Abstract
Metatranscriptomics generates large volumes of sequence data about transcribed genes in natural environments. Taxonomic annotation of these datasets depends on availability of curated reference sequences. For marine microbial eukaryotes, current reference libraries are limited by gaps in sequenced organism diversity and barriers to updating libraries with new sequence data, resulting in taxonomic annotation of about half of eukaryotic environmental transcripts. Here, we introduce Marine Functional EukaRyotic Reference Taxa (MarFERReT), a marine microbial eukaryotic sequence library designed for use with taxonomic annotation of eukaryotic metatranscriptomes. We gathered 902 publicly accessible marine eukaryote genomes and transcriptomes and assessed their sequence quality and cross-contamination issues, selecting 800 validated entries for inclusion in MarFERReT. Version 1.1 of MarFERReT contains reference sequences from 800 marine eukaryotic genomes and transcriptomes, covering 453 species- and strain-level taxa, totaling nearly 28 million protein sequences with associated NCBI and PR2 Taxonomy identifiers and Pfam functional annotations. The MarFERReT project repository hosts containerized build scripts, documentation on installation and use case examples, and information on new versions of MarFERReT.
Subject terms: Classification and taxonomy, Microbial genetics, Transcriptomics
Background & Summary
Microbial eukaryotes perform essential ecological functions in marine ecosystems as phototrophs, predators, and parasites1. This evolutionarily diverse group of organisms collectively possesses hundreds of millions of taxonomically distinct genes encoding metabolic processes that shape global biogeochemical cycles2. Eukaryotic metatranscriptomes are a sample of the nucleotides that result from community-wide transcriptional patterns and provide a window into how different members of the community function in situ. Metatranscriptome samples have been collected and annotated for studies of marine microbial eukaryotes across the world, including the global-scale Tara Oceans expedition2. Identifying the taxonomic origin of these sequences depends on the quality and depth of the reference sequence library used for annotation. Early metatranscriptome analyses were limited by sparsity of reference sequences among marine protists, with many lineages lacking any sequenced representatives. The sequence landscape improved dramatically in 2014 with development of the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP), a community-wide undertaking that resulted in public availability of 678 assembled transcriptomes derived from over 340 marine eukaryotic strains3. The MMETSP increased the number of sequenced marine protist species by approximately one order of magnitude, greatly enhancing the breadth and quality of environmental sequence annotation.
Since the release of the MMETSP, a variety of new marine eukaryote-focused reference libraries have arisen that build upon MMETSP by aggregating additional marine microbial genomes and transcriptomes from different sources (Table 1). The PhyloDB reference library4 was last updated publicly in 2015 and contains microbial eukaryotes, bacteria, archaea, and viruses isolated from both marine and non-marine environments. MARMICRODB5 is a prokaryote-focused database that includes a subset of eukaryotic reference sequences from the MMETSP. The EukZoo protein database6 of aquatic microbial eukaryotes was released and updated in 2018 and added 61 genomes and transcriptomes to the MMETSP dataset. The METdb repository7 was assembled in 2019 and included MMETSP re-assemblies8 in addition to 34 marine protist transcriptomes generated from cultures within the Roscoff Culture Collection (RCC). Most recently, the EukProt database9 was released in 2022 and incorporates genome-scale predicted proteins from a broad spectrum of eukaryotic phyla, including a high proportion of sequences from terrestrial plants, animals, and fungi along with 272 of the 678 MMETSP transcriptomes. These culture-driven sequencing and database efforts have contributed significantly to the landscape of available reference sequences, but coverage gaps in taxonomic representation persist, and approximately half of assembled marine metatranscriptome transcripts from the Tara Oceans expedition and other environmental sequencing studies have no significant similarity to any known sequence in reference sequences libraries2,10.
Table 1.
Comparison of reference sequence library features.
| Library Name | Latest Release | Total Entries | Domain Focus | Reviewed Publication | Code | Data DOI |
|---|---|---|---|---|---|---|
| MMETSP3 | 2014 | 678 transcriptomes | Marine microbial eukaryotes | Yes | No | No |
| PhyloDB4 | 2015 | 19,962 viral, 230 archaeal, 4910 bacterial, 894 eukaryotic taxa (409 from MMETSP) | General | No | No | No |
| EukZoo6 | 2018 | 739 genomes, transcriptomes (678 from MMETSP) | Aquatic microbial eukaryotes | No | Yes | Yes |
| METdb7 | 2019 | 464 transcriptomes (410 from MMETSP) | Marine microbial eukaryotes | Poster | Yes | No |
| MMETSP re-assemblies8 | 2019 | 678 MMETSP transcriptomes | Marine microbial eukaryotes | Yes | Yes | Yes17 |
| EukProt9 | 2022 | 993 species (272 from MMETSP) | General eukaryotes | Yes | No | Yes |
| MarFERReT | 2023 | 902 total genomes & transcriptomes (678 from MMETSP), 800 QC-validated entries | Marine microbial eukaryotes | Yes | Yes299 | Yes298 |
Library Name: shorthand name of the database. Latest Release: most recent release of the database or publication. Total Entries: number of included references, broken down by provenance. Domain Focus: taxonomic emphasis of the database. Publication: peer-reviewed publication accompanies the database. Code: availability of an accessible codebase complete with documentation.
The emergence of single-cell amplified genomes and transcriptomes (SAGs and SATs) now allows for targeted sequencing of single cells from environmental samples without the need for culturing11, opening the window of sequencing opportunities for the uncultured majority of marine eukaryote species. Taxonomic identification of a eukaryotic SAG is assigned, when possible, from analysis of 18 S rDNA generated during the amplification step, although as with other annotations, 18 S rDNA-based taxonomy is also dependent on reference sequences. SAGs from MAST-3 and MAST-4 clades of marine stramenopiles and the Chrysophyte H1 and H2 clades are publicly available12. The availability of SAGs from uncultured organisms in combination with the continued isolation and sequencing of marine eukaryote strains from diverse environments such as Antarctic coastal waters13, deep waters14, and the oligotrophic open ocean15 improves the ability to provide taxonomic affiliation to previously unannotated sequences.
As additional cultured and uncultured reference transcriptomes and genomes from different sources continue to become publicly available, their ongoing integration into open and reproducible reference sequence libraries remains vital to enhancing environmental sequence annotation. Alongside the static database release, publishing documented code for generating the database product ensures reproducibility and transparency in the release of future library versions. Furthermore, open-source development of a library codebase allows researchers to expand the core codebase and dependencies and fork the codebase independently as desired, thus helping to ensure the resource continues to grow. This also allows researchers to add new reference sequences for targeted research questions without dependence on centralized library releases. Reference libraries currently available for marine microbes either do not meet these criteria (Table 1) or they lack the necessary strain- or species-level diversity to annotate marine microbial eukaryote metatranscriptomes. For instance, the objective of the recently released EukProt9 reference database is designed to include at least one representative genera from sequenced lineages for broad eukaryotic diversity, and leave out sequenced species in well-sequenced lineages to balance the total database size. Although useful for broad annotation purposes, this does not meet the specific research needs of metatranscriptome studies that investigate species- and strain-level sequence differences in diverse marine eukaryote lineages15,16.
Here, we introduce the Marine Functional EukaRyotic Reference Taxa (MarFERReT), an updated open-source marine eukaryote reference protein sequence library with a reproducible framework allowing for community-supported expansion over time. MarFERReT was created out of the need for a reference sequence library that 1) focuses on species and strain-level representation of marine microbial eukaryotes, 2) is represented by a stable and accessible publication and/or DOI, 3) captures recent advances in published sequence data, 4) has transparent and replicable code, and 5) can expand over time with documented releases. MarFERReT v1 was designed to be a comprehensive marine microbial eukaryote reference library for the taxonomic annotation of environmental metatranscriptome data that facilitates access to marine eukaryote reference sequences and helps further our understanding of the diverse functional potential of marine protists.
A total of 902 candidate reference entries considered for inclusion into MarFERReT were collected online from public-access sequencing projects released between 2002 and 2021, with a predominant focus on marine microbial eukaryotes (Fig. 1a). The reference sequences were gathered from 754 assembled transcriptomes12,14,17–53 (including 2 single-cell amplified transcriptomes), and 148 genomes (including 8 single-cell amplified genomes)54–291. Sequences were collected from sources as nucleotides or translated peptides. Predicted protein sequences were annotated with Pfam 34.0292 to provide uniform functional prediction. Each reference entry is linked to an NCBI Taxonomy293 identifier (NCBI taxID) reflecting the latest changes in the NCBI Taxonomy. For 885 candidate entries we have also included matching identifiers from the PR2 database ecosystem294,295 of protist ribosomal reference sequences. The PR2 identifiers294 and the addition of 9 classification levels from the community curated PR2 database295 complement the NCBI Taxonomy classifications, and facilitate cross-comparison with 18 S RNA metabarcoding studies. We use NCBI taxIDs as the primary taxonomic identifier in most of our downstream processes owing to its widespread community adoption, interoperability with last common ancestry (LCA) taxonomic annotation software, and strain-level taxonomic resolution.
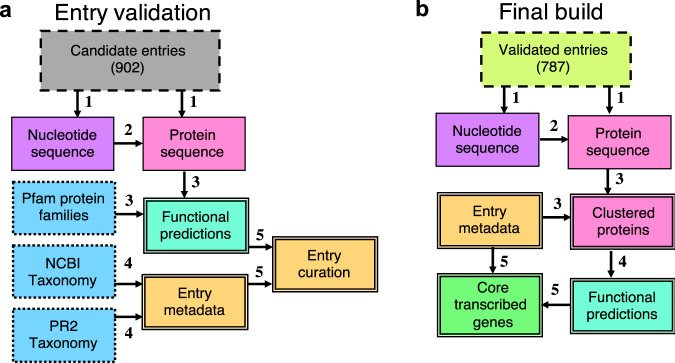
Fig. 1.
Diagrammatic overview of MarFERReT validation and build processes. Boxes represent the data sets involved in building MarFERReT and the border style indicates the data type: external sequence inputs (dashed line), external taxonomic and functional annotation resources (dotted lines), internal data products (single solid line) and output MarFERReT data products (double lines). Arrows indicate processes. (a) Candidate entry and NCBI taxID validation: (1) Candidate entries were identified from primary data sources and downloaded as nucleotide and protein reference sequences; (2) six-frame translation301 and frame-selection of nucleotide sequences into protein sequences; (3) functional annotation of protein sequences with Pfam292 protein families using HMMER 3.3302; (4) curation of NCBI Taxonomy293 IDs (taxIDs) for MarFERReT candidate entries and additional incorporation of matched IDs and classification from the PR2 Taxonomy ecosystem294,295; (5) candidate entries are assessed with evidence from external studies and by taxonomic analysis of ribosomal protein sequences for potential cross-contamination. Validated entries accepted for the quality-controlled build are recorded in the entry metadata. (b) Quality-controlled MarFERReT build with validated entries. For the set of 800 validated entries, the same methods used in 1a were used for (1) aggregating nucleotide and protein data and (2) translating nucleotide to protein sequences; (3) intra-taxa clustering at the strain or species level: protein sequence data sharing the same NCBI taxIDs are pooled together and clustered307 at 99% identity using updated taxIDs contained in the metadata; (4) Final Pfam annotation of the clustered protein sequences; (5) identification of core transcribed genes from functional annotations of transcriptome-derived entries.
We include quality control steps to validate candidate entries for inclusion into a final, quality-controlled library build. An analyses of ribosomal protein taxonomies was applied to all MarFERReT candidate entries to assess sequence data for potential cross-contamination. We also incorporated cross-contamination analyses of MMETSP transcriptome entries8 derived from two previous studies296,297 (see Technical Validation section). In total, we identified 102 candidate entries with sample contamination or sequence quality issues. The remaining 800 candidate entries were included in the final MarFERReT v1.1 data products298 (Fig. 1b), encompassing 453 species or strain-level taxa. Sequence redundancy was reduced within each taxon by combining sequences with the same NCBI taxID and clustering them at a 99% protein sequence identity threshold, resulting in a total of 27,951,013 translated and clustered protein sequences (Fig. 2, see Data Records for full description of data).
