Abstract
Background
Squamous cell carcinoma (SCC) is the most common oral malignancy, and somatic mutations in some driver genes have been implicated in SCC development. Clear cell SCC (CCSCC) is a rare histological variant of SCC, and various clear cell neoplasms must be considered in the differential diagnosis of CCSCC in the oral cavity. Based on a limited number of CCSCC cases reported in the oral cavity, CCSCC is considered an aggressive variant of SCC with a poor prognosis; however, its genetic characteristics remain unknown.
Methods
A maxillary gingival tumor in an 89-year-old female was described and investigated using immunohistochemical staining, special staining, fluorescence in situ hybridization, and next-generation sequencing (NGS) with a custom panel of driver genes, including those associated with SCC and clear cell neoplasm development.
Results
Histopathological examination revealed a proliferation of atypical epithelial cells with abundant clear cytoplasm and enlarged and centrally placed round nuclei. The tumor was exophytic with deep, penetrating proliferation. The atypical clear cells were continuous with the conventional SCC cells. Immunohistochemical analysis showed that the clear cells were positive for CK AE1/AE3 and CK5/6 and nuclear-positive for p63. In contrast, the clear cells were negative for αSMA, S100, HMB45, Melan-A, CD10, and p16. p53 immunoreactivity exhibited a wild-type expression pattern. Additionally, the clear cells were positive for periodic acid-Schiff (PAS) and negative for diastase-PAS, mucicarmine, and Alcian blue. Based on these results, the diagnosis of CCSCC was confirmed. Molecular analysis of the clear cells identified PIK3CA p.E542K (c.1624G>A) and HRAS p.G12A (c.35 G>C) somatic mutations classified as oncogenic. No pathogenic variants were identified in TP53, EWSR1, AKT1, PTEN, BRAF, KRAS, NRAS, RASA1, or MAML2.
Conclusions
We report a case of CCSCC of the oral cavity with PIK3CA and HRAS mutations. The identification of PIK3CA and/or HRAS mutations is rare in SCC; however, both mutations are important potential targets for antitumor therapy. A detailed analysis of gene mutations in CCSCC may lead to a better understanding of its biological behavior and an improved prognosis, as well as a differential diagnosis from other clear cell neoplasms.
Keywords: Squamous cell carcinoma, Clear cell squamous cell carcinoma, Oral tumor, PIK3CA, HRAS
Introduction
Squamous cell carcinoma (SCC) is the most common malignancy of the oral cavity [1]. Oral SCC (OSCC) is SCC that arises from the oral mucosal epithelium and has different histological subtypes, including basaloid, verrucous, spindle cell, papillary, adenosquamous, acantholytic, and caniculatum variants [1]. Clear cell SCC (CCSCC) is a rare histological variant of SCC and is characterized by the presence of abundant clear cytoplasm [2]. Kuo first described CCSCC of the skin [3], and Frazier et al. reported CCSCC of the oral cavity [4]. Clear cell change occurs extremely rarely in mucosal SCC, and only 12 cases of CCSCC developed in the oral cavity have been reported to date, including the present case [4–16] (Table 1). Based on a limited number of CCSCC cases reported in the oral cavity, CCSCC has been purported to be an aggressive variant of SCC that has a poor prognosis [9, 11–13] (Table 1).
Table 1.
