Highlights
-
•
N-terminal modification improves the catalytic activity of 1,4-α-glucan branching enzymes.
-
•
Two mutants exhibited enhanced specificity activities (up to 1.28-fold), compared to the wild-type GBE.
-
•
The α-1,6-glycosidic linkage ratios of maltodextrin samples modified by L25R and L25A were significantly increased compared to wild-type GBE.
Keywords: 1,4-α-glucan branching enzyme; N-terminal domain; Enzymatic activity; Salt bridge; Carbohydrate-binding module 48
Abstract
The 1,4-α-glucan branching enzyme (GBE, EC 2.4.1.18) has garnered considerable attention for its ability to increase the degree of branching of starch and retard starch digestion, which has great industrial applications. Previous studies have reported that the N-terminal domain plays an important role in the expression and stability of GBEs. To further increase the catalytic ability of Gt-GBE, we constructed five mutants in the N-terminal domain: L19R, L19K, L25R, L25K, and L25A. Specific activities of L25R and L25A were increased by 28.46% and 23.46%, respectively, versus the wild-type Gt-GBE. In addition, the α-1,6-glycosidic linkage ratios of maltodextrin samples treated with L25R and L25A increased to 5.71%, which were significantly increased by 19.96% compared with that of the wild-type Gt-GBE. The results of this study suggest that the N-terminal domain selective modification can improve enzyme catalytic activity, thus further increasing the commercial application of enzymes in food and pharmaceutical industries.
Introduction
The 1,4-α-glucan branching enzyme (GBE, EC 2.4.1.18) catalyzes the cleavage of α-1,4-glycosidic linkages in starch to form short glucan chains and short chains attach to the acceptor chains via α-1,6-glycosidic linkages to form new branches, which increases the degree of branching of starch (Abad et al., 2002, Feng et al., 2016, Hayashi et al., 2017). GBEs have significant potential of improving the slow digestion and anti-digestive properties of starch with their unique transglycosylation (Jo et al., 2016, Ren et al., 2020, Ming-Mao et al., 2011, Park et al., 2018, Tetlow and Emes, 2014, Yu et al., 2021). However, most GBEs have problems in the process of application, such as large amount of enzyme addition and long reaction time. To overcome these drawbacks, it has become a key research point to improve the catalytic activity of GBEs to expand the application in food and pharmaceutical industries.
Most GBEs belong to glycoside hydrolase family 13 (GH13) and possess 3 common domains: a carboxyl-terminal domain, a central (β/α)8-barrel catalytic domain, and an amino-terminal domain (Abad et al., 2002, Feng et al., 2016, Suzuki and Suzuki, 2016). Among them, the N-terminal domain, which is an important structural and functional domain of proteins, plays an important role in maintaining the structural stability and functional properties of proteins. Currently, a series of N-terminal domain structural modification strategies have been explored to improve the catalytic activity of enzymes. For example, Ni et al. found that truncation of the N-terminal domain of Limosilactobacillus reuteri 121, Lactobacillus jensenii JV-V16, L. johnsonii NCC533, L. gasseri DSM20604 and L. reuteri LTH5448 fructansucrases showed an obvious effect on the catalytic activity without altering the product specificity (Ni et al., 2021). Zheng et al. truncated the first 10N-terminal amino acid residues of xylanase, and the mutant exhibited a 1.36-fold increase in enzyme activity (Zheng et al., 2016). Xiong et al. constructed a recombinant mutant PTxA-DB by replacing five residues (T10Y, N11H, N12D, Y15F, N30L), combined with an additional disulfide bridge (T2C-T29C) in the N-terminal region of xylanase PjxA. The specific activity of PTxA-DB was increased by 1.72-fold, compared with the wild-type (Xiong et al., 2019). Feng et al. mutated the highly flexible residues Ala26 and Gly29 of variant L6 of l-asparaginase to obtain the variant L6-A26N/G29F. The variant showed a 3.44-fold increase in specific activity in contrast to the wild-type (Feng et al., 2019).