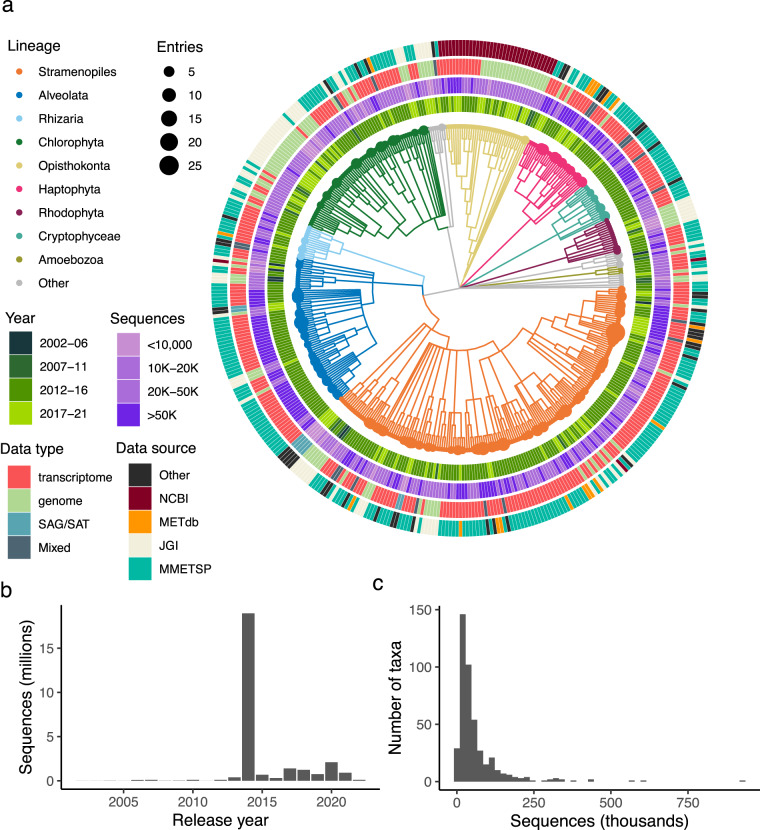
Fig. 2.
Cladogram of 800 validated MarFERReT entries and summary of reference metadata. (a) Cladogram of hierarchical taxonomic ranks of marine eukaryotes within the NCBI Taxonomy framework293 using the NCBI CommonTree tool. Each tip is a unique taxon included in MarFERReT, defined by its NCBI taxID identifier. Branches are colored by taxonomic lineage with size of the closed circle at each tip proportional to the number of validated entries in each taxon. Concentric rings describe metadata and statistics for each taxon. From innermost ring outward: year of publication or data release for sequence data (average year of release for multiple entries), number of clustered sequences in taxon, raw input format of sequence data: transcriptome, transcriptome shotgun assembly; genome, genome-derived gene models; SAG, single-cell amplified genome; SAT, single-cell amplified transcriptome; or a combination of types (mixed), and source of data: NCBI293, METdb7, JGI PhycoCosm300, or MMETSP3. (b) Number of clustered sequences in MarFERReT build by year of data release, and (c) Histogram showing distribution of clustered sequence count for 453 taxa in the final build.
The implementation of this workflow consists of a combination of Python, Bash, and R scripts, and is freely available as part of the MarFERReT repository. All core database processing steps have been containerized to maximize reproducibility, reduce issues related to software dependencies, and lower technical barriers to use. Documentation included in the repository details the steps required for users to replicate the construction of MarFERReT or to build derivative versions, as well as example use cases. Additionally, supporting code infrastructure is designed to facilitate the addition of new curated reference sequence material and annotation products by users and in future releases. The project repository containing code and documentation can be found here299. MarFERReT v1.1 data products are available online through the Zenodo repository (link)298, and contain all data files necessary to begin using MarFERReT for sequence annotation (see Data Records and Usage Notes sections for more detail).
Methods
Collation of sequence data
First (arrow 1, Fig. 1a), candidate entries were collected from publicly available web resources in the form of genomic, transcriptomic, and single-cell-amplified genome (SAG) and transcriptome (SAT) data. Reference sequences were collected from primary sources as either nucleotides (5,986,735 sequences) or translated peptides (30,826,074 sequences). Four projects were major sources of sequence information for MarFERReT. The Marine Microbial Eukaryote Sequence Project3 (MMETSP) is the largest contributor to MarFERReT with 678 candidate entries. The MMETSP sequences were collected as peptide translations from Version 2 of the MMETSP re-assemblies8,17. A total of 116 entries54–252 were collected from JGI Phycocosm300 as translated protein predictions of gene models from assembled genomes. A total of 41 transcriptomes from Roscoff Culture Collection18 isolates were collected from the METdb database7 as nucleotide transcriptome assemblies. To provide classification breadth to metazoan (animal) sequences consistently captured in protist-focused metatranscriptomes2, sequences from 36 Metazoan species were included from NCBI GenBank, including nucleotide transcriptome assemblies from 14 copepod transcriptomes26–47 and translated gene models from 22 marine species selected for broad coverage of 12 metazoan phyla254–292. The remaining 23 candidate entries were collected from other sequencing projects including translated gene models from 8 single-cell amplified genomes of uncultured stramenopiles12, ten assembled transcriptomes of diatom isolates from Antarctic and North Atlantic coastal waters13, two single-cell amplified assembled transcriptomes from early-branching dinoflagellates14, and translated transcriptome assemblies from open-ocean isolates of haptophytes and diatoms of the North Pacific15,25. Information for each candidate entry is available in the data repository in MarFERReT.v1.metadata.csv298 (see the Data Records section), including the web link to the primary data, the original file name, and the associated publication and DOI (link).
Six-frame translation and frame selection
Second (arrow 2, Fig. 1a), nucleotide sequences were translated in six frames with transeq vEMBOSS:6.6.0.059301 using Standard Genetic Code, to bring all reference sequence material into translated amino acid sequence space. The translation frame containing the longest open reading frame sequence was retained for downstream analysis.
Functional annotation of protein sequences
Third (arrow 3, Fig. 1a), candidate entry protein sequences were annotated against the Pfam 34.0292 collection of 19,179 protein family Hidden Markov Models (HMMs) using HMMER 3.3302. The highest-stringency cutoff score (‘trusted cutoff’) assigned by Pfam to each hmm profile was used as a minimum score threshold. The best scoring Pfam annotation (highest bitscore) was used if the protein received more than one Pfam match. A total of 12,554,711 candidate entry sequences received annotation with 12,549 of the 19,179 total possible profiles in Pfam 34.0. The raw Pfam annotations and best-scoring annotations are available online in the MarFERReT Zenodo repository (see Data Records).
Curation of sequence metadata
Fourth (arrow 4, Fig. 1a), NCBI Taxonomy293 hierarchical relationships were determined for the NCBI taxIDs in MarFERReT using the ‘taxtastic’ package303 v0.9.2. Prior to this, an NCBI taxID for each entry was determined through source metadata or manually assigned. All entries were manually updated if necessary to reflect changes and additions to the NCBI Taxonomy database, as of October 11th, 2022. For 761 of the 902 candidate entries; an NCBI Taxonomy was provided with the source metadata. The original taxIDs were unchanged for 497 entries, and for 264 entries, the taxID was updated to reflect the best-possible taxonomic match in NCBI Taxonomy as of October 11th, 2022. For the remaining 141 entries without an NCBI taxID from the data source, organism name and strain information were used to find the most specific match in NCBI Taxonomy, searching for strain-specific matches, then species-specific matches if strain was not available, followed by unclassified species in the genus. The complete record of taxID curation for every entry is available online (see ‘MarFERReT.v1.entry_curation.csv’ in Data Records).
Each entry is also associated with its closest match within the PR2 protist ribosomal reference database294 and the accompanying community-curated classification scheme of the PR2 database ecosystem295. A custom python script was used with the updated NCBI taxIDs associated with MarFERReT entries to identify the 18 S sequence tags in the PR2 database that share the same NCBI taxID. We used RapidFuzz304, a rapid fuzzy string matching algorithm to help identify the PR2 sequence tags matching MarFERReT entries at the strain-level wherever possible, as the strain identities are not always captured in the NCBI Taxonomy framework and the PR2 taxonomy levels only descend to the species level, even if strain information is included in the ID description. Each entry was manually reviewed to identify the most accurate strain-level match, proceeding to species-level matches if no strain match was found, and genus-level if no species-level match exists in the PR2 database. The associated PR2 ID and the full PR2 lineage classification are also available in the entry metadata on Zenodo298 (‘MarFERReT.v1.metadata.csv’ in the Data Records section), code documentation is available here (link). For subsequent steps, we use the NCBI taxID for strain-level identification and interoperability with key software.
Validation of candidate entries
Fifth (arrow 5, Fig. 1a), we assessed the 902 candidate entries for potential quality and contamination issues to identify a set of validated entries to include in a quality-controlled MarFERReT build (Fig. 3). Five metrics were used to flag candidate entries for exclusion from the final build. First, we flagged 24 entries with less than 1,200 total sequences, and second we flagged an additional 4 entries with less than 500 total assigned Pfam domains (Fig. 3b) from the functional annotation step. For candidate entries from the MMETSP re-assemblies8,17, we incorporated cross-contamination estimates derived from two independent studies. Lasek-Nesselquist and Johnson296 examined 26 ciliate MMETSP entries for potential contamination and identified 18 samples with an estimated 25 to 86% contamination; these 18 MMETSP entries were flagged in our assessment. Van Vlierberghe et al.297 investigated all 678 MMETSP entries for potential cross-contamination through taxonomic analysis of ribosomal protein sequences; we flagged 30 entries with over 50% sequence cross-contamination reported here. The estimates from van Vlierberghe et al.297 for 8 entries with 100% reported contamination were not flagged as the wrong NCBI taxID was used in their determination (entry IDs 378, 379, 380, 381, 504, 505, 506 and 507).
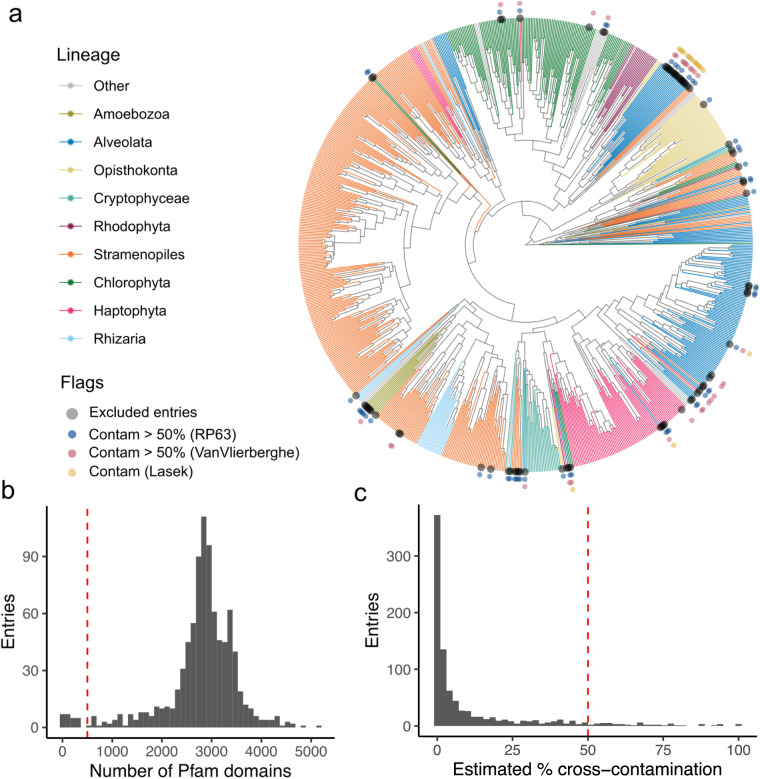
Fig. 3.
Validation of candidate entry sequences for cross-contamination. (a) Circular tree from hierarchical clustering of a binary distance matrix, constructed from the presence/absence of approximately 12,000 Pfam protein families in 874 candidate entries; entries with low sequence or pfam flags were excluded. Grey points at the tip indicate one of 102 entries excluded from the final build, with the other points marking the flag(s) for excluded entries. Contam > 50% (RP63), cross-contamination estimates over 50% from this study; Contam > 50% (VanVlierberghe), cross-contamination estimates over 50% from van Vlierberghe et al.297; Contam (Lasek), reported contamination for ciliate entries from Lasek-Nesselquist and Johnson296. (b) Histogram of Pfam domains in annotated candidate entry sequences; red dotted line indicates the 500 Pfam minimum threshold for inclusion. (c) Histogram of estimated cross-contamination in entries from ribosomal protein analysis; red dotted line indicates 50% cutoff threshold for exclusion.
For the fifth metric, we calculated potential cross-contamination estimates for all candidate entries (Fig. 3c) by deploying an approach similar to the method used to assess MMETSP transcriptomes by van Vlierberghe et al.297. These estimates were calculated for the 874 candidate entries without the low sequence or low Pfam flags noted above. The approach utilizes taxonomic annotation of ribosomal sequences matched to one of 63 Pfam protein domain IDs specific to ribosomal proteins and present in over 90% of candidate entries (referred to in this manuscript as ‘RP63’). The full set of UniProtKB reference sequences associated with each Pfam domain305 was downloaded and used to create a DIAMOND reference database for the DIAMOND306 sequence alignment software using the ‘diamond makedb’ command with default parameters and the NCBI database, as scripted in build_diamond_db.sh. Ribosomal protein sequences from MarFERReT candidate entries were retrieved based on Pfam functional annotations (arrow 3, Fig. 1a) and taxonomically identified by Last Common Ancestor estimation using the ‘diamond blastp’ command (parameters: -b 100 -c 1 -e 1e-5 –top 10 -f 102), with results reported as an NCBI taxID. For each entry, we determined the number of ribosomal proteins with an annotation within the pre-specified lineages used by van Vlierberghe et al.297: Amoebozoa, Ciliophora, Colpodellida, Cryptophyceae, Dinophyceae, Euglenozoa, Glaucocystophyceae, Haptophyta, Heterolobosea, Palpitomonas, Perkinsozoa, Rhizaria, Rhodophyta, Stramenopiles, and Viridiplantae. We also include the broad taxonomic lineages of Metazoa, Bacteria, Archaea, and Viruses to identify potential contamination by these groups. An estimate of potential contamination for each entry was generated by calculating the percentage of ribosomal sequences identified outside of the expected lineage. Based on this analysis, we flagged 53 entries with over 50% estimated contamination (Fig. 3c). These RP63 results are provided in the ‘MarFERReT.v1.QC_estimates.csv’ file described in the Data Records section.