Clinical characteristics of clear cell squamous cell carcinoma cases in the oral cavity origin
| Case | Author | Age/sex | Location | Recurrent | Metastasis | Follow-up |
|---|---|---|---|---|---|---|
| 1 | Frazier et al. [4] | 59/F | Mandibular gingiva | N/A | N/A | Lost |
| 2 | Kumar et al. [7] | 70/F | Anterior maxilla and right mandibular (2 sites) | N/A | LN | Died within 2 months |
| 3 | Nainani et al. [8] | 52/M | Buccal mucosa | N/A | LN | Died within 3 months |
| 4 | Kaliamoorthy et al. [9] | 35/F | Lateral tongue and lingual vestibule | N/A | No | N/A |
| 5 | Khoury et al. [10] | 66/F | Tongue to the floor of the mouth | N/A | Lung (3 months later) | N/A |
| 6 | Devi et al. [11] | 55/M | Maxillary alveolar ridge | N/A | LN | Alive 5 months |
| 7 | Katoti et al. [12] | 59/M | Upper jaw | N/A | N/A | N/A |
| 8 | Ramani et al. [13] | 42/F | Mandibular alveolar mucosa | + (6 months later) | N/A | Lost |
| 9 | Hasegawa et al. [14] | 70/M | Tongue | + | LN and Lung (3 months later) | N/A |
| 10 | Mukkanwar et al. [15] | 60/M | Posterolateral border of the tongue | N/A | N/A | Lost |
| 11 | Mahamad Apandi et al. [16] | 65/M | Floor of the mouth | + (26 months later) | Lung (34 months later), LN (38 months later) | N/A |
| 12 | Present case | 89/F | Maxillary alveolar ridge | + (3 months later) | LN (8 months later) | Died within 8 months |
F female, M male, + positive, − negative, N/A data not available, LN lymph node
Activation of phosphatidylinositol 3-kinase (PI3K)/AKT and RAS/RAF signaling pathways, which regulate cell proliferation and growth, apoptosis, autophagy, invasion, and migration, is observed in various malignancies, including SCC [17, 18]. Mutations in the genes involved in signaling pathways are closely related to cancer development and prognosis [17, 18]. Currently, efforts are focused on understanding the molecular and cellular consequences of these mutations and the opportunities for targeted therapies [18–21]. Detailed analysis of genetic mutations in pathways involving potential targets for antitumor therapy may lead to an improved prognosis for CCSCC. However, no reports identifying genetic mutations in CCSCC exist, and the genetic characteristics are still unknown. Moreover, various clear cell neoplasms may be found in the oral cavity, which must be considered in the differential diagnosis of CCSCC [6]. These clear cell neoplasms that are independent of SCC have well-defined genetic profiles that may help specify diagnoses in difficult cases (Table 2). Here, we report a case of CCSCC of the maxillary gingiva with PIK3CA and HRAS mutations and review the literature on CCSCC of the oral cavity.
Table 2.
Staining panel and molecular findings for the differential diagnosis of clear cell neoplasms in the oral cavity
| CK | p63 | SMA | S100 | Melan-A | CD10 | d-PAS | Molecular findings | |
|---|---|---|---|---|---|---|---|---|
| Clear MEC | + | + | − | − | − | − | + | CRTC1/3::MAML2 |
| Clear MEca | + | + | + | + | − | − | − |
EWSR1 rearrangement PLGA1 rearrangement |
| HCCC | + | + | − | − | − | − | − |
EWSR1::ATF1 EWSR1::CREM |
| CCOC | + | + | − | − | − | − | − | EWSR1::ATF1, EWSR1::CREM |
| Malignant melanoma | − | − | + | + | + | − | − | BRAF mutation |
| CCRCC | + | − | − | − | − | + | − | |
| Present case | + | + | − | − | − | − | − |
PIK3CA mutation HRAS mutation |
CK, cytokeratin; SMA, smooth muscle actin; d-PAS, diastase-periodic acid-Schiff; MEC, mucoepidermoid carcinoma; CRTC1/3, CREB regulated transcription coactivator 1/3; MAML2, mastermind-like transcriptional coactivator 2; MEca, myoepithelial carcinoma; EWSR1: Ewing sarcoma breakpoint region 1; PLGA1, pleomorphic adenoma gene 1; HCCC, hyalinizing clear cell carcinoma; ATF1, activating transcription factor 1; CREM, cAMP responsive element modulator; CCOC, clear cell odontogenic carcinoma; BRAF, B-Raf proto-oncogene, serine/threonine kinase; CCRCC, clear cell renal cell carcinoma; PIK3CA, phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha; HRAS, HRAS proto-oncogene, GTPase
Case Report
Clinical Summary
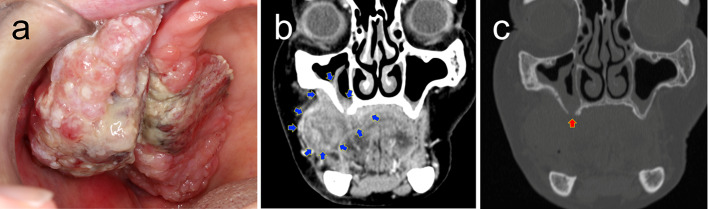
An 89-year-old female was referred to the Osaka University Dental Hospital for painful swelling in the upper right gingival region. The patient had noticed a gingival mass for 1 month and had no significant medical history. Intraoral examination revealed an approximately 60 × 40 mm lobulated mass with an ulcerative surface on the right maxillary posterior gingiva extending to the buccal mucosa (Fig. 1a). Computed tomography (CT) revealed an infiltrative lesion with maxillary bone resorption (Fig. 1b, c). With a provisional diagnosis of SCC, an incisional biopsy was performed (Fig. 2) followed by segmental maxillectomy (Fig. 3). The tumor recurred after 3 months, and the patient died 8 months after surgery due to complications related to disease recurrence (Table 1).