GBE from Geobacillus thermoglucosidans STB02 (Gt-GBE) has good thermostability with an optimum temperature of 60°C (Ban et al., 2018). Compared with other GBEs, Gt-GBE is more capable of meeting high temperature requirement of starch gelatinization process in practical applications. However, Gt-GBE has problems such as large enzyme addition in production process, which limits their application in industries. Therefore, the catalytic activity of Gt-GBE needs to be enhanced by molecular modification. In the study of constructing a series of progressive N-terminal domain truncation mutants of Gt-GBE, it was determined that the N-terminal domain plays an important role in maintaining the catalytic activity and thermostability, especially the region where the first 10 N-terminal amino acids are located (Xin et al., 2019). Based on this finding, we selected the region containing the first 10 N-terminal amino acids of Gt-GBE for further molecular modification. Sequence analysis indicated that the surface of Gt-GBE contains many charged residues, which is conducive to the formation of electrostatic interactions (salt bridges). Thus, we analyzed the structure of the region containing the first 10 N-terminal amino acids and their adjacent regions (<5 Å) to identify potential amino acids for targeted mutagenesis to form intramolecular salt bridges. To reduce the blindness of constructing salt bridges, we searched for appropriate amino acid residue sites to mutate into charged amino acid residues to form salt bridges with the original charged amino acid residues of Gt-GBE. According to the analysis of the three-dimensional (3D) simulated structure of Gt-GBE, two amino acid sites, 19 and 25, were selected as target amino acids for the construction of salt bridges. In the present study, we constructed five GBE variants (L19K, L19R, L25R, L25K, and L25A) in the N-terminal domain to increase the specific activity and catalytic efficiency.
Materials and methods
Strains, plasmids and materials
The DNA sequence of 1,4-α-glucan branching enzyme gene Gt-gbe was obtained from NCBI (GenBank: KJ660983.1). The Gt-gbe/pET-20b(+) was constructed in our laboratory. Escherichia coli JM109 was used as the cloning host, while E. coli BL21(DE3) was used for protein expression of the wild-type Gt-GBE and mutants. Isoamylase (I5284, EC 3.2.1.68, 10,000,000 U/mg protein) and potato amylopectin were purchased from Sigma Chemical Co. (St. Louis, MO, USA). High-fidelity DNA polymerase was purchased from Vazyme Biotechnology Co. (Nanjing, China). All other chemicals were obtained from Sinopharm Chemical Reagent Co. (Shanghai, China).
Construction of mutants
The recombinant plasmid Gt-gbe/pET-20b(+) was used as the template to generate mutated enzyme genes by one-step PCR method. The PCR primers required for each mutation were recognized by two complementary oligonucleotides, one for the sence and another for the antisence strand. Their sequences are presented in Supplementary Table 1. PCR-amplified products were incubated with DpnI overnight at 37°C and then transformed into E. coli JM109 competent cells.
Production and purification of enzymes
Production of GBE proteins were performed as previously described, with minor modifications (Liu et al., 2017). A single colony of E. coli BL21(DE3) cells harboring recombinant Gt-gbe/pET-20b(+) or its mutant plasmid was inoculated into Luria-Bertani (LB) medium containing 100 μg/mL ampicillin and incubated at 37°C for 6–8 h until the absorbance at 600 nm reached approximately 0.6. And then, 1% portion of LB culture was added to Terrific Broth (TB) medium and incubated on a rotary shaker at 30°C for 48 h. Subsequently, the supernatant (crude enzyme) was collected after centrifugation at 10,000×g for 15 min at 4°C. In practice, we found that many proteins could be expressed without IPTG. Therefore, we expressed GBE without adding IPTG for cost-saving purposes.
The crude enzyme solution was purified by His-tag affinity chromatography as previously described (Ban et al., 2020). The crude enzyme solution was loaded onto the 5 mL HisTrap™ HP Column (GE Healthcare) equilibrated with buffer A (20 mM imidazole, 500 mM NaCl, 50 mM Tris-HCl, pH 7.5). The bound enzyme was eluted with 60% buffer B (500 mM imidazole, 500 mM NaCl, 50 mM Tris-HCl, pH 7.5). The eluate was pooled and stored at −80 °C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Bradford protein assay were performed for analyzing the protein purity and the total protein concentration.
Enzyme activity assay
The activities of GBEs were measured using iodine assay as previously described (Liu et al., 2017). Potato amylopectin type III (Sigma, USA) was used as the substrate and dissolved in 50 mM sodium phosphate buffer (pH 7.5). Then, 50 μL of the enzyme extract was added to 950 μL of the substrate solution (2.5 mg/mL) and incubated at 50°C for 15 min. The reaction was aborted by boiling for 30 min. The resultant reaction mixture (300 µL) was subsequently mixed with 5 mL of the iodine reagent at room temperature in the dark, followed by the measurement of the absorbance at 530 nm (A530) after 15 min. One unit of enzyme activity was defined as the amount of branching enzyme that caused a 1% decrease in absorbance value per min (Ban et al., 2016, Palomo et al., 2009).