A total of 102 entries failed at least one of the five quality control metrics described above. The metrics and flags are provided in the file MarFERReT.v1.curation.csv as described in the Data Records section. Python scripts and documentation for these analyses are available in the code repository here: (link).
Final build of MarFERReT with accepted candidate entries
A total of 800 validated candidate entries were accepted for inclusion into the final MarFERReT build, after excluding 102 entries with flags identified during the entry validation step. From these validated entries, protein sequences sharing the same NCBI taxIDs were combined into bins, thus preserving strain-level sequence diversity where possible. Sequence redundancy was reduced (arrow 3, Fig. 1b) by pooling protein sequences within NCBI taxID bins and clustering at the 99% amino acid sequence identity threshold with MMseqs. 2307. A representative sequence was retained for each cluster, and all sequences were renamed with a unique identifier. Pfam functional annotations for the 27,951,013 intra-species clustered protein sequences were propagated from the functional annotation step of the validation protocol (arrow 4, Fig. 1b). Clustered protein sequences, taxonomy and entry data tables, Pfam annotations, and other sequence-level protein data are available online (see Data Records for description of all files).
Identification of Core Transcribed Genes
The Pfam annotations of MarFERReT protein sequences were used to identify a set of cross-taxa core transcribed genes (CTGs; arrow 5, Fig. 1b) that serve as corollary of the BUSCO genome completeness metric308 oriented towards marine eukaryotic metatranscriptomes. For any given high-level taxonomic lineage, the CTGs are operationally defined here as the set of Pfam families observed in translated transcriptomes of at least 95% of the species within the given lineage. Only validated entries were used for this analysis. The CTG inventories were identified based on the Pfam 34.0 annotation of 7,514,355 proteins translated from the 654 validated transcriptome and SAT entries (the 146 validated genomic and SAG-sourced entries were not included), and a presence-absence matrix of Pfam functions was generated from the functional annotations of 332 taxa. We derived CTG inventories for all eukaryotes as a whole group, and for nine major marine lineages with at least 10 transcriptomic reference taxa each: Bacillariophyta (diatoms), Ochrophyta (excluding the Bacillariophyta subclade), Dinophyceae, Chlorophyta, Haptophyta, Cryptophyceae, Opisthokonta, Rhizaria and Amoebozoa. Bacillariophyta are listed separately from the Ochrophyta parent clade because this is a well-studied ochrophyte lineage with a large number of sequenced taxa. As an example, the ‘Haptophyta’ CTGs are comprised of the set of 1,074 Pfam domains observed in at least 28 of the 29 haptophyte taxa with transcriptomes in MarFERReT v1.1. The list of Pfam IDs and their detection frequencies are available online for these 9 major marine lineages and for Eukaryotes as a whole298 (see the Data Records section, MarFERReT.core_genes.v1.csv), and documented in the code repository (link)299.
Data Records
The final MarFERReT v1.1 data products are available online through a Zenodo repository (link)298 and contain the data files necessary to begin using MarFERReT for sequence annotation. The raw source data for the 902 candidate entries considered for MarFERReT v1.1, including the 800 QC validated entries, are available for download from their respective online repositories. Links to the entry source website and associated publications for the entries is listed in MarFERReT.v1.metadata.csv on Zenodo298, and detailed instructions and code for downloading the raw sequence data from source are available in the MarFERReT code repository299 (link). Sequence source URL locations and linked publications are also available. The permanent DOI representing all MarFERReT versions is 10.5281/zenodo.7055911.
The following files are available in the MarFERReT data repository:
MarFERReT.v1.metadata.csv
This CSV file contains descriptors of each of the 902 database entries, including data source, taxonomy, and sequence descriptors. Data fields are as follows:
entry_id: Unique MarFERReT sequence entry identifier
accepted: Acceptance into the final MarFERReT build (Y/N). The Y/N values can be adjusted to customize the final build output according to user-specific needs
marferret_name: A human and machine friendly string derived from the NCBI Taxonomy293 organism name; maintaining strain-level designation wherever possible
tax_id: The NCBI Taxonomy ID (taxID)
pr2_accession: Best-matching PR2 accession ID294 associated with entry (see Methods)
pr2_rank: The lowest shared rank between the entry and the pr2_accession
pr2_taxonomy: PR2 Taxonomy classification295 scheme of the pr2_accession
data_type: Type of sequence data; transcriptome shotgun assemblies (TSA), gene models from assembled genomes (genome), and single-cell amplified genomes (SAG) or transcriptomes (SAT)
data_source: Online origin of sequence data; from a Zenodo data repository (Zenodo), a datadryad.org repository (datadryad.org), MMETSP re-assemblies on Zenodo (MMETSP)17, NCBI GenBank (NCBI), JGI Phycocosm (JGI-Phycocosm), the TARA Oceans portal on Genoscope (TARA), or entries from the Roscoff Culture Collection through the METdb database repository (METdb)
source_link: URL where the original sequence data and/or metadata was collected
pub_year: Year of data release or publication of linked reference
ref_link: Pubmed URL directs to the published reference for entry, if available
ref_doi: DOI of entry data from source, if available
source_filename: Name of the original sequence file name from the data source
seq_type: Entry sequence data retrieved in nucleotide (nt) or amino acid (aa) alphabets
n_seqs_raw: Number of sequences in the original sequence file
source_name: Full organism name from entry source
original_taxID: Original NCBI taxID from entry data source metadata, if available
alias: Additional identifiers for the entry, if available
MarFERReT.v1.entry_curation.csv
This CSV file contains curation and quality-control information on the 902 candidate entries considered for incorporation into MarFERReT v1.1, including curated NCBI Taxonomy IDs and entry validation statistics. Data fields are as follows:
entry_id: Unique MarFERReT sequence entry identifier
marferret_name: Organism name in human and machine friendly format, including additional NCBI taxonomy strain identifiers if available
tax_id: Verified NCBI taxID used in MarFERReT
taxID_status: Status of the final NCBI taxID (Assigned, Updated, or Unchanged)
taxID_notes: Notes on the original_taxID
n_seqs_raw: Number of sequences in the original sequence file
n_pfams: Number of Pfam domains identified in protein sequences
qc_flag: Early validation quality control flags for the following: LOW_SEQS; less than 1,200 raw sequences; LOW_PFAMS; less than 500 Pfam domain annotations
flag_Lasek: Flag notes from Lasek-Nesselquist and Johnson296; contains the flag ‘FLAG_LASEK’ indicating ciliate samples reported as contaminated in this study
VV_contam_pct: Estimated contamination reported for MMETSP entries in van Vlierberghe et al.297
flag_VanVlierberghe: Flag for a high level of estimated contamination, from ‘flag_VanVlierberghe’ values over 50%: FLAG_VV
rp63_npfams: Number of ribosomal protein Pfam domains out of 63 total
rp63_contam_pct: Percent of total ribosomal protein sequences with an inferred taxonomic identity in any lineage other than the recorded identity, as described in the Technical Validation section from analysis of 63 Pfam ribosomal protein domains
flag_rp63: Flag for a high level of estimated contamination, from ‘rp63_contam_pct’ values over 50%: FLAG_RP63
flag_sum: Count of the number of flag columns (‘qc_flag’, ‘flag_Lasek’, ‘flag_VanVlierberghe’, and ‘flag_rp63’). All entries with one or more flags are nominally rejected (‘accepted’ = N); entries without any flags are validated and accepted (‘accepted’ = Y)
accepted: Acceptance into the final MarFERReT build (Y or N)
MarFERReT.v1.RP63_QC_estimates.csv
This CSV file contains the results of the ‘RP63’ cross-contamination check using ribosomal proteins (see Methods). The lineage bin columns are the taxonomic categories that define whether a query sequence is placed within or outside the expected lineage.
entry_handle: Human-readable tag concatenating the MarFERReT ‘entry_id’ with the ‘marferret_name’ (from MarFERReT.v1.metadata.csv)
entry_id: Unique MarFERReT sequence entry identifier
tax_id: The NCBI Taxonomy ID (taxID)
n_seqs: Number of protein sequences annotated as a Pfam ribosomal protein family
n_pfams: Number of unique Pfam protein families
tax_group: The expected lineage of this entry sample from the ‘predefined lineage’ categories below
contam_pct: The percentage of ribosomal protein sequences identified in a lineage other than the expected ‘tax_group’ lineage
[lineage bins]: Series of 21 columns Amoebozoa, Ciliophora, Colpodellida, Cryptophyceae, Dinophyceae, Euglenozoa, Glaucocystophyceae, Haptophyta, Heterolobosea, Opisthokonta, Palpitomonas, Perkinsozoa, Rhizaria, Rhodophyta, Stramenopiles, Viridiplantae, Bacteria, Archaea, Viruses, Other, Unknown
MarFERReT.v1.proteins.faa.gz
This Gzip-compressed FASTA file contains the 27,951,013 final translated and clustered protein sequences for all 800 accepted MarFERReT entries. The FASTA sequence definition line (‘defline’) contains the unique identifier for the sequence and its reference (mftX, where ‘X’ is a ten-digit integer value).
MarFERReT.v1.taxonomies.tab.gz
This Gzip-compressed tab-separated file is formatted for interoperability with the DIAMOND306 protein alignment tool commonly used for downstream analyses (see Usage Notes) and contains some columns without any data. Each row contains an entry for one of the MarFERReT protein sequences in MarFERReT.v1.proteins.faa.gz. Note that ‘accession.version’ and ‘taxid’ are populated columns while ‘accession’ and ‘gi’ have NA values; the latter columns are required for back-compatibility as input for the DIAMOND alignment software and LCA analysis.
The columns in this file contain the following information:
accession: (NA)
accession.version: The unique MarFERReT sequence identifier (‘mftX’)
taxid: The NCBI Taxonomy ID associated with this reference sequence
gi: (NA)
MarFERReT.v1.proteins_info.tab.gz
This Gzip-compressed tab-separated file contains a row for each final MarFERReT protein sequence with the following columns:
aa_id: The unique identifier for each MarFERReT protein sequence
entry_id: The unique numeric identifier for each MarFERReT entry
source_defline: The original, unformatted sequence identifier
MarFERReT.candidate_entry_Pfam_annotations.tar.gz
This Gzip-compressed archive contains the raw HMMER3302 output from the search of Pfam 34.0292 HMM profiles against the full set of protein sequences from candidate entries. The archive contains files for each entry with the suffix ‘.Pfam34.domtblout.tab’ and prefixed with the ‘entry_id’ and ‘marferret_name’ values from MarFERReT.v1.metadata.csv. The ‘domtblout.tab’ files are the output from hmmsearch using the–domtblout parameter containing 3 header and 10 footer rows beginning with ‘#’ and rows for each hmmsearch match with 22 whitespace-delimited fields and a target sequence description (see here for more information on the hmmsearch output file formats). The ‘target name’ (original sequence identifier from MarFERReT.v1.proteins_info.tab.gz), ‘query name’ (Pfam name), ‘accession’ (Pfam ID), ‘E-value’ and ‘score’ (full sequence match scores) are retained in downstream data products.
MarFERReT.v1.best_pfam.csv.gz
This Gzip-compressed CSV file contains the best-scoring Pfam annotation for intra-species clustered protein sequences from the 800 final MarFERReT entries; derived from the raw hmmsearch annotations in MarFERReT.candidate_entry_Pfam_annotations.tar.gz. This file contains the following fields:
aa_id: The unique MarFERReT protein sequence ID (‘mftX’)
entry_id: Unique MarFERReT sequence entry identifier
source_defline: Original FASTA sequence identifier
pfam_name: The shorthand Pfam protein family name
pfam_id: The Pfam identifier
MarFERReT.v1.entry_pfam_sums.csv.gz
This Gzip-compressed CSV file contains a reduced version of MarFERReT.v1.best_pfam.csv.gz; grouped by ‘entry_id’ and ‘pfam_id’ to summarize the number of sequences (‘n_seqs’) with each unique entry_id-pfam_id pair. Contains the ‘entry_id’, ‘pfam_id’, ‘pfam_name’ and ‘n_seqs’ columns.
MarFERReT.v1.core_genes.csv
This CSV file contains the core transcribed gene (CTG) catalog derived from MarFERReT transcribed reference sequence data (see Methods) to be used in environmental metatranscriptome analysis in conjunction with other MarFERReT data products. The columns contain the following values:
lineage: Name of major marine microbial eukaryote lineage
n_taxa: Number of species- and strain-level taxa this Pfam observed in
pfam_id: Pfam protein family identifier
frequency: Proportion of species (n_species) in lineage where pfam_id is observed
Technical Validation
MarFERReT was developed for the primary application of marine metatranscriptome annotation. We used several independent criteria to assess the 902 candidate genome and transcriptome entries from different sources, and flagged 102 entries for exclusion (Fig. 3a) and retained 800 validated entries for the quality-controlled build as detailed in the Methods subsection, Validation of candidate entries (Fig. 1a). These flags and related metrics are reported for entries in MarFERReT.v1.entry_curation.csv (see Data Records section).