Fig. 1.
Clinical presentation. a Intraoral finding. Representative coronal computed tomography (CT) images with bone window (b) and with contrast-enhanced (c). The blue arrows indicate a large tumor extension (b). The red arrow indicates a bone penetration at the alveolar process of the right maxilla (c)
Fig. 2.
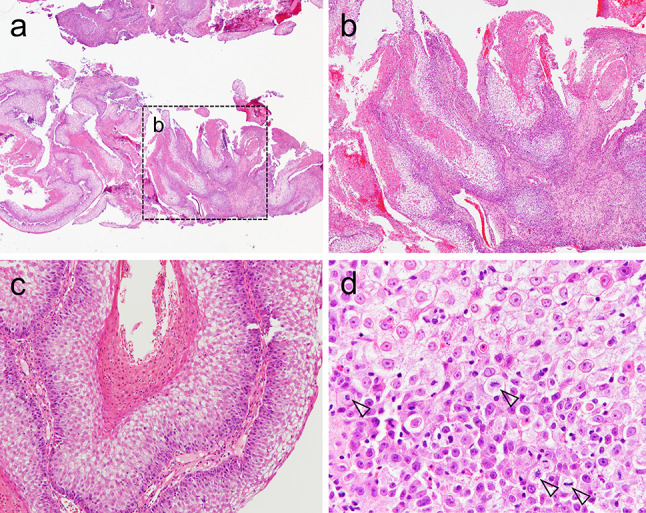
Histological findings of the biopsy. The exophytic tumor mass showed a penetrating growth pattern, resulting in several deep crypts filled with keratin debris (b was the black dotted-box area in a). c The tumor cells featured abundant clear cytoplasm, especially from the parabasal cells to the surface epithelium. d Clear cells exhibited enlarged and centrally placed round nuclei and nuclear and cellular atypia. Arrowheads indicate mitotic figures
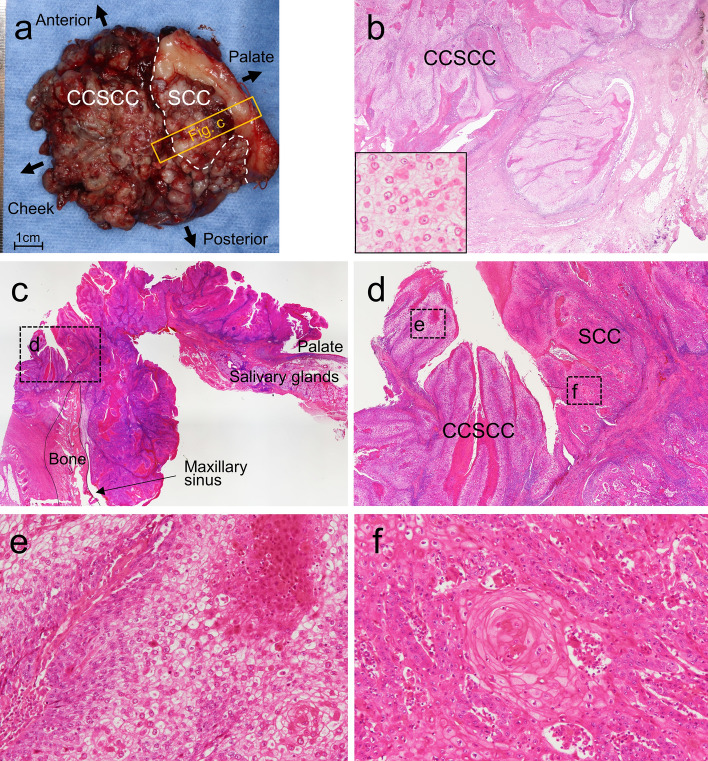
Fig. 3.
