Abstract
Target identification studies are a major hurdle in probe and drug discovery pipelines due to the need to chemically modify small molecules of interest, which can be time intensive and have low throughput. Here, we describe a versatile and scalable method for attaching chemical moieties to a small molecule, isocyanate-mediated chemical tagging (IMCT). By preparation of a template resin with an isocyanate capture group and a cleavable linker, nucleophilic groups on small molecules can be modified with an enforced one-to-one stoichiometry. We demonstrate a small molecule substrate scope that includes primary and secondary amines, thiols, phenols, benzyl alcohols, and primary alcohols. Cheminformatic analyses predict that IMCT is reactive with more than 25% of lead-like compounds in publicly available databases. To demonstrate that the method can produce biologically active molecules, we generated FKBP12 photoaffinity labeling (PAL) compounds with a wide range of affinities and showed that purified and crude cleavage products can bind to and label FKBP12. This method could be used to rapidly modify small molecules for many applications, including the synthesis of PAL probes, fluorescence polarization probes, pull-down probes, and degraders.
Target identification studies are critical in probe and drug discovery, particularly for compounds arising from high-throughput screening. Assay cascades may permit varying degrees of mechanistic inference, but definite identification of on- and off-target interactions is challenging and may require probe development by modification of the lead compound with enrichment handles, fluorescent labels, or photoaffinity-labeling (PAL) groups.1 Modification also requires an understanding of the compound’s structure–activity relationship (SAR) to ensure that any modification site does not impact bioactivity. Therefore, comprehensive target identification typically occurs late in the lead discovery pipeline, despite the critical information it provides. To address the expanding need for chimeric molecules, generalized strategies for chemical tagging have been developed. Current methods to rapidly modify small molecules include photo-cross-linking,2,3 click chemistry,4,5 or reactions targeted toward prevalent but specific functional groups. Photo-cross-linking and click chemistry-based methods require multiple synthetic steps to introduce diazirine, alkyne, or azide groups to either the moiety to be added and/or the small molecule of interest. As a result, an upfront time investment is needed to synthesize these components. NHS esters and maleimide groups can selectively react with amines and thiols, respectively, but reactivity with a single nucleophilic group limits the application of these approaches to a small portion of the available chemical space.
Here, we demonstrate a generalizable, rapid method to add chemical moieties to small molecules, which we refer to as isocyanate-mediated chemical tagging (IMCT). IMCT (1) employs chemistries that will react with most drug-like small molecules, (2) reacts with one-to-one stoichiometry, and (3) utilizes mild conditions that should not alter functional groups outside of the expected scope.
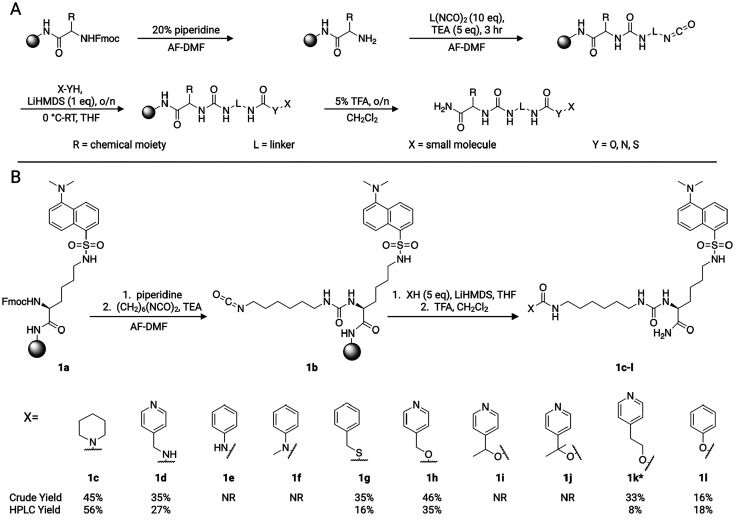
Inspired by isocyanate-based small molecule microarrays (SMMs),6−8 the method uses an electrophilic isocyanate group to modify a variety of nucleophilic groups8,9 (Scheme 1). Reaction of isocyanates with amines, alcohols, and thiols leads to the formation of ureas, carbamates, and S-thiocarbamates, respectively. These groups have been found to be stable under certain biologic conditions,10−13 although there are exceptions, such as the use of phenolic carbamates as prodrugs.14,15 The isocyanate reaction occurs on the surface of the resin, where physical separation of reactants enforces a one-to-one stoichiometry and prevents excessive reactivity. For many compounds, the broad reactivity of isocyanate will result in a mixture of conjugation sites, reducing the risk that the conjugation will occlude binding.8 Additional benefits of coupling the compound to beads include the ability to obtain a desired concentration of modified small molecules with minimal starting material and the ease of purification. Our implementation of the IMCT strategy involves synthesis of a chemical tag on resin, preparation with commercial diisocyanate linkers, and subsequent capture of nucleophilic groups on the target small molecule in the presence of lithium bis(trimethylsilyl)amide (LiHMDS) (Scheme 1A).
