Abstract
Background
Hypertrophic cardiomyopathy (HCM) is an autosomal dominant genetic disorder, characterized by cardiomyocyte hypertrophy, cardiomyocyte disarray and fibrosis, which has a prevalence of ∼1: 200–500 and predisposes individuals to heart failure and sudden death. The mechanisms through which diverse HCM-causing mutations cause cardiac dysfunction remain mostly unknown and their identification may reveal new therapeutic avenues. MicroRNAs (miRNAs) have emerged as critical regulators of gene expression and disease phenotype in various pathologies. We explored whether miRNAs could play a role in HCM pathogenesis and offer potential therapeutic targets.
Methods and results
Using high-throughput miRNA expression profiling and qPCR analysis in two distinct mouse models of HCM, we found that miR-199a-3p expression levels are upregulated in mutant mice compared to age- and treatment-matched wild-type mice. We also found that miR-199a-3p expression is enriched in cardiac non-myocytes compared to cardiomyocytes. When we expressed miR-199a-3p mimic in cultured murine primary cardiac fibroblasts and analyzed the conditioned media by proteomics, we found that several extracellular matrix (ECM) proteins (e.g., TSP2, FBLN3, COL11A1, LYOX) were differentially secreted (data are available via ProteomeXchange with identifier PXD042904). We confirmed our proteomics findings by qPCR analysis of selected mRNAs and demonstrated that miR-199a-3p mimic expression in cardiac fibroblasts drives upregulation of ECM gene expression, including Tsp2, Fbln3, Pcoc1, Col1a1 and Col3a1. To examine the role of miR-199a-3p in vivo, we inhibited its function using lock-nucleic acid (LNA)-based inhibitors (antimiR-199a-3p) in an HCM mouse model. Our results revealed that progression of cardiac fibrosis is attenuated when miR-199a-3p function is inhibited in mild-to-moderate HCM. Finally, guided by computational target prediction algorithms, we identified mRNAs Cd151 and Itga3 as direct targets of miR-199a-3p and have shown that miR-199a-3p mimic expression negatively regulates AKT activation in cardiac fibroblasts.
Conclusions
Altogether, our results suggest that miR-199a-3p may contribute to cardiac fibrosis in HCM through its actions in cardiac fibroblasts. Thus, inhibition of miR-199a-3p in mild-to-moderate HCM may offer therapeutic benefit in combination with complementary approaches that target the primary defect in cardiac myocytes.
Keywords: Hypertrophic cardiomyopathy, miRNA, Fibrosis, Hypertrophy, miR-199a-3p
Graphical abstract
Created with BioRender.com
Highlights
-
•
microRNA miR-199a-3p expression levels are upregulated in 3 different mouse models of cardiomyopathy.
-
•
miR-199a-3p mimic expression induces expression of several extracellular matrix genes and proteins in cardiac fibroblasts.
-
•
In vivo inhibition of miR-199a-3p attenuates cardiac fibrosis in mild-to-moderate hypertrophic cardiomyopathy in mice.
1. Introduction
Hypertrophic cardiomyopathy (HCM), often referred to as a “disease of the sarcomere”, is the most common cause of sudden death in young athletes and has a prevalence of 1:200 to 1:500 in the general population [[1], [2], [3]]. HCM is characterized by diastolic dysfunction, which along with cardiomyocyte hypertrophy, disarray, and fibrosis, leads to progressively worsening disease over decades and precipitate in atrial fibrillation, heart failure or sudden cardiac death. Mutations in genes encoding human beta myosin heavy chain (βMHC/MYH7) and cardiac myosin binding protein C (MYBPC3) represent about 50–75 % of all HCM-causing mutations known to date [4,5]. Despite identification of causal mutations in HCM, the cellular pathways that are aberrantly regulated due to these mutations and lead to the development of HCM pathology remain largely unknown. We have previously shown that there are global gene expression changes in hearts isolated from an HCM mouse model and identified non-myocyte activation of TGFβ1 mediated pathways as a key mechanism for increased fibrosis in HCM [6,7].
Over the last two decades, microRNAs (miRNAs), 21–25-nucleotide non-coding RNAs, have emerged as critical regulators of gene expression, capable of targeting a vast number of mRNAs and in turn, modulating varied physiological and pathological processes. miRNAs have been identified as regulators of cardiac regeneration [8] as well as cardiac hypertrophy and fibrosis in various cardiac disease models [9]. Furthermore, targeting of miRNAs has proven to offer a therapeutic benefit in various animal models of disease, with several miRNA inhibitors currently in Phase 1/2 clinical trials [10]. miRNAs have also been identified to be differentially expressed in HCM patient samples [[11], [12], [13]] and HCM mouse models [[14], [15], [16]]; however, whether their dysregulation plays a causal role in HCM pathogenesis remains largely unknown.
There is a pressing need to explore the mechanisms of cardiac fibrosis and hypertrophy to develop novel therapies for HCM, as conventional strategies (e.g. β-blockers, calcium channel blockers) [17] only delay progression or offer symptomatic relief. Mavacamten, an allosteric inhibitor of myosin, has been found to provide therapeutic benefit in a subset of patients with symptomatic obstructive HCM [18,19] and recent preclinical studies with genome-editing to correct the disease-causing mutations in βMHC/MYH7 have yielded promising results [20,21]. Nevertheless, complementary strategies that could prevent or slow down cardiac fibrosis progression are likely to provide additional benefits to HCM patients.
In our prior work, we have reported aberrant gene expression related to pro-fibrotic signalling in cardiac non-myocytes isolated from HCM mouse hearts [7]. In this study, we aimed to determine the potential of miRNA-based therapies for the prevention of fibrosis and hypertrophy in an HCM mouse model. Thus, we hypothesized that dysregulated expression of miRNAs in cardiac non-myocytes may play a causal role in HCM pathogenesis through (1) the regulation of a pro-fibrotic gene programme, and (2) the promotion of cardiac fibrosis and hypertrophy. To test our hypothesis, we used heterozygous knock-in mice bearing a human βMHC (MYH7) R719W or R403Q mutation in murine αMhc (αMHC403/+) and (αMHC719/+). These mouse HCM models have been previously characterized and while the homozygous knock-in mice are lethal around postnatal day 7, heterozygous mice have been shown to recapitulate the hallmarks of human HCM [6,7,22,23]. Our experiments have identified miR-199a-3p as an upregulated miRNA in two mouse models of HCM. Using in vitro and in vivo approaches, we show that miR-199a-3p regulates extracellular matrix (ECM) gene and protein expression in cardiac fibroblasts and its inhibition in mild-to-moderate disease attenuates fibrotic remodeling, which may offer therapeutic benefit.
2. Results
2.1. Expression of miR-199a-3p is upregulated in mouse HCM hearts
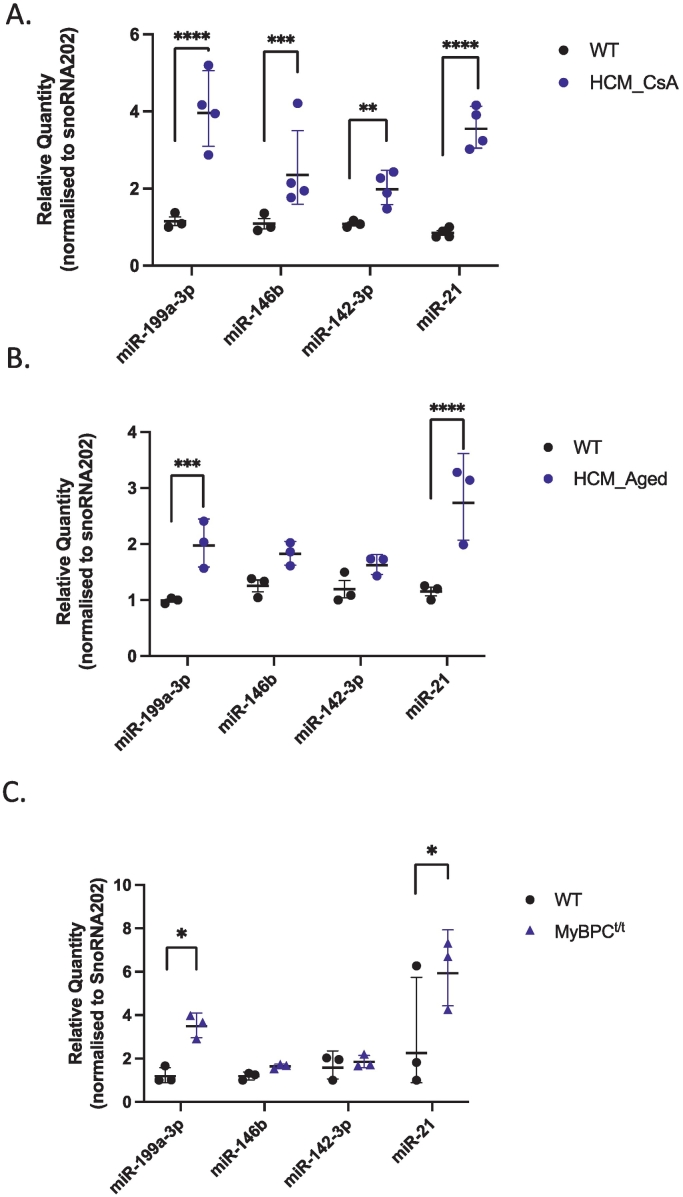
To identify and interrogate miRNA(s) that may regulate gene expression in HCM and thus have a role in determining disease phenotype, we performed RNA-seq to profile miRNA expression in two different mouse models of HCM. Heterozygous HCM mice (αMHC403/+ and αMHC719/+) display minimal HCM pathology early in life but develop age-dependent cardiac hypertrophy and fibrosis that becomes evident at >35 weeks of age [6] (referred to as HCM_Aged in this study). Previous work has shown that treating young pre-hypertrophic heterozygous mice (6–10 weeks old) with Cyclosporin A (CsA) accelerates manifestation of the HCM phenotype, with marked cardiac hypertrophy and fibrosis after 2–3 weeks of CsA treatment [7,23] (referred to as HCM_CsA in this study). Using RNA-seq, we profiled miRNAs from left ventricle (LV) of hypertrophic HCM_CsA, HCM_Aged and treatment- and age-matched wild-type (WT) littermate control mice. Our results show 54 differentially expressed miRNAs in hypertrophic HCM_CsA mice (9 miRNAs ≥1.5x down-regulated and 45 miRNAs ≥1.5x up-regulated, p < 0.05, Supp Table 1). In addition, we identified 21 differentially expressed miRNAs in hypertrophic HCM_Aged mice (7 miRNAs ≥1.5-fold down-regulated and 14 miRNAs ≥1.5-fold up-regulated, p < 0.05, Supp Table 2). Our analyses show that miR-142-3p, miR-146b, miR-199a-5p, miR-199a-3p, miR-21-3p, miR-214 and miR-341 are differentially expressed, common to both HCM_CsA and HCM_Aged mice models (≥1.5-fold-change, p < 0.05, Supp Table 3). To validate these observations, we quantified the selected miRNAs by quantitative PCR (qPCR) and confirmed the differential expression of miR-199a-3p (3.5-fold, p < 0.0001) and miR-21 (4.2-fold, p < 0.0001) in HCM_CsA mice hearts (Fig. 1A). Similarly, expression levels of miR-199a-3p (2.0-fold, p = 0.0006) and miR-21 (2.4-fold, p < 0.0001) were significantly increased in HCM_Aged mice hearts (Fig. 1B). The differential expression of miR-146b (2.2-fold, p = 0.0007) and miR-142-3p (1.8-fold, p = 0.0085) was confirmed only in HCM_CsA mice hearts (Fig. 1A), and they both showed a trend towards an increase in HCM_Aged mice hearts (1.5-fold p = 0.0577 for miR-146b, 1.4-fold p = 0.1303 for miR-142-3p) (Fig. 1B). Supporting our findings, up-regulation of these miRNAs was also reported in a transgenic mouse model of severe HCM [14].