Histological findings of the surgical specimen. a Gross view of the surgical specimen. b Region of clear cell squamous cell carcinoma (CCSCC). CCSCC infiltrated deep connective tissue (inset: cytology of CCSCC). c, d Transitional area between CCSCC (left side) and conventional squamous cell carcinoma (SCC) (right side). c shows the orange lined-box area in a. d shows the black dotted-box area in c. e, f Region of CCSCC or SCC in the transitional area (both from the black dotted-box area in d)
Pathological Findings
Biopsy showed an infiltrative neoplasm (Fig. 2a). The tumor was primarily exophytic but with a pattern of deep, pushing invasion with keratin debris in crypt spaces (Fig. 2a, b). Tumor cells had prominent clear cytoplasm, especially from the parabasal cells to the surface epithelium (Fig. 2c). Clear cells exhibited enlarged, centrally placed, round nuclei (Fig. 2d). Nuclear and cellular atypia of tumor cells were observed (Fig. 2d). Based on these observations, a clear-cell variant of SCC was suspected, and subsequent segmental maxillectomy was performed (Fig. 3a).
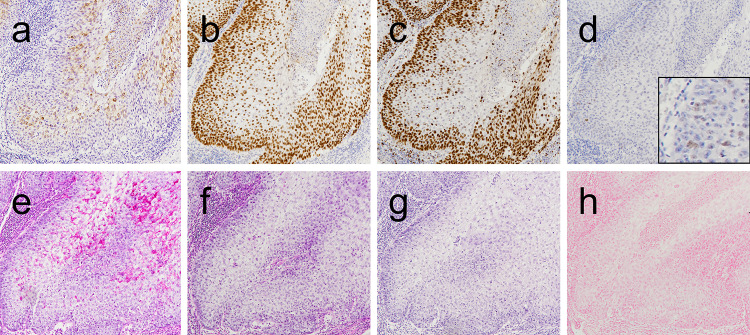
Histopathological examination of the surgical specimen affirmed biopsy findings. Additionally, sheets and islands of atypical clear cells infiltrated deep connective tissues (Fig. 3b). The clear cells were contiguous with conventional SCC cells connected to the normal epithelium (Fig. 3c–f), indicating the clear cells to be a component of the SCC. Clear cells accounted for a majority (80%) of tumor cells (Fig. 3a). Conventional SCC extended from the oral surface to the maxillary sinus with bone resorption (Fig. 3c). We did not perform immunohistochemical staining, special staining, or molecular analysis of the conventional SCC since the conventional SCC was observed only in formalin-fixed paraffin-embedded (FFPE) samples containing bone and decalcified by formic acid (Fig. 3c). Immunohistochemical analysis showed that the clear cells were positive for both CK AE1/AE3 (Fig. 4a) and CK5/6 mainly in the upper half of the epithelial layers, and were nuclear-positive for p63 in all epithelial layers (Fig. 4b). In contrast, the clear cells were negative for αSMA, S100, HMB45, Melan-A, CD10, and p16. The clear cells exhibited a p53 wild-type expression pattern (negative to weakly positive) (Fig. 4d). Immunohistochemical evaluation of Ki-67 revealed nuclear staining, mostly in the basal and parabasal cells of the atypical epithelium (Fig. 4c). Additionally, the cytoplasm of the clear cells was positive for periodic acid-Schiff (PAS) and negative for diastase-PAS, mucicarmine, and Alcian blue, suggesting accumulation of glycogen in the cytoplasm of the atypical clear cells (Fig. 4e–h).
Fig. 4.