Scheme 1. (A) Synthesis of Chimeric Molecules with IMCT. (B) Reactivity Screen of Various Nucleophile Groups with IMCT.
TEA = triethylamine. AF-DMF = amine free-dimethylformamide. TFA = trifluoroacetic acid. CH2Cl2 = dichloromethane. NR = no reaction. * indicates crude yield and HPLC yield were obtained from the same reaction mixture.
First, we tested the reactivity of IMCT with aromatic derivatives across various nucleophilic groups and substitution patterns. Reaction yields were calculated from the weight of the product isolated after purification and from crude cleavage material using standard curves of purified product with liquid chromatography mass spectrometry (LCMS). Purified product was obtained either from the same reaction mixture as crude material or from an independent reaction. The expected product was formed with primary (27–35%) and secondary amine (45–56%), thiol (16–35%), phenol (16–18%), benzyl alcohol (35–46%), and primary alcohol (8–33%) groups. Yields obtained from each method are largely comparable and consistent with the nucleophilicity of each group (Scheme 1B). LiHMDS has been shown to promote the addition of alcohols over amines,16 which may explain the similar reactivities of amines and primary alcohols. A mass corresponding to an amine cleavage product was observed for secondary and tertiary alcohols (data not shown). This is consistent with the acid-sensitivity of other sterically hindered carbamates, such as the tert-butyloxycarbonyl (Boc) protecting group.17,18 No reaction product was observed with aniline or methyl aniline.
A major side product, 1m, from the reactivity studies was isolated and identified to inform future reaction optimization conditions. Product 1m was detected in each reaction and corresponds to the diisocyanate linker reacting with two molecules of starting material (Scheme S1). In a reaction generating 1c, 1m consumed 46% of the starting material and made up 23% of the final reaction mixture by mole. Because this side reaction can generate competitive small molecules in certain biological assays, we undertook a preliminary characterization of the effects of reaction parameters on the 1m yield. While resin loading and bead swelling did not improve 1c:1m contamination, increasing the amount of hexamethylene diisocyanate significantly reduced 1m contamination (Table S1). We hypothesize that this is achieved through the faster or more complete saturation of reactive sites. Further increasing the diisocyanate concentration or multiparameter optimization may lead to better product purity.
To predict the reactivity of IMCT with publicly available lead-like libraries,19−24 we used SMILES arbitrary target specification (SMARTS) queries25 (Table S2), which describe chemical substructures using extensions of SMILES, to determine what percent of compounds had primary and secondary amines, primary and aromatic alcohols, and thiol groups (Table 1). In accordance with reactivity results (Scheme 1), compounds containing aniline, methyl aniline, and secondary and tertiary alcohols were not counted as reactive with IMCT in the computational analysis. We found that greater than 25% of these small molecule libraries contain at least one nucleophile that we expect to be reactive with the method.
