Abstract
Mating pair stabilization occurs during conjugative DNA transfer whereby the donor and recipient cells form a tight junction which requires pili as well as TraN and TraG in the donor cell. The role of the outer membrane protein, TraN, during conjugative transfer was examined by introduction of a chloramphenicol resistance cassette into the traN gene on an F plasmid derivative, pOX38, to produce pOX38N1::CAT. pOX38N1::CAT was greatly reduced in its ability to transfer DNA, indicating that TraN plays a greater role in conjugation than previously thought. F and R100-1 traN were capable of complementing pOX38N1::CAT transfer equally well when wild-type recipients were used. F traN, but not R100-1 traN, supported a much lower level of transfer when there was an ompA mutation or lipopolysaccharide (LPS) deficiency in the recipient cell, suggesting receptor specificity. The R100-1 traN gene was sequenced, and the gene product was found to exhibit 82.3% overall similarity with F TraN. The differences were mainly located within a central region of the proteins (amino acids 162 to 333 of F and 162 to 348 of R100-1). Deletion analysis of F traN suggested that this central portion might be responsible for the receptor specificity displayed by TraN. TraN was not responsible for TraT-dependent surface exclusion. Thus, TraN, and not the F pilus, appears to interact with OmpA and LPS moieties during conjugation, resulting in mating pair stabilization, the first step in efficient mobilization of DNA.
Bacterial conjugation is the mechanism whereby DNA is transferred from a donor to a recipient cell through a complex apparatus that is encoded by a transfer system. One of the most studied conjugative systems is the F plasmid of Escherichia coli K-12, and the entire sequence of its transfer region of 40 genes (tra) is known (reviewed in reference 12). Five classes of genes have been identified so far: those for (i) pilus synthesis, (ii) surface exclusion, (iii) mating pair stabilization (MPS), (iv) regulation, and (v) origin of transfer (oriT) nicking and DNA mobilization. About 20 genes encoded within the tra region are absolutely required for conjugation to occur since mutations in these genes completely abrogate transfer. In other cases, mutations within tra genes cause transfer levels to decrease by several orders of magnitude. In this report, we demonstrate that traN, whose gene product is involved in MPS, is more important for transfer efficiency than previously thought.
The interactions that occur between a donor cell carrying the F plasmid and a recipient cell trigger plasmid transfer and lead to the establishment of an F plasmid in both, resulting in the formation of two donor cells. Very little is known about the molecular basis of the initial events preceding the transfer of DNA, but three distinct steps are thought to occur: (i) pilus attachment to the recipient cell, (ii) pilus retraction, and (iii) eventual stabilization of the contact established between the cells (26). Fifteen proteins are required for pilus assembly; mutations in these genes yield nonpiliated cells, yet a distinct mechanism of pilus assembly or retraction has not been delineated (12). MPS is comparatively simpler and involves two proteins, TraN and TraG (26). Nonpiliated cells do not transfer DNA, showing that the pilus is absolutely required for DNA transfer, while mutations in traN or traG drastically reduce transfer efficiency but do not completely abolish it (12). Together, TraN and TraG are thought to stabilize mating pairs through an as yet unknown mechanism.
TraN is expressed as a protein of 602 amino acids (aa) that is cleaved at the N terminus to the mature form of 584 aa and targeted to the outer membrane, where it apparently resides (25). Its presence in the outer membrane suggests that it might mediate interactions with the recipient cell during conjugation. TraG, an inner membrane protein, was shown to be bifunctional since mutations in the proximal half of the gene affect pilus assembly while mutations in the distal portion of traG affect MPS (26). A stable 487-aa periplasmic product termed TraG*, possibly resulting from proteolysis at a putative peptide cleavage site found in the inner membrane protein TraG, has been observed (11). It is tempting to speculate that the primary function of TraG*, which is the C-terminal segment of TraG, is MPS.
Mutations in traN and traG affect the formation of contact-specific, multicellular mating aggregates, seen in mixtures of donor and recipient cells by electron microscopy (3). These mating aggregates are resistant to shear forces and appear to play a role in conjugation in liquid culture. The amber mutant traN548, as well as some traG mutants that express F pili, mate more efficiently on a solid surface, whereas mutations in other transfer genes do not have this property (3, 26). It has been shown that stable mating aggregates contain electron-dense regions at the site of contact between the outer membranes of the mating cells, and it has been proposed that this electron-dense region is due to the accumulation of proteins at the contact site. TraN, being an outer membrane protein (OMP), is a likely component of this junction, where it associates with recipient cell components known to be required for efficient mating (see below) (10). In addition, stable mating aggregates accumulated at the nonpermissive temperature in studies using a temperature-sensitive mutant of traD, which encodes the purported DNA pump (34). These results imply that DNA transfer proceeds with greater efficiency after MPS has occurred.
By isolating mutants that are poor recipients (Con−), two major types of mutations which lower the mating efficiency of the F plasmid have been found. The first type are mutations in genes (rfa) which affects biosynthesis of the inner core of the lipopolysaccharide (LPS), specifically mutations within the gene rfaP, which encodes a protein responsible for pyrophosphorylethanolamine (PPEA) addition to the heptose I residue and extension of the heptose II residue by heptose III in the inner core of LPS (4). The second type are mutations in the gene for one of the major porins, ompA (3, 4). These mutations decrease mating efficiency by 2 to 3 logs when present in the recipient cell but have no detrimental effects on mating efficiency when they are present in the donor cell (4). Requirements for OmpA in the recipient cell are completely alleviated if the mating is allowed to occur on a solid surface (3). LPS and OmpA may thus be contributed by the recipient cell to the electron-dense region mentioned above and are postulated to act as receptors during conjugation.
R100-1, also known as NR1, is a closely related plasmid that belongs to the incompatibility group FII and encodes a number of antibiotic resistance determinants (reviewed in reference 47). Transfer of R100-1 is not affected by the recipient cell mutations in rfa or ompA, suggesting that R100-1 requires an alternate component of the recipient cell (4). Initially, it was thought that OmpA and LPS were receptors for the pilus tip during recipient cell recognition, but experiments demonstrated that both F traA and R100-1 traA complemented a null mutant of the pilin gene, traA, to equal levels, irrespective of the recipient cell used, indicating that the pilin subunit is not the donor component that interacts with OmpA or LPS (4).