Immunohistochemical and special stains. a CK AE1/AE3, b p63, c Ki-67, and d p53 immunostains. e Periodic acid-Schiff (PAS), f PAS with diastase, g mucicarmine, and h Alcian blue stains
Postoperative positron emission tomography (PET) CT was performed to assess distant metastasis. No signs of tumors were detected in other organs. Therefore, the final diagnosis of CCSCC of the maxillary gingiva was established.
Molecular Analysis
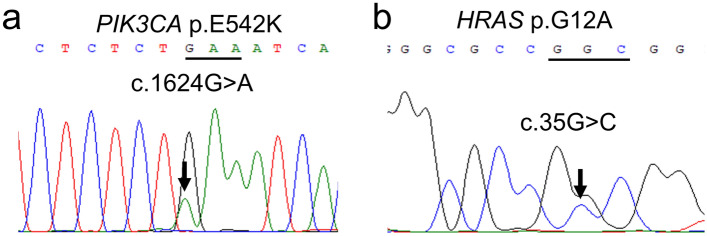
To further investigate the genetic profile of CCSCC, targeted next-generation sequencing (NGS) was performed using a custom panel as previously described [22]. The gene panel was designed using SureDesign (https://earray.chem.agilent.com/suredesign) to cover the whole EWSR1 gene (coverage 90.91%), and entire exons of TP53 gene or genes associated with the PI3K/AKT and RAS/RAF signaling pathways (PIK3CA, AKT1, PTEN, BRAF, KRAS, NRAS, HRAS, and RASA1). FFPE samples, in which tumor cells comprised approximately 60% of the total cells, were selected, and DNA was obtained from the sample. Polymerase chain reaction (PCR) assays and direct sequencing were performed to confirm gene mutations. Sequencing identified PIK3CA p.E542K (c.1624G>A) and HRAS p.G12A (c.35G>C) somatic mutations classified as oncogenic (Fig. 5a, b). No pathogenic variants were identified in TP53, EWSR1, AKT1, PTEN, BRAF, KRAS, NRAS, or RASA1. Moreover, MAML2 rearrangement was not detected by fluorescence in situ hybridization (Z-2014-50, ZyoVision, Bremerhaven, Germany) (data not shown).
Fig. 5.
Molecular analysis. Direct gene sequencing shows chromatograms for PIK3CA p.E542K (c.1624G>A) (a) and HRAS p.G12A (c.35G>C) (b)
Discussion
CCSCC of the oral cavity is rare, and its genetics relative to conventional SCC are unclear. To date, only 12 cases of CCSCC in the oral cavity have been reported, including our case [4, 7–16]. Previous reports have described that, histologically, CCSCC cells show abundant clear cytoplasm along with enlarged and centrally placed round nuclei [7, 9–11, 14]. It has been suggested that the proportion of clear cells required to define CCSCC is > 25% [2, 5]. Our case was consistent with the definition of CCSCC, both in the cytopathological findings and the proportion of clear cells in the lesion. Thus, our case is the first case of CCSCC, to our knowledge, in which a gene mutation has been described.
In the oral cavity, it is necessary to distinguish CCSCC from other tumors composed of clear cells: salivary gland carcinomas (clear cell variant mucoepidermoid carcinoma, clear cell myoepithelial carcinoma, and hyalinizing clear cell carcinoma [HCCC]), odontogenic carcinoma (clear cell odontogenic carcinoma [CCOC]), malignant melanoma, and metastatic carcinoma [6, 23] (Table 2). The lack of intracellular mucin, confirmed by d-PAS, mucicarmine, and Alcian blue staining results, excluded the diagnosis of mucoepidermoid carcinoma [6]. The lack of myoepithelial markers (such as SMA and S100) excluded clear-cell myoepithelial carcinoma [6]. The tumor location on the oral surface and the histology of squamous differentiation excluded HCCC and CCOC [6, 23]. The lack of S100, Melan-A, and HMB45 immunoreactivity excluded malignant melanoma [6, 23]. Metastatic tumors, such as clear cell renal cell carcinoma (CCRCC), were excluded because there were no signs of tumors in the other organs and CD10 immunoreactivity was absent [6]. These pathological and clinical findings led to the diagnosis of CCSCC in our case. Besides, our case of CCSCC was genetically different from other clear cell neoplasms in that MAML2 rearrangement (characteristic of mucoepidermoid carcinoma), EWSR1 rearrangement/translocation (characteristics of clear cell variants of myoepithelial carcinoma, HCCC, and CCOC), and BRAF mutations (detected in malignant melanoma and odontogenic tumors) were not detected (Table 2) [6, 23]. On the other hand, both PIK3CA (p.E542K) and HRAS (p.G12A) oncogenic mutations were detected, which are hardly detected in SCC (Fig. 5a, b) [1]. Thus, genetically, CCSCC may be an entity of SCC and distinct from other clear cell neoplasms in the oral cavity.