Table 1. Reactivity of IMCT with Publicly Available, Lead-Like Libraries.
| Chemical Library | Library Size | Reactive with IMCT (%) | Reactive Amines (%) | Reactive Alcohols (%) |
|---|---|---|---|---|
| BindingDb | 1,035,478 | 34 | 25 | 13 |
| Broad Repurposing | 6,761 | 38 | 23 | 19 |
| MLPCN | 514,841 | 32 | 10 | 24 |
| ChEMBL | 1,970,684 | 31 | 20 | 15 |
| REAL Diverse | 25,270,741 | 25 | 17 | 10 |
Although greater than 25% of small molecules are predicted to be reactive, modification of molecules at a nucleophilic site will cause loss of activity if the nucleophilic site confers function and if there are no other nucleophilic sites on the molecule. However, if multiple nucleophilic groups exist on a molecule, then IMCT will generate a mixture of compounds modified at different nucleophilic sites, which improves the probability that an active molecule is synthesized. For molecules with multiple nucleophilic groups, the proportion of molecules modified at each conjugation site in the final reaction mixture will depend on steric hindrance and nucleophilicity of the conjugation site. By immobilization of the isocyanate group on resin, physical separation should ensure that only one site is modified on each small molecule. However, for sufficiently large compounds, it is possible that multiple conjugations occur on the same molecule. This hypothetical situation might be addressed by decreasing the resin loading or increasing the compound concentration. Overall, the broad reactivity of IMCT and enforced one-to-one stoichiometry of reactants due to bead immobilization reduce the likelihood that the activity of a small molecule will be lost after modification.
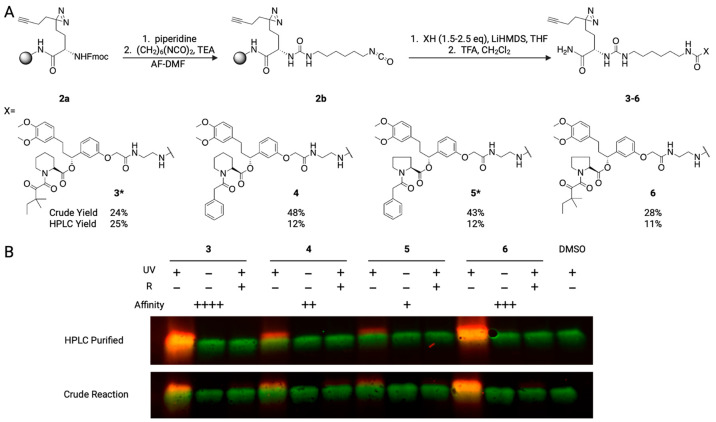
We then tested whether the IMCT method could be used to synthesize PAL probes. Early implementation of IMCT for target identification and validation in high-throughput screening pipelines would radically de-risk lead discovery efforts. Therefore, we synthesized amine analogues of well-established FKBP12 probes, which have a broad range of affinities resulting from small changes to a common scaffold (Kd = 25.7, 800, 5170, and 44000 nM).8,26,27 These analogues were then used to synthesize PAL probes with IMCT (Figure 1A). The crude reaction yields were consistent (24–48%) with the primary amines in the reactivities screen, with the FKBP12 binders with alkyl appendages (24–28%) having lower yields than the molecules with aromatic groups (43–48%).
Figure 1.
(a) Schematic of the PAL synthesis of FKBP12 probes with IMCT. (b) Fluorescent imaging of FKBP12 (green) labeled with PAL probes (10 μM) with or without UV irradiation or rapamycin (10 μM, R) treatment. Alexa Fluor 647 picolyl azide (red) was installed on covalently modified FKBP12 by using CuAAC. ++++ = highest affinity for FKBP12. + = lowest affinity for FKBP12. * indicates crude yield and HPLC yield were obtained from the same reaction mixture.