In this study, we examined the MPS gene, traN, of F and R100-1. A null mutation of traN was constructed by inserting a chloramphenicol resistance (Cmr) cassette into the coding region of traN to generate pOX38N1::CAT. We ascertained the potential effect of Con− recipient cells by complementing this traN null mutation with either F or R100-1 traN in the donor cell. Assays for mating efficiencies to LPS-deficient or ompA recipient cells, as well as to recipient cells expressing F TraT, a protein involved in surface exclusion, were done to demonstrate plasmid or allelic specificity. Finally, the sequence of R100-1 TraN is reported for comparison to that of F.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general reagents.
Bacterial strains and plasmids used in this study are described in Table 1. Cells were grown in Luria-Bertani media (LB; 1% Difco tryptone, 0.5% Difco yeast extract, 1% NaCl [BDH Inc.]) at 37°C on a tube roller or shaker. Glucose was added to cultures to a final concentration of 100 mM to repress expression from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter or as a general growth enhancer. Antibiotics (Sigma Chemical Co.) were added to final concentrations as follows: ampicillin, 50 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 25 μg/ml; spectinomycin, 100 μg/ml; streptomycin, 200 μg/ml; tetracycline, 10 μg/ml; and nalidixic acid, 16 μg/ml. IPTG (Sigma) was added as an inducer, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Sigma) was added as an indicator where needed.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant phenotype | Reference(s) or source |
|---|---|---|
| Strains | ||
| ED24 | Spcr F− Lac− | 1 |
| XK100 | Spcr derivative of BL21(DE3) carrying T7 RNA polymerase under lacUV5 control in the chromosome | 41 |
| XK1200 | Nalr F−lacΔU124 Δ(nadA aroG gal attL bio gyrA) | 31; K. Ippen-Ihler |
| DH5α | NalrsupE44 ΔlacU169 (φ80 lacZΔ80M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 14; lab stock |
| MV1193 | Smr Δ(lac-proAB) rpsL thi endA sbcB15 hsdR4 Δ(srl-recA)306::Tn10(Tetr) F′[traD36 proAB+ lacIqlacZΔM15] | 48; lab stock |
| JE2571-1 | Smr Nalrleu thr Flac Pil− λ+ | David Bradley |
| RD17 | Δ(pro-lac)XIII λ−recA56 rel-1 supE44 thi-1 | 43 |
| MC4100 | SmraraD139 Δ(argF-lac)U169 rpsL150 relA1 flbB3501 deoC1 ptsF25 rbsR | C. Manoil; lab stock |
| JC3272 | Smr F−lacΔX74 gal his trp lys rpsL tsx | 1; lab stock |
| HB101 | NalrsupE44 hsdS20 recA13 ara-14 proA1 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 9; spontaneous Nalr in 22 |
| ompA and LPS mutants | ||
| CC102 | SmrgalE28 of JC3272 | C. Manoil |
| CS180-2 | Smrthr leuB6 proA argE his thi galK lacY trpE mtl xyl ara-14; CS180 reisolated rpsL Su+ | 7; C. Schnaitman |
| CC277 | KmrompA902::Tn5 of CC102 | 28; C. Manoil |
| CS1834 | SmrrecB21 recC22 sbcB15 sbcC201 argE3 his-4 leuB6 proA2 thr-1 ara-14 galK2 mtl-1 xyl-5 thi-1 rpsL32 supE44 Bx-33Δ(argF-lac)U169 Δ(trpEA)2 | 35; C. Schnaitman |
| CS1999 | Δlac of CS180 Mur | 19; C. Schnaitman |
| CS2198 | KmrrfaJ19::TnlacZ of CS1999 | 35; C. Schnaitman |
| CS2193 | KmrrfaP23::TnlacZ of CS1834 | C. Schnaitman |
| Plasmids | ||
| Conjugative plasmids and derivatives | ||
| pOX38 | IncFI Tra+ RepFIA+, f1 HindIII fragment of F | 13 |
| pOX38::Tc | Tcr, pOX38::mini-Tn10 | 4 |
| FlacN548 | traN amber mutant of Flac | 29 |
| FN548::Tc | Tcr Lac−, mini-Tn10 in lac | 4 |
| R100-1 | IncFII Tra+ Cmr Far Smr Spr Sur Tcr | 21, 46 |
| pRS29 | Tcr, 15-kb partial EcoRI fragment of F containing traN, cloned into pSC101 | 2 |
| pRS29N548 | traN amber mutant of pRS29 | K. Ippen-Ihler |
| pOX38N1::CAT | Cmr, traN1::CAT of pOX38 (see Materials and Methods) | This study |
| pBS derivatives | ||
| pBS KS+ or SK+ | Ampr, cloning vector, 3.0 kb | Stratagene |
| F traN | ||
| pKI375 | Ampr, 3.0-kb Asp7001 fragment of F, trbC+ traN+ trbE+ | 25; K. Ippen-Ihler |
| pKI375N1::CAT | Ampr Cmr, same as pKI375 except replacement of a BbrPI/EcoRV fragment of pKI375 with a CAT cassette (see Materials and Methods) | This study |
| pS55, pS53, pS84, pS63, pS62, pS83, pS52, pS82 | Ampr, 3′ deletions of traN in pKI375 (see Fig. 1) | 25; K. Ippen-Ihler |
| pBK2K90T | Ampr, K90T mutation in putative ATP-binding motif of traN in pKI375, trbC+ traN+ trbE+ | This study |
| R100-1 traN | ||
| pBK7 | Ampr, 6.2-kb SacI/SalI fragment of R100-1 in pBS SK+ containing trbI, traW, traU, orfF, trbC, traN, trbE, and traF | This study |
| pBK8 | Ampr, 3.5-kb NsiI fragment of pBK7 in pBS SK+/PstI, traN+ trbE+ traF+ | This study |
| pBK8-2818 | Ampr, 2.8-kb, 3′ deletion of pBK8, traN+ trbE+ | This study |
| Miscellaneous | ||
| pK194 | Kmr, 2.4-kb cloning vector, p15a replicon | 18 |
| pK194T | Kmr, 1.1-kb PCR-amplified traT+ fragment in pK194 | 4 |
| pUC4C | Ampr Cmr, 3.6-kb end-filled AsuII fragment of Cmr cassette (pBR327) cloned into PstI-digested pUC4K (to remove Kmr cassette) | 24 |
Cloning.
General techniques were performed as described elsewhere (6) unless otherwise indicated. All DNA modification enzymes were from Boehringer Mannheim. Vent polymerase (proofreading+) was from New England Biolabs. Large-scale plasmid preparations were done as specified by Qiagen (36a) except that after resuspension of the DNA pellet in Tris-EDTA buffer, the DNA was subjected to two phenol extractions, and the aqueous phase was collected and ethanol precipitated as usual. Small-scale plasmid preparations were performed as previously described (8) or by using QIAquick miniprep kits.