Fig. 1.
Validation of differentially regulated miRNAs in HCM mouse models by Taqman qPCR analysis. Total RNA was isolated, reverse transcribed and qPCR was performed with specific Taqman miRNA assays (Applied Biosystems). Relative quantity of each miRNA was calculated using the 2-ΔΔCT method (snoRNA202 used as a normalization control). miRNA expression levels were quantified for the following mice models: (A) HCM_CsA vs. WT_CsA mice (n = 3–4); (B) HCM_Aged vs. WT_Aged mice (n = 3); (C) Mybpc3 homozygous mutant (MyBPCt/tvs. WT mice (n = 3). Data are represented as geometric mean of relative quantity ±SD, Statistics: 2way ANOVA with Sidak's multiple comparisons test. Adjusted p value **** p < 0.0001, ***p < 0.001, ** p = 0.0085, *p < 0.05.
We also explored whether these differentially expressed miRNAs displayed similar expression patterns in an additional mouse model of cardiomyopathy (MyBPCt/t), which expresses a truncated peptide of cardiac myosin binding protein C (MyBPC), similar to that found in human HCM. Homozygous Mybpc3 mutant (MyBPCt/t) mice display myocyte hypertrophy, disarray and fibrosis [24]. We found that the expression levels of both miR-199a-3p (2.9-fold, p = 0.0181) and miR-21 (2.6-fold, p = 0.0361) were significantly upregulated in MyBPCt/t mice hearts relative to WT littermate controls, whereas we did not detect any significant differences in the expression levels of miR-146b and miR-142-3p (Fig. 1C). Altogether, these observations suggest that diverse disease-causing mutations share similar phenotypes and perturbations in miRNA expression; however, whether such changes are causal to the HCM phenotype remains to be determined.
In our previous work, we reported cardiac non-myocyte mediated signalling and TGFβ1 pathway as key regulators of fibrosis and hypertrophy in HCM mouse models [7]. To investigate the possible role(s) of miRNAs in promoting HCM pathology, we used qPCR to quantify the expression levels of miR-199a-3p, miR-21, miR-146b and miR-142-3p in myocytes vs non-myocytes isolated from adult mouse hearts. We found an increased expression level of all 4 miRNAs in non-myocytes, suggesting a possible role in cardiac non-myocyte mediated signalling (Supp Fig. 1). Next, using Ingenuity Pathway Analysis (IPA, Qiagen), we analyzed the putative targets of each miRNA (predicted by TargetScan Mouse Version 7.2 (https://www.targetscan.org/mmu_72/) to identify the potential molecular pathways regulated by these miRNAs. Interestingly, target pathway analysis of miR-199a-3p was most notable, as IPA analysis revealed TGFβ1 as the top upstream regulator (Suppl Table 4A) and Rac/actin cytoskeletal signalling as two of the top canonical pathways of predicted targets (Suppl Table 4B). As ECM protein deposition associated with fibrosis is tightly regulated by actin-cytoskeletal signalling in response to external cues such as TGFβ1 [25], we focused our studies on the role of miR-199a-3p in cardiac fibroblasts.
2.2. miR-199a-3p overexpression induces differential ECM protein and gene expression
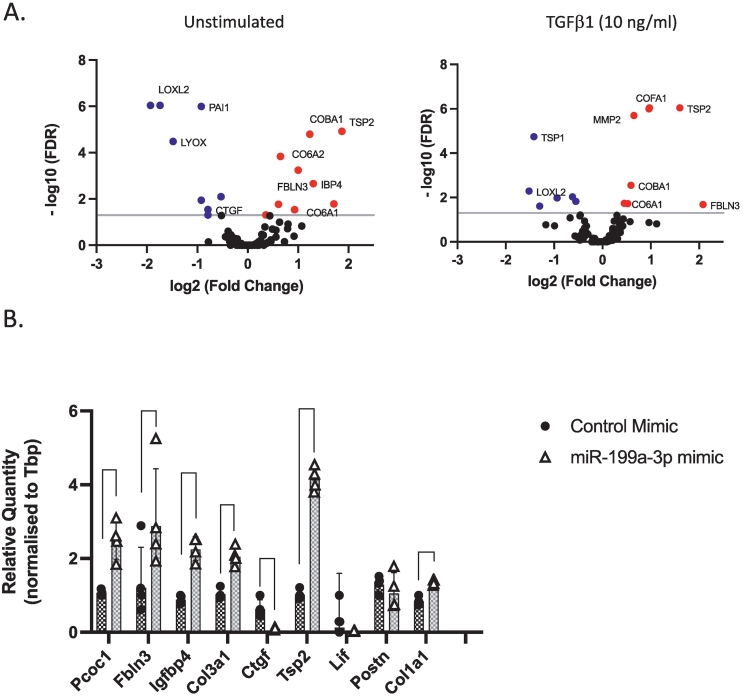
To explore whether miR-199a-3p may regulate ECM protein deposition, which is one of the hallmarks of tissue fibrosis, we took an unbiased proteomics approach. We transfected the primary cardiac fibroblasts with miR-199a-3p or control mimics and incubated the cells in either serum-free media supplemented with vehicle or TGFβ1 (10 ng/ml) for 48 h. We then processed the conditioned media for secretome analysis as previously described [26,27]. Our results show that in unstimulated miR-199a-3p mimic transfected cells, the levels of normalized spectral counts of several secreted ECM proteins, including Thrombospondin 2 (TSP2), Collagen 6a2 (COL6A2), Collagen 11a1 (COL11A1/COBA1), Fibulin 3 (FBLN3), Insulin growth factor binding protein 4 (IGFBP4/IBP4), Lysyl oxidase like 2 (LOXL2), Plasminogen activator inhibitor 1 (PAI1) and Lysyl oxidase (LYOX), are significantly altered (Fig. 2A, left graph and Supp Table 5). Additionally, in miR-199a-3p mimic transfected cells stimulated with TGFβ1, the levels of several ECM proteins including TSP2, matrix metalloproteinase 2 (MMP2), collagen 15a1 (COL15a1/COFA1), are significantly upregulated (Fig. 2A, right graph and Supp Table 5). Increased levels of Collagen1-alpha-1 (COL1A1) and Collagen 3-alpha-1 (COL3A1) are often associated with fibrosis and our results show that their levels are also slightly upregulated (but not statistically significant). We also assessed the protein expression levels of COL1A1 as measured by ELISA (Supp Fig. 2A) and COL3A1 as measured by Western blotting (Supp Fig. 2B) and found that the protein expression levels of both collagens are increased in miR-199a-3p mimic expressing cells stimulated with TGFβ1.
Fig. 2.
miR-199a-3p mimic expression regulates expression of ECM. (A) Adult murine cardiac fibroblasts were transfected with control or miR-199a-3p mimics and conditioned media was collected and processed for mass-spectrometric analysis. Volcano plots depicting proteomics analysis in basal unstimulated conditions (left graph) and in TGFβ1 stimulated conditions (right graph). Blue dots indicate significantly down-regulated proteins and red dots indicate significantly up-regulated proteins in miR-199a-3p mimic expressing cells as compared to control-mimic expressing cells (B) Validation of proteomics data was conducted by corresponding gene expression levels through quantitative PCR. Relative quantity of each gene was calculated using the 2-ΔΔCT method (Tbp used as a normalization control). Data are represented as geometric mean ± SD (n = 4 experiments). Statistics: Paired 2-tailed t-test with multiple comparisons-FDR, p value **p < 0.01, ***p < 0.001, ****p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To confirm our proteomics findings, we transfected cardiac fibroblasts with miR-199a-3p or control mimics and using qPCR, quantified mRNA expression level of Tsp2, Fbln3, Igfbp4, Col1a1 and Col3a1. Our results indicate that the expression levels of these ECM genes are all significantly upregulated (Fig. 2B). Altogether, our observations suggest that miR-199a-3p may regulate pathways involved in ECM gene and protein expression in cardiac fibroblasts.
2.3. Impact of miR-199a-3p inhibition in HCM mouse model
Guided by our findings in cultured cardiac fibroblasts, we sought to test the hypothesis that upregulation of miR-199a-3p in cardiac tissue may promote ECM protein deposition and have adverse impact in vivo during disease development. When we examined the expression level of miR-199a-3p in young heterozygous HCM mice hearts (4–8 weeks old, referred to as HCM_Young), our results revealed a significant upregulation as compared to that in young WT mice hearts (Supp Fig. 3A), suggesting that the expression of miR-199a-3p may be induced early on during HCM disease development. To assess whether miR-199a-3p has any causal role in promoting disease pathology, we evaluated whether the inhibition of miR-199a-3p activity in vivo can prevent cardiac hypertrophy and fibrosis in HCM mouse model. After body weight and echocardiographic measurements were collected at baseline, young HCM mice (4 weeks old) were injected subcutaneously with locked nucleic acid (LNA)-antimiR-199a-3p (N = 9) or control oligo (N = 10) (12.5 mg/kg) at days 0, 2 and 4, based on previous studies [28,29] and then monthly onwards until the end-point measurements were taken at 45–47 weeks old. Tissues were collected for gravimetric, RNA, protein, and histological analyses (Supp Fig. 3B). In parallel, we also collected cardiac tissues from untreated WT_Aged and HCM_Aged mice (>45 weeks old) to serve as controls. During the experiment, one mouse from LNA-control group and two mice from LNA-antimiR-199a-3p group had to be culled prematurely due to developing dermatitis (Supp Fig. 3C) and these mice were excluded from the study.