The ‘LOW_SEQS’ flag was given to 24 transcriptome entries from the MMETSP dataset with less than 1,200 sequences; these low counts were also noted by van Vlierberghe et al.297. This threshold cutoff is far less than the median sequence count of approximately 31,000 across all entries. We assessed the number of Pfam annotations to candidate entry sequences to ensure that entries properly contained protein-coding sequence content. Over 90% of entries have between 1,500 and 4,500 Pfam domains, and we assigned the ‘LOW_PFAMS’ flag to 4 entries with less than 500 Pfams if they were not already flagged with ‘LOW_SEQS’ (Fig. 3c).
We investigated entries for potential sequence cross-contamination through taxonomic analysis of 63 ribosomal protein sequences (referred to here as ‘RP63’), flagging 53 entries with over 50% estimated cross-contamination (Fig. 3c). This approach is similar to the one described by van Vlierberghe et al.297, except that it utilizes sequences from the publicly-available Pfam resource305 and DIAMOND306 Lowest Common Ancestor analysis instead of manually-curated alignments to provide expanded representation, and also identifies entries with cross-kingdom contamination from bacteria (see MarFERReT.v1.QC_estimates.csv in Data Records). The RP63 flags were added to 30 entries with over 50% contamination from van Vlierberghe et al.297 for the MMETSP transcriptomes and for 18 ciliate MMETSP entries reported as contaminated from Lasek-Nesselquist & Johnson296.
Together, we flagged a total of 102 candidate entries (Fig. 3) with sequence quality issues or high levels of potential contamination, and these are not included in the final MarFERReT build (Fig. 2). We recognize that predetermined value cutoffs for entry inclusion have the potential to eliminate valuable and diverse entries from the final product. The Usage Notes section describes how users can create a derivative version of the database by adding or removing entries included in the build process. The final build of MarFERReT v1 data products includes 800 validated MarFERReT entries that encompass 453 taxa and approximately 30 million clustered protein sequences (Fig. 2).
Usage Notes
The code for reproducing MarFERReT data products from primary source sequence is available in a public repository299, along with documentation. The software used for critical database assembly steps is packaged in stable, version-controlled containers, and scripts for pulling these containers from public repositories are included in the repository. Users can make use of pre-built MarFERReT v1.1 data products298 or create their own development version of MarFERReT using the containerized workflow described below. Code, documentation, and tutorials for this project are available on the MarFERReT repository: https://github.com/armbrustlab/marferret.
To use the processed MarFERReT protein data and associated data products directly, proceed to step 1 below (Using MarFERReT data products directly). To replicate the MarFERReT build process or create a derivative reference library with the containerized pipeline using the raw source data, skip to step 2 below (Cloning the MarFERReT repository).
Using MarFERReT data products directly
Finalized MarFERReT data products include over 27 million intra-species clustered protein sequences, metadata with curated taxonomy identifiers, Pfam protein annotations, core transcribed gene catalogs for marine microbial eukaryote lineages, and other supporting data298. The URLs are provided for the sources of the individual sequences, and the compiled, translated, and clustered sequences are available for download through the public repository, Zenodo (link).
If downloaded directly, steps 2 (Cloning the MarFERReT repository) through 7 (Building the Core Transcribed Gene catalog) can be skipped. MarFERReT can be combined with other protein sequence reference libraries or new reference sequence material for expanded phylogenetic coverage. For an example of combining MarFERReT with other databases or new reference sequence entries, see step 8 (Combining MarFERReT with other reference sequences). An example workflow for using these data to annotate environmental metatranscriptome is described in subsection 9 below (Using MarFERReT to annotate environmental metatranscriptomes).
Cloning the MarFERReT repository
The first step is to copy the MarFERReT pipeline code and cloning the repository into a suitable directory where the database will be built.
Collecting and organizing inputs
We do not host the primary raw data in the final MarFERReT data products; the original links, raw filenames, and other source reference information for all candidate entries are available in the MarFERReT entry curation table on Zenodo298 (MarFERReT.v1.entry_curation.csv). Two sets of input files are required to replicate the MarFERReT build: 1) the source reference sequences and 2) a corresponding metadata file (MarFERReT.v1.metadata.csv). The source reference sequences will need to be downloaded from their various public locations, and their file names will need to match the entries in the metadata table. The metadata file entitled MarFERReT.v1.metadata.csv contains information on the source data public URL, original file names, URL link to associated publication, and associated data object DOI for all reference sources considered for use in the first MarFERReT build. The ‘accepted’ field indicates entries that were accepted or excluded from the final quality-controlled version of the protein sequence library. These values are given here as a default suggestion and can be toggled to include (“Y”) or exclude (“N”) individual entries to meet the needs of specific research questions. See the full description of this file under Data Records. Detailed instructions for finding and downloading the source reference sequences used to build MarFERReT v1.1 can be found in this document. Once the MarFERReT repository is cloned onto the target machine, a new directory called ‘source_seqs’ must be created under the data directory. All FASTA files of the source reference sequences should be deposited into this directory. Before running the MarFERReT pipeline, all FASTA files should be unzipped.
Building software containers
The MarFERReT database construction pipeline is containerized to obviate concerns with software dependencies. Additionally, MarFERReT supports both Singularity and Docker containerization, depending on user preference. The necessary containers can be built in two steps:
Install either Singularity or Docker on target machine.
Navigate to the containers directory and run either the build_singularity_images.sh or build_docker_images.sh script from the command line.
Running MarFERReT database construction pipeline
Once the input source reference sequences have been collected, metadata has been organized, and the software containers have been built, the MarFERReT database construction pipeline is ready. Navigate to the scripts directory and run the assemble_marferret.sh script from the command line. The user will be prompted to enter either 1 or 2 depending on whether Singularity or Docker containerization is used. The pipeline will take several hours to run, depending on individual computer system specifications. When it is done, the following outputs in the data directory will be available:
MarFERReT.v1.proteins.faa.gz–MarFERReT protein library
MarFERReT.v1.taxonomies.tab.gz–taxonomy mapping file required as input for building diamond database
MarFERReT.v1.proteins_info.tab.gz–mapping file connecting each MarFERReT protein to its originating reference sequence
/aa_seq–directory with translated & standardized amino acid sequences
/taxid_grouped–directory with amino acid sequences grouped by taxid
/clustered–directory with amino acid sequences clustered within taxid
The three.gz files listed above can also be downloaded directly from the Zenodo repository298.
Annotating MarFERReT database sequences
Information on the functions of the proteins included in MarFERReT can be added by annotating the sequences with one of the many bioinformatic tools available for functional inference. In this repository we have included a script for annotating the database with Pfam292 (now included as a part of the InterPro consortium305).
To annotate MarFERReT, first download a copy of the Pfam database of HMM profiles. Make a new directory named ‘pfam’ under the ‘data’ directory. Download into this directory the latest version of Pfam from the Pfam ftp site.
Once the Pfam HMM database has been downloaded, navigate to the ‘scripts’ directory and run the pfam_annotate.sh script from the command line. In addition to the ‘data/pfam/Pfam-A.hmm’ HMM database, this script requires the ‘data/MarFERReT.v1.proteins.faa.gz’ file as an input. The complete set of Pfam annotations can also be found on Zenodo302 (see Data Records for full description).
Building the Core Transcribed Gene catalog
After the MarFERReT protein sequences have been functionally annotated, sets of CTGs can be derived from the RNA-derived data for specific marine lineages or for all eukaryotes. Selecting the Pfam IDs that are present in at least 95% of species of a given lineage allows us to define a set of functions that can be reasonably expected to be found in a relatively complete transcriptome. These CTG catalogs can be used downstream of environmental sequence annotation with MarFERReT to assess the coverage of environmental taxon bins, as demonstrated in Case Study 2. Documentation and code for generating and using the CTG catalogs from Pfam annotations for user-defined lineages are found here (link).
Combining MarFERReT with other reference sequences
MarFERReT can be combined with other domain-focused reference sequence libraries or new reference sequence transcriptomes and genomes to expand taxonomic coverage. In the Case Studies, we show an example combining MarFERReT with a filtered version of the prokaryote-focused MARMICRODB library5,309. Both libraries use NCBI Taxonomy identifiers as their primary classification framework, facilitating compatible annotation approaches. After downloading or building the MarFERReT protein sequence database, bacterial sequences can be downloaded from the MARMICRODB Zenodo repository309 and the libraries concatenated together for use in downstream processes.
MarFERReT can also be combined with individual reference sequence transcriptomes and genomes that have just been released, or are not incorporated in current reference libraries, or to add representation for specific research needs. This also requires that every sequence entry has an NCBI Taxonomy identifier. Instruction and code for combining MarFERReT with other large reference libraries like MARMICRODB or with sets of individual reference sequence entries can be found on the codebase repository here: c ombining_marferret_and_other_references.md.
Using MarFERReT to annotate environmental metatranscriptomes
Two case studies, with accompanying code, are provided to illustrate how MarFERReT can be used in a command-line environment for common analyses, either by itself or in conjunction with other protein sequence libraries to assign taxonomic identity to environmental sequences, and to approximate sequencing coverage within environmental taxonomic bins (Fig. 4).
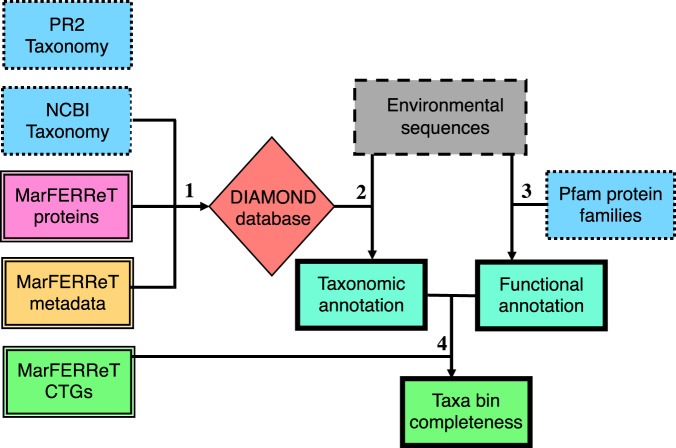
Fig. 4.
Schematic of case study 1 and 2 use of MarFERReT for annotation of environmental metatranscriptomes. Example workflow showing how MarFERReT data products can be used to annotate unknown assembled sequences and assess taxonomic bins. Boxes indicate datasets; box borders indicate environmental contig sequence data (dashed line), taxonomic and functional annotation from external resources (dotted lines), MarFERReT data products (double lines) and taxonomic and functional annotation results (bold lines). The red diamond indicates a user-constructed DIAMOND306 database for lowest common ancestor determination; prior to this step the user could combine MarFERReT with other libraries for expanded taxonomic coverage as shown in case study 1. Arrows represent processes: (1) construction of DIAMOND306 database using MarFERReT proteins and data, and taxonomy files from NCBI Taxonomy; matching PR2 Taxonomy294,295 identifiers and classifications are also provided in the metadata for alternative classification approaches. (2) Taxonomic annotation of environmental contigs using a DIAMOND306 database built from MarFERReT proteins; (3) Functional annotation of environmental contigs with HMMER302 3.3 on Pfam292 protein family hmm profiles; (4) Completeness assessment of taxonomically- and functionally-annotated metatranscriptome bins using MarFERReT core transcribed genes.
Case Study 1 shows how MarFERReT can be used to annotate unknown environmental sequences using the DIAMOND303 fast protein-alignment tool. In summary, a DIAMOND-formatted database is created from sequence data and NCBI Taxonomy293 information, and subsequently used to annotate unknown environmental reads (Fig. 4, arrow 1 and 2).
Case Study 2 documents how to identify core transcribed genes for user-defined lineages using MarFERReT protein sequences, and provides an example on how to estimate the completeness of environmental transcriptome bins with taxonomic annotation (from Case Study 1) and functional annotation with Pfam 34.0292 (now included as a part of the InterPro consortium). The example shown here uses ‘species-level’ annotations (or lower) for enhanced taxonomic specificity. In summary, the taxonomic and functional annotations are aggregated together, and the percentage of lineage-specific CTGs is determined for each species-level environmental taxon bin (Fig. 4, arrow 3 and 4).
While the case studies discussed above demonstrate the practicality of using MarFERReT for annotating environmental metatranscriptomes, MarFERReT can also be used to supplement other analyses that depend on reference proteins, including the analysis of marine metagenomes and metaproteomes.