Construction of pOX38N1::CAT.
A null mutant of traN was constructed by inserting the Cmr (chloramphenicol acetyltransferase [CAT]) cassette from pUC4C into traN. To isolate the cassette, pUC4C was digested with BamHI to give the desired fragment of 0.86 kb, which was excised from a 1.5% agarose gel and purified by the GlassMAX DNA isolation matrix system (Gibco BRL). The ends were filled in with Klenow enzyme to blunt the fragment, and the entire cassette (978 bp) was ligated to pKI375 digested with EcoRV and BbrPI (which removes 860 bp and creates blunt ends) to generate pKI375N1::CAT. To generate pOX38N1::CAT, a triparental mating was conducted (32). Donor cells containing pOX38 (RD17/pOX38) and recipient cells containing pKI375N1::CAT (XK100/pKI375N1::CAT) were grown to mid-log phase in LB with appropriate antibiotics, the cells were pelleted and washed to remove antibiotics, resuspended in fresh LB, then mixed at equal volumes, and incubated for 1 h at 37°C to allow plasmid transfer and recombination. Recipient HB101 cells at mid-log phase were added at a fourfold excess volume and incubated for a further hour at 37°C to allow the newly generated pOX38N1::CAT to be transferred. Desired recombinants were Nalr Cmr and Amps. PCR amplification using primers SRNF and SRNR, and subsequent digestion with EcoRI, demonstrated that pKI375N1::CAT had correctly recombined with the traN sequence on pOX38.
Mating assays.
Mating assays were performed as previously described (4). Briefly, cultures were grown to mid-log to late log phase in LB with appropriate antibiotics, and glucose was added to all donor cultures to a final concentration of 100 mM. Cells were pelleted and washed once with cold LB to remove antibiotics. One hundred microliters each of donors and recipients was mixed with 800 μl of cold LB, vortexed briefly, and allowed to mate for 30 min at 37°C. Matings were terminated by vortexing vigorously and putting the cells on ice to prevent further mating events. After serial dilutions in cold LB, 10 μl of each dilution was plated separately on selective plates for donors and for transconjugants. Colonies were counted after overnight incubation, and the number of transconjugants per 100 donor cells was calculated. Typically 107 to 108 donor cells/ml were used in each mating assay. Multiple experiments were performed with separate and multiple cultures in each experiment; each culture was started from a single isolated colony. Plasmids containing mutations were always compared to the wild-type plasmid within the same experiment. Surface exclusion indices were as previously described (4). Cold minimal saline solution caused loss of donor cell viability when used as a diluent; therefore, cold LB was used instead. The donor cultures required the addition of glucose to optimize the growth rates. Since pKI375 and related plasmids were not stably maintained in cells, only fresh transformants were used for all mating assays, and glycerol stocks were not used except for E. coli ED24 containing pOX38::Tc, pOX38::Km, or FlacN548::Tc, E. coli JE2571-1 or XK1200 containing R100-1, and all recipient strains.
DNA synthesis and sequencing.
DNA synthesis was performed on an Applied Biosystems 391 or 381A DNA synthesizer (PCR-MATE). DNA sequencing was performed by the Sanger dideoxy method, using an Applied Biosystems 373 DNA Sequencer (STRETCH) or a Sequenase 2.0 kit (United States Biochemical). Oligonucleotides used in this study were SRNF (5′-GATCCGCAGGTGGCTCATGATTTAC-3′), SRNR (5′-GAATTCTTACCGTATCCCACGACC-3′), LFR53 (5′-ACCGGAACAACCATC-3′), LFR55 (5′-CACAGGAGAGTATCC-3′), LFR56 (5′-TTCATCAGGTCTTCG-3′), LFR59 (5′-ACTCCGACACTGACGGTCAG-3′), LFR60 (5′-AGTACTGCACACGGACTGCC-3′), and LFR61 (5′-CAGACAGGTTCCCTCAGCGG-3′). Sequence analysis was performed with the Wisconsin Package sequence analysis software (Genetics Computer Group Inc. [GCG], Madison, Wis.).
Site-directed mutagenesis.
Single-stranded DNA was produced as described elsewhere (44). Site-directed mutagenesis was performed as specified by Zoller and Smith (48), using the two-primer method and primers LFR53 and SRNF. Heteroduplex DNA was then transformed into E. coli DH5α, and the colonies were screened by colony hybridization. Colony lifts were done as described previously (39) on nitrocellulose membranes. The DNA was cross-linked to the membrane by using a Bio-Rad GS Gene Linker UV chamber, and γ-32P-labeled LFR53 was hybridized in 5× Denhardt’s solution (2.5× SSC [20× SSC is 3 M NaCl plus 0.3 M sodium citrate · 2H2O, pH 7.0], 0.5% sodium dodecyl sulfate [SDS], 90 mM Tris [pH 7.5], 0.9 M NaCl, 6 mM EDTA, 100 μg of heat-denatured calf thymus DNA [Sigma] per ml) overnight at 37°C. Membranes were washed in 50 ml of 6× SSC–0.1% SDS for 15-min intervals initially at room temperature and then with increasingly stringent conditions (increasing temperature) until signal from potentially positive colonies could be distinguished above background. The membranes were autoradiographed with Kodak X-Omat AR film at −70°C, using an intensifying screen. DNA was extracted from potentially positive colonies, and the presence of the mutation was confirmed by automated sequencing.
Generation of 3′ deletions of pBK8.
Deletion of the 3′ end of pBK8 was done with the Erase-a-Base system (Promega) (36). Approximately 10 μg of plasmid DNA (pBK8) was digested with BamHI and SacI, and deletions were generated by using exonuclease III. The first three time points were screened for deletions of the appropriate size by EcoRI restriction digests of small-scale plasmid preparations. The exact endpoint of the deletion was determined by sequencing using Sequenase and the reverse sequencing primer of plasmid pBluescript (pBS) SK+.
RESULTS
Construction of pOX38N1::CAT.