PIK3CA and RAS (KRAS, NRAS, and HRAS) mutations activate the PI3K/AKT and RAS/RAF pathways, respectively [17, 18]. Both pathways are critical drivers of tumorigenesis and potential targets for antitumor therapy [17–21]. Oncogenic mutations in PIK3CA and RAS have been identified in various malignancies, and both occasionally coexist [24]. However, in head and neck SCC (HNSCC), including OSCC, most genetic mutations are associated with tumor suppressor genes such as TP53, and genetic mutations in the PI3K/AKT or RAS/RAF pathways are rare [1, 25–32]. Kobayashi et al. reported that the most frequently mutated gene among 284 HNSCC cases was TP53 (67%), followed by PIK3CA (8%), AKT1 (4%), and HRAS (3%) [27]. Among HNSCC cases, only one had both PIK3CA and HRAS mutations [27]. In OSCC, the mutation frequency of PIK3CA ranges from 0 to 13.92% [30–32]. No significant correlation was found between PIK3CA mutations and survival rates in HNSCC and OSCC [27, 30–32]. Mutation frequencies of HRAS in OSCC range from 5 to 17.4% [30–32]. Carrying an HRAS mutation is considered a high-risk factor for poor prognosis and survival in HNSCC and OSCC [31–33]. HNSCC with HRAS mutations shows poor clinical outcomes with a high recurrence rate following primary definitive treatment (50–67% recurrence within 6 months), short disease-free survival (4.0 months; 95% CI 1.0 to 36.0), and overall survival (15.0 months; 95% CI 6.0 to 52.0) [33]. In this context, CCSCC recurred 3 months after primary resection in our patient, who had clear surgical margins at the time of resection, and the patient subsequently died 8 months later from complications related to tumor recurrence. Further studies are required to determine the association between HRAS mutations and poor prognoses in other CCSCC cases.
Several PI3K/AKT and RAS/RAF targeting agents are currently undergoing clinical trials, and molecular profiling of these targets needs to be investigated [17–21]. A recent study demonstrated that tipifarnib, a farnesyltransferase inhibitor that disrupts HRAS function, dramatically improved clinical outcomes in patients with HRAS-mutant HNSCC [21]. Moreover, PI3K inhibitors have demonstrated antiproliferative, pro-apoptotic, and antitumor activities in a range of preclinical cancer models as a single agent or in combination with other anticancer therapies [19, 20]. CCSCC of the oral cavity is considered an SCC variant [8, 11, 13–15]. Thus, the PI3K/AKT and RAS/RAF pathways may be important potential targets for future therapeutic options in patients with CCSCC.
Conclusion
In conclusion, we report a case of CCSCC in the oral cavity associated with PIK3CA and HRAS mutations, potential targets for antitumor therapy. A detailed analysis of gene mutations in CCSCC may lead to a better understanding of its biological behavior and an improved prognosis, as well as distinguish CCSCC from other clear cell neoplasms.