Future MarFERReT releases
MarFERReT was designed to be updated as new microbial eukaryote functional reference sequences are publicly released, with releases identified either through literature reviews, updates to public repositories, or through user nominations. Additionally, users can create their own database derivatives using this framework. Suggestions for new additions or changes to future versions of MarFERReT can be submitted via the ‘Issues’ request function in this code repository (link). When submitting an organism request for future MarFERReT versions, the following information is required:
Full scientific name of the organism (with strain name if possible)
An NCBI taxID of the organism (as specific as possible, e.g. strain-level)
A URL to the location of the assembled source data, with additional instructions if necessary
Brief justification for why this organism should be included, e.g. “New SAGs from a clade of marine haptophytes”.
A citation or publication for the data, if available.
New entries will be processed through the workflow described for candidate entries and validated through phylogenetic analysis of ribosomal protein families (see Methods and Technical Validation). Future versions of MarFERReT will be documented in a changelog in the code repository (link), describing any additions or modifications to the library composition. The changelog will detail updates to the MarFERReT code and MarFERReT files hosted on Zenodo298, including revisions to the scripts, metadata files, functional annotation protocols, protein sequence library, DIAMOND databases, and Core Transcribed Gene inventories.
Acknowledgements
This work was supported by grants from the Simons Foundation (Award IDs 723795, 721244, and 549945FY22 to E.V.A.).
Author contributions
R.D.G. and E.V.A. conceived and designed the study, with input from S.B. and S.C. R.D.G. and S.C. contributed to the acquisition of data and curation of metadata. R.D.G. and S.B. developed, wrote, and documented the code used in the study, and S.B. developed the containerized pipeline. R.D.G. drafted the manuscript, and S.B., S.C. and E.V.A. provided critical revisions. All authors reviewed and approved the final version of the manuscript for submission.
Code availability
Code, documentation, and tutorials for this project are available on the MarFERReT repository299: https://github.com/armbrustlab/marferret. This repository has also been archived for the v.1.1 release310 and the archived code is available on Zenodo here: https://zenodo.org/records/10278540. Information on the software versions and parameters used in this publication are included in the MarFERReT containerized build on the repository and in the archived code.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
R. D. Groussman, Email: rgrous83@uw.edu
E. V. Armbrust, Email: armbrust@uw.edu
References
- 1.Caron DA, et al. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 2017;15:6–20. doi: 10.1038/nrmicro.2016.160. [DOI] [PubMed] [Google Scholar]
- 2.Carradec Q, et al. A global ocean atlas of eukaryotic genes. Nat. Commun. 2018;9:373. doi: 10.1038/s41467-017-02342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A.E. Allen Lab. PhyloDB, version 1.075. https://github.com/allenlab/PhyloDB (2015).
- 5.Becker JW, Hogle SL, Rosendo K, Chisholm SW. Co-culture and biogeography of Prochlorococcus and SAR11. The ISME journal. 2019;13:1506–1519. doi: 10.1038/s41396-019-0365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, Hu S, Caron D. 2018. EukZoo, an aquatic protistan protein database for meta-omics studies. (0.2) [Data set] Zenodo. [DOI]
- 7.Niang, G. et al. METdb: A genomic reference database for marine species. F1000Research910.7490/f1000research.1118000.1 (2020).
- 8.Johnson, L. K., Alexander, H. & Brown, C. T. Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. Gigascience 8, giy158 10.5281/zenodo.746048 (2019). [DOI] [PMC free article] [PubMed]
- 9.Richter, D. J. et al. EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotes. Peer Community Journal, 2 (2022).
- 10.Groussman, R. D., Coesel, S. N., Durham, B. P. & Armbrust, E. V. Diel-Regulated Transcriptional Cascades of Microbial Eukaryotes in the North Pacific Subtropical Gyre. Front. Microbiol. 12 (2021). [DOI] [PMC free article] [PubMed]
- 11.Roy RS, et al. Single cell genome analysis of an uncultured heterotrophic stramenopile. Sci. Rep. 2014;4:1–8. doi: 10.1038/srep04780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeleuthner Y, et al. Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nature Communications. 2018;9:310. doi: 10.1038/s41467-017-02235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guajardo M, Jimenez V, Vaulot D, Trefault N. 2021. Transcriptomes from Thalassiosira and Minidiscus diatoms from English Channel and Antarctic coastal waters (Version 1) [Data set] Zenodo. [DOI]
- 14.Cooney EC, et al. Single-cell transcriptomics of Abedinium reveals a new early-branching dinoflagellate lineage. Genome Biol. Evol. 2020;12:2417–2428. doi: 10.1093/gbe/evaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert BS, et al. The dynamic trophic architecture of open-ocean protist communities revealed through machine-guided metatranscriptomics. Proc. Natl. Acad. Sci. 2022;119:e2100916119. doi: 10.1073/pnas.2100916119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coesel SN, et al. Diel transcriptional oscillations of light-sensitive regulatory elements in open-ocean eukaryotic plankton communities. Proc. Natl. Acad. Sci. 2021;118:e2100235118. doi: 10.1073/pnas.2011038118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson LK, Alexander H, Brown CT. 2017. MMETSP re-assemblies [Data set] Zenodo. [DOI]
- 18.Niang G, 2020. METdb: a genomic reference database for marine species [Data set] Zenodo. [DOI]
- 19.Guajardo M, Jimenez V, Vaulot D, Trefault N. 2021. (Assemblies) Transcriptomes from Thalassiosira and Minidiscus diatoms from English Channel and Antarctic coastal waters (Version 1) [Data set] Zenodo. [DOI]
- 20.Janouškovec J, et al. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. Elife. 2019;8:e49662. doi: 10.7554/eLife.49662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NCBI GenBank. TSA: Cephaloidophora cf. communis isolate WS-2016. https://www.ncbi.nlm.nih.gov/nuccore/GHVH00000000.1 (2016).
- 22.NCBI GenBank. Malassezia globosa strain CBS 7966 EST library. https://www.ncbi.nlm.nih.gov/biosample/SAMN01758921 (2007).
- 23.Urushihara H, et al. Comparative genome and transcriptome analyses of the social amoeba Acytostelium subglobosum that accomplishes multicellular development without germ-soma differentiation. BMC Genomics. 2015;16(1):80. doi: 10.1186/s12864-015-1278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.NCBI GenBank. full-length enriched Acytostelium cDNA library. https://www.ncbi.nlm.nih.gov/biosample/SAMN02905743 (2015).
- 25.Groussman RD, 2022. Transcriptome assemblies of three diatom and three prymnesiophyte isolates from station ALOHA and Kaneohe Bay (1.0) [Data set] Zenodo. [DOI]
- 26.Roncalli V, Cieslak MC, Passamaneck Y, Christie AE, Lenz PH. Glutathione S-transferase (GST) gene diversity in the crustacean Calanus finmarchicus–contributors to cellular detoxification. PLoS One. 2015;10(5):e0123322. doi: 10.1371/journal.pone.0123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.NCBI GenBank. Calanus finmarchicus. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA236528 (2014).
- 28.Maas AE, Blanco-Bercial L, Lo A, Tarrant AM, Timmins-Schiffman E. Variations in Copepod Proteome and Respiration Rate in Association with Diel Vertical Migration and Circadian Cycle. Biol Bull. 2018;235(1):30–42. doi: 10.1086/699219. [DOI] [PubMed] [Google Scholar]
- 29.NCBI GenBank. Calanus glacialis. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA237014 (2015).
- 30.Roncalli V, Cieslak MC, Sommer SA, Hopcroft RR, Lenz PH. De novo transcriptome assembly of the calanoid copepod Neocalanus flemingeri: A new resource for emergence from diapause. Mar Genomics. 2018;37:114–119. doi: 10.1016/j.margen.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 31.NCBI GenBank. TSA: Neocalanus flemingeri, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GFUD00000000/ (2018).
- 32.NCBI GenBank. TSA: Acartia tonsa, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GFWY00000000/ (2017).
- 33.NCBI GenBank. TSA: Eurytemora carolleeae sequence, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GEAN00000000/ (2016).
- 34.Roncalli V, et al. A deep transcriptomic resource for the copepod crustacean Labidocera madurae: A potential indicator species for assessing near shore ecosystem health. PLoS One. 2017;12(10):e0186794. doi: 10.1371/journal.pone.0186794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCBI GenBank. TSA: Labidocera madurae, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GFWO00000000/ (2017).
- 36.Barreto FS, Pereira RJ, Burton RS. Hybrid dysfunction and physiological compensation in gene expression. Mol Biol Evol. 2015;32:613–622. doi: 10.1093/molbev/msu321. [DOI] [PubMed] [Google Scholar]
- 37.NCBI GenBank. TSA: Tigriopus californicus, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GBSZ00000000/ (2015).
- 38.Kim HS, et al. De novo assembly and annotation of the Antarctic copepod (Tigriopus kingsejongensis) transcriptome. Mar Genomics. 2016;28:37–39. doi: 10.1016/j.margen.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 39.NCBI GenBank. TSA: Tigriopus sp. 1 SL-2012, transcriptome shotgun assembly.https://www.ncbi.nlm.nih.gov/nuccore/GDFW01000000/ (2015).
- 40.Kim HS, et al. Identification of xenobiotic biodegradation and metabolism-related genes in the copepod Tigriopus japonicus whole transcriptome analysis. Mar Genomics. 2015;24(Pt 3):207–208. doi: 10.1016/j.margen.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 41.NCBI GenBank. TSA: Tigriopus japonicus, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GCHA00000000/ (2015).
- 42.Lee BY, et al. RNA-seq based whole transcriptome analysis of the cyclopoid copepod Paracyclopina nana focusing on xenobiotics metabolism. Comp Biochem Physiol Part D Genomics Proteomics. 2015;15:12–19. doi: 10.1016/j.cbd.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 43.NCBI GenBank. TSA: Paracyclopina nana, transcriptome shotgun assembly.https://www.ncbi.nlm.nih.gov/nuccore/GCJT01000000/ (2015).
- 44.NCBI GenBank. TSA: Eucyclops serrulatus, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GARW01000000/ (2014).
- 45.NCBI GenBank. TSA: Lepeophtheirus salmonis, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/HACA00000000/ (2015).
- 46.NCBI GenBank. TSA: Caligus rogercresseyi, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GAZX00000000/ (2014).
- 47.NCBI GenBank. TSA: Pleuromamma xiphias, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GFCI00000000/ (2018).
- 48.Onyshchenko A, Roberts WR, Ruck EC, Lewis JA, Alverson AJ. The genome of a nonphotosynthetic diatom provides insights into the metabolic shift to heterotrophy and constraints on the loss of photosynthesis. New Phytol. 2021;232(4):1750–1764. doi: 10.1111/nph.17673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCBI GenBank. TSA: Nitzschia sp. Nitz4, transcriptome shotgun assembly. https://www.ncbi.nlm.nih.gov/nuccore/GIQR00000000/ (2020).
- 50.Mars Brisbin M, Mitarai S. Differential gene expression supports a resource-intensive, defensive role for colony production in the bloom-forming haptophyte, Phaeocystis globosa. J Eukaryot Microbiol. 2019;66(5):788–801. doi: 10.1111/jeu.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mars Brisbin M, Mitarai S. 2018. Phaeocystis globosa colonial gene expression. Zenodo. https://zenodo.org/record/1476491
- 52.Seeleuthner et al. Tara Oceans SAGs. http://www.genoscope.cns.fr/tara/, Tara Oceans10.1038/s41467-017-02235-3 (2018).
- 53.Cooney, E. et al. Single cell transcriptomics of Abedinium reveals a new early-branching dinoflagellate lineage, Dryad, Dataset10.5061/dryad.pg4f4qrk0 (2020). [DOI] [PMC free article] [PubMed]
- 54.John U, et al. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci Adv. 2019;5:eaav1110. doi: 10.1126/sciadv.aav1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.JGI PhycoCosm. Amoebophrya ceratii AT5.2. https://phycocosm.jgi.doe.gov/Amoce1/Amoce1.home.html (2019).
- 56.JGI PhycoCosm. Aplanochytrium kerguelense PBS07 v1.0.https://phycocosm.jgi.doe.gov/Aplke1/Aplke1.home.html (2013).
- 57.JGI PhycoCosm. Aurantiochytrium limacinum ATCC MYA-1381 v1.0.https://phycocosm.jgi.doe.gov/Aurli1/Aurli1.home.html (2012).
- 58.JGI PhycoCosm. Aureococcus anophagefferens clone 1984 v1.0. https://phycocosm.jgi.doe.gov/Auran1/Auran1.home.html
- 59.Gao C, et al. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics. 2014;15:582. doi: 10.1186/1471-2164-15-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.JGI PhycoCosm. Auxenochlorella protothecoides 0710. https://phycocosm.jgi.doe.gov/Auxeprot1/Auxeprot1.home.html (2014).
- 61.Vogler BW, et al. Characterization of plant carbon substrate utilization by Auxenochlorella protothecoides. Algal Research. 2018;34:37–48. doi: 10.1016/j.algal.2018.07.001. [DOI] [Google Scholar]
- 62.JGI PhycoCosm. Auxenochlorella protothecoides UTEX 25. https://phycocosm.jgi.doe.gov/Auxpr25_1/Auxpr25_1.home.html (2018).
- 63.Moreau H, et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012;13(8):R74. doi: 10.1186/gb-2012-13-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.JGI PhycoCosm. Bathycoccus prasinos RCC1105. https://phycocosm.jgi.doe.gov/Batpra1/Batpra1.home.html (2012).