The amber mutant FlacN548::Tc (4) was initially used for complementation experiments; however, a high level of reversion resulted in loss of the amber mutation, possibly due to recombination between the wild-type traN sequences on pKI375 and the N548 amber mutation on Flac. Therefore, a promotorless CAT cassette was inserted in traN on the pOX38 F plasmid derivative to generate a null mutation. This would have allowed the Cmr marker to be followed easily during mating assays as well as enabling a determination of the relative expression levels of traN. Unfortunately, Cmr was never observed, suggesting that insufficient traN expression was generated from the main promoter of the F plasmid, PY, which is approximately 11 kb upstream of the traN start codon. To ensure that Cmr was expressed, a CAT cassette with its own promoter was isolated from plasmid pUC4C and ligated into BbrPI/EcoRV-digested pKI375, resulting in replacement of approximately one-half of the coding sequence of traN with the CAT cassette to generate pKI375N1::CAT (Fig. 1). A triparental mating was used to cross this traN null mutation into pOX38, and the desired recombinants were selected on the basis of resistance to nalidixic acid and chloramphenicol and sensitivity to ampicillin. Sensitivity to phage f1 infection demonstrated proper pilus assembly and therefore uninterrupted expression of the tra genes downstream of traN which are required for pilus synthesis.
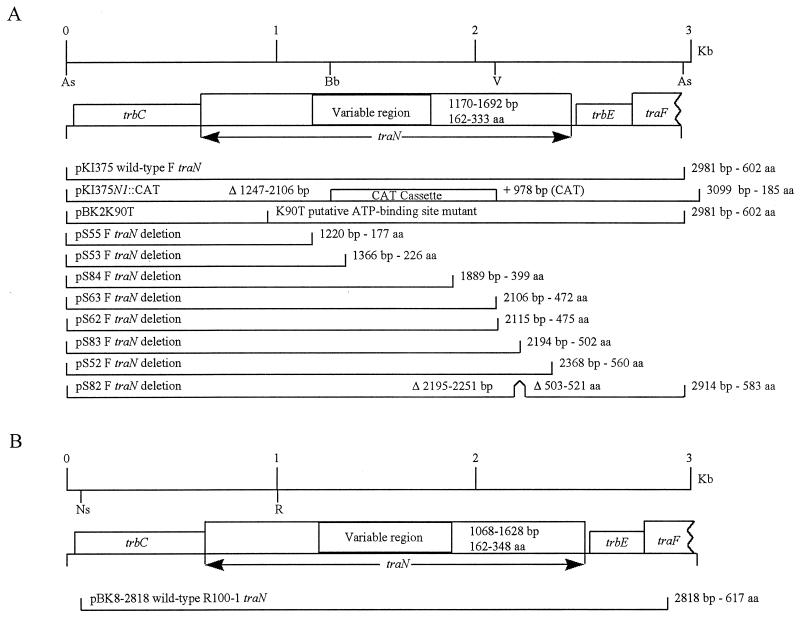
FIG. 1.
Physical maps of the various F constructs used in this study. (A) F plasmid derivatives. (B) R100-1 plasmid derivatives. The region corresponding to the F or R100-1 plasmid from which each of these constructs is generated is shown; sizes are marked at the top. Restriction enzymes: As, Asp7001; Bb, BbrPI; Ns, NsiI; R, EcoRI; V, EcoRV. tra and trb genes are boxed. The variable region in TraN is also boxed, and the range of amino acids that this region represents in F and R100-1 TraN is shown. Each construct is shown approximately to scale, and the length of the insert in pBS is shown along with the number of expected amino acids remaining in TraN. The F plasmid deletions are all derivatives of the initial clone of F traN, pKI375, which is a 3-kb Asp7001 fragment of the F plasmid cloned into pBS KS+/EcoRV (25). The R100-1 traN clone is a derivative of a 3.5-kb NsiI fragment of R100-1 cloned into pBS SK+/PstI and subsequently treated with exonuclease to generate a deletion. pS82 is an in-frame deletion of 18 aa. T7 RNA polymerase-directed transcription proceeds from left to right in both diagrams. The region replaced by the CAT cassette in pKI375N1::CAT is boxed. The K90T mutation is a Lys-to-Thr mutation at residue 90 in TraN.
EcoRI digestion of the product of PCR amplification using primers SRNF and SRNR, which amplify the entire traN gene, further confirmed that correct recombination and replacement of the wild-type traN sequence on pOX38 with pKI375N1::CAT had occurred. The null mutation was constructed such that CAT and traN transcription are both in the same direction as transcription from the PY promoter. Complementation with pRS29 (traN+) restored wild-type levels of conjugation (average of 11 transconjugants per 100 donor cells), while a similar clone containing an amber mutation of traN, pRS29N548, gave a 600-fold reduction (average of 0.018 transconjugants per 100 donor cells), indicating that the deficiency in conjugation of pOX38N1::CAT was specific to traN and that expression of downstream genes was not affected.
Cloning and sequencing of R100-1 traN.
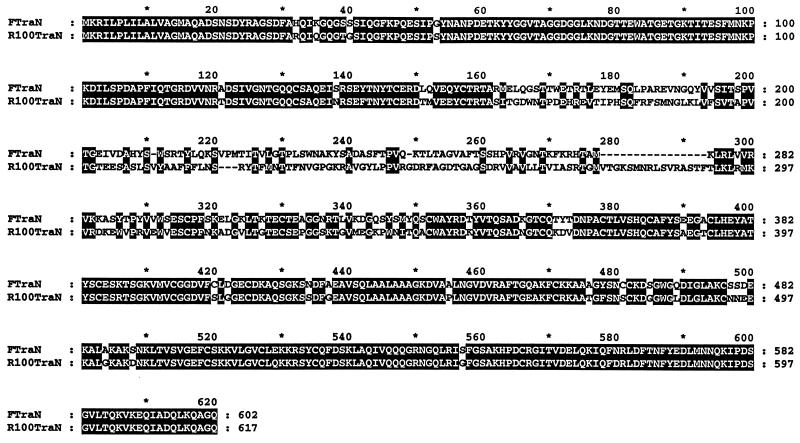
R100-1 and ColB2 are plasmids closely related to F; their transfer proteins are almost identical in sequence and function to those of F (4, 5, 20). The differences in R100-1 and ColB2 have been exploited as reservoirs of naturally occurring alleles of transfer genes. PCR amplification of traN using primers SRNF and SRNR demonstrated that a band of the same size could be generated by using F, R100-1, or ColB2 template DNA, indicating that a coding region corresponding to traN exists in all of these plasmids (data not shown). traN of R100-1 was sequenced with primers SRNF and SRNR after PCR amplification. Primers were generated as needed to sequence across the entire R100-1 traN open reading frame on both strands. The predicted amino acid sequence yielded a protein that was very similar to F TraN (73.6% identity; 82.3% similarity [Fig. 2]). The central region of R100-1 TraN from aa 162 to 348 is remarkably dissimilar to that of the corresponding region of F TraN (aa 162 to 333) and includes 15 extra amino acids. This sequence divergence in the central portion of TraN may indicate specificity of receptor interaction, while the conserved N and C termini might imply preserved functional domains. Preliminary sequence analysis of ColB2 TraN showed that it was virtually identical to F TraN in the central region and was not studied further.
