in Human Cells. Dev. Cell, 17: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, Y., Yoshiba, S., Gupta, A., Watanabe, K., and Kitagawa, D.. 2016. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat. Commun., 7: 12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans, S.J., Wong, M.L., and O’Farrell, P.H.. 1999. Mitotic Regulators Govern Progress through Steps in the Centrosome Duplication Cycle. J. Cell Biol., 147: 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, J., Cole, R., and Rieder, C.. 1993. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J. Cell Biol., 122: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S., Pimenta-Marques, A., and Bettencourt-Dias, M.. 2017. Maintaining centrosomes and cilia. J. Cell Sci., 130: 3789–3800. [DOI] [PubMed] [Google Scholar]
- Whitehead, C.M. and Rattner, J.B.. 1998. Expanding the role of HsEg5 within the mitotic and post-mitotic phases of the cell cycle. J. Cell Sci., 111: 2551–2561. [DOI] [PubMed] [Google Scholar]
- Wilde, A. 1999. Stimulation of Microtubule Aster Formation and Spindle Assembly by the Small GTPase Ran. Science, 284: 1359–1362. [DOI] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola, J.V., Davis, R.L., Yoon, M., Motamedi, A., Kroll, A., Seo, C.P., Hsia, J.E., Kim, S.K., Mitchell, J.W., Mitchell, B.J., Desai, A., Gahman, T.C., Shiau, A.K., and Oegema, K.. 2015. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science, 348: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., de Heus, C., Liu, Q., Bouchet, B.P., Noordstra, I., Jiang, K., Hua, S., Martin, M., Yang, C., Grigoriev, I., Katrukha, E.A., Altelaar, A.F.M., Hoogenraad, C.C., Qi, R.Z., Klumperman, J., and Akhmanova, A.. 2016. Molecular Pathway of Microtubule Organization at the Golgi Apparatus. Dev. Cell, 39: 44–60. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Hughes, M., and Clarke, P.R.. 1999. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J. Cell Sci., 112: 2453–2461. [DOI] [PubMed] [Google Scholar]