- 65.Curtis BA, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- 66.JGI PhycoCosm. Bigelowiella natans CCMP2755 v1.0. https://phycocosm.jgi.doe.gov/Bigna1/Bigna1.home.html (2012).
- 67.Denoeud F, et al. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011;12:R29. doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.JGI PhycoCosm. Blastocystis hominis Singapore isolate B (sub-type 7). https://phycocosm.jgi.doe.gov/Blahom1/Blahom1.home.html (2011).
- 69.Browne DR, et al. Draft nuclear genome sequence of the liquid hydrocarbon-accumulating green microalga Botryococcus braunii Race B (Showa) Genome Announc. 2017;5(16):e00215–17. doi: 10.1128/genomeA.00215-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.JGI PhycoCosm. Botryococcus braunii Showa v2.1.https://phycocosm.jgi.doe.gov/Botrbrau1/Botrbrau1.home.html (2017).
- 71.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23(15):1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 72.JGI PhycoCosm. Breviolum minutum. https://phycocosm.jgi.doe.gov/Bremi1/Bremi1.home.html (2013).
- 73.Arimoto A, et al. A siphonous macroalgal genome suggests convergent functions of homeobox genes in algae and land plants. DNA Res. 2019;26:183–192. doi: 10.1093/dnares/dsz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.JGI PhycoCosm. Caulerpa lentillifera. https://phycocosm.jgi.doe.gov/Caulen1/Caulen1.home.html (2019).
- 75.Nishiyama T, et al. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell. 2018;174(2):448–464.e24. doi: 10.1016/j.cell.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 76.JGI PhycoCosm. Chara braunii S276. https://phycocosm.jgi.doe.gov/Chabra1/Chabra1.home.html (2018).
- 77.Hirooka S, et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA. 2017;114:E8304–E8313. doi: 10.1073/pnas.1707072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.JGI PhycoCosm. Chlamydomonas eustigma NIES-2499. https://phycocosm.jgi.doe.gov/Chleu1/Chleu1.home.html (2017).
- 79.Craig RJ, et al. Comparative genomics of Chlamydomonas. Plant Cell. 2021;33:1016–1041. doi: 10.1093/plcell/koab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.JGI PhycoCosm. Chlamydomonas incerta SAG 7.73. https://phycocosm.jgi.doe.gov/Chlin1/Chlin1.home.html (2021).
- 81.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.JGI PhycoCosm. Chlamydomonas reinhardtii CC-503 v5.6. https://phycocosm.jgi.doe.gov/Chlre5_6/Chlre5_6.home.html (2007).
- 83.JGI PhycoCosm. Chlamydomonas schloesseri CCAP 11/173. https://phycocosm.jgi.doe.gov/Chlsc1/Chlsc1.home.html (2021).
- 84.Hamada M, et al. Metabolic co-dependence drives the evolutionarily ancient Hydra-Chlorella symbiosis. Elife. 2018;7:e35122. doi: 10.7554/eLife.35122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.JGI PhycoCosm. Chlorella sp. A99.https://phycocosm.jgi.doe.gov/ChloA99_1/ChloA99_1.home.html (2018).
- 86.JGI PhycoCosm. Chlorella sorokiniana DOE1412. https://phycocosm.jgi.doe.gov/ChloDOE1412_1/ChloDOE1412_1.home.html (2018).
- 87.JGI PhycoCosm. Chlorella sorokiniana UTEX 1230. https://phycocosm.jgi.doe.gov/Chloso1230_1/Chloso1230_1.home.html (2018). [DOI] [PMC free article] [PubMed]
- 88.Arriola MB, et al. Genome sequences of Chlorella sorokiniana UTEX 1602 and Micractinium conductrix SAG 241.80: implications to maltose excretion by a green alga. Plant J. 2018;93:566–586. doi: 10.1111/tpj.13789. [DOI] [PubMed] [Google Scholar]
- 89.JGI PhycoCosm. Chlorella sorokiniana UTEX 1602. https://phycocosm.jgi.doe.gov/Chloso1602_1/Chloso1602_1.home.html (2018).
- 90.JGI PhycoCosm. Chlorella sorokiniana str. 1228. https://phycocosm.jgi.doe.gov/Chloso1228_1/Chloso1228_1.home.html (2018).
- 91.JGI PhycoCosm. Chlorella variabilis NC64A v1.0. https://phycocosm.jgi.doe.gov/ChlNC64A_1/ChlNC64A_1.home.html (2010).
- 92.Wang S, et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat Plants. 2020;6(2):95–106. doi: 10.1038/s41477-019-0560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.JGI PhycoCosm. Chlorokybus atmophyticus CCAC 0220. https://phycocosm.jgi.doe.gov/Chlat1/Chlat1.home.html (2020).
- 94.Lemieux C, Turmel M, Otis C, Pombert JF. A streamlined and predominantly diploid genome in the tiny marine green alga Chloropicon primus. Nat Commun. 2019;10(1):4061. doi: 10.1038/s41467-019-12014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.JGI PhycoCosm. Chloropicon primus CCMP1205.https://phycocosm.jgi.doe.gov/Chlpri1/Chlpri1.home.html (2019).
- 96.Collén J, et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. 2013;110:5247–5252. doi: 10.1073/pnas.1221259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.JGI PhycoCosm. Chondrus crispus Stackhouse.https://phycocosm.jgi.doe.gov/Chocri1/Chocri1.home.html (2013).
- 98.Roth MS, et al. Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc Natl Acad Sci USA. 2017;114(21):E4296–E4305. doi: 10.1073/pnas.1619928114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.JGI PhycoCosm. Chromochloris zofingiensis SAG 211-14 v5.0.https://phycocosm.jgi.doe.gov/Chrzof1/Chrzof1.home.html (2017).
- 100.JGI PhycoCosm. Chrysochromulina parva Lackey. https://phycocosm.jgi.doe.gov/Chrpa1/Chrpa1.home.html (2019).
- 101.Hovde BT, et al. Genome sequence and transcriptome analyses of Chrysochromulina tobin: metabolic tools for enhanced algal fitness in the prominent order Prymnesiales (Haptophyceae) PLoS Genet. 2015;11:e1005469. doi: 10.1371/journal.pgen.1005469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.JGI PhycoCosm. Chrysochromulina tobin CCMP291.https://phycocosm.jgi.doe.gov/Chrsp_1/Chrsp_1.home.html (2015).
- 103.Liu H, et al. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun Biol. 2018;1:95. doi: 10.1038/s42003-018-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.JGI PhycoCosm. Cladocopium goreaui SCF055-01.https://phycocosm.jgi.doe.gov/Clago1/Clago1.home.html (2018).
- 105.Nishitsuji K, et al. A draft genome of the brown alga, Cladosiphon okamuranus, S-strain: a platform for future studies of ‘mozuku’ biology. DNA Res. 2016;23(6):561–570. doi: 10.1093/dnares/dsw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.JGI PhycoCosm. Cladosiphon okamuranus S strain. https://phycocosm.jgi.doe.gov/Claok1/Claok1.home.html (2016).
- 107.Blanc G, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.JGI PhycoCosm. Coccomyxa subellipsoidea C-169 v3.0.https://phycocosm.jgi.doe.gov/Cosub3/Cosub3.home.html (2012).
- 109.Dorrell RG, et al. Convergent evolution and horizontal gene transfer in Arctic Ocean microalgae. Life Sci Alliance. 2022;6(3):e202201833. doi: 10.26508/lsa.202201833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.JGI PhycoCosm. Cryptophyceae sp. CCMP2293 v1.0.https://phycocosm.jgi.doe.gov/Crypto2293_1/Crypto2293_1.home.html (2017).
- 111.Rossoni AW, et al. The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. Elife. 2019;8:e45017. doi: 10.7554/eLife.45017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.JGI PhycoCosm. Cyanidioschyzon merolae Soos.https://phycocosm.jgi.doe.gov/CyamerSoos_1/CyamerSoos_1.home.html (2019).
- 113.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.JGI PhycoCosm. Cyanidioschyzon merolae strain 10D. https://phycocosm.jgi.doe.gov/Cyamer1/Cyamer1.home.html (2007).
- 115.Price DC, et al. Analysis of an improved Cyanophora paradoxa genome assembly. DNA Res. 2019;26(4):287–299. doi: 10.1093/dnares/dsz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.JGI PhycoCosm. Cyanophora paradoxa CCMP329.https://phycocosm.jgi.doe.gov/Cyapar1/Cyapar1.home.html (2019).
- 117.Traller JC, et al. Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol Biofuels. 2016;9:258. doi: 10.1186/s13068-016-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.JGI PhycoCosm. Cyclotella cryptica CCMP332. https://phycocosm.jgi.doe.gov/Cyccr1/Cyccr1.home.html (2016).
- 119.Polle JEW, et al. Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina Strain CCAP19/18. Genome Announc. 2017;5(43):e01105–17. doi: 10.1128/genomeA.01105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.JGI PhycoCosm. Dunaliella salina CCAP19/18. https://phycocosm.jgi.doe.gov/Dunsal1/Dunsal1.home.html (2017).
- 121.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 122.JGI PhycoCosm. Ectocarpus siliculosus Ec 32. https://phycocosm.jgi.doe.gov/Ectsil1/Ectsil1.home.html (2010).
- 123.JGI PhycoCosm. Edaphochlamys debaryana CCAP 11/70. https://phycocosm.jgi.doe.gov/Edade1/Edade1.home.html (2021).
- 124.Read, BA, et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499(7457):209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 125.JGI PhycoCosm. Emiliania huxleyi CCMP1516 v1.0.https://phycocosm.jgi.doe.gov/Emihu1/Emihu1.home.html (2013).
- 126.JGI PhycoCosm. Enallax costatus CCAP 276/31 v1.0.https://phycocosm.jgi.doe.gov/Enacos1/Enacos1.home.html (2018).
- 127.Tanaka T, et al. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell. 2015;27(1):162–176. doi: 10.1105/tpc.114.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.JGI PhycoCosm. Fistulifera solaris JPCC DA0580.https://phycocosm.jgi.doe.gov/Fisso1/Fisso1.home.html (2015).
- 129.JGI PhycoCosm. Flechtneria rotunda SEV3-VF49 v1.0.https://phycocosm.jgi.doe.gov/Flerot1/Flerot1.home.html (2018).
- 130.Mock T, et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature. 2017;541(7638):536–540. doi: 10.1038/nature20803. [DOI] [PubMed] [Google Scholar]
- 131.JGI PhycoCosm. Fragilariopsis cylindrus CCMP 1102. https://phycocosm.jgi.doe.gov/Fracy1/Fracy1.home.html (2017).
- 132.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350(6261):691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 133.JGI PhycoCosm. Fugacium kawagutii CCMP2468. https://phycocosm.jgi.doe.gov/Fugka2468_1/Fugka2468_1.home.html (2015).
- 134.JGI PhycoCosm. Galdieria phlegrea Soos.https://phycocosm.jgi.doe.gov/Galph1/Galph1.home.html (2019).
- 135.JGI PhycoCosm. Galdieria sulphuraria 002. https://phycocosm.jgi.doe.gov/Gsu002_1/Gsu002_1.home.html (2019).
- 136.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317(5846):1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 137.JGI PhycoCosm. Giardia intestinalis ATCC 50803. https://phycocosm.jgi.doe.gov/Giaint1/Giaint1.home.html (2007).
- 138.Hanschen ER, et al. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 2016;7:11370. doi: 10.1038/ncomms11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.JGI PhycoCosm. Gonium pectorale NIES-2863.https://phycocosm.jgi.doe.gov/Gonpec1/Gonpec1.home.html (2016).
- 140.Lee J, et al. Analysis of the Draft Genome of the Red Seaweed Gracilariopsis chorda Provides Insights into Genome Size Evolution in Rhodophyta. Mol Biol Evol. 2018;35(8):1869–1886. doi: 10.1093/molbev/msy081. [DOI] [PubMed] [Google Scholar]
- 141.JGI PhycoCosm. Gracilariopsis chorda isolate SKKU-2015. https://phycocosm.jgi.doe.gov/Graco1/Graco1.home.html (2018).
- 142.JGI PhycoCosm. Guillardia theta CCMP2712 v1.0.https://phycocosm.jgi.doe.gov/Guith1/Guith1.home.html (2012).
- 143.Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.JGI PhycoCosm. Hyaloperonospora arabidopsidis Emoy2 v2.0.https://phycocosm.jgi.doe.gov/Hyaar1/Hyaar1.home.html (2010).
- 145.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.JGI PhycoCosm. Klebsormidium nitens NIES-2285. https://phycocosm.jgi.doe.gov/Klenit1/Klenit1.home.html (2014).
- 147.JGI PhycoCosm. Mesostigma viride CCAC 1140.https://phycocosm.jgi.doe.gov/Mesovir1/Mesovir1.home.html (2020).