Next, we tested whether the purified and crude cleavage PAL analogues were able to engage pure protein. The product yields of crude products were estimated using a standard curve of the purified product. From these yields, stock solutions were prepared to have a fixed concentration of the active compound. FKBP12 was treated with a small molecule (10 μM) and UV-irradiated (365 nm) to induce the covalent linkage between the small molecule and protein. Alexa Fluor 647 picolyl azide was installed on the covalently modified protein through the alkyne handle using copper-catalyzed azide–alkyne Huisgen 1,3-dipolar cycloaddition (CuAAC). Probe labeled proteins and FKBP12 in each sample were detected via gel electrophoresis (Figure 1B). Samples that were not irradiated served as negative controls, and samples that were pretreated with rapamycin (10 μM) demonstrated binding selectivity.
For purified and crude probes, we detected engagement of the PAL analogues with FKBP12 and demonstrated that rapamycin prevented interaction with the PAL probes. Additional competition experiments with parent inhibitors suggest that IMCT does not abrogate the affinity of the original molecule (Figure 2A). The fluorescent intensity of the bands correlated with the affinity of the parent molecules for FKBP12 (Kd = 5 > 4 > 6 > 3), except for crude 6, which was brighter than crude 3 (Tables S3 and S4). We hypothesize that 6 may have improved aqueous solubility or stability or undergoes a higher efficiency CuAAC reaction than 3, which resulted in marginally lower levels of FKBP12 labeling with purified 6 and higher levels of FKBP12 labeling with crude 6 compared to 3.
Figure 2.
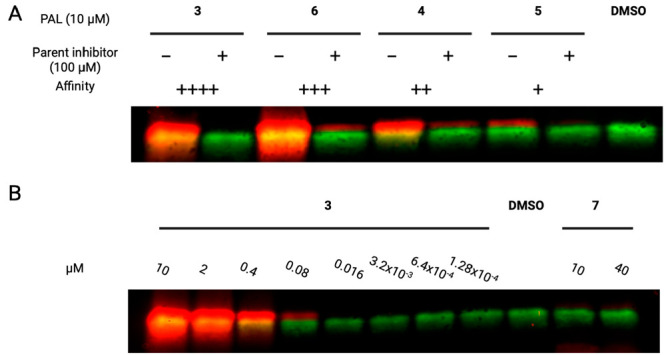
(a) Fluorescent imaging of FKBP12 (green) labeled with PAL probes (10 μM) with or without parent inhibitor (100 μM) treatment. (b) Fluorescent imaging of FKBP12 (green) labeled with 3 in an 8-point, 5-fold dilution series or linker 7 (10 or 40 μM). All samples were exposed to UV irradiation. Alexa Fluor 647 picolyl azide (red) was installed on covalently modified FKBP12 using CuAAC. ++++ = highest affinity for FKBP12. + = lowest affinity for FKBP12.
To demonstrate the specificity of the PAL probes, FKBP12 was treated with 3 in an 8-point, 5-fold dilution series (Figure 2B). Dose-dependent labeling of FKBP12 was observed (Table S5). PAL linker 7 was also included as a negative control. Crude 7 was generated on a bead and tested at two concentrations (10 and 40 μM), which were calculated based on resin loading. Nonspecific labeling of FKBP12 was not seen with linker 7 only. Thus, for the FKBP12 interaction, IMCT byproducts do not impair photoaffinity labeling nor do they result in spurious labeling. Overall, these experiments demonstrate that IMCT generated active PAL probes in a facile synthesis that does not require further purification.