Enzyme thermostability assay
The effects of different temperatures on the activities of the wild-type Gt-GBE and mutants were determined at temperature ranging from 30 to 70°C. The activities of GBEs were measured at each temperature, and the one exhibiting the highest enzymatic activity was defined as the optimum temperature for the enzymatic reaction.
The thermostability of the wild-type Gt-GBE and mutants were determined by pre-incubating the enzymes for 2 h at 60°C. Each pre-incubation mixture was sampled several times at appropriate intervals, and the residual activity was measured (Palomo et al., 2009). The half-life of the wild-type Gt-GBE and its mutants were obtained when half of activity was lost (Ban et al., 2020).
Proton nuclear magnetic resonance hydrogen-1 spectroscopic analysis
Proton nuclear magnetic resonance hydrogen-1 (1H NMR) facilitates the identification of the α-1,4- and α-1,6-glycosidic linkages of starch samples. The differences in the catalytic ability of different GBE mutants could be characterized based on the amounts of α-1,4- and α-1,6-glycosidic linkages. The analysis was performed as described elsewhere (Li et al., 2016), albeit with some modifications. The control was treated without Gt-GBE. Freeze-dried maltodextrin samples were dissolved in D2O (final concentration of 20 mg/mL (W/V)) and then boiled for pasting. Analysis was performed using the 1H NMR spectra. The chemical shifts in the anomeric protons of the α-1,4 and α-1,6-glycosidic linkages were detected at 5.37 ppm and 4.96 ppm, respectively. The α-1,6 glycosidic linkage ratio was calculated by dividing the area of α-1,6 glycosidic linkage peak by the total area of α-1,4 glycosidic linkage and α-1,6 glycosidic linkage peaks.
Chain length distribution
The chain length distribution of maltodextrin samples was analyzed as previously described, with slight modifications (Kong et al., 2018). Maltodextrin was incubated with the wild-type Gt-GBE and mutants (L25R, L25K, L25A) at 50°C, respectively. The reaction mixture was boiled for 30 min to stop the reaction. The treated maltodextrin samples were then dried by vacuum freeze dryer. Each lyophilized maltodextrin sample (10 mg) was dissolved in 2 mL of sodium acetate buffer (50 mM, pH 3.5) and incubated with isoamylase (100 U/mg substrate) at 40°C for 24 h to hydrolyze α-1,6-glycosidic linkages, followed by boiling for 30 min to terminate the reaction. Maltodextrin treated without GBE served as the control. Debranched samples were analyzed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using an HPAEC system (ICS-5000, Dionex Co., USA) equipped with a pulsed amperometric detector (PAD) and a CarboPac PA200 column (ThermoFisher Scientific, Waltham, MA, USA). The gradient elution procedure was reported in a previous study (Ren et al., 2017). The chain length distribution was characterized as a percentage of the total peak area (Tian et al., 2016).
Statistical analysis
The values are presented as the means of triplicate observations. Data from the survey were analyzed using the SPSS software. Data results causing a difference of P < 0.05 were considered to indicate statistical significance.
Structure modeling
Based on the amino acid sequence of Gt-GBE, the crystal structure of E. coli GBE (PDB: 1M7X), which has high sequence identity (44.83%) of Gt-GBE, was selected as the template to simulate the homology model of GBE mutants. The SWISS-MODEL server was used to simulate the homology models. PyMOL was used to analyze the 3D structures and visualize the interactions between different amino acid residues.
Results
Expression, production, and purification of the wild-type Gt-GBE and its mutants
Wild-type Gt-GBE and five mutants, L19K, L19R, L25R, L25K and L25A, were inserted into the pET-20b(+) plasmid and cloned in E. coli BL21(DE3). Using potato amylopectin as the substrate, the enzyme activities were assayed as described in the “Materials and methods” section. All GBEs showed the ability to form branches. SDS-PAGE analysis also showed that the five mutants were highly expressed in E. coli BL21(DE3). No significant difference was observed in the expression of the wild-type Gt-GBE and mutants (data not shown).
GBEs were purified by Histidine-tag (His-tag) affinity chromatography using the HisTrap™ HP column as described above. In SDS-PAGE analysis, all GBEs showed a single band, which was consistent with their theoretical molecular weights (Supplementary Fig. 1).