FIG. 2.
Comparison of the protein sequences of F and R100-1 TraN. F TraN sequence is derived from nucleotides 12997 to 14802 of the F tra operon and corrected from a previously published sequence (12 [GenBank accession no. U01159]). R100-1 traN was sequenced as described in Materials and Methods. The sequence of R100-1 traN is contained within a larger fragment of R100-1 sequence (nucleotides 8469 to 10322; GenBank accession no. AF005044). The sequences were aligned with the GCG Pileup program and shaded with the Genedoc program (http://www.cris.com/∼ketchup/genedoc.shtml). Single-letter amino acid codes are used; conserved amino acids are shaded in black, with amino acid residues corresponding to either F or R100-1 TraN numbered on the right. Dashes are used to indicate gaps in the sequence generated during computer alignment.
Complementation with R100-1 traN.
R100-1 traN was initially cloned on a SacI/SalI fragment in pBS SK+ (pBK7, 6.2 kb). A smaller NsiI fragment of pBK7 was subcloned into pBS SK+/PstI (pBK8, 3.5 kb). A further deletions in pBK8 resulted in the elimination of R100-1 traF, a gene which in the F plasmid is involved in pilus assembly and is essential for transfer. Exonuclease III and S1 nuclease were used to generate a 3′ deletion of pBK8, which was characterized by DNA sequencing. This plasmid, pBK8-2818, carries a 2,818-bp fragment of R100-1 which encodes traN and trbE but not traF. It was assumed that R100-1 trbE would not interfere in complementation assays since a null mutation of F trbE had no effect on mating efficiency, F-pilus expression, or phage infection (25).
TraN expression in all of these plasmids, including the pKI375 derivatives and the R100-1 traN clones was oriented in the same direction as the T7 promoter and opposite the lac promoter of the pBS vector (Fig. 1). In all complementation experiments, basal levels of expression from an unknown or plasmid-driven promoter, in the absence of T7 RNA polymerase, were sufficient for full complementation to wild-type levels (see below). Thus, a source of the T7 RNA polymerase gene was not required in mating assays. R100-1 traN, from plasmid pBK8-2818, complemented pOX38N1::CAT to the same levels as F traN, suggesting conservation of function (see below). TraN was specifically labeled from plasmids pKI375 and pBK8-2818 by induction of T7 RNA polymerase in E. coli XK100 and by addition of rifampin to inhibit host protein synthesis; radioactive labeling and separation on SDS-polyacrylamide gels demonstrated that a protein of the expected size for TraN was expressed from each of these plasmids (data not shown).
Interactions of F TraN and OmpA.
Since TraN is an OMP in the donor cell and appears to be involved in MPS, it might interact with exposed moieties on the surface of the recipient cell such as OmpA or LPS which have previously been implicated in F conjugation (4). CC277 is a strain which carries a null mutation in ompA. As shown in Table 2, depending on the nature of the recipient cell (CC277 versus MC4100; ompA versus ompA+), pOX38N1::CAT transfer is complemented to different levels with plasmids carrying F or R100-1 traN. When pOX38N1::CAT was complemented with F traN, mating efficiency decreased to 0.025% of wild-type levels with CC277 compared to MC4100 (ompA versus wild type; 0.04 versus 160 transconjugants per 100 donor cells), while R100-1 traN mating efficiency decreased to 56% of wild-type levels (55 versus 99 transconjugants per 100 donors). This finding suggests that F TraN, but not R100-1 TraN, may interact with OmpA during conjugation to bring about MPS. These results have been confirmed with strains CC650 and CC651, both of which carry ompA point mutations that are known to affect conjugation (data not shown and reference 27). Since the two TraN proteins exhibit dissimilar sequences for at least a third of their lengths, it is possible that this region is important for specific interactions with OMPs in the recipient cell during conjugation.
TABLE 2.
Complementation analysis of pOX38N1::CAT with F or R100-1 traN
| Plasmid | traN genotype | Mating efficiencya
|
|
|---|---|---|---|
| ompA | Wild type | ||
| pOX38::Tc | F | 0.12 | 67 |
| R100-1 | R100-1 | 233 | 200b |
| pOX38N1::CAT/pKI375 | F | 0.040 | 160 |
| pOX38N1::CAT/pBK8-2818 | R100-1 | 56 | 99 |
| pOX38N1::CAT/pBS KS+ | Vector control | 0.00038 | 0.00043 |
Number of transconjugants per 100 donor cells. Typically, the number of donors was 107 to 108 cells/ml. If more than one assay was done, the results were averaged. The ompA mutant is a null mutant, strain CC277, a derivative of JC3272. The wild-type strain is MC4100. Similar results were obtained with JC3272.
The wild-type strain is CS2198; since R100-1 is Smr, MC4100 cannot be used as a wild-type control.
Effects of LPS-deficient recipient cells on traN complementation.
It has been demonstrated that F and R100-1 mating efficiencies are differentially affected by various mutations in biosynthetic genes for LPS (4). Similar to the assays with the ompA mutations described above, complementation was assayed by using rfa recipients, which are deficient in various moieties in the inner core of the LPS, and donor cells carrying pOX38N1::CAT and plasmids expressing either of the two traN alleles.
Table 3 indicates that as in the case of ompA recipients, F traN complementation levels were affected by LPS-defective recipients (28-fold reduction; 130 versus 4.7 transconjugants per 100 donor cells; CS2193 versus CS2198; rfaP+ versus rfaP), while R100-1 traN levels were not (1.6-fold reduction; 73 versus 45 transconjugants per 100 donor cells; CS2193 versus CS2198). The decrease in mating efficiency was not as great for F traN in rfaP null strains (3.6%) as for ompA null strains (0.025%). It is very clear, however, that an rfaP mutation had little effect on R100-1 traN-mediated transfer or on the transfer of the wild-type R100-1 plasmid.
TABLE 3.
Effects of LPS-deficient recipients on complementation ability of F and R100 traN
| Plasmid | traN genotype | Mating efficiencya
|
|
|---|---|---|---|
| rfaP | Wild type | ||
| pOX38::Tc | F | 6 | 80 |
| R100-1 | R100-1 | 188 | 200 |
| pOX38N1::CAT/pKI375 | F | 4.7 | 130 |
| pOX38N1::CAT/pBK8-2818 | R100-1 | 31 | 73 |
| pOX38N1::CAT/pBS KS+ | Vector control | 0.0024 | 0.00023 |
Defined as in Table 2, footnote a. The rfaP mutant is strain CS2193. The wild-type strain is CS2198, which is rfaJ19::TnlacZ (Kmr) but does not affect mating efficiency.