In conclusion, IMCT can conjugate functional chemical moieties onto target molecules at primary and secondary amines, thiols, and aromatic and primary alcohols with yields between 8 and 56%. By cheminformatic analysis of compound screening libraries and binding databases, we estimate that our method can modify more than 25% of the relevant chemical space directly, and we hypothesize that this is an underestimation, as compounds can be substituted with nearly identical analogues in most applications. Our method is generalizable and does not require prior substrate modification as long as the substrate contains a reactive nucleophile. For molecules with multiple nucleophilic groups, IMCT is predicted to produce a mixture of molecules modified at different conjugation sites, reducing the likelihood that modification will abrogate activity. We have also demonstrated that small molecules modified by IMCT preserve their target-binding properties. IMCT-generated analogues of FKBP12 ligands show protein labeling activity across a range of lead-like affinities, and the IMCT/PAL workflow is sufficiently sensitive that compound purification is unimportant or even entirely dispensable. Theoretically, after preparation of the template resin, the entire IMCT workflow can be completed in a two-day period for 96 compounds, using existing parallel synthesis set-ups.28,29 This would permit throughput-constrained assay formats to be implemented earlier in the lead discovery process, accelerating target identification studies. We envision possible applications of IMCT to include PAL analogues, fluorescence polarization probes, affinity pull-down probes, and chimeric inducers of target proximity such as proteolysis targeting chimeras.
Acknowledgments
We would like to thank Keisuke Motoyoma, Huarui Cui, Maddy Henley, and the Pentelute lab for scientific discussions that supported this work. We would also like to thank Dr. Mohanraja Kumar in the MIT Department of Chemistry Instrumentation Facility for assisting with analytical chemistry. The Table of Contents graphic, Scheme 1, Figure 1, Figure 2, and Scheme S1 were created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.3c00352.
Detailed experimental procedures and compound characterizations (PDF)
Author Present Address
% Cullinan Oncology, Cambridge, MA, 02142, United States
Author Present Address
# Dana Farber Cancer Institute, Boston, MA, 02215, United States
Author Present Address
$ Flagship Pioneering, Cambridge, MA, 02142, United States
Author Present Address
& Neurocrine Biosciences, San Diego, CA, 92130, United States
The project was supported by the National Cancer Institute (P30-CA14051 and U54 project number 5U54CA143874-05), the National Science Foundation (NSF Career Award 1845464), Burroughs Wellcome Fund Career Award at the Scientific Interface, the MIT Center for Precision Cancer Medicine, and the Koch Institute Frontier Research Program. C.C.H. was supported by the Ludwig Center for Molecular Oncology and the NSF. J.A.K. was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG). R.M.W. was supported by the Ludwig Center for Molecular Oncology and the MIT Center for Precision Cancer Medicine.
The authors declare the following competing financial interest(s): C.C.H., J.A.K., C.C., C.M.W., and A.N.K. have filed a patent (US 20220089537) related to this project.
Supplementary Material
References