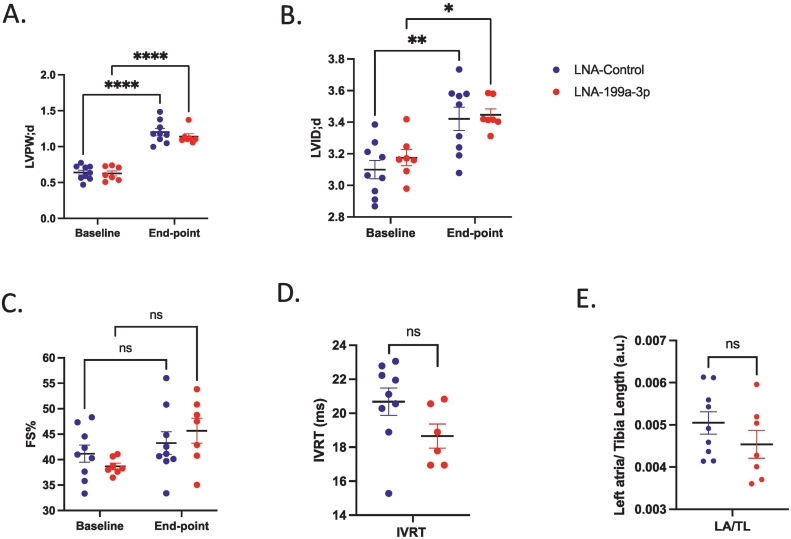
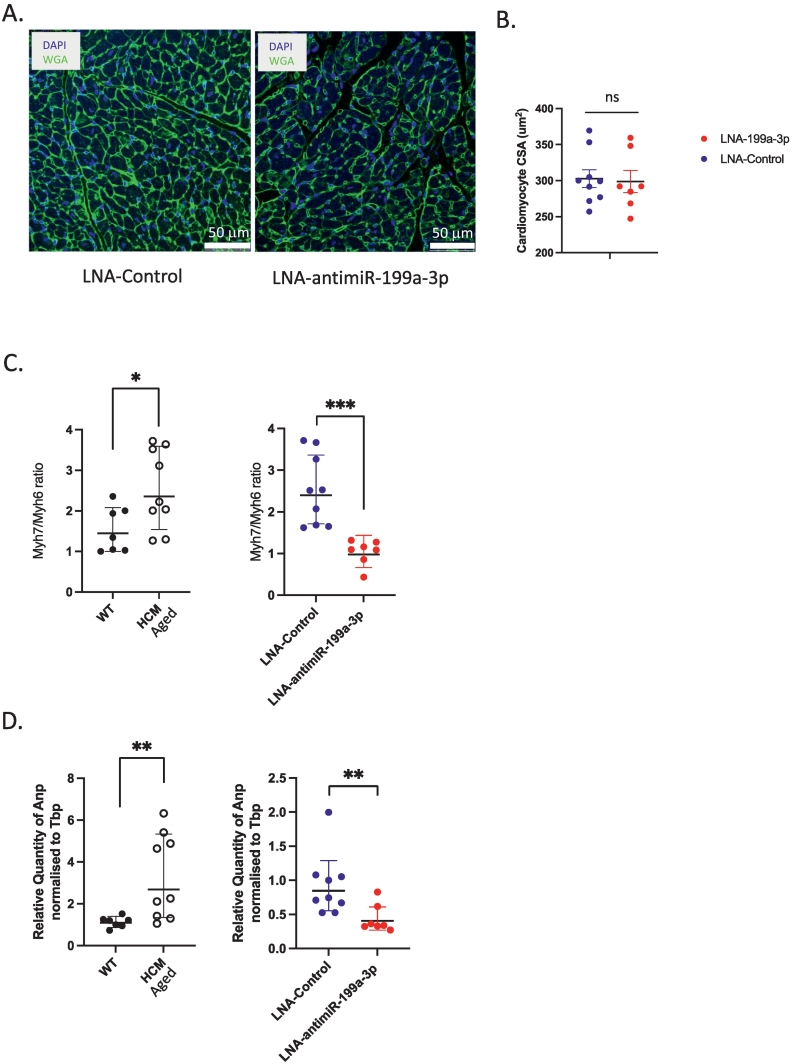
As previously reported [7,22,30] untreated HCM_Aged mice developed significantly increased diastolic left ventricular wall thickness (LVPW thickness;d) and fractional shortening (FS%) and a decreased diastolic left ventricular internal diameter (LVID; d) as compared to WT_Aged mice (Supp Fig. 4A, B, C). In comparison, there were no significant differences in end-point diastolic LVPW thickness (Fig. 3A), LVID (Fig. 3B) or FS% (Fig. 3C) between LNA-control and LNA-antimiR-199a-3p treated mice, despite significant inhibition of miR-199a-3p levels (91.8 % inhibition, p < 0.0001, Supp Fig. 3D). Furthermore, upon post-mortem gravimetric analysis, while left atrial weight/tibial length ratio was significantly increased, as expected [6] in untreated HCM_Aged mice compared to WT_Aged mice (Supp Fig. 4D), we did not observe any significant differences in left atrial weight/tibial length ratio between LNA-control and antimiR-199a-3p treated HCM mice (Fig. 3E). To determine if inhibition of miR-199a-3p affected diastolic function, isovolumetric relaxation time (IVRT) was measured by mitral Doppler echocardiography. Impairment of relaxation can be detected by a prolongation of the IVRT, which typically increases in diastolic dysfunction [31]. While there was a trend towards a decrease of IVRT in LNA antimiR-199a-3p treated mice compared to LNA control group, this was not statistically significant (Fig. 3D). In line with these results, we did not detect any significant differences in cross-sectional area of cardiomyocytes in heart sections from HCM mice treated with LNA-control or LNA antimiR-199a-3p (Fig. 4A, B). Altogether, these results indicate that miR-199a-3p does not contribute to cardiac hypertrophy associated with HCM, as assessed by echocardiographic and morphologic measurements.
Fig. 3.
Effects of in vivo miR-199a-3p inhibition on echocardiography and gravimetric parameters of HCM mice. HCM mice were treated with either LNA-control (n = 9, blue-filled circles or LNA-antimiR-199a-3p (n = 7, red-filled circles) (12.5 mg/kg monthly). (A) diastolic left ventricular posterior wall thickness (LVPW;d) (B) diastolic left ventricular internal dimension (LVID;d) and (C) fractional shortening (FS) % at baseline and end-point of the study. Data are expressed as mean ± SEM. Statistics: 2way ANOVA with Sidak's multiple comparisons test. P value: *p = 0.0457, **p = 0.0082, ****p < 0.0001. (D) Isovolumetric relaxation time (IVRT) at end-point. (E) Gravimetric analysis shows left atrial /tibia length ratio (LA/TL). Data are expressed as mean ± SEM, Statistics: Unpaired two-tailed t-test. P value p = ns (not significant). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Effects of in vivo miR-199a-3p inhibition on cardiomyocyte response in HCM mice. (A) Micrographs depict 5 μm paraffin-embedded heart sections from LNA-control and LNA-antimiR-199a-3p treated mice, stained with fluorescent-labelled wheat-germ agglutinin (WGA; green) and DAPI (blue) to demarcate cardiomyocyte borders and identify nuclei, respectively. (B) Graph shows quantification of cardiomyocyte cross-sectional area (CSA, um2) as measured by using NIH Image J image analysis software. Statistics: Unpaired two-tailed t-test. There is no statistical difference (ns) between LNA-control (n = 9, blue filled circles) or LNA-antimiR-199a-3p (n = 7, red filled circles). mRNA levels of cardiac stress markers (C) Myh6, Myh7 and (D) Anp were quantified by real time-quantitative PCR in left ventricular heart tissue obtained from aged untreated WT (n = 7, black-filled circles), age-matched HCM (n = 9, empty circles), and aged HCM mice treated with LNA-control (n = 9, blue filled circles) or LNA-antimiR-199a-3p (n = 7, red filled circles). Relative quantity of each gene was calculated using the 2-DDCT method (Tbp was used as a normalization control). Data are expressed as geometric mean ± SD. Statistics: Unpaired two-tailed t-test with Welch's correction. P value * p < 0.05, ** p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
During pathologic cardiac remodeling, in response to cardiomyocyte stress, expression of several fetal genes is induced leading to increased Myh7/Myh6 ratio (Fig. 4C, left graph) and Anp expression (Fig. 4D, left graph) in HCM_Aged mice hearts as compared to WT_Aged mice hearts. Interestingly, we found that the treatment of aging HCM mice with LNA-antimiR-199a-3p significantly reduced the Myh7/Myh6 ratio (Fig. 4C, right graph) and Anp mRNA expression (Fig. 4D, right graph). We also found that the levels of miR-199a-3p significantly correlated with the Myh7/Myh6 ratio (r = 0.75, p < 0.001) and Anp levels (r = 0.87, p < 0.0001). These results indicate that at a molecular level, miRNA-199a-3p inhibition significantly attenuates the activation of fetal gene program; however, such inhibition appears not to be sufficient to improve HCM-associated changes in cardiomyocyte morphology or function.
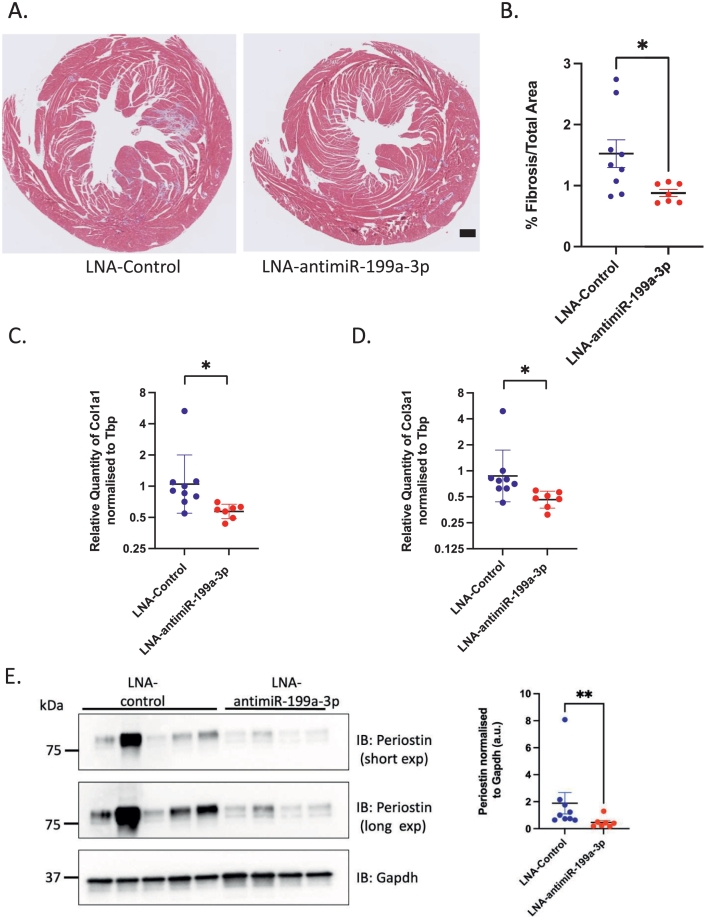
Next, we assessed whether the inhibition of miR-199a-3p activity in HCM mice had any effect on cardiac fibrosis. As previously reported for this mouse model [6,22], untreated HCM_Aged mice developed significant myocardial fibrosis as shown by Masson's Trichrome staining and quantification of fibrotic area percentage (Supp Fig. 5A, B), as well as increased fibrotic gene expression (Col1a1 and Col3a1) as measured by qPCR (Supp Fig. 5C and D). Furthermore, our results demonstrate that the treatment of aging HCM mice with LNA-antimiR-199a-3p significantly reduced the amount of cardiac fibrosis as compared to LNA-control treated mice (Fig. 5A, 5B, 1.53 % vs 0.88 %, p = 0.03). In support of these observations, our results also show a small but significant decrease in Col1a1 and Col3a1 mRNA levels in hearts of HCM mice treated with LNA-antimiR-199a-3p (Fig. 5C, D) as compared to control mice. Moreover, the levels of miR-199a-3p significantly correlated with Col1a1 (r = 0.88, p < 0.0001) and Col3a1 levels (r = 0.88, p < 0.0001). We have previously shown that pro-fibrotic protein periostin (POSTN) is upregulated in HCM mice hearts [7]. Thus, we assessed the expression levels of periostin and found that periostin expression is decreased after LNA-antimiR-199a-3p treatment of HCM mice, as compared to control-treated HCM mice (Fig. 5E). Altogether, our results suggest that miR-199a-3p may promote fibrosis by modulating the expression of pro-fibrotic genes and upregulating ECM protein expression in HCM mouse hearts.
Fig. 5.