Surface exclusion.
Complementation assays were again performed with pOX38N1::CAT and the two traN alleles to determine if TraN is important for the surface exclusion function of TraT, an OMP. Surface exclusion can be measured as a ratio of the mating efficiency of a recipient cell expressing TraT compared to a wild-type recipient cell (no TraT expressed), which is called the surface exclusion index (Sfx) (15). A positive Sfx, therefore, indicates surface exclusion and, since surface exclusion is plasmid specific, would indicate specificity of interactions of TraT with donor cell components. As shown in Table 4, the Sfx is independent of traN. pOX38N1::CAT in the presence of F traN exhibited an Sfx of 245, while R100-1 traN had an Sfx of 284. The wild-type plasmids pOX38::Tc and R100-1 had Sfxs of 77 and 1.5 respectively, indicating that pOX38N1::CAT is surface excluded to approximately the same degree as pOX38::Tc, regardless of which TraN protein is expressed. Thus, TraT appears not to interfere with TraN function during mating pair formation, and surface exclusion as mediated by TraT is not specific to TraN but may be due to some other plasmid-specific protein.
TABLE 4.
Sfxs of different alleles of traN
| Plasmid(s) | traN genotype | Mating efficiencya
|
Sfxb | |
|---|---|---|---|---|
| pK194T/ JC3272 | pK194/ JC3272 | |||
| pOX38::Tc | F | 0.13 | 10 | 77 |
| R100-1 | R100-1 | 60 | 91 | 1.5 |
| pOX38N1::CAT/pKI375 | F | 0.28 | 61 | 245 |
| pOX38N1::CAT/pBK8-2818 | R100-1 | 0.037 | 10 | 284 |
| pOX38N1::CAT/pBS KS+ | Vector control | <0.00025c | 0.0024 | >9.5 |
Defined as in Table 2, footnote a. pK194T expresses TraT from its own promoter; pK194 is the vector control.
Mating efficiency to the vector control (pK194) divided by mating efficiency of TraT-expressing cells (pK194T).
There were no colonies present; therefore, 0.00025 is the theoretical upper limit if only one colony existed at the lowest dilution.
Complementation with F traN 3′ deletions.
To further examine the function of TraN, we obtained a number of 3′ deletions of F traN (25) and tested them for the ability to complement pOX38N1::CAT (Table 5). All of these constructs express a stable form of TraN from the T7 promoter in XK100 cells, as determined previously (25) (Fig. 1).
TABLE 5.
Complementation analysis of F traN mutations
| Plasmid | TraN expresseda | Mating efficiency, wild-typeb | % of wild-type mating efficiency (deletion/pKI375) | Mating efficiency, ompAb |
|---|---|---|---|---|
| pKI375 | F | 160 | 100 | 0.040 |
| pS55 | aa 1–177 | 0.0011 | 0.00052 | ND |
| pS53 | aa 1–226 | 0.0019 | 0.00089 | ND |
| pS84 | aa 1–399 | 0.83 | 1.46 | 0.023 |
| pS63 | aa 1–472 | <0.0078c | <0.012 | <0.0052 |
| pS62 | aa 1–475 | 0.16 | 0.15 | <0.00023 |
| pS83 | aa 1–502 | 0.011 | 0.013 | 0.008 |
| pS82 | aa 1–583 | 0.0060 | 0.015 | <0.0023 |
| pBK2K90T | K90T mutation | 162 | 90 | ND |
The 3′ deletions were tested in assays using wild-type or ompA recipients in order to delimit the region of TraN which potentially interacts with OmpA. As shown in Table 5, many of the deletions (pS55, pS53, pS63, pS83, and pS82) complemented pOX38N1::CAT poorly, usually at only 1 order of magnitude above the vector control (range of 0.001 to 0.0087 transconjugants per 100 donor cells). Interestingly, some deletions were able to complement pOX38N1::CAT to a higher degree (pS84 and pS62; 0.83 and 0.16 transconjugants per 100 donor cells, respectively). Complementation with the plasmid pS52, which carries a deletion of the last 40 aa of the C terminus of TraN, could not be carried out because cells containing the plasmid were inviable.
Since pS84 is the largest deletion which is still affected by an ompA mutation in recipient cells to a degree similar to that of the wild-type plasmid (pKI375), a region between the N terminus and residue 399 appears to be important for interaction with OmpA. This finding suggests that this region, which encompasses the dissimilar region between F and R100-1 TraN, might be important for OmpA recognition by F TraN.
DISCUSSION
Previously, OmpA and LPS were thought to be the receptors for the F pilus during the initial contact between the donor and recipient cells (4). Our work demonstrates that the outer membrane protein TraN is responsible for interactions with OmpA and LPS, and that this interaction enables stable mating pair formation which leads to a much more efficient transfer of DNA. The receptor in the recipient cell for the pilus tip, as well as for R100-1 TraN, remains unknown.
TraN receptor in the recipient cell.
Our results indicate that F TraN interacts either directly or indirectly with OmpA and LPS to enable MPS. The LPS component is potentially a PPEA moiety in the inner core (4). In contrast, R100-1 TraN probably recognizes a distinct moiety during conjugation, as it gives wild-type levels of plasmid transfer when either Con− mutation (rfaP or ompA) exists in the recipient cell. Two previous R100-1 Con− mutants have been previously isolated; however, the nature of the mutation which specifically decreased R100-1 transfer efficiency but not F transfer efficiency is unknown (16). Sequence analysis of R100-1 TraN revealed a high degree of similarity to F TraN at the N and C termini, while a central dissimilar region indicated significant sequence mosaicism which might reflect allele-specific receptor interactions. Radioactive labeling indicated that a protein of the expected size was expressed from pBK8-2818, and we concluded that this was R100-1 TraN. Preliminary sequence analysis showed that the ColB2 traN central region was identical to that of F traN (data not shown). We expected sequence conservation between F and ColB2 traN since ColB2 and F have similar requirements for moieties in the recipient cell during conjugation (4).
It is not known whether the rfa locus is directly important for conjugation or whether mutations within the gene decrease transfer efficiency due to secondary effects on OmpA, by affecting the insertion or the conformation of OmpA in the outer membrane. A decrease of 30 to 50% in OmpA concentration has been reported for LPS-defective cells; however, it is not known if this would cause the specific decrease in F plasmid-mediated transfer (4). As well, addition of purified phosphorylethanolamine, thought to mimic PPEA, decreases F and R100-1 but not ColB2 mating efficiency (4). LPS may interact with a donor component such as the pilus tip or TraN that is separate from its effect on OmpA.