- Schenone M.; Dančík V.; Wagner B. K.; Clemons P. A. Target Identification and Mechanism of Action in Chemical Biology and Drug Discovery. Nat. Chem. Biol. 2013, 9 (4), 232–240. 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasova I.; Bulat B.; Jung N.; Bräse S. Synthesis of Diaziridines and Diazirines via Resin-Bound Sulfonyl Oximes. Org. Lett. 2017, 19 (1), 34–37. 10.1021/acs.orglett.6b03252. [DOI] [PubMed] [Google Scholar]
- Sannino A.; Gironda-Martínez A.; Gorre E. M. D.; Prati L.; Piazzi J.; Scheuermann J.; Neri D.; Donckele E. J.; Samain F. Critical Evaluation of Photo-Cross-Linking Parameters for the Implementation of Efficient DNA-Encoded Chemical Library Selections. ACS Comb. Sci. 2020, 22 (4), 204–212. 10.1021/acscombsci.0c00023. [DOI] [PubMed] [Google Scholar]
- Meng G.; Guo T.; Ma T.; Zhang J.; Shen Y.; Sharpless K. B.; Dong J. Modular Click Chemistry Libraries for Functional Screens Using a Diazotizing Reagent. Nature 2019, 574 (7776), 86–89. 10.1038/s41586-019-1589-1. [DOI] [PubMed] [Google Scholar]
- Srinivasan R.; Tan L. P.; Wu H.; Yang P. Y.; Kalesh K. A.; Yao S. Q. High-Throughput Synthesis of Azide Libraries Suitable for Direct “Click” Chemistry and in Situ Screening. Org. Biomol. Chem. 2009, 7 (9), 1821–1828. 10.1039/b902338k. [DOI] [PubMed] [Google Scholar]
- Bradner J. E.; McPherson O. M.; Koehler A. N. A Method for the Covalent Capture and Screening of Diverse Small Molecules in a Microarray Format. Nat. Protoc. 2006, 1 (5), 2344–2352. 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- Duffner J. L.; Clemons P. A.; Koehler A. N. A Pipeline for Ligand Discovery Using Small-Molecule Microarrays. Curr. Opin. Chem. Biol. 2007, 11 (1), 74–82. 10.1016/j.cbpa.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Bradner J. E.; McPherson O. M.; Mazitschek R.; Barnes-Seeman D.; Shen J. P.; Dhaliwal J.; Stevenson K. E.; Duffner J. L.; Park S. B.; Neuberg D. S.; et al. A Robust Small-Molecule Microarray Platform for Screening Cell Lysates. Chem. Biol. 2006, 13 (5), 493–504. 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Bailey M. E.; Kirss V.; Spaunburgh R. G. Reactivity of Organic Isocyanates. Ind. Eng. Chem. 1956, 48 (4), 794–797. 10.1021/ie50556a035. [DOI] [Google Scholar]
- Ghosh A. K.; Brindisi M. Urea Derivatives in Modern Drug Discovery and Medicinal Chemistry. J. Med. Chem. 2020, 63 (6), 2751–2788. 10.1021/acs.jmedchem.9b01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacondio F.; Silva C.; Lodola A.; Fioni A.; Rivara S.; Duranti A.; Tontini A.; Sanchini S.; Clapper J. R.; Piomelli D.; Mor M.; Tarzia G.; et al. Structure-Property Relationships of a Class of Carbamate-Based Fatty Acid Amide Hydrolase (FAAH) Inhibitors: Chemical and Biological Stability. Chem. Med. Chem. 2009, 4 (9), 1495–1504. 10.1002/cmdc.200900120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Brindisi M. Organic Carbamates in Drug Design and Medicinal Chemistry. J. Med. Chem. 2015, 58 (7), 2895–2940. 10.1021/jm501371s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. L.; Shi Y.; Kang J. S.; Oelschlaeger P.; Yang K. W. Amino Acid Thioester Derivatives: A Highly Promising Scaffold for the Development of Metallo-β-Lactamase L1 Inhibitors. ACS Med. Chem. Lett. 2015, 6 (6), 660–664. 10.1021/acsmedchemlett.5b00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y.; Yanagisawa E.; Ohshima T.; Takeda S.; Aburada M.; Miyamoto K. I. Synthesis and Evaluation of Carbamate Prodrugs of a Phenolic Compound. Chem. Pharm. Bull. 2007, 55 (2), 328–333. 10.1248/cpb.55.328. [DOI] [PubMed] [Google Scholar]
- Saari W. S.; Schwering J. E.; Lyle P. A.; Smith S. J.; Engelhardt E. L. Cyclization-Activated Prodrugs. Basic Carbamates of 4-Hydroxyanisole. J. Med. Chem. 1990, 33 (1), 97–101. 10.1021/jm00163a016. [DOI] [PubMed] [Google Scholar]