- 148.Cheng S, et al. Genomes of Subaerial Zygnematophyceae provide insights into land plant evolution. Cell. 2019;179:1057–1067.e14. doi: 10.1016/j.cell.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 149.JGI PhycoCosm. Mesotaenium endlicherianum SAG 12.97. https://phycocosm.jgi.doe.gov/Mesen1/Mesen1.home.html (2019).
- 150.JGI PhycoCosm. Micractinium conductrix SAG 241.80.https://phycocosm.jgi.doe.gov/Micco1/Micco1.home.html (2018). [DOI] [PubMed]
- 151.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324(5924):268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 152.JGI PhycoCosm. Micromonas commoda NOUM17 (RCC 299).https://phycocosm.jgi.doe.gov/MicpuN3v2/MicpuN3v2.home.html (2009).
- 153.JGI PhycoCosm. Micromonas pusilla CCMP1545. https://phycocosm.jgi.doe.gov/MicpuC3v2/MicpuC3v2.home.html (2009).
- 154.JGI PhycoCosm. Minidiscus variabilis CCMP495 v1.0. https://phycocosm.jgi.doe.gov/Mintr2/Mintr2.home.html (2020).
- 155.Bogen C, et al. Reconstruction of the lipid metabolism for the microalga Monoraphidium neglectum from its genome sequence reveals characteristics suitable for biofuel production. BMC Genomics. 2013;14:926. doi: 10.1186/1471-2164-14-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.JGI PhycoCosm. Monoraphidium neglectum SAG 48.87.https://phycocosm.jgi.doe.gov/Monneg1/Monneg1.home.html (2013).
- 157.JGI PhycoCosm. Naegleria gruberi v1.0. https://phycocosm.jgi.doe.gov/Naegr1/Naegr1.home.html
- 158.Corteggiani Carpinelli E, et al. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 2014;7:323–335. doi: 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- 159.JGI PhycoCosm. Nannochloropsis gaditana B-31. https://phycocosm.jgi.doe.gov/Nangad1/Nangad1.home.html (2014).
- 160.Vieler A, et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012;8(11):e1003064. doi: 10.1371/journal.pgen.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.JGI PhycoCosm. Nannochloropsis oceanica CCMP1779 v1.0. https://phycocosm.jgi.doe.gov/Nanoce1779/Nanoce1779.home.html (2017).
- 162.Ohan JA, et al. Nuclear Genome Assembly of the Microalga Nannochloropsis salina CCMP1776. Microbiol Resour Announc. 2019;8(44):e00750–19. doi: 10.1128/MRA.00750-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.JGI PhycoCosm. Nannochloropsis salina CCMP1776.https://phycocosm.jgi.doe.gov/Nansal1776_1/Nansal1776_1.home.html (2019).
- 164.Nishitsuji K, et al. Draft genome of the brown alga, Nemacystus decipiens, Onna-1 strain: Fusion of genes involved in the sulfated fucan biosynthesis pathway. Sci Rep. 2019;9(1):4607. doi: 10.1038/s41598-019-40955-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.JGI PhycoCosm. Nemacystus decipiens Onna-1. https://phycocosm.jgi.doe.gov/Nemde1/Nemde1.home.html (2019).
- 166.Oliver A, et al. Diploid genomic architecture of Nitzschia inconspicua, an elite biomass production diatom. Sci Rep. 2021;11(1):15592. doi: 10.1038/s41598-021-95106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.JGI PhycoCosm. Nitzschia inconspicua GAI-293 v2.0.https://phycocosm.jgi.doe.gov/Nithil2/Nithil2.home.html (2021).
- 168.JGI PhycoCosm. Ochromonadaceae sp. CCMP2298 v1.0.https://phycocosm.jgi.doe.gov/Ochro2298_1/Ochro2298_1.home.html (2017).
- 169.JGI PhycoCosm. Ochromonas sp. CCMP1393 v1.4.https://phycocosm.jgi.doe.gov/Ochro1393_1_4/Ochro1393_1_4.home.html (2020).
- 170.JGI PhycoCosm. Ostreococcus sp. RCC809. https://phycocosm.jgi.doe.gov/OstRCC809_2/OstRCC809_2.home.html (2014).
- 171.Blanc-Mathieu R, et al. Population genomics of picophytoplankton unveils novel chromosome hypervariability. Sci Adv. 2017;3:e1700239. doi: 10.1126/sciadv.1700239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.JGI PhycoCosm. Ostreococcus tauri RCC1115 v1.0.https://phycocosm.jgi.doe.gov/Ostta1115_2/Ostta1115_2.home.html (2017).
- 173.Blanc-Mathieu R, et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genomics. 2014;15:1103. doi: 10.1186/1471-2164-15-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.JGI PhycoCosm. Ostreococcus tauri RCC4221 v3.0.https://phycocosm.jgi.doe.gov/Ostta4221_3/Ostta4221_3.home.html (2014).
- 175.Swart EC, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11(1):e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.JGI PhycoCosm. Oxytricha trifallax JRB310. https://phycocosm.jgi.doe.gov/Oxytri1/Oxytri1.home.html (2013).
- 177.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 178.JGI PhycoCosm. Paramecium tetraurelia d4_2. https://phycocosm.jgi.doe.gov/Partet1/Partet1.home.html (2006).
- 179.JGI PhycoCosm. Paraphysomonas imperforata CCMP1604 v1.4. https://phycocosm.jgi.doe.gov/Parimp1_4/Parimp1_4.home.html (2020).
- 180.JGI PhycoCosm. Pavlovales sp. CCMP2436 v1.0.https://phycocosm.jgi.doe.gov/Pavlov2436_1/Pavlov2436_1.home.html (2016).
- 181.JGI PhycoCosm. Pelagophyceae sp. CCMP2097 v1.0.https://phycocosm.jgi.doe.gov/Pelago2097_1/Pelago2097_1.home.html (2017).
- 182.JGI PhycoCosm. Phaeocystis antarctica CCMP1374 v2.2.https://phycocosm.jgi.doe.gov/Phaant1/Phaant1.home.html (2019).
- 183.JGI PhycoCosm. Phaeocystis globosa Pg-G v2.3.https://phycocosm.jgi.doe.gov/Phaglo1/Phaglo1.home.html (2019).
- 184.JGI PhycoCosm. Phaeodactylum tricornutum CCAP 1055/1 v2.0.https://phycocosm.jgi.doe.gov/Phatr2/Phatr2.home.html (2008).
- 185.Lamour KH, et al. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact. 2012;25(10):1350–1360. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.JGI PhycoCosm. Phytophthora capsici LT1534 v11.0.https://phycocosm.jgi.doe.gov/Phyca11/Phyca11.home.html (2012).
- 187.JGI PhycoCosm. Phytophthora cinnamomi var cinnamomi v1.0.https://phycocosm.jgi.doe.gov/Phyci1/Phyci1.home.html (2012).
- 188.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 189.JGI PhycoCosm. Phytophthora infestans T30-4.https://phycocosm.jgi.doe.gov/Phyinf1/Phyinf1.home.html (2009).
- 190.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 191.JGI PhycoCosm. Phytophthora sojae v3.0.https://phycocosm.jgi.doe.gov/Physo3/Physo3.home.html (2006).
- 192.Dahlin LR, et al. Development of a high-productivity, halophilic, thermotolerant microalga Picochlorum renovo. Commun. Biol. 2019;2:388. doi: 10.1038/s42003-019-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.JGI PhycoCosm. Picochlorum renovo. https://phycocosm.jgi.doe.gov/Picre1/Picre1.home.html (2019).
- 194.Gonzalez-Esquer CR, et al. Nuclear, chloroplast, and mitochondrial genome sequences of the prospective microalgal biofuel strain Picochlorum soloecismus. Genome Announc. 2018;6:e01498–17. doi: 10.1128/genomeA.01498-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.JGI PhycoCosm. Picochlorum soloecismus DOE101. https://phycocosm.jgi.doe.gov/Picsp_1/Picsp_1.home.html (2018).
- 196.Junkins EN, et al. Draft Genome Sequence of Picocystis sp. Strain ML, Cultivated from Mono Lake, California. Microbiol Resour Announc. 2019;8:e01353–18. doi: 10.1128/MRA.01353-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.JGI PhycoCosm. Picocystis sp. ML. https://phycocosm.jgi.doe.gov/Pico_ML_1/Pico_ML_1.home.html (2019).
- 198.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.JGI PhycoCosm. Plasmodium falciparum 3D7. https://phycocosm.jgi.doe.gov/Plafal1/Plafal1.home.html (2002).
- 200.Brawley SH, et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta) Proc Natl Acad Sci USA. 2017;114:E6361–E6370. doi: 10.1073/pnas.1703088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.JGI PhycoCosm. Porphyra umbilicalis isolate 4086291. https://phycocosm.jgi.doe.gov/Porumb1/Porumb1.home.html (2017).
- 202.Li L, et al. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat Ecol Evol. 2020;4(9):1220–1231. doi: 10.1038/s41559-020-1221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.JGI PhycoCosm. Prasinoderma coloniale CCMP1413.https://phycocosm.jgi.doe.gov/Praco1/Praco1.home.html (2020).
- 204.JGI PhycoCosm. Pseudo-nitzschia multiseries CLN-47.https://phycocosm.jgi.doe.gov/Psemu1/Psemu1.home.html (2012).
- 205.Nakamura Y, et al. The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis) PLoS One. 2013;8(3):e57122. doi: 10.1371/journal.pone.0057122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.JGI PhycoCosm. Pyropia yezoensis U-51.https://phycocosm.jgi.doe.gov/Pyrye1/Pyrye1.home.html (2013).
- 207.Suzuki S, Yamaguchi H, Nakajima N, Kawachi M. Raphidocelis subcapitata (Pseudokirchneriella subcapitata) provides an insight into genome evolution and environmental adaptations in the Sphaeropleales. Sci Rep. 2018;8(1):8058. doi: 10.1038/s41598-018-26331-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.JGI PhycoCosm. Raphidocelis subcapitata NIES-35.https://phycocosm.jgi.doe.gov/Rapsub1/Rapsub1.home.html (2018).
- 209.Glöckner G, et al. The genome of the foraminiferan Reticulomyxa filosa. Curr Biol. 2014;24(1):11–18. doi: 10.1016/j.cub.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 210.JGI PhycoCosm. Reticulomyxa filosa. https://phycocosm.jgi.doe.gov/Retfil1/Retfil1.home.html (2014).
- 211.Ye N, et al. Saccharina genomes provide novel insight into kelp biology. Nat Commun. 2015;6:6986. doi: 10.1038/ncomms7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.JGI PhycoCosm. Saccharina japonica str. Ja. https://phycocosm.jgi.doe.gov/Sacja1/Sacja1.home.html (2015).
- 213.Jiang RH, et al. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet. 2013;9:e1003272. doi: 10.1371/journal.pgen.1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.JGI PhycoCosm. Saprolegnia parasitica CBS 223.65. https://phycocosm.jgi.doe.gov/Sappar1/Sappar1.home.html (2013).
- 215.JGI PhycoCosm. Scenedesmus obliquus EN0004 v1.0.https://phycocosm.jgi.doe.gov/SceoblEN4_1/SceoblEN4_1.home.html (2020).
- 216.JGI PhycoCosm. Scenedesmus obliquus UTEX 393. https://phycocosm.jgi.doe.gov/Sobl393_1/Sobl393_1.home.html (2017).
- 217.Starkenburg SR, et al. Draft nuclear genome, complete chloroplast genome, and complete mitochondrial genome for the biofuel/bioproduct feedstock species Scenedesmus obliquus strain DOE0152z. Genome Announc. 2017;5(32):e00617–17. doi: 10.1128/genomeA.00617-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.JGI PhycoCosm. Scenedesmus obliquus UTEX B 3031. https://phycocosm.jgi.doe.gov/Sceobl1/Sceobl1.home.html (2017).
- 219.Calhoun S, et al. A multi-omic characterization of temperature stress in a halotolerant Scenedesmus strain for algal biotechnology. Commun. Biol. 2021;4:333. doi: 10.1038/s42003-021-01859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.JGI PhycoCosm. Scenedesmus sp. NREL 46B-D3 v1.0.https://phycocosm.jgi.doe.gov/Scesp_1/Scesp_1.home.html (2021).
- 221.JGI PhycoCosm. Schizochytrium aggregatum ATCC 28209 v1.0.https://phycocosm.jgi.doe.gov/Schag1/Schag1.home.html (2013).
- 222.Osuna-Cruz CM, et al. The Seminavis robusta genome provides insights into the evolutionary adaptations of benthic diatoms. Nat Commun. 2020;11(1):3320. doi: 10.1038/s41467-020-17191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.JGI PhycoCosm. Seminavis robusta D6. https://phycocosm.jgi.doe.gov/Semro1/Semro1.home.html (2020).
- 224.JGI PhycoCosm. Symbiochloris reticulata Spain reference genome v1.0. https://phycocosm.jgi.doe.gov/Dicre1/Dicre1.home.html (2016).
- 225.Aranda M, et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.JGI PhycoCosm. Symbiodinium microadriaticum CCMP2467. https://phycocosm.jgi.doe.gov/Symmic1/Symmic1.home.html (2016).