Critical region important for receptor interactions.
In an attempt to determine exactly which region of TraN is important for its function, we used a number of deletion derivatives of F traN and assayed for their ability to complement pOX38N1::CAT transfer to wild-type and ompA recipients. The minimal traN sequence that gave reasonable levels of complementation was a truncated TraN of 399 aa (pS84) that presumably contains sufficient information for TraN function. Since complementation by pS84 was affected by an ompA mutation in the recipient cell, the possibility exists that the recognition domain for OmpA is in the first 399 aa.
No pattern of complementation was seen with the smaller deletions in traN (pS62, pS63, pS83, pS82, and pS52), possibly due to effects on the transmembrane segments of TraN. Compared to deletion of an entire transmembrane segment, partial deletions could have more deleterious effects as a result of improper insertion of the protein into the membrane (17). This may explain why two closely related deletions, pS62 and pS63, exhibit such a marked difference in complementation ability and why the construct pS52 resulted in cell inviability.
Surface exclusion.
Redundant transfer of plasmid DNA from a donor to a recipient cell is prevented by a process known as surface exclusion, which is mediated by two proteins, TraT in the outer membrane and TraS in the inner membrane. In conjunction, they inhibit the level of F+ to F+ transfer by several hundred-fold. TraT is plasmid specific; that is, F TraT inhibits F transfer, while R100-1 TraT inhibits R100-1 transfer (reviewed in reference 42). This specificity has been correlated with a single amino acid change in TraT (G120A) which changes the specificity from F to R100-1 (15). It has been reported that TraT interferes with infection of E. coli by bacteriophages which are specific for OmpA, suggesting that TraT blocks binding of F plasmid-encoded products to OmpA in a similar manner (38). We tested the possibility that TraT inhibits TraN-mediated MPS during donor-to-donor conjugation and found that TraT-dependent surface exclusion was independent of the allele of traN expressed in the donor cell. Therefore, TraT cannot block TraN-OmpA interactions and might interfere with some other aspect of transfer, possibly F pilus recognition of the recipient cell.
Two functions for TraN?
The results presented here suggest that TraN has two functions: (i) recognition of a receptor and (ii) another function, possibly interactions with other plasmid-specific components. This is most apparent when one compares the effects on transfer efficiency when either ompA or traN is mutated. The lack of OmpA in the recipient cell leads to a 100- to 1,000-fold decrease in transfer efficiency. When traN is mutated in the donor cell, however, transfer efficiency is decreased 1 million-fold, suggesting that other conjugative functions are dependent on TraN. Similarly, mating on solid surfaces relieves the requirement for OmpA in the recipient cell, while traN mutations are only partially suppressed on a solid support. The MPS pathway may, therefore, be more complex and involve interactions with other transfer proteins as a prelude to DNA transport. TraG is potentially a partner in MPS (11, 26). Since MPS, a process that stabilizes two cells during conjugation, requires both an OMP and an inner membrane or periplasmic protein, this process might involve formation of a complex in the donor cell envelope.
Computer analysis of TraN.
An analysis of the topology of TraN within the outer membrane could provide clues as to which portions of TraN are responsible for interactions with OmpA. Sequence comparison of TraN to the porin superfamily showed no similarity of TraN to the conserved transmembrane segments (17). Membrane topology experiments using epitope insertions are currently being done to identify those regions of TraN which are exposed on the cell surface and those which reside within the periplasm.
Several other proteins known to bind OmpA are the tail proteins (gp38) of the T-even-type phages, TuII*, K3, and Ox2, all of which bind OmpA as a step in their infective processes (33). Analyses of ompA revealed that mutations in sequences coding for several extracellular loops affected phage binding, while the sole mutation in ompA found to affect F plasmid conjugation is a G154D substitution in the fourth extracellular loop of OmpA; no other mutation which affects F-mediated transfer has been found in ompA (37). A multiple sequence alignment with these gp38 proteins found no significant homology with TraN.
TraN contains a sequence (84-ATGETGKT-91) which resembles an ATP-binding loop or Walker box (25, 45). Mutation of the conserved lysine residue at position 90 to a threonine, which is important for interactions with the phosphate residues of ATP, resulted in a construct that was fully functional in complementing pOX38N1::CAT (Table 5) (23, 40).
Interestingly, F and R100-1 TraN contain 20 conserved cysteine residues, primarily in the C-terminal half of the proteins, which may be important for intra- or intermolecular disulfide bonds. The periplasm contains a highly oxidizing environment, directed by several proteins (reviewed in reference 30), resulting in the formation of disulfide bridges between apposed cysteine residues which may assist in protein folding. The role of these cysteines in the function of TraN is currently under investigation.