- Uesugi S.; Li Z.; Yazaki R.; Ohshima T. Chemoselective Catalytic Conjugate Addition of Alcohols over Amines. Angew. Chemie - Int. Ed. 2014, 53 (6), 1611–1615. 10.1002/anie.201309755. [DOI] [PubMed] [Google Scholar]
- Lundt B. F.; Johansen N. L.; Vølund A.; Markussen J. Removal of t-Butyl and t-Butoxycarbonyl Protecting Groups with Trifluoroacetic Acid. Int. J. Pept. Protein Res. 1978, 12 (5), 258–268. 10.1111/j.1399-3011.1978.tb02896.x. [DOI] [PubMed] [Google Scholar]
- Agami C.; Couty F. The Reactivity of the N-Boc Protecting Group: An Underrated Feature. Tetrahedron 2002, 58 (14), 2701–2724. 10.1016/S0040-4020(02)00131-X. [DOI] [Google Scholar]
- Gilson M. K.; Liu T.; Baitaluk M.; Nicola G.; Hwang L.; Chong J. BindingDB in 2015: A Public Database for Medicinal Chemistry, Computational Chemistry and Systems Pharmacology. Nucleic Acids Res. 2016, 44 (D1), D1045–D1053. 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S. M.; Bittker J. A.; Liu Z.; Gould J.; McCarren P.; Hirschman J. E.; Johnston S. E.; Vrcic A.; Wong B.; Khan M.; et al. The Drug Repurposing Hub: A Next-Generation Drug Library and Information Resource. Nat. Med. 2017, 23 (4), 405–408. 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S. L.; Kotz J. D.; Li M.; Aubé J.; Austin C. P.; Reed J. C.; Rosen H.; White E. L.; Sklar L. A.; Lindsley C. W.; et al. Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes. Cell 2015, 161 (6), 1252–1265. 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea T. I.; Bologa C. G.; Boyer S.; Curpan R. F.; Glen R. C.; Hopkins A. L.; Lipinski C. A.; Marshall G. R.; Martin Y. C.; Ostopovici-Halip L.; et al. A Crowdsourcing Evaluation of the NIH Chemical Probes. Nat. Chem. Biol. 2009, 5 (7), 441–447. 10.1038/nchembio0709-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–7. 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. C.; Sanders J. M.; Gao Y.-D.; Tudor M.; Haidle A. M.; Klein D. J.; Converso A.; Lesburg C. A.; Zang Y.; Wood H. B. Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery. J. Chem. Inf. Model. 2020, 60 (9), 4144–4152. 10.1021/acs.jcim.0c00113. [DOI] [PubMed] [Google Scholar]
- SMARTS - A Language for Describing Molecular Patterns. https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html (accessed September 15, 2022).
- Ong S. E.; Schenone M.; Margolin A. A.; Li X.; Do K.; Doud M. K.; Mani D. R.; Kuai L.; Wang X.; Wood J. L.; et al. Identifying the Proteins to Which Small-Molecule Probes and Drugs Bind in Cells. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (12), 4617–4622. 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt D. A.; Luengo J. I.; Yamashita D. S.; Oh H. J.; Konialian A. L.; Yen H. K.; Rozamus L. W.; Brandt M.; Bossard M. J.; Levy M. A.; et al. Design, Synthesis, and Kinetic Evaluation of High-Affinity FKBP Ligands and the x-Ray Crystal Structures of Their Complexes with FKBP12. J. Am. Chem. Soc. 1993, 115 (22), 9925–9938. 10.1021/ja00075a008. [DOI] [Google Scholar]
- Murray J. K.; Gellman S. H. Parallel Synthesis of Peptide Libraries Using Microwave Irradiation. Nat. Protoc. 2007, 2 (3), 624–631. 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
- Weller H. N.; Rubin A. E.; Moshiri B.; Ruediger W.; Li W. J.; Allen J.; Nolfo J.; Bertok A.; Rosso V. W. Development and Commercialization of the MiniBlock Synthesizer Family: A Historical Case Study. J. Lab. Autom. 2005, 10 (1), 59–71. 10.1016/j.jala.2004.07.002. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.