- 227.Featherston J, et al. The 4-Celled Tetrabaena socialis nuclear genome reveals the essential components for genetic control of cell number at the origin of multicellularity in the Volvocine lineage. Mol. Biol. Evol. 2018;35:855–870. doi: 10.1093/molbev/msx332. [DOI] [PubMed] [Google Scholar]
- 228.JGI PhycoCosm. Tetrabaena socialis NIES-571. https://phycocosm.jgi.doe.gov/Tetso1/Tetso1.home.html (2018).
- 229.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.JGI PhycoCosm. Tetrahymena thermophila SB210. https://phycocosm.jgi.doe.gov/Tetthe1/Tetthe1.home.html (2006).
- 231.Lommer M, et al. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 2006;13(7):R66x. doi: 10.1186/gb-2012-13-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.JGI PhycoCosm. Thalassiosira oceanica CCMP1005.https://phycocosm.jgi.doe.gov/Thaoce1/Thaoce1.home.html (2012).
- 233.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 234.JGI PhycoCosm. Thalassiosira pseudonana CCMP 1335 v3.0.https://phycocosm.jgi.doe.gov/Thaps3/Thaps3.home.html (2004).
- 235.Kissinger JC, Gajria B, Li L, Paulsen IT, Roos DS. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31(1):234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.JGI PhycoCosm. Toxoplasma gondii ME49. https://phycocosm.jgi.doe.gov/Toxgon1/Toxgon1.home.html (2003).
- 237.Greshake Tzovaras B, et al. What is in Umbilicaria pustulata? A metagenomic approach to reconstruct the holo-genome of a lichen. Genome Biol. Evol. 2020;12:309–324. doi: 10.1093/gbe/evaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.JGI PhycoCosm. Trebouxia sp. A1-2.https://phycocosm.jgi.doe.gov/TrebA12_1/TrebA12_1.home.html (2020).
- 239.Mahan KM, et al. Annotated genome sequence of the high-biomass-producing yellow-green alga Tribonema minus. Microbiol Resour Announc. 2021;10(24):e0032721. doi: 10.1128/MRA.00327-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.JGI PhycoCosm. Tribonema minus UTEX B 3156 v1.0. https://phycocosm.jgi.doe.gov/Trimin1/Trimin1.home.html (2021).
- 241.Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.JGI PhycoCosm. Trichomonas vaginalis G3.https://phycocosm.jgi.doe.gov/Trivag1/Trivag1.home.html (2007).
- 243.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 244.JGI PhycoCosm. Trypanosoma brucei brucei TREU927. https://phycocosm.jgi.doe.gov/Trybru1/Trybru1.home.html (2005).
- 245.De Clerck O, et al. Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol. 2018;28:2921–2933.e5. doi: 10.1016/j.cub.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 246.JGI PhycoCosm. Ulva mutabilis Foyn. https://phycocosm.jgi.doe.gov/Ulvmu1/Ulvmu1.home.html (2018).
- 247.Shan T, et al. First genome of the brown alga Undaria pinnatifida: Chromosome-level assembly using PacBio and Hi-C technologies. Front Genet. 2020;11:140. doi: 10.3389/fgene.2020.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.JGI PhycoCosm. Undaria pinnatifida M23.https://phycocosm.jgi.doe.gov/Undpi1/Undpi1.home.html (2020).
- 249.Woo YH, et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife. 2015;4:e06974. doi: 10.7554/eLife.06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.JGI PhycoCosm. Vitrella brassicaformis CCMP3155. https://phycocosm.jgi.doe.gov/Vitbras1/Vitbras1.home.html (2015).
- 251.Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329(5988):223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.JGI PhycoCosm. Volvox carteri v2.1.https://phycocosm.jgi.doe.gov/Volca2_1/Volca2_1.home.html (2010).
- 253.Schmitt P, Gueguen Y, Desmarais E, Bachère E, de Lorgeril J. Molecular diversity of antimicrobial effectors in the oyster Crassostrea gigas. BMC Evol Biol. 2010;10:23. doi: 10.1186/1471-2148-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.NCBI GenBank. Crassostrea gigas genome assembly cgigas_uk_roslin_v1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_902806645.1/ (2020).
- 255.Albertin CB, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524(7564):220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.NCBI GenBank. Octopus bimaculoides genome assembly Octopus_bimaculoides_v2_0. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_001194135.1/ (2015).
- 257.Knudsen B, Kohn AB, Nahir B, McFadden CS, Moroz LL. Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica: conservation of the gene order in Euthyneura. Mol Phylogenet Evol. 2006;38(2):459–469. doi: 10.1016/j.ympev.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 258.NCBI GenBank. Aplysia californica genome assembly AplCal3.0.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000002075.1/ (2013).
- 259.Swart E, et al. Species-specific transcriptomic responses in Daphnia magna exposed to a bio-plastic production intermediate. Environ Pollut. 2019;252(Pt A):399–408. doi: 10.1016/j.envpol.2019.05.057. [DOI] [PubMed] [Google Scholar]
- 260.NCBI GenBank. Daphnia magna genome assembly ASM2063170v1.1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_020631705.1/ (2021).
- 261.Polinski JM, et al. The American lobster genome reveals insights on longevity, neural, and immune adaptations. Sci Adv. 2021;7(26):eabe8290. doi: 10.1126/sciadv.abe8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.NCBI GenBank. Homarus americanus genome assembly GMGI_Hamer_2.0.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_018991925.1/ (2021).
- 263.Denoeud F, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.NCBI GenBank. Oikopleura dioica genome assembly ASM20953v1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_000209535.1/ (2010).
- 265.NCBI GenBank. Hippoglossus stenolepis genome assembly HSTE1.2. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_022539355.2/ (2021).
- 266.NCBI GenBank. Tursiops truncatus genome assembly mTurTru1.mat.Y https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_011762595.1/ (2020).
- 267.NCBI GenBank. Dibothriocephalus latus genome assembly D_latum_Geneva_0011_upd. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_900617775.1/ (2018).
- 268.Young ND, et al. The Opisthorchis viverrini genome provides insights into life in the bile duct. Nat Commun. 2014;5:4378. doi: 10.1038/ncomms5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.NCBI GenBank. Opisthorchis viverrini genome assembly OpiViv1.0.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000715545.1/ (2014).
- 270.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.NCBI GenBank. Capitella teleta genome assembly Capca1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_000328365.1/ (2013).
- 272.NCBI GenBank. Helobdella robusta genome assembly Helobdella robusta v1.0. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000326865.1/ (2012).
- 273.NCBI GenBank. Anisakis simplex genome assembly A_simplex_0011_upd. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_900617985.1/ (2018).
- 274.NCBI GenBank. Trichuris trichiura genome assembly TTRE2.1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_000613005.1/ (2014).
- 275.Simakov O, et al. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci Adv. 2022;8(5):eabi5884. doi: 10.1126/sciadv.abi5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.NCBI GenBank. Hydra vulgaris genome assembly Hydra_105_v3.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_022113875.1/ (2021).
- 277.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 278.NCBI GenBank. Nematostella vectensis genome assembly ASM20922v1. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000209225.1/ (2007).
- 279.Zhang X, et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017;15(10):e2003790. doi: 10.1371/journal.pbio.2003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.NCBI GenBank. Apostichopus japonicus genome assembly ASM275485v1.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_002754855.1/ (2017).
- 281.Sodergren E, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314(5801):941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.NCBI GenBank. Strongylocentrotus purpuratus genome assembly Spur_5.0. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000002235.5/ (2019).
- 283.Rayko M, et al. Draft genome of Bugula neritina, a colonial animal packing powerful symbionts and potential medicines. Sci Data. 2020;7(1):356. doi: 10.1038/s41597-020-00684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.NCBI GenBank. Bugula neritina genome assembly ASM1079987v2. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_010799875.2/ (2020).
- 285.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466(7307):720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.NCBI GenBank. Amphimedon queenslandica genome assembly v1.0.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000090795.1/ (2010).
- 287.Luo YJ, et al. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun. 2015;6:8301. doi: 10.1038/ncomms9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.NCBI GenBank. Lingula anatina genome assembly LinAna2.0. https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_001039355.2/ (2019).
- 289.NCBI GenBank. Adineta ricciae genome assembly Ar_ARIC003_reference_genomic_v1.https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCA_905250025.1/ (2021).
- 290.Hulatt CJ, Wijffels RH, Posewitz MC. The Genome of the Haptophyte Diacronema lutheri (Pavlova lutheri, Pavlovales): A Model for Lipid Biosynthesis in Eukaryotic Algae. Genome Biol Evol. 2021;13:evab178. doi: 10.1093/gbe/evab178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.NCBI GenBank. Diacronema lutheri strain:NIVA-4/92. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA725470/ (2021).
- 292.Mistry J, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Guillou L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012;41(D1):D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.del Campo J, et al. EukRef: Phylogenetic curation of ribosomal RNA to enhance understanding of eukaryotic diversity and distribution. PLOS Biology. 2018;16:e2005849. doi: 10.1371/journal.pbio.2005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Lasek-Nesselquist E, Johnson MD. A phylogenomic approach to clarifying the relationship of Mesodinium within the Ciliophora: a case study in the complexity of mixed-species transcriptome analyses. Genome Biology and Evolution. 2019;11(11):3218–3232. doi: 10.1093/gbe/evz233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Van Vlierberghe M, Di Franco A, Philippe H, Baurain D. Decontamination, pooling and dereplication of the 678 samples of the Marine Microbial Eukaryote Transcriptome Sequencing Project. BMC Res Notes. 2021;14(1):306. doi: 10.1186/s13104-021-05717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Groussman RD, Blaskowski S, Coesel SN, Armbrust EV. 2023. MarFERReT: an open-source, version-controlled reference library of marine microbial eukaryote functional genes (1.1) [Data set] Zenodo. [DOI] [PMC free article] [PubMed]
- 299.Groussman, R. D., Blaskowski, S., Coesel, S. N. Marine Functional EukaRyotic Reference Taxa (Version 1.1) [Computer software]. https://github.com/armbrustlab/marferret (2023). [DOI] [PMC free article] [PubMed]
- 300.Grigoriev IV, et al. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res. 2021;49:D1004–D1011. doi: 10.1093/nar/gkaa898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 302.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.FHCRC Computational Biology. taxtastic. Githubhttps://github.com/fhcrc/taxtastic (2022).
- 304.Bachmann M. maxbachmann/RapidFuzz: Release 1.8.0 [Computer software]. 2021 doi: 10.5281/zenodo.5584996. [DOI] [Google Scholar]
- 305.Paysan-Lafosse T, et al. InterPro in 2022. Nucleic acids research. 2023;51(D1):D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 307.Steinegger M, Söding J. Clustering huge protein sequence sets in linear time. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-04964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 309.Hogle SL. 2019. MARMICRODB database for taxonomic classification of (marine) metagenomes (1.0.0) [Data set] Zenodo. [DOI]
- 310.Groussman RD. 2023. Codebase and documentation for MarFERReT microbial eukaryote reference sequence library (Version 1.1) [Software] Zenodo. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Liu Z, Hu S, Caron D. 2018. EukZoo, an aquatic protistan protein database for meta-omics studies. (0.2) [Data set] Zenodo. [DOI]
- Guajardo M, Jimenez V, Vaulot D, Trefault N. 2021. Transcriptomes from Thalassiosira and Minidiscus diatoms from English Channel and Antarctic coastal waters (Version 1) [Data set] Zenodo. [DOI]
- Johnson LK, Alexander H, Brown CT. 2017. MMETSP re-assemblies [Data set] Zenodo. [DOI]
- Niang G, 2020. METdb: a genomic reference database for marine species [Data set] Zenodo. [DOI]
- Guajardo M, Jimenez V, Vaulot D, Trefault N. 2021. (Assemblies) Transcriptomes from Thalassiosira and Minidiscus diatoms from English Channel and Antarctic coastal waters (Version 1) [Data set] Zenodo. [DOI]
- Groussman RD, 2022. Transcriptome assemblies of three diatom and three prymnesiophyte isolates from station ALOHA and Kaneohe Bay (1.0) [Data set] Zenodo. [DOI]
- Mars Brisbin M, Mitarai S. 2018. Phaeocystis globosa colonial gene expression. Zenodo. https://zenodo.org/record/1476491
- Groussman RD, Blaskowski S, Coesel SN, Armbrust EV. 2023. MarFERReT: an open-source, version-controlled reference library of marine microbial eukaryote functional genes (1.1) [Data set] Zenodo. [DOI] [PMC free article] [PubMed]
- Hogle SL. 2019. MARMICRODB database for taxonomic classification of (marine) metagenomes (1.0.0) [Data set] Zenodo. [DOI]
- Groussman RD. 2023. Codebase and documentation for MarFERReT microbial eukaryote reference sequence library (Version 1.1) [Software] Zenodo. [DOI]
Data Availability Statement
Code, documentation, and tutorials for this project are available on the MarFERReT repository299: https://github.com/armbrustlab/marferret. This repository has also been archived for the v.1.1 release310 and the archived code is available on Zenodo here: https://zenodo.org/records/10278540. Information on the software versions and parameters used in this publication are included in the MarFERReT containerized build on the repository and in the archived code.