Effects of in vivo miR-199a-3p inhibition on cardiac remodeling in HCM mice. (A) Micrographs depict 5 μm paraffin-embedded heart sections from LNA-control and LNA-antimiR-199a-3p treated mice stained with Masson's Trichrome (MT). Blue staining depicts collagen deposition areas. Scale bar = 0.5 mm (B) Graph shows quantification of fibrosis in heart sections using NIH Image J Software in LNA-control (blue-filled circles) and LNA-antimiR-199a-3p (red-filled circles) treated mice hearts. Data are expressed as mean ± SEM. Statistics: Unpaired 2-tailed t-test with Welch's correction, *p-value <0.05. mRNA levels of collagens (C) Col1a1 and (D) Col3a1 were quantified by real time-quantitative PCR in left ventricular heart tissue obtained from LNA-control (n = 9, blue filled circles) or LNA-antimiR-199a-3p treated HCM mice (n = 7, red filled circles). Relative quantity of each gene was calculated using the 2-DDCT method (Tbp was used as a normalization control). Data are expressed as geometric mean ± SD. Statistics: Unpaired 2-tailed t-test with Welch's correction. P value * p < 0.05 (E) Representative immunoblots show Periostin and Gapdh protein expression (left) and densitometric analysis of Periostin normalized to Gapdh levels (LNA-control n = 9 or LNA-antimiR-199a-3p treated n = 7 HCM mice). Data are presented mean ± SEM. Statistics: Unpaired Mann-Whitney test. P value ** p = 0.005. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. miR-199a-3p targets Cd151/Itga3 and inhibits AKT signalling in cardiac fibroblasts
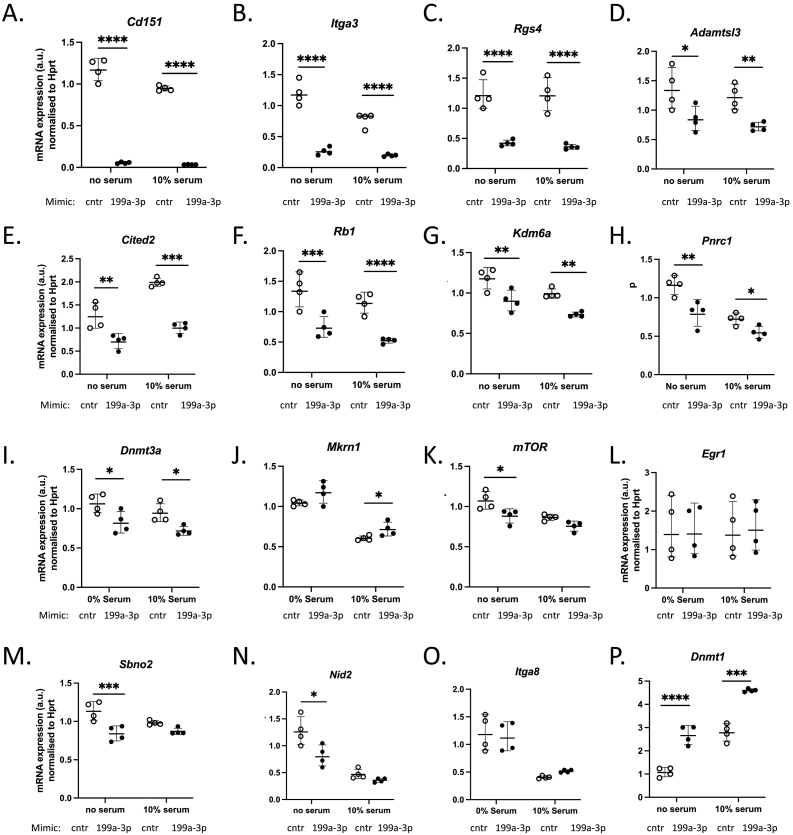
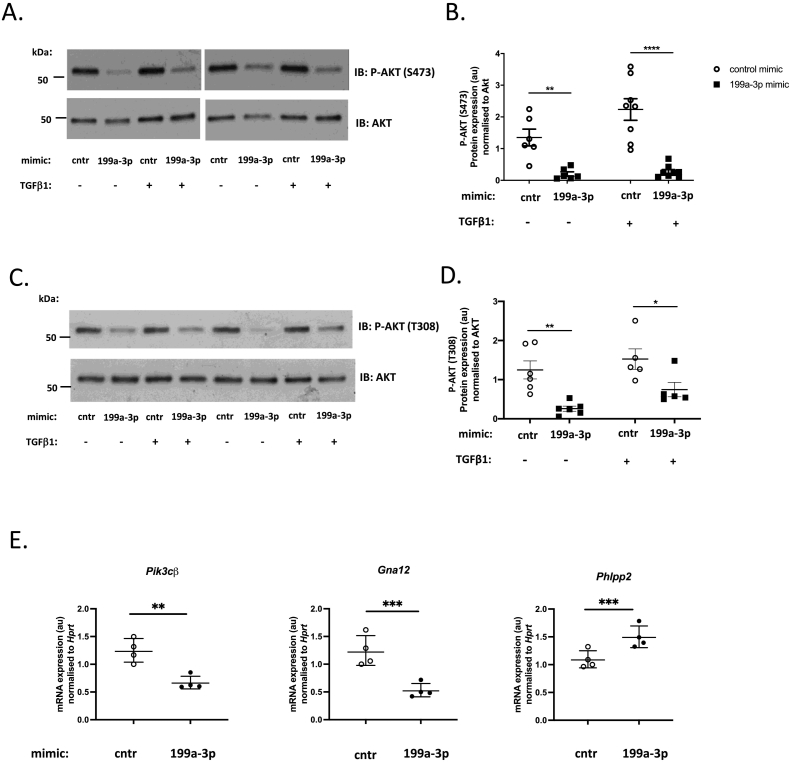
To identify the relevant miR-199a-3p target(s) which may be involved in regulation of ECM gene and protein expression and cardiac fibrosis during HCM development, we used the miRNA target prediction algorithm, TargetScan [32,105]. Since there are hundreds of potential targets, we chose to assess the predicted mRNAs based on their molecular functions and selected those with wider downstream effects, such as cell surface receptors, transcription factors and regulators. Our qPCR analysis identified several targets which were significantly downregulated in cells expressing miR-199a-3p mimic as compared to control cells (Fig. 6A-O). Among the most significantly down-regulated mRNA targets, we identified Cd151, which encodes a transmembrane protein belonging to the tetraspanin superfamily (Fig. 6A) and Itga3, which encodes a laminin binding integrin a3 (Fig. 6B). These genes have previously been reported as the most significant targets of miR-199a-3p in NIH3T3 cells [33]. Interestingly, CD151 and ITGA3 proteins are known to associate together to regulate both ECM remodeling and downstream outside-inside signalling pathways [[34], [35], [36]]. To determine the impact of inhibition of Cd151 and Itga3 expression, we interrogated the known downstream pathways. While MAPK ERK 1/2 activation is not affected (data not shown), we observed a significant attenuation of AKT signalling, as shown by reduction in AKT Ser473 (Fig. 7A, B) and Thr308 (Fig. 7C, D) phosphorylation levels, in cells overexpressing miR-199a-3p as compared to control cells cultured with or without TGFβ1. We have also investigated additional targets that may be involved in down-regulation of AKT activation and identified several upstream AKT regulators: Pik3cβ and Gna12 as direct targets and Phlpp2 as an indirect target of miR-199a-3p (Fig. 7E). Taken together, these results suggest that through targeting of Cd151 and Itga3 (and additional AKT upstream regulators), miR-199a-3p may regulate PI3K/AKT pathway in cardiac fibroblasts.
Fig. 6.
Identification of direct targets of miR-199a-3p in cultured adult mouse cardiac fibroblasts. mRNA levels of computationally predicted miR-199a-3p targets were quantified by real time-quantitative PCR in cardiac fibroblasts, cultured in serum-free media or 10 % FBS/DMEM, transfected with either control mimic (cntr) or miR-199a-3p mimic (199a-3p). (A) Cd151, (B) Itga3, (C) Rgs4, (D) Adamtsl3, (E) Cited2, (F) Rb1 (G) Kdm6a (H) Pnrc1 (I) Dnmt3a (J) Mkrn1 (K) mTor (L) Egr1, (M) Sbno2, (N) Nid2, (O) Itga8 (P) Dnmt1. Relative quantity of each gene was calculated using the 2-DDCT method (Hprt was used as a normalization control, n = 4 per group). Data are presented geometric mean ± SD. Statistical analysis was performed by two-way ANOVA with Sidak's multiple comparisons test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Fig. 7.
miR-199a-3p mimic inhibits AKT phosphorylation in cardiac fibroblasts. Cardiac fibroblasts were transfected with control mimic (cntr) or miR-199a-3p mimic (199a-3p) and cultured with or without TGFβ1 (10 ng/ml) for 48 h. (A) Representative immunoblots show phospho-AKT (Ser473) (upper panels) and total AKT (lower panels) (B) Densitometric analysis of phospho-Akt (Ser473) normalized to total Akt (n = 8 per group). Data are presented as mean ± SEM. Statistical analysis was performed by two-way ANOVA with Sidak's multiple comparisons test, ** p < 0.01, **** p < 0.0001. (C) Representative immunoblots show phospho-AKT(Thr308) (upper panel) and total AKT (lower panel) (D) Densitometric analysis of phospho-AKT (Thr308) normalized to total AKT (n = 5–6 per group). Data is represented as mean ± SEM. Statistical analysis was performed by two-way ANOVA with Sidak's multiple comparisons test, * p < 0.05, ** p < 0.01 (E) mRNA expression levels of miR-199a-3p targets Pik3cβ, Gna12 and Phlpp2. Relative quantity of each gene was calculated using the 2-DDCT method (Hprt was used as a normalization control, n = 4 per group). Data are presented as geometric mean ± SD. Statistical analysis was performed by paired two-tailed t-test. *** p < 0.001, ** p < 0.01.
3. Discussion
In this study, we show that (1) the cardiac expression of miR-199a-3p is significantly upregulated in three different mouse models with HCM-causing mutations in sarcomeric protein-encoding genes; (2) overexpression of miR-199a-3p in cardiac fibroblasts modifies ECM RNA and protein expression; and (3) the inhibition of miR-199a-3p activity in vivo using a specific LNA-antimiR oligonucleotide attenuates cardiac fibrosis in an HCM mouse model without affecting cardiac myocyte hypertrophy.
Our results showing increased cardiac expression of miR-199a-3p in HCM mouse models complement previous evidence of miR-199a-3p upregulation in distinct mouse models of cardiac hypertrophy and heart failure [14,37,38] as well as in HCM patients' plasma and tissues [39,40]. While upregulation of miR-199a-3p may be a compensatory response to cardiac stress, our results indicate that miR-199a-3p regulates ECM gene and protein expression in cardiac fibroblasts and contributes to cardiac fibrotic remodeling in an HCM mouse model.
One of our initial observations was that expression levels of miR-199a-3p is significantly higher in non-myocytes as compared to myocytes isolated from adult mouse hearts, in agreement with studies reporting similar findings [41]. To determine whether increased miR-199a-3p expression in HCM hearts is due to non-myocytes, we quantified miR-199a-3p in freshly isolated cardiac non-myocytes and myocytes from aged HCM and WT hearts (Supp Fig. 6); however, we did not detect any significant differences in its expression. It is worth noting that due to highly variable nature of HCM phenotype, our small sample size (n = 3) is presumably not sufficient to draw conclusions and further work is needed. Alternatively, increased miR-199a-3p expression in HCM hearts maybe due to increased numbers of cardiac fibroblasts, given that miR-199a-3p (and miR-199a-5p) are reported to be highly expressed in mouse cardiac fibroblasts (second most abundant miRNAs following let-7 family) [42].
miR-199a is expressed in connective tissue of various organs [41,43] and its increased expression has been reported to accompany fibrosis in liver, lung and kidney [[44], [45], [46]], suggesting a possible association between fibrosis and miR-199a-3p. In this study, we found significant changes in the expression levels of several ECM RNAs and proteins in cardiac fibroblasts overexpressing miR-199a-3p as compared to control cells, suggesting a role in regulating ECM remodeling. Among the most significantly upregulated ECM genes/proteins, we identified angiogenic factor thrombospondin 2 (TSP2), which is involved in ECM reorganisation and endothelial cell proliferation [[47], [48], [49], [50]]. Notably, increased circulating TSP2 levels in heart failure patients is associated with a poor prognosis [[51], [52], [53]]. While Tsp2 mRNA is not a direct target of miR-199a-3p, it is reported to be negatively regulated by AKT1-mediated pathways [54] and in line with this, we found that overexpression of miR-199a-3p in cardiac fibroblasts significantly inhibited AKT activation (Fig. 7), which can presumably lead to Tsp2 upregulation.