Thermostability of the wild-type Gt-GBE and its mutants
The increase in enzyme catalytic activity is often accompanied by an increase in enzyme structure flexibility, which is more conducive to substrate binding. On the contrary, the thermostability of enzymes requires a decrease in the flexibility of the enzyme structure, which makes it more resistant to the thermal effects. Therefore, an increase in the catalytic activity may lead to a decrease in thermostability (Zhong et al., 2009, Lonhienne et al., 2000).
Firstly, we determined the thermostability of the wild-type Gt-GBE and five mutant crude enzymes at 60°C. The thermostability assay showed that among these mutants, L19K and L19R lost most of their activities after 20 min of incubation, while L25R, L25A and L25K exhibited relatively high thermostability compared with the wild-type Gt-GBE (Supplementary Fig. 2). Thus, we selected these three mutants, L25R, L25A, and L25K for further studies.
We next determined the optimum temperature of the wild-type Gt-GBE and its mutants in the temperature range of 30–70°C using amylopectin as the substrate. As shown in Fig. 1A, the mutants L25R, L25A and L25K showed the highest specific activity at 60°C, which was similar to the wild-type Gt-GBE. Meanwhile, we determined the thermostability of GBEs at 60°C, the results are presented in Table 1 and Fig. 1B. The half-life at 60°C (t (1/2, 60°C)) of L25R and L25A were not significantly different from that of the wild-type Gt-GBE. Also, the wild-type Gt-GBE, L25R and L25A maintained over 40% of residual activity after incubating at 60 °C for 50 min. These results indicated that the mutation of Leu25 to Arg and Ala did not significantly decrease the thermostability.
Fig. 1.
Characteristics of the wild-type Gt-GBE and its mutants. A: Optimum temperature of the wild-type Gt-GBE and mutants. B: Relative activity of GBEs after incubation of 50 min at 60°C. C: 1H NMR of maltodextrin treated with wild-type Gt-GBE and mutants L25R, L25A, and L25K. Significant differences are indicated by different letters (P < 0.05). D: Chain length distribution analysis of maltodextrin before and after modification by wild-type Gt-GBE or mutant GBEs. E: 1H NMR spectrum of maltodextrin samples treated with GBEs.
Table 1.
Thermostability parameters of GBEs.
| Protein samples | t (1/2, 60°C) (min)a |
|---|---|
| WT | 46.1 ± 0.2b |
| L25A | 48.2 ± 0.1b |
| L25R | 45.0 ± 0.1b |
| L25K | 41.1 ± 0.5a |
a Data with different superscript letters within a column are significantly different (P < 0.05).
Enzyme activity of the wild-type Gt-GBE and its mutants
The specific activities of GBEs were determined at 50°C and pH 7.5 using iodine stain assay. The specific activities of the wild-type Gt-GBE and its mutants are shown in Table 2. Among the three mutants, L25R and L25A exhibited significantly higher specific activities compared with the wild-type Gt-GBE, with a 1.28-fold and 1.23-fold increase in specific activity, respectively. In addition, the enzymatic activity of L25K was similar to that of the wild-type Gt-GBE.
Table 2.
Specific activities of GBEs.
| Protein samples | Specific activity (U/mg)a | Relative enzyme activity (%) |
|---|---|---|
| WT | 1790.16 ± 55.60b | 100.00 |
| L25R | 2299.58 ± 54.35d | 128.46 |
| L25A | 2210.13 ± 117.28c | 123.46 |
| L25K | 1716.23 ± 19.29a | 95.87 |
a Values are presented as means ± standard deviation (n = 3). Data with different superscript letters within a column are significantly different (P < 0.05).
1H NMR analysis
GBE catalyze the hydrolysis of α-1,4-glycosidic linkages of maltodextrin and form new α-1,6-glycosidic linkages by transglycosylation. Thus, compared with unmodified maltodextrin, GBE-modified maltodextrin contains more α-1,6-glycosidic linkages. To further characterize the changes in catalytic properties of the GBE variations, we measured the α-1,6-glycosidic linkage ratios of the products modified by wild-type Gt-GBE and its mutants using 1H NMR. The 1H NMR spectrum of maltodextrin samples treated with GBEs is shown in Fig. 1E, and the α-1,6-glycosidic linkage ratios of each sample, which were further calculated, are shown in Fig. 1C. The maltodextrin sample treated with the wild-type Gt-GBE exhibited an α-1,6-glycosidic linkage ratio of 4.76%. The α-1,6-glycosidic linkage ratios of mutants, L25R and L25A, were 5.71%, which were considerably higher than that of the wild-type Gt-GBE. However, the mutant L25K showed the same α-1,6-glycosidic linkage ratio as the wild-type Gt-GBE. These results indicated that the mutants L25R and L25A have higher specific enzyme activities than wild-type Gt-GBE.