Abbreviations
- AKT1
AKT serine/threonine kinase 1
- BRAF
B-Raf proto-oncogene, serine/threonine kinase
- HCCC
Hyalinizing clear cell carcinoma
- CCOC
Clear cell odontogenic carcinoma
- CCRCC
Clear cell renal cell carcinoma
- CCSCC
Clear cell squamous cell carcinoma
- CT
Computed tomography
- EWSR1
Ewing sarcoma breakpoint region 1
- HNSCC
Head and neck squamous cell carcinoma
- HRAS
HRAS proto-oncogene, GTPase
- KRAS
KRAS proto-oncogene, GTPase
- MAML2
Mastermind-like transcriptional coactivator 2
- NGS
Next-generation sequencing
- NRAS
NRAS proto-oncogene, GTPase
- OSCC
Oral squamous cell carcinoma
- PAS
Periodic acid-Schiff
- PCR
Polymerase chain reaction
- PET-CT
Positron emission tomography-computed tomography
- PI3K
Phosphatidylinositol 3-kinase
- PIK3CA
Phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha
- PTEN
Phosphatase and tensin homolog
- RASA1
RAS P21 protein activator 1
- SCC
Squamous cell carcinoma
Author Contributions
All the authors contributed to this study. KH designed the study. KH, TS, AT, YU, SO, KO, YH, EM, and ST interpreted the H&E staining and immunohistochemical findings. KH and DM performed molecular analysis and assembled the data. KH, YH, and ST contributed to the manuscript’s writing. NU, YI, and SM reviewed clinical and radiological data. All authors have reviewed and approved the manuscript for submission.
Funding
Open access funding provided by Osaka University. The authors have no funding or financial relationships.
Data Availability
The surgical materials and datasets analyzed in the current study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
All authors state that they have no conflicts of interest.
Ethical Approval
This case report was approved by the Ethical Review Board of the Graduate School of Dentistry, Osaka University (No. R1-E46) and performed in accordance with committee guidelines and regulations.
Informed Consent
The requirement for informed consent was waived by the Ethical Review Board.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ, editors. World Health Organization classification of head and neck tumours. 4. Lyon: IARC; 2017. pp. 108–111. [Google Scholar]
- 2.Elder DE, Massi D, Scolyer RA, Willemze R, editors. World Health Organization classification of skin tumours. 4. Lyon: IARC; 2018. pp. 43–44. [Google Scholar]
- 3.Kuo T. Clear cell carcinoma of the skin. A variant of the squamous cell carcinoma that simulates sebaceous carcinoma. Am J Surg Pathol. 1980;4:573–583. doi: 10.1097/00000478-198012000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Frazier JJ, Sacks H, Freedman PD. Primary glycogen-rich clear cell squamous cell carcinoma of the mandibular gingiva. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114:e47–e51. doi: 10.1016/j.oooo.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Corbalán-Vélez R, Ruiz-Macia JA, Brufau C, López-Lozano JM, Martínez-Barba E, Carapeto FJ. Clear cells in cutaneous squamous cell carcinoma. Actas Dermosifiliogr. 2009;100:307–316. doi: 10.1016/S1578-2190(09)70068-X. [DOI] [PubMed] [Google Scholar]
- 6.Cipriani NA, Kakkar A. Top 10 clear cell head and neck lesions to contemplate. Head Neck Pathol. 2023;17:33–52. doi: 10.1007/s12105-022-01518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar K, Shetty DC, Wadhwan V, Gupta P. Synchronous oral squamous cell carcinomas with unusual histopathological feature. J Oral Maxillofac Pathol. 2012;16:420–424. doi: 10.4103/0973-029X.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nainani P, Singh HP, Paliwal A, Nagpal N. A rare case report of clear cell variant of oral squamous cell carcinoma. J Clin Diagn Res. 2014;8:QD07–QD09. doi: 10.7860/JCDR/2014/11536.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaliamoorthy S, Sethuraman V, Ramalingam SM, Arunkumar S. A rare case of clear cell variant of oral squamous cell carcinoma. J Nat Sci Biol Med. 2015;6:245–247. doi: 10.4103/0976-9668.149209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoury ZH, Bugshan A, Lubek JE, Papadimitriou JC, Basile JR, Younis RH. Glycogen-rich clear cell squamous cell carcinoma originating in the oral cavity. Head Neck Pathol. 2017;11:552–560. doi: 10.1007/s12105-017-0812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devi A, Kamboj M, Singh V, Singh S. Clear-cell variant of squamous cell carcinoma in maxilla as primary lesion: a rare case. J Oral Maxillofac Pathol. 2017;21:425–428. doi: 10.4103/jomfp.JOMFP_180_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakoti LM, Mahanta D, Sharma JD, Chowdhury Z. Clear-cell squamous cell carcinoma: an uncommon variant of very common malignancy in the head and neck. Int J Oral Health Sci. 2018;8:136–139. doi: 10.4103/ijohs.ijohs_23_18. [DOI] [Google Scholar]