To determine whether the upregulation of miR-199a-3p is causal to HCM pathology in the mouse model, we utilized LNA-based antimiRs to inhibit miRNA-199a-3p activity. Even though inhibition of miR-199a-3p activity did not have any significant impact on cardiac function parameters; interestingly, we found that the gene expression levels of cardiomyocyte stress markers (Anp, Myh7/Myh6 ratio) were significantly downregulated. Notably, our results reveal that the inhibition of miR-199a-3p activity significantly attenuated the gene expression levels of pro-fibrotic Col1a1 and Col3a1 and the development of cardiac fibrosis in aging HCM mice. Our observations are consistent with a recent study which reported that miR-199a-3p mimic expression increased the expression of Anp and Myh7 in neonatal mouse cardiomyocytes and Col1a1 and Col3a1 in cardiac fibroblasts, exacerbating fibrosis associated with Ang-II infusion in a mouse model [55]. Supporting these findings, miR-199a-3p has been proposed to regulate both hepatic [56] and renal fibrosis [57] in mouse models.
It is worth noting that LNA-antimiR-199a-3p treatment does not appear to prevent the development of cardiac fibrosis in more severe HCM cases, as significant fibrosis was observed in mice that had to be culled early (prior to study endpoint) due to the development of spontaneous dermatitis (n = 2 in LNA-antimiR-199a-3p group). While these mice were excluded from further analysis, we noted that one of the hearts displayed significant fibrosis despite being treated with LNA-antimiR-199a-3p (data not shown). These results suggest that the inhibition of miR-199a-3p is not sufficient to prevent the development of fibrosis in mice with severe HCM development. In fact, this is in line with our observations that inhibition of miR-199a-3p in an HCM mouse model, where Cyclosporin A (CsA) is used to accelerate disease in young HCM mice, had no effect on cardiac stress marker and fibrotic gene/protein expression (unpublished). Altogether, these results indicate that inhibition of miR-199a-3p is beneficial in mild-to-moderate disease development. Considering that miRNAs are fine tuners of gene expression, multiple miRNAs likely act in concert to modulate disease progression. Indeed, others have shown that simultaneous inhibition of miR-199a-5p and miR-214 has a stronger therapeutic impact in a pressure overload-induced heart failure model than the inhibition of individual miRNAs [37]. Future studies will be needed to determine whether concomitant inhibition of miR-199a-3p and other miRNAs, (e.g., miR-199a-5p and miR-214, which are all transcribed from the same pri-miRNA), could provide a greater therapeutic benefit.
To explore the pathways that miR-199a-3p may interact with to regulate ECM gene and protein expression, we utilized a miRNA target prediction database (TargetScan) and identified Cd151, Itga3, Pik3cβ and Gna12, which have all been reported to regulate AKT signalling [36,[58], [59], [60]] as direct-targets of miR-199a-3p. Moreover, we have shown that the miR-199a-3p overexpression inhibits AKT activation in cardiac fibroblasts. While further studies are needed to pinpoint the miR-199a-3p target(s) that regulate AKT signalling and ECM protein expression in cardiac fibroblasts, the potential targeting of CD151 is particularly interesting. CD151 has been reported to regulate ECM deposition through its association and/or regulation of MMPs [[61], [62], [63], [64]] and is involved in adhesion, invasion, angiogenesis and vascular permeability through the regulation of cytoskeletal reorganisation and activation of various signalling molecules including AKT, eNOS, RAC, CDC42 and RHO pathways [36,58,65]. CD151 overexpression has also been shown to promote vascularisation via the AKT pathway in a rat myocardial infarction model [66]. Given that AKT signalling is a well-known regulator of endothelial cell function and nitric oxide (NO) release [67] and has cardioprotective roles [[68], [69], [70], [71]], its inhibition by miR-199a-3p (through possible targeting of Cd151) has implications for vascular functions such as angiogenesis and capillary permeability. In line with this, in an in vitro model of diabetic retinopathy miR-199a-3p was shown to inhibit angiogenesis [72] and regulate NO bioavailability [73] via the PI3K/AKT pathway. Interestingly, microvascular dysfunction is often observed in HCM patients [74,75] and reduced capillary density has also been proposed as an underlying factor in disease progression into end-stage heart failure in HCM [76]. Increased capillary permeability in the heart has recently been shown to induce diastolic dysfunction independent of fibrosis or cardiomyocyte dysfunction in a mouse model [77]. Future studies will be needed to determine whether the attenuation of fibrotic remodeling in the HCM mouse model upon inhibition of miR-199a-3p could be partially due to improvement of cardiac fibroblast and/or endothelial/vascular functions via AKT-mediated signalling.
Our results support the increasing evidence in literature that miR-199a (which is processed to give rise to miR-199a-3p and miR-199a-5p) has critical functions in cardiac biology. Circulating miR-199a-3p levels were found to be differentially expressed and investigated as a possible biomarker in various cardiac pathologies [78], with increased levels reported in coronary heart disease [79] and acute myocardial infarction [80], decreased levels correlated with increasing acuity of heart failure [81,82].
Studies have shown that cardiac specific expression of miR-199a plays a role in cardiomyocyte cell size regulation [83] and induces cardiac hypertrophy [38,84] and increases expression of fetal and profibrotic genes [84]. Similarly, both miR-199a-3p and miR-199a-5p mimics were found to increase AngII induced cardiac fibrosis, COL1A1, COL3A1, ANP and β-MHC levels in mouse myocardium [55]. Furthermore, induced expression of miR-199a in cardiomyocytes causes endothelial dysfunction via NO-dependent paracrine effects [85] and inhibition of miR-199a-3p or -5p were both associated with increased NO bioavailability, regulating angiogenesis and vascular tone [73].
While above studies point to a “pathologic” function of miR-199a (both -3p and -5p), others have shown “cardioprotective” roles. Induced expression of miR-199a-3p in cardiomyocytes has been reported to promote cardiomyocyte proliferation (via targeting HopX, Homer [86], Cd151 [87]) and cardiac regeneration [86] and provide cardiac repair in mouse [88] and pig models of myocardial infarction (MI) [89]. miR-199a was shown to enhance the potency of human IPS-cell-derived cardiomyocytes for myocardial repair [90], protect cardiomyocytes from simulated ischemia-reperfusion injury [91] and doxorubicin-induced cell death [92] and promote proliferation of mouse cardiac c-kit+ cells [93]. In addition, miR-199a-3p inhibitors promoted stem cell differentiation into cardiac myocytes [94] and it was shown that loss of long non-coding RNA CRRL (which acts as a sponge for miR-199a-3p) promotes cardiomyocyte regeneration and improves cardiac repair [95]. However, it is important to note that sustained expression of miR-199a-3p in myocytes lead to sudden arrhythmic death in treated pigs [89], indicating that expression of miR-199a-3p needs to be tightly regulated.
Our results that inhibition of miR-199a-3p attenuates fibrosis in HCM mice, without affecting cardiac function, may be due to its cell- and context-dependent effects as discussed above and our findings should be interpreted considering some limitations discussed below:
4. Limitations
-
(1)
One of the limitations of our study is the challenge of measuring the potency of antimiRs. Apparent inhibition of miR-199a-3p determined by qPCR at the end of the study (Supp Fig. 3D) may not represent the actual degree of inhibition in vivo. While qPCR assessment of miRNA levels has been widely used to measure the extent of inhibition [29,96,97], this method may also produce inconsistent results depending on the miRNA being measured [98]. An alternative approach is to measure the expression level of miRNA targets to assess the potency of miRNA inhibitor. However, the magnitude of derepression of the target maybe subtle (e.g. 1.2× fold) and distribution of mRNA targets can differ across cells types [99]. When we quantified mRNA expression of Hopx, a miR-199a-3p target identified in cardiomyocytes [86], we did not detect its derepression in LNA-antimiR-199a-3p treated mice hearts. We also tested the additional mRNA targets (Cd151, Itga3, Rgs4, Rb1 and Dnmt3a) we identified in cultured cardiac fibroblasts and did not detect any derepression. Given that there are often cell-type specific effects on mRNA targets, whole tissue RNA extracts used for quantification may have masked any possible differences. It is also possible that miRNA-199a-3p is involved in co-targeting networks with other miRNAs (e.g. miR-199a-5p [73]) and combined inhibition may be a better approach.
-
(2)
Another limitation is a smaller sample size of our in vivo study, and it is possible that we did not have sufficient statistical power to detect subtle differences in cardiac structure and function, given the subtle effects on cardiac fibrosis.
-
(3)
Last but not the least, our study did not evaluate the mechanisms by which miR-199a-3p may modulate fibrotic gene expression and cardiac fibrosis. While we used mimic expression approach in cultured cardiac fibroblasts to identify multiple targets including Cd151/Itga3 and AKT pathways, we recognize that this approach can be biased due to the high levels of miRNA. Therefore, further studies will be needed to elucidate the role, if any, of these targets in regulation of distinct fibrotic pathways and their relevance in vivo.
5. Conclusions
Our in vitro studies in cultured cardiac fibroblasts have shown that miR-199a-3p mimic expression induces perturbations in ECM gene/protein expression and inhibits AKT activation. Moreover, our in vivo studies have shown that the inhibition of miR-199a-3p activity by LNA-antimiR attenuates fibrosis associated with mild-to-moderate HCM, without impacting the development of hypertrophy. Altogether, our findings suggest that targeting of miR-199a-3p to limit fibrosis may offer a complementary therapeutic approach to newly emerging therapies that target disease-causing mutations in sarcomeric protein genes to treat HCM. Furthermore, as miR-199a-3p has been identified as a promising tool to induce cardiomyocyte proliferation [100] [86,88] when overexpressed in cardiac tissue, understanding its molecular targets and biological functions in other cardiac cells will be critical for further exploration of specific and safe therapeutic strategies.
6. Methods
6.1. Mouse models
Heterozygous knock-in HCM mice carrying human βMHC (MYH7) R719W or R403Q mutation in murine αMhc gene (αMHC403/+) and (αMHC719/+) recapitulate the hallmarks of human HCM and have been previously generated and characterized [6,7,22,23] by the Seidman Laboratory. Cyclosporin A-treated young mutant HCM mice have been shown to display accelerated HCM phenotype as reported previously [7,23] and referred to as HCM_CsA in this study. Preparation and administration of mouse chow containing CsA has been described previously [7]. HCM mice develop overt hypertrophy and fibrosis phenotype as they age (>35 weeks) and are referred to as HCM_Aged in this study. Homozygous mice carrying Mybpc3 truncation mutation (MyBPCt/t) have also been previously described [24]. In vivo experiments with LNA-antimiR-199a-3p have been carried out in heterozygous αMHC403/+ mice (referred to as HCM mice, unless indicated otherwise), which were gifted by the Seidman Laboratory and rederived by King's College Biological Services to ensure pathogen-free lineage. Female 129SVE-F mice used for re-derivation was obtained from Taconic (USA). Mice were housed in pathogen-free, individually ventilated cages with a 12-hour light-dark cycle, controlled humidity, and temperature (20–22C), fed standard mouse chow and water ad libitum. All mice were maintained and studied using protocols approved by the Animal Care and Use Committee of Harvard Medical School and in compliance with the Home Office regulations, as detailed in the Home Office Guidance on the operation of the Animals (Scientific Procedures) Act 1986. Only male heterozygous mice and gender-matched wild type littermates were used for the studies, as female heterozygous mice do not consistently develop cardiac hypertrophy or fibrosis [6].
6.2. miRNA library preparation and analysis
miRNA libraries were prepared from total RNA isolated from left ventricles of hypertrophic Cyclosporin A treated αMHC719/+ mice (referred to as HCM_CsA) and aged >35 weeks αMHC403/+ mice (referred to as HCM_Aged) as described in detail previously [101,102]. All sequencing data has been submitted to NCBI-Gene Expression Omnibus (GEO Accession numbers: GSE234772 (HCM_Aged) and GSE234773 (HCM_CsA).