REFERENCES
- 1.Achtman M, Willetts N, Clark A J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971;106:529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman M, Skurray R A, Thompson R, Helmuth R, Hall S, Beutin L, Clark A J. Assignment of tra cistrons to EcoRI fragments of F sex factor DNA. J Bacteriol. 1978;133:1383–1392. doi: 10.1128/jb.133.3.1383-1392.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achtman M, Morelli G, Schwuchow S. Cell-cell interactions in conjugating Escherichia coli: role of the F pili and fate of mating aggregates. J Bacteriol. 1978;135:1053–1061. doi: 10.1128/jb.135.3.1053-1061.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony K G, Sherburne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 5.Anthony K G, Kathir P, Moore D, Ippen-Ihler K, Frost L S. Analysis of the traLEKBP sequence and the TraP protein from three F-like plasmids: F, R100-1, and ColB2. J Bacteriol. 1996;178:3194–3200. doi: 10.1128/jb.178.11.3194-3200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl J A, editors. Current protocols in molecular biology, and supplements. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7.Austin E A, Graves J F, Hite L A, Parker C T, Schnaitman C A. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990;172:5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid and alkaline procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer H W, Roulland-Dussoix D. A comparative analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;170:61–91. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 10.Durrenberger M B, Villiger W, Bachi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 11.Firth N, Skurray R. Characterization of the F plasmid bifunctional gene, traG. Mol Gen Genet. 1992;232:145–143. doi: 10.1007/BF00299147. [DOI] [PubMed] [Google Scholar]
- 12.Frost L A, Ippen-Ihler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guyer M S, Reed R R, Steitz J W, Low K B. Identification of a sex-factor affinity site in E. coli as γδ. Cold Spring Harbor Symp Quant Biol. 1980;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Harrison J L, Taylor I M, Platt K, O’Connor C D. Surface exclusion specificity of the TraT lipoprotein is determined by single alterations in five-amino-acid region of the protein. Mol Microbiol. 1992;6:2825–2832. doi: 10.1111/j.1365-2958.1992.tb01462.x. [DOI] [PubMed] [Google Scholar]
- 16.Havekes L, Hoekstra W, Kempen H. Relation between, F, R100 and R144 Escherichia coli K-12 donor strains in mating. Mol Gen Genet. 1977;155:185–189. doi: 10.1007/BF00393158. [DOI] [PubMed] [Google Scholar]
- 17.Jeanteur D, Lakey J H, Pattus F. The porin superfamily: diversity and common features. In: Ghuysen J-M, Hakenback R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 363–380. [Google Scholar]
- 18.Jobling M G, Holmes R K. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZα and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990;18:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klena J D, Ashford II R S, Schnaitman C A. Role of Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J Bacteriol. 1992;174:7297–7307. doi: 10.1128/jb.174.22.7297-7307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klimke W A, Fekete R, Manchak J, Anthony K, Frost L S. Plasmid specificity and interaction: the similarities and differences between the transfer regions of two compatible plasmids, F and R100-1. In: Syvanen M, Kado C, editors. Horizontal gene transfer. New York, N.Y: Chapman and Hall; 1997. pp. 23–39. [Google Scholar]
- 21.Lawn A M, Meynell E, Meynell G G, Datta N. Sex pili and the classification of sex factors in the Enterobacteriaceae. Nature. 1967;216:343–346. doi: 10.1038/216343a0. [DOI] [PubMed] [Google Scholar]
- 22.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan K M, Knight K L. Mutagenesis of the P-loop motif in the ATP binding site of the RecA protein from Escherichia coli. J Mol Biol. 1993;232:1048–1059. doi: 10.1006/jmbi.1993.1459. [DOI] [PubMed] [Google Scholar]
- 24.Maneewannakul K, Maneewannakul S, Ippen-Ihler K. Sequence alteration affecting F plasmid transfer gene expression: a conjugation system dependent on transcription by the RNA polymerase phage T7. Mol Microbiol. 1992;6:2961–2973. doi: 10.1111/j.1365-2958.1992.tb01755.x. [DOI] [PubMed] [Google Scholar]
- 25.Maneewannakul S, Kathir P, Ippen-Ihler K. Characterization of the F plasmid mating aggregation gene traN and of a new F transfer region locus trbE. J Mol Biol. 1992;225:299–311. doi: 10.1016/0022-2836(92)90923-8. [DOI] [PubMed] [Google Scholar]
- 26.Manning P A, Morelli G, Achtman M. TraG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc Natl Acad Sci USA. 1981;78:7487–7491. doi: 10.1073/pnas.78.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoil C. A genetic approach to defining the sites of interaction of a membrane protein with different external agents. J Mol Biol. 1983;169:507–519. doi: 10.1016/s0022-2836(83)80063-1. [DOI] [PubMed] [Google Scholar]
- 28.Manoil C, Rosenbusch J P. Conjugation-deficient mutants of Escherichia coli distinguish classes of functions of the outer membrane OmpA protein. Mol Gen Genet. 1982;187:148–156. doi: 10.1007/BF00384398. [DOI] [PubMed] [Google Scholar]
- 29.Miki T, Horiuchi T, Willetts N S. Identification and characterization of four new tra cistrons on the E. coli K12 sex factor F. Plasmid. 1978;1:316–323. doi: 10.1016/0147-619x(78)90048-3. [DOI] [PubMed] [Google Scholar]
- 30.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore D, Sowa B A, Ippen-Ihler K. The effect of tra mutations on the synthesis of the F-pilin membrane polypeptide. Mol Gen Genet. 1981;184:260–264. doi: 10.1007/BF00272914. [DOI] [PubMed] [Google Scholar]
- 32.Moore D, Wu J H, Kathir P, Hamilton C M, Ippen-Ihler K. Analysis of transfer genes and gene products within the traB-traC region of the Escherichia coli fertility factor, F. J Bacteriol. 1987;169:3994–4001. doi: 10.1128/jb.169.9.3994-4002.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona R, Kramer C, Henning U. Bacteriophage receptor area of outer membrane protein OmpA of Escherichia coli K-12. J Bacteriol. 1985;165:539–543. doi: 10.1128/jb.164.2.539-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panicker M M, Minkley E G., Jr DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J Bacteriol. 1985;162:584–590. doi: 10.1128/jb.162.2.584-590.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pradel E, Parker C T, Schnaitman C A. Structures of the rfaB, rfaI, rfaJ, and rfaS genes of Escherichia coli K-12 and their roles in assembly of the lipopolysaccharide core. J Bacteriol. 1992;174:4736–4745. doi: 10.1128/jb.174.14.4736-4745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Promega Corporation. Promega protocols and applications guide. 2nd ed. Madison, Wis: Promega Corporation; 1991. [Google Scholar]
- 36a.Qiagen. Qiagen plasmid handbook. Chatsworth, Calif: Qiagen; 1995. [Google Scholar]
- 37.Ried G, Henning U. A unique amino acid substitution in the outer membrane protein OmpA causes conjugation deficiency in Escherichia coli K-12. FEBS Lett. 1987;223:387–390. doi: 10.1016/0014-5793(87)80324-1. [DOI] [PubMed] [Google Scholar]
- 38.Riede I, Eschbach M L. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986;205:241–245. doi: 10.1016/0014-5793(86)80905-x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Story R M, Steitz T A. Structure of the RecA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 41.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 42.Sukupolvi S, O’Connor C D. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol Rev. 1990;54:331–341. doi: 10.1128/mr.54.4.331-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai M-M, Wong R Y-P, Hoang A, Deonier R. Transposition of Tn1000: in vivo properties. J Bacteriol. 1987;169:5556–5562. doi: 10.1128/jb.169.12.5556-5562.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 45.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willetts N, Maule J. Specificities of the IncF plasmid conjugation genes. Genet Res (Cambridge) 1986;47:1–11. doi: 10.1017/s0016672300024447. [DOI] [PubMed] [Google Scholar]
- 47.Womble D D, Rownd R H. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic-resistant plasmids. Microbiol Rev. 1988;52:433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. Methods Enzymol. 1987;154:329. doi: 10.1016/0076-6879(87)54083-6. [DOI] [PubMed] [Google Scholar]