- 13.Ramani P, Gheena S, Karunagaran M, Hannah R. Clear-cell variant of oral squamous cell carcinoma: a rare entity. J Oral Maxillofac Pathol. 2021;25:S22–S26. doi: 10.4103/jomfp.JOMFP_295_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa K, Fujii S, Kurppa KJ, Maehara T, Oobu K, Nakamura S, et al. Clear cell squamous cell carcinoma of the tongue exhibits characteristics as an undifferentiated squamous cell carcinoma. Pathol Res Pract. 2022;235:153909. doi: 10.1016/j.prp.2022.153909. [DOI] [PubMed] [Google Scholar]
- 15.Mukkanwar RN, Palaskar S, Pawar R, Shah DR. Clear cell variant of oral squamous cell carcinoma: case report and review. Autops Case Rep. 2022;12:e2021388. doi: 10.4322/acr.2021.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahamad Apandi NI, Ramanathan A, Ismail SM, Ranganathan K. Do clear cell changes in oral squamous cell carcinoma warrant it being recognised as a variant? Cureus. 2022;14:e25057. doi: 10.7759/cureus.25057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyssonnaux C, Eychène A. The Raf/MEK/ERK pathway: new concepts of activation. Biol Cell. 2001;93:53–62. doi: 10.1016/s0248-4900(01)01125-x. [DOI] [PubMed] [Google Scholar]
- 19.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 20.Marquard FE, Jücker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem Pharmacol. 2020;172:113729. doi: 10.1016/j.bcp.2019.113729. [DOI] [PubMed] [Google Scholar]
- 21.Ho AL, Brana I, Haddad R, Bauman J, Bible K, Oosting S, et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J Clin Oncol. 2021;39:1856–1864. doi: 10.1200/JCO.20.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori Y, Hirose K, Ozeki M, Hata K, Motooka D, Tahara S, et al. PIK3CA mutation correlates with mTOR pathway expression but not clinical and pathological features in Fibfibroipose vascular anomaly (FAVA) Diagn Pathol. 2022;17:19. doi: 10.1186/s13000-022-01199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose K, Usami Y, Kohara M, Sato S, Iwamoto Y, Murakami S, et al. Clear cell carcinoma of palatal minor salivary gland harboring a novel EWSR1-ATF1 fusion gene: report of a case and review of the literature. Head Neck Pathol. 2021;15:676–681. doi: 10.1007/s12105-020-01211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS ONE. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi K, Yoshimoto S, Matsumoto F, Ando M, Murakami N, Omura G, et al. All-exon TP53 sequencing and protein phenotype analysis accurately predict clinical outcome after surgical treatment of head and neck squamous cell carcinoma. Ann Surg Oncol. 2019;26:2294–2303. doi: 10.1245/s10434-019-07287-x. [DOI] [PubMed] [Google Scholar]
- 28.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starzyńska A, Sejda A, Adamska P, Marvaso G, Sakowicz-Burkiewicz M, Adamski Ł, et al. Prognostic value of the PIK3CA, AKT, and PTEN mutations in oral squamous cell carcinoma: literature review. Arch Med Sci. 2020;17:207–217. doi: 10.5114/aoms.2020.100780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SJ, Liu H, Liao CT, Huang PJ, Huang Y, Hsu A, et al. Ultra-deep targeted sequencing of advanced oral squamous cell carcinoma identifies a mutation-based prognostic gene signature. Oncotarget. 2015;6:18066–18080. doi: 10.18632/oncotarget.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batta N, Pandey M. Mutational spectrum of tobacco associated oral squamous carcinoma and its therapeutic significance. World J Surg Oncol. 2019;17:198. doi: 10.1186/s12957-019-1741-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman N, Marcelo KL, Hopkins JF, Khan NI, Du R, Hong L, et al. HRAS mutations define a distinct subgroup in head and neck squamous cell carcinoma. JCO Precis Oncol. 2023;7:e2200211. doi: 10.1200/PO.22.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The surgical materials and datasets analyzed in the current study are available from the corresponding author upon reasonable request.