6.3. Isolation, culture, and transfection of murine adult cardiac fibroblasts
Adult mouse cardiac fibroblasts were isolated from young 8–10 week old male C57BL6/J mice (Charles River, UK) by Collagenase II-based (Worthington Biochem, LS004176) serial digestion, as described in detail previously [26,27] Cells isolated from each individual mouse heart were treated as an individual biological replicate for the subsequent experiments and cultured separately. Isolated cells were first plated onto T25 culture flasks coated with 0.2 % gelatin (Sigma-Aldrich, cat G1393-100 ml) and cultured at 37 °C with 5 % CO2 until confluency and trypsinized and passaged onto larger gelatin-coated T75 culture flasks after 3–4 days (passage 1). At passage 2, cells were seeded at a density 10–12 × 104 cells/well into gelatin coated 6-well plates for transfection next day (at 60–70 % confluency). Cardiac fibroblasts were transfected with 50 nM final concentration of miRIDIAN miRNA control mimic (CN-001000-01-05, Dharmacon) or miR-199a-3p mimic (C-310455-07-0005, Dharmacon) with Lipofectamine RNAiMAX (InvitrogenTM cat. 13778150), according to manufacturer's instructions. Briefly, transfection mixes for miRNA mimic were prepared in Opti-MEM (reduced serum medium, Gibco cat. 31985070) and added to cells in fresh DMEM. Next day, the media was replaced with fresh media. For experiments with TGFβ-stimulation, cells were either cultured in DMEM alone or with DMEM supplemented with 10 ng/ml TGFβ1 (Peprotech) for 48 h before conditioned media and cell lysates were collected for proteomics and RNA/protein analysis, respectively. For studies to identify direct targets of miR-199a-3p, the cells were transfected and cultured in DMEM supplemented with 10 % Fetal Bovine Serum (FBS, Merck F7524-500ML) or no serum, as indicated, and cell lysates were collected 24 h post-transfection for downstream RNA and protein analyses. Adult cardiac non-myocytes were isolated, as previously described [7] and immediately processed for RNA isolation for the experiments described in Supp Fig. 1.
6.4. Preparation of miRNA mimics for in vitro studies
miRNA mimics, supplied in lyophilized form, were resuspended in 1x siRNA resuspension buffer (5x Buffer, Horizon, cat B-002000-UB-100) at a concentration of 50 μM and stored in single use aliquots at —80C freezer. For transfections, mimic stocks were diluted to 10 uM working concentration.
6.5. Secretome analysis
Conditioned medium collected from fibroblasts transfected with miR-199a-3p mimic and control mimic with or without TGFβ stimulation was processed for proteomics analysis as described in detail previously [27]. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD042904 and 10.6019/.
6.6. Preparation of cell and tissue lysates
Transfected cells were first washed with ice-cold phosphate buffered saline (PBS) and then lysed in Triton Lysis Buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4, 0.2 % Triton-X100, 2 mM sodium orthovanadate (NaV), 10 mM sodium fluoride (NaF), 1x EDTA-free protease inhibitor table (Roche)) or RIPA Lysis Buffer (150 mM NaCl, 50 mM Tris-HCL pH 7.4, 1 % Triton X-100, 5 mM EDTA, 2 mM sodium orthovanadate (NaV), 10 mM sodium fluoride (NaF), 1x EDTA-free protease inhibitor tablet (Roche), 0.5 % sodium deoxycholate, 0.1 % sodium dodecyl sulphate (SDS)). After incubation at 4 °C for 20 min on a rotating platform, cell lysates were centrifuged at 4 °C for 10 min at 14000 rpm to obtain the soluble fraction and protein concentration was quantified by Bradford assay (Bio-Rad) or BCA Assay. Lysates were diluted to the same concentration using lysis buffer, mixed with 4xLaemmli buffer (250 mM Tris-HCl pH 6.8, 40 % glycerol, 8 % SDS, 20 % β-mercaptoethanol (BME), 0.008 % bromophenol blue), boiled at 95 °C for 5 min and stored at -20 °C. Frozen ventricular heart tissues were processed and lysed, as described previously [104].
6.7. SDS-PAGE and Immunoblotting
Proteins were resolved by SDS-PAGE (4–20 % Mini-PROTEAN® 10 well 30 μl, cat. 4568093, Biorad) according to manufacturer's instructions. Precision plus protein dual colour standard (Bio-Rad, cat. 1610374) was loaded on each gel for estimation of protein size. Proteins were transferred onto 0.45 μm polyvinylidene difluoride (PVDF) membrane (Amersham, cat. 88518) by electrophoretic transfer in the Mini Trans-Blot® cell at 100 V for 60 min in transfer buffer (25 mM Tris, 192 uM glycine, 20 % (v/v) methanol). Following transfer, membranes were blocked with 5 % (w/v) of milk or bovine serum albumin (BSA) in TBS-T (20 mM Tris-HCl, pH 7.6, 137 uM NaCl, 0.1 % Tween) at room temperature for 1 h with gentle shaking. Membranes were incubated with primary antibodies overnight at 4 °C on a rotating platform. Antibody suppliers and dilutions used are listed in Supp Table 6. Next day membranes were washed 4 × 5 min in TBS-T and incubated at room temperature for 1 h with secondary antibody (anti-rabbit HRP, Cell Signalling 7074 1:2000; anti-mouse HRP, Dako Agilent P0447 1:2000) conjugated with horseradish peroxidase (HRP) in 5 % milk/TBST, followed by another 4 × 5 min washes in TBS-T. Proteins were detected using ECL Prime (GE Healthcare cat. 12316992) or SuperSignalTM West Femto (ThermoFisher cat. 34094) reagents, according to manufacturer's instructions. Chemiluminescence was captured on Amersham Hyperfilm™ (GE Healthcare, cat. 28-9068-37) and was visualized by processing film in the X-ray film processor (RG II Fuji). Protein band intensities were quantified by Quantity One software using GS-800 calibrated densitometer. Protein expression of each sample was normalized to the expression of endogenous Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or total AKT, as indicated. Where data from different membranes was combined to be analyzed as a group, protein expression was normalized to the sum of all data points within the same membrane, rather than to the chosen calibrator sample. For periostin immunoblots (Fig. 5E), chemiluminescence was captured on iBright Imaging System (Thermofisher) and densitometric analyses were performed using NIH Image J software.
6.8. Pro-collagen 1α1 enzyme-linked immunosorbent assay
Expression of pro-collagen 1α1 in cell lysates was quantified in duplicates with mouse pro-collagen 1α1 SimpleStep ELISA® kit (Abcam, ab210579), according to manufacturer's instructions. End-point optical density was measured on microplate reader (BioTek EL808) at wavelength 450 nm and concentration of pro-collagen 1a1 of the samples was calculated by interpolating against the standard curve.
6.9. Cell/Tissue Processing for RNA isolation
Cardiac fibroblasts were washed with ice-cold PBS and lysed with 700 μl QIAzol lysis reagent (Qiagen, cat. 79306). Snap-frozen mouse heart tissue (LV) samples were pulverized using a pre-chilled mortar and pestle before lysing in 700 μl of QIAzol. Samples were homogenized by processing in TissueLyser II for 2 × 2 min at 20 Hz using 5 mm stainless steel beads (Qiagen, cat. 69989). Total RNA was isolated with miRNeasy Mini miRNA isolation kit (Qiagen, 217004), according to manufacturer's instructions. Concentration of RNA was measured by NanoDrop™ 2000C Spectrophotometer (Thermo Scientific). Quality of isolated RNA was analyzed with Agilent RNA 6000 Nano kit on Agilent 2100 Bioanalyzer system to ensure RNA Integrity Numbers (RIN) were 8 or above.
6.10. Reverse transcription and qPCR
Total RNA was reverse transcribed using SuperScript IV (SSIV) reverse transcription (RT) kit by Invitrogen (ThermoFisher, cat. 18091050) for gene quantification or TaqMan miRNA Reverse Transcription (RT) kit (ThermoFisher cat. 4366596) for miRNA quantification, according to manufacturers' instructions. Primer sequences for quantitative polymerase chain reaction (qPCR) with SYBR Green assay were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (Supp Table 7) and UPL-probes and primer sequences were obtained from Roche (Supp Table 7). Gene expression was quantified by qPCR using Power SYBR mix (Applied Biosystems, ThermoFisher cat. 4367659) with 5 ng cDNA per reaction in 384-well plates in triplicates. The plates were run in ABI Prism 7900HT cycler (Applied Biosystems) with the following cycling conditions: 95 °C 10 min, (95 °C 15 s, 60 °C 1 min) x 40 cycles. For miRNA qPCR, reactions were prepared using TaqMan Universal PCR master mix II (Applied Biosystems, ThermoFisher cat. 4440043), and TaqMan miRNA assay (Supp Table 8 for list of assays) according to manufacturer's instructions with the following cycling conditions: 50 °C 2 min, 95 °C 10 min, (95 °C 15 s, 60 °C 1 min) X 40 cycles. qPCR reactions were set up in triplicates and run on a StepOnePlus Real-Time PCR system (ThermoFisher) or in HT7900 (ABI) or QuantiStudio 7 (ABI). The relative quantity (RQ) of the investigated genes or miRNAs was calculated using the 2 -△△CT method [103]. Hypoxanthine Guanine Phosphoribosyltransferase (Hprt), Peptidylprolyl Isomerase A (Ppia), TATA-Box Binding Protein (Tbp) and Glyceraldehyde-3- Phosphate Dehydrogenase (Gapdh) mouse genes were used as endogenous normalization controls, depending on the experimental setup. SnoRNA202 was used as an endogenous normalization control for miRNA quantification.
6.11. Preparation and administration of in vivo LNA antimiR inhibitors
LNA antimiR-199a-3p and LNA control inhibitors were custom-made and supplied in lyophilized form by Qiagen (199900_166641_Negative control: ACGTCTATACGCCCA; 199900_166640_hsa-miR-199a-3p: ATGTGCAGACTACTG) and resuspended in sterile phosphate buffered saline (PBS, ThermoFisher cat. 10010023) to a final concentration of 2.5 mg/ml and stored in −80 °C until use. 4-week-old HCM heterozygous male mice (αMHC403/+) were randomly assigned into LNA antimiR-199a-3p (n = 9) and LNA control (n = 10) groups. Injection volumes were calculated based on the weight of mice. LNA antimiR-199a-3p and LNA control inhibitors were administered by 3 subcutaneous injections (12.5 mg/kg/day) at monthly intervals until mice reached 45–47 weeks of age.
6.12. Echocardiography
VisualSonics Vevo 3100 imaging system (Scanhead transducer MX400, 30 MHz, resolution 50 um, cardiac mouse) was used to acquire echocardiography data.
During the procedure, anesthesia was maintained via nose cone inhalation of 1.5–2.0 % isoflurane and oxygen flow rate of 1 L/min. Heart rate of 400–500 beats per minute (bpm) and body temperature of 37.5 ± 0.5 °C, was maintained throughout. Eye lubricant was applied to each eye to prevent drying of the corneas. Fur was removed from the chest area using electric hair clipper and depilator cream (Veet sensitive) prior to applying pre-warmed ultrasound gel (Aquasonic 100) to the chest. Two-dimensional images (B-mode) and M-mode recordings were acquired in both parasternal long axis (PLAX) and short axis (SAX) views at the mid-ventricular (papillary muscle) level. ECG and respiration gating were used to minimize imaging artifacts. Echocardiography data was analyzed with FUJIFILM VisualSonics Vevo Lab (v2.1.0) software using standard cardiovascular packages. PLAX one-dimensional M-mode projection was used to measure end-diastolic and systolic interventricular septum (IVS) thickness, LV internal diameter (LVID) and LV posterior wall (LVPW) thickness. LV anterior wall (LVAW) thickness and LVPW were also measured in SAX one-dimensional M-mode projection. All measurements were averaged from 3 heart cycles in diastole and systole. Functional parameters including cardiac output, stroke volume, ejection fraction (EF) and fractional shortening (FS) were obtained from calculations based on LV M-mode tracing. All echocardiographic studies were blinded for both genotype and treatment during the measurement and analysis stages.
6.13. Dissection and gravimetric data collection
Mice were anaesthetized by inhalation of 3–4 % isoflurane and sacrificed by cervical dislocation. After dissecting, weights of whole heart, left atrium (LA) and ventricles were measured and the heart was transversely cut at mid-papillary level with the base processed for histology (fixed overnight at 4 °C in 4 % paraformaldehyde (PFA)) and the apex (for RNA and protein analysis) snap frozen in liquid N2 and stored at −80 °C. Tibia was also isolated as described previously [104] and used to normalize gravimetric data.
6.14. Masson's trichrome staining
Dissected heart tissue was fixed with 4 % PFA for 24 h at 4 °C, processed in the automated tissue processor (Leica TP1020) with successive alcohol/xylene incubations and embedded in paraffin. Tissues embedded in paraffin were sectioned into 5 μm sections (Leica RM 2125RTS) and mounted onto microscope slides (SuperFrost Plus, WVR cat. 631-0108). Masson's Trichrome (MT) kit (Sigma cat. HT15-1KT) was used to stain heart sections, according to manufacturer's instructions, and the sections were mounted with Eukitt quick-hardening medium (Sigma-Aldrich, cat 03989). The slides were scanned, and images were acquired with Hamamatsu NanoZoomer Scanner S360 (UCL-IQPath histology service) and the amount of fibrosis in sections was quantified with ImageJ software using colour-threshold method and expressed as percentage of total heart tissue area.
6.15. Myocyte cross-sectional area quantification
5 μm paraffin tissue sections were de-paraffinized, rehydrated and incubated with Alexa-488 conjugated wheat germ agglutinin (WGA-Alexa 488, 1:500, Thermofisher cat W11261) and DAPI (4′, 6-diamidino-2-phenylindole, Sigma-Aldrich, cat 10236276001, 1:500) diluted in TBS-T at room temperature for 1 h in a humidified chamber protected from light. After staining, slides were washed 3 × 5 min in TBS-T and were mounted with 10 μl of mounting medium (ProLong Gold Antifade, Invitrogen, Thermofisher P36935) according to manufacturer's instructions. Images of WGA-stained heart sections were acquired using 63 x objective of inverted confocal microscope (TCS SP5 system, Leica) and transverse-cut myocytes with central nuclei were traced manually using NIH ImageJ software. As a minimum 200 myocytes were analyzed for each experimental animal.
6.16. In silico miRNA target prediction
miRNA target prediction database TargetScanMouse 7.2 (https://www.targetscan.org/vert_72) [105] was used to search for computationally predicted targets of miR-199a-3p. A list of over 200 potential targets was closely examined, searches of scientific literature performed to identify the most relevant targets, which were shortlisted for experimental validation. STRING database (https://string-db.org/) [106] was used to establish connection between targets, e.g. those that belong to the same signalling pathway.
6.17. Statistics
GraphPad Prism 9.5.0 software or Microsoft Excel was used for statistical analysis of quantitative data. Statistics for each experiment is indicated in corresponding figure legend. Depending on experimental set up, data were analyzed by paired or unpaired 2-tailed t-test, 1way or 2way ANOVA, followed by Tukey's or Sidak's multiple comparison tests or unpaired Mann-Whitney test, as appropriate. Statistical analysis for gel-based MS data was performed as described previously [27]. For correlation analysis, Pearson r was calculated. Differences were considered significant when p-value < 0.05.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Prof James Clark, Dr. Hannah Lewis and Mr. Norman Catibog at King's College London for expert training and technical advice on echocardiography and Dr. Shiney Reji at King's College London for technical assistance throughout the project. SE is grateful to Professors Christine and Jon Seidman (Harvard Medical School) for supporting her generation of miRNA-Seq expression data at the start of this project and subsequently providing αMHC403/+ mice to her laboratory at King's College London, and Dr. Danos Christodolou (previously in the Seidman group) for support with computational analysis of miRNA-Seq expression data.
Disclosures
None.
Funding
This work was supported by a BHF Intermediate Fellowship (FS/16/21/31860) to S Eminaga and a BHF Interdisciplinary PhD studentship to T Barwari. Dr. Barallobre-Barreiro is a British Heart Foundation Intermediate Fellow (FS/19/33/34328). M Mayr is a British Heart Foundation (BHF) Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/16/14/32397, RG/F/21/110053).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmccpl.2023.100056.
Appendix A. Supplementary data
Supplementary material
References
- 1.Botvinick E.H., et al. Hypertrophic cardiomyopathy in the young: another form of ischemic cardiomyopathy? J Am Coll Cardiol. 1993;22(3):805–807. doi: 10.1016/0735-1097(93)90194-6. [DOI] [PubMed] [Google Scholar]
- 2.Maron B.J., et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 3.Semsarian C., et al. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65(12):1249–1254. doi: 10.1016/j.jacc.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Garfinkel A.C., Seidman J.G., Seidman C.E. Genetic pathogenesis of hypertrophic and dilated cardiomyopathy. Heart Fail Clin. 2018;14(2):139–146. doi: 10.1016/j.hfc.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marian A.J., Braunwald E. Hypertrophic cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res. 2017;121(7):749–770. doi: 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisterfer-Lowrance A.A., et al. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272(5262):731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 7.Teekakirikul P., et al. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010;120(10):3520–3529. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braga L., et al. Non-coding RNA therapeutics for cardiac regeneration. Cardiovasc Res. 2021;117(3):674–693. doi: 10.1093/cvr/cvaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barwari T., Joshi A., Mayr M. MicroRNAs in cardiovascular disease. J Am Coll Cardiol. 2016;68(23):2577–2584. doi: 10.1016/j.jacc.2016.09.945. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty C., et al. Therapeutic advances of miRNAs: a preclinical and clinical update. J Adv Res. 2021;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ntelios D., et al. A critical approach for successful use of circulating microRNAs as biomarkers in cardiovascular diseases: the case of hypertrophic cardiomyopathy. Heart Fail Rev. 2022;27(1):281–294. doi: 10.1007/s10741-021-10084-y. [DOI] [PubMed] [Google Scholar]
- 12.Luo F., Liu W., Bu H. MicroRNAs in hypertrophic cardiomyopathy: pathogenesis, diagnosis, treatment potential and roles as clinical biomarkers. Heart Fail Rev. 2022;27(6):2211–2221. doi: 10.1007/s10741-022-10231-z. [DOI] [PubMed] [Google Scholar]
- 13.Thottakara T., et al. A novel miRNA screen identifies miRNA-4454 as a candidate biomarker for ventricular fibrosis in patients with hypertrophic cardiomyopathy. Biomolecules. 2021;11(11) doi: 10.3390/biom11111718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagnall R.D., et al. Global microRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0044744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., et al. Differences in microRNA-29 and pro-fibrotic gene expression in mouse and human hypertrophic cardiomyopathy. Front Cardiovasc Med. 2019;6:170. doi: 10.3389/fcvm.2019.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vakrou S., et al. Allele-specific differences in transcriptome, miRNome, and mitochondrial function in two hypertrophic cardiomyopathy mouse models. JCI Insight. 2018;3(6) doi: 10.1172/jci.insight.94493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marian A.J. Contemporary treatment of hypertrophic cardiomyopathy. Tex Heart Inst J. 2009;36(3):194–204. [PMC free article] [PubMed] [Google Scholar]
- 18.Spertus J.A., et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): health status analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2021;397(10293):2467–2475. doi: 10.1016/S0140-6736(21)00763-7. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler M.T., et al. Effects of mavacamten on measures of cardiopulmonary exercise testing beyond peak oxygen consumption: a secondary analysis of the EXPLORER-HCM randomized trial. JAMA Cardiol. 2023;8(3):240–247. doi: 10.1001/jamacardio.2022.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichart D., et al. Efficient in vivo genome editing prevents hypertrophic cardiomyopathy in mice. Nat Med. 2023;29(2):412–421. doi: 10.1038/s41591-022-02190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chai A.C., et al. Base editing correction of hypertrophic cardiomyopathy in human cardiomyocytes and humanized mice. Nat Med. 2023;29(2):401–411. doi: 10.1038/s41591-022-02176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf C.M., et al. Somatic events modify hypertrophic cardiomyopathy pathology and link hypertrophy to arrhythmia. Proc Natl Acad Sci U S A. 2005;102(50):18123–18128. doi: 10.1073/pnas.0509145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fatkin D., et al. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106(11):1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell B.K., et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104(9):1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melchionna R., et al. Actin cytoskeleton and regulation of TGFbeta signaling: exploring their links. Biomolecules. 2021;11(2) doi: 10.3390/biom11020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abonnenc M., et al. Extracellular matrix secretion by cardiac fibroblasts: role of microRNA-29b and microRNA-30c. Circ Res. 2013;113(10):1138–1147. doi: 10.1161/CIRCRESAHA.113.302400. [DOI] [PubMed] [Google Scholar]
- 27.Barwari T., et al. Inhibition of profibrotic microRNA-21 affects platelets and their releasate. JCI Insight. 2018;3(21) doi: 10.1172/jci.insight.123335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patrick D.M., et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120(11):3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardo B.C., et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A. 2012;109(43):17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McConnell B.K., et al. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res. 2001;88(4):383–389. doi: 10.1161/01.res.88.4.383. [DOI] [PubMed] [Google Scholar]
- 31.Schnelle M., et al. Echocardiographic evaluation of diastolic function in mouse models of heart disease. J Mol Cell Cardiol. 2018;114:20–28. doi: 10.1016/j.yjmcc.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeary S.E., et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366(6472) doi: 10.1126/science.aav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werfel S., et al. Preferential microRNA targeting revealed by in vivo competitive binding and differential Argonaute immunoprecipitation. Nucleic Acids Res. 2017;45(17):10218–10228. doi: 10.1093/nar/gkx640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemler M.E. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 35.Kazarov A.R., et al. An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology. J Cell Biol. 2002;158(7):1299–1309. doi: 10.1083/jcb.200204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda Y., et al. Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood. 2007;109(4):1524–1532. doi: 10.1182/blood-2006-08-041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.el Azzouzi H., et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab. 2013;18(3):341–354. doi: 10.1016/j.cmet.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 38.van Rooij E., et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roncarati R., et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2014;63(9):920–927. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 40.Leptidis S., et al. A deep sequencing approach to uncover the miRNOME in the human heart. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0057800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider M., et al. Cell-specific detection of microRNA expression during cardiomyogenesis by combined in situ hybridization and immunohistochemistry. J Mol Histol. 2011;42(4):289–299. doi: 10.1007/s10735-011-9332-8. [DOI] [PubMed] [Google Scholar]
- 42.Ramanujam D., et al. MicroRNA-21-dependent macrophage-to-fibroblast signaling determines the cardiac response to pressure overload. Circulation. 2021;143(15):1513–1525. doi: 10.1161/CIRCULATIONAHA.120.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vacchi-Suzzi C., et al. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0052442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami Y., et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lino Cardenas C.L., et al. miR-199a-5p is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9(2) doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C.G., et al. Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology. 2012;142(5):1206–1217 e7. doi: 10.1053/j.gastro.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calabro N.E., Kristofik N.J., Kyriakides T.R. Thrombospondin-2 and extracellular matrix assembly. Biochim Biophys Acta. 2014;1840(8):2396–2402. doi: 10.1016/j.bbagen.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopes N., et al. Thrombospondin 2 regulates cell proliferation induced by Rac1 redox-dependent signaling. Mol Cell Biol. 2003;23(15):5401–5408. doi: 10.1128/MCB.23.15.5401-5408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong L.C., et al. Thrombospondin 2 inhibits microvascular endothelial cell proliferation by a caspase-independent mechanism. Mol Biol Cell. 2002;13(6):1893–1905. doi: 10.1091/mbc.E01-09-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krady M.M., et al. Thrombospondin-2 modulates extracellular matrix remodeling during physiological angiogenesis. Am J Pathol. 2008;173(3):879–891. doi: 10.2353/ajpath.2008.080128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanatani S., et al. Circulating thrombospondin-2 reflects disease severity and predicts outcome of heart failure with reduced ejection fraction. Circ J. 2014;78(4):903–910. doi: 10.1253/circj.cj-13-1221. [DOI] [PubMed] [Google Scholar]
- 52.Kimura Y., et al. High serum levels of thrombospondin-2 correlate with poor prognosis of patients with heart failure with preserved ejection fraction. Heart Vessel. 2016;31(1):52–59. doi: 10.1007/s00380-014-0571-y. [DOI] [PubMed] [Google Scholar]
- 53.Zhang K., et al. Role of thrombospondin1 and thrombospondin2 in cardiovascular diseases (review) Int J Mol Med. 2020;45(5):1275–1293. doi: 10.3892/ijmm.2020.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bancroft T., et al. Up-regulation of thrombospondin-2 in Akt1-null mice contributes to compromised tissue repair due to abnormalities in fibroblast function. J Biol Chem. 2015;290(1):409–422. doi: 10.1074/jbc.M114.618421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng N., et al. Diverging targets mediate the pathological roleof miR-199a-5p and miR-199a-3p by promoting cardiac hypertrophy and fibrosis. Mol Ther Nucleic Acids. 2021;26:1035–1050. doi: 10.1016/j.omtn.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X., et al. Twist1-induced miR-199a-3p promotes liver fibrosis by suppressing caveolin-2 and activating TGF-beta pathway. Signal Transduct Target Ther. 2020;5(1):75. doi: 10.1038/s41392-020-0169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang R., et al. p53 induces miR199a-3p to suppress SOCS7 for STAT3 activation and renal fibrosis in UUO. Sci Rep. 2017;7:43409. doi: 10.1038/srep43409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Detchokul S., et al. Tetraspanins as regulators of the tumour microenvironment: implications for metastasis and therapeutic strategies. Br J Pharmacol. 2014;171(24):5462–5490. doi: 10.1111/bph.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arcucci S., et al. Organismal roles for the PI3Kalpha and beta isoforms: their specificity, redundancy or cooperation is context-dependent. Biochem J. 2021;478(6):1199–1225. doi: 10.1042/BCJ20210004. [DOI] [PubMed] [Google Scholar]
- 60.Yoo E.J., et al. Galpha(12) facilitates shortening in human airway smooth muscle by modulating phosphoinositide 3-kinase-mediated activation in a RhoA-dependent manner. Br J Pharmacol. 2017;174(23):4383–4395. doi: 10.1111/bph.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanez-Mo M., et al. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112(8):3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 62.Yue S., Mu W., Zoller M. Tspan8 and CD151 promote metastasis by distinct mechanisms. Eur J Cancer. 2013;49(13):2934–2948. doi: 10.1016/j.ejca.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 63.Shi G.M., et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology. 2010;52(1):183–196. doi: 10.1002/hep.23661. [DOI] [PubMed] [Google Scholar]
- 64.Devbhandari R.P., et al. Profiling of the tetraspanin CD151 web and conspiracy of CD151/integrin beta1 complex in the progression of hepatocellular carcinoma. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang F., et al. Tetraspanin CD151 maintains vascular stability by balancing the forces of cell adhesion and cytoskeletal tension. Blood. 2011;118(15):4274–4284. doi: 10.1182/blood-2011-03-339531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Z., Liu Z. CD151 gene delivery activates PI3K/Akt pathway and promotes neovascularization after myocardial infarction in rats. Mol Med. 2006;12(9–10):214–220. doi: 10.2119/2006-00037.Zheng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ackah E., et al. Akt1/protein kinase Balpha is critical for ischemic and VEGF-mediated angiogenesis. J Clin Invest. 2005;115(8):2119–2127. doi: 10.1172/JCI24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villarreal F., et al. Regulation of cardiac fibroblast collagen synthesis by adenosine: roles for Epac and PI3K. Am J Phys Cell Phys. 2009;296(5):C1178–C1184. doi: 10.1152/ajpcell.00291.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., et al. Calycosin as a novel PI3K activator reduces inflammation and fibrosis in heart failure through AKT-IKK/STAT3 axis. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.828061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee M.Y., et al. Endothelial Akt1 mediates angiogenesis by phosphorylating multiple angiogenic substrates. Proc Natl Acad Sci U S A. 2014;111(35):12865–12870. doi: 10.1073/pnas.1408472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su F., et al. Cardioprotection by PI3K-mediated signaling is required for anti-arrhythmia and myocardial repair in response to ischemic preconditioning in infarcted pig hearts. Lab Investig. 2015;95(8):860–871. doi: 10.1038/labinvest.2015.64. [DOI] [PubMed] [Google Scholar]
- 72.Wang L., Liu W.X., Huang X.G. MicroRNA-199a-3p inhibits angiogenesis by targeting the VEGF/PI3K/AKT signalling pathway in an in vitro model of diabetic retinopathy. Exp Mol Pathol. 2020;116 doi: 10.1016/j.yexmp.2020.104488. [DOI] [PubMed] [Google Scholar]
- 73.Joris V., et al. MicroRNA-199a-3p and MicroRNA-199a-5p take part to a redundant network of regulation of the NOS (NO synthase)/NO pathway in the endothelium. Arterioscler Thromb Vasc Biol. 2018;38(10):2345–2357. doi: 10.1161/ATVBAHA.118.311145. [DOI] [PubMed] [Google Scholar]
- 74.Cecchi F., et al. Coronary microvascular dysfunction and prognosis in hypertrophic cardiomyopathy. N Engl J Med. 2003;349(11):1027–1035. doi: 10.1056/NEJMoa025050. [DOI] [PubMed] [Google Scholar]
- 75.Petersen S.E., et al. Evidence for microvascular dysfunction in hypertrophic cardiomyopathy: new insights from multiparametric magnetic resonance imaging. Circulation. 2007;115(18):2418–2425. doi: 10.1161/CIRCULATIONAHA.106.657023. [DOI] [PubMed] [Google Scholar]
- 76.Nijenkamp L., et al. Sex-specific cardiac remodeling in early and advanced stages of hypertrophic cardiomyopathy. PLoS One. 2020;15(5) doi: 10.1371/journal.pone.0232427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abelanet A., et al. Increased capillary permeability in heart induces diastolic dysfunction independently of inflammation, fibrosis, or cardiomyocyte dysfunction. Arterioscler Thromb Vasc Biol. 2022;42(6):745–763. doi: 10.1161/ATVBAHA.121.317319. [DOI] [PubMed] [Google Scholar]
- 78.Wronska A. The role of microRNA in the development, diagnosis, and treatment of cardiovascular disease: recent developments. J Pharmacol Exp Ther. 2023;384(1):123–132. doi: 10.1124/jpet.121.001152. [DOI] [PubMed] [Google Scholar]
- 79.Su M., et al. Circulating microRNA profiles based on direct S-Poly(T)Plus assay for detection of coronary heart disease. J Cell Mol Med. 2020;24(11):5984–5997. doi: 10.1111/jcmm.15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong Z., et al. Expression profiling and bioinformatics analysis of circulating microRNAs in patients with acute myocardial infarction. J Clin Lab Anal. 2020;34(3) doi: 10.1002/jcla.23099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ovchinnikova E.S., et al. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail. 2016;18(4):414–423. doi: 10.1002/ejhf.332. [DOI] [PubMed] [Google Scholar]
- 82.Vegter E.L., et al. Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int J Cardiol. 2016;224:231–239. doi: 10.1016/j.ijcard.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 83.Song X.W., et al. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol. 2010;225(2):437–443. doi: 10.1002/jcp.22217. [DOI] [PubMed] [Google Scholar]
- 84.Li Z., et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017;24(7):1205–1213. doi: 10.1038/cdd.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haghikia A., et al. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J. 2011;32(10):1287–1297. doi: 10.1093/eurheartj/ehq369. [DOI] [PubMed] [Google Scholar]
- 86.Eulalio A., et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 87.Tao Y., et al. miR-199a-3p promotes cardiomyocyte proliferation by inhibiting Cd151 expression. Biochem Biophys Res Commun. 2019;516(1):28–36. doi: 10.1016/j.bbrc.2019.05.174. [DOI] [PubMed] [Google Scholar]
- 88.Lesizza P., et al. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ Res. 2017;120(8):1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 89.Gabisonia K., et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569(7756):418–422. doi: 10.1038/s41586-019-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bian W., et al. miR-199a overexpression enhances the potency of human induced-pluripotent stem-cell-derived cardiomyocytes for myocardial repair. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.673621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park K.M., et al. Carvedilol-responsive microRNAs, miR-199a-3p and -214 protect cardiomyocytes from simulated ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2016;311(2):H371–H383. doi: 10.1152/ajpheart.00807.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J.Y., et al. Mesenchymal stem cell-derived small extracellular vesicles protect cardiomyocytes from doxorubicin-induced cardiomyopathy by upregulating survivin expression via the miR-199a-3p-Akt-Sp1/p53 signaling pathway. Int J Mol Sci. 2021;22(13) doi: 10.3390/ijms22137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu J., Wang Y., Cui J., et al. miR199a-3p regulates P53 by targeting CABLES1 in mouse cardiac c-kit+ cells to promote proliferation and inhibit apoptosis through a negative feedback loop. Stem Cell Res Ther. 2017;8:127. doi: 10.1186/s13287-017-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen H.P., et al. MiR-199a-3p inhibition facilitates cardiomyocyte differentiation of embryonic stem cell through promotion of MEF2C. J Cell Physiol. 2019;234(12):23315–23325. doi: 10.1002/jcp.28899. [DOI] [PubMed] [Google Scholar]
- 95.Chen G., et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J Mol Cell Cardiol. 2018;122:152–164. doi: 10.1016/j.yjmcc.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 96.Montgomery R.L., et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elmen J., et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Androsavich J.R., et al. Polysome shift assay for direct measurement of miRNA inhibition by anti-miRNA drugs. Nucleic Acids Res. 2016;44(2) doi: 10.1093/nar/gkv893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clark P.M., et al. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep. 2014;4:5947. doi: 10.1038/srep05947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torrini C., et al. Common regulatory pathways mediate activity of MicroRNAs inducing cardiomyocyte proliferation. Cell Rep. 2019;27(9):2759–2771 e5. doi: 10.1016/j.celrep.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vigneault F., et al. High-throughput multiplex sequencing of miRNA. Curr Protoc Hum Genet. 2012 doi: 10.1002/0471142905.hg1112s73. (Chapter 11: p. Unit 11 12 1-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eminaga S., et al. Quantification of microRNA expression with next-generation sequencing. Curr Protoc Mol Biol. 2013 doi: 10.1002/0471142727.mb0417s103. (Chapter 4: p. Unit 4 17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 104.Lewis H.R., et al. Phosphorylation at serines 157 and 161 is necessary for preserving cardiac expression level and functions of Sarcomeric Z-disc protein telethonin. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.732020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Agarwal V., et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szklarczyk D., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material