Branched chain length distribution of GBE-treated maltodextrin
To investigate whether changes in the transglycosylation pattern of mutants, we determined the chain length distribution of maltodextrins after incubating with the wild-type Gt-GBE and its mutants. The chain lengths were divided into the degree of polymerization (DP) 1–12, DP 13–24, DP 25–36, and DP > 36. HPAEC-PAD was used to analyze the products after debranching with isoamylase enzyme. As shown in Table 3, compared with non-modified maltodextrin, the amount of short chains (DP 1–12) of maltodextrin modified by wild-type Gt-GBE was significantly higher than that of other chain lengths, with an increase of 4.45% (from 67.04% to 71.49%). The result indicated that during the modification of maltodextrin, Gt-GBE hydrolyzed the longer chains and generated more short chains (DP 1–12).
Table 3.
Chain length distribution of maltodextrins treated with the wild-type Gt-GBE and its mutants L25R, L25A, and L25K.
| Protein samples | Branch chain length distribution (%)a |
|||
|---|---|---|---|---|
| DP 1–12 | DP 13–24 | DP 25–36 | DP > 36 | |
| Control | 67.04 ± 0.11a | 26.06 ± 0.02c | 5.58 ± 0.03e | 1.33 ± 0.01c |
| WT | 71.49 ± 0.02b | 25.21 ± 0.04a | 2.98 ± 0.09d | 0.33 ± 0.05b |
| L25R | 72.13 ± 0.04c | 25.26 ± 0.15a | 2.12 ± 0.02a | 0.04 ± 0.01a |
| L25A | 72.01 ± 0.02d | 25.07 ± 0.01a | 2.89 ± 0.05c | 0.04 ± 0.01a |
| L25K | 71.76 ± 0.13e | 25.48 ± 0.12b | 2.70 ± 0.02b | 0.06 ± 0.02a |
a Data with different superscript letters within a column are significantly different (P < 0.05).
As shown in Table 3 and Fig. 1D, maltodextrins incubated with GBE variations showed a similar transglycosylation pattern as the wild-type Gt-GBE. Of these, compared with the wild-type Gt-GBE, maltodextrins incubated with L25R or L25A showed an increase in the amount of short chains and a decrease in the amount of long chains. This was probably caused by the fact that the mutation site was far from the catalytic center, without changing the polar environment of catalytic center, and the change in amino acid side-chain length did not affect the interactions near binding sites.
Structural analysis
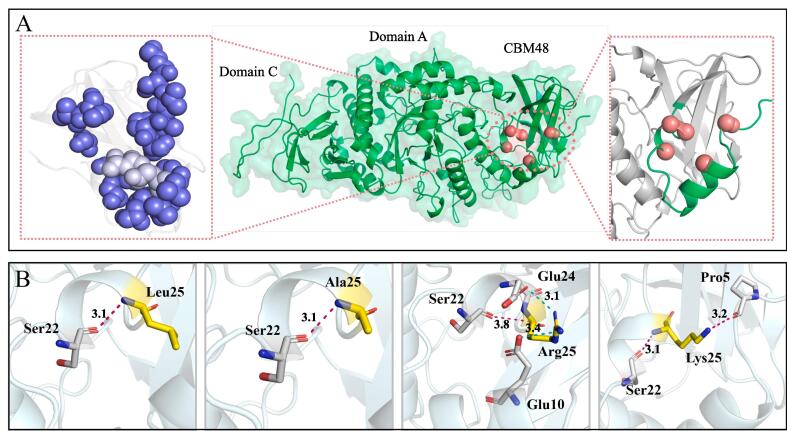
The crystal structure of GBE from E. coli, which shares 44.83% sequence identity with Gt-GBE. Based on this crystal structure, we modelled the 3D structure of Gt-GBE using the SWISS-MODEL homology modelling server (Fig. 2). As shown in Fig. 2A, Gt-GBE consists of three horizontally aligned domains. Through multiple sequence alignment, Gt-GBE contains a carbohydrate-binding module 48 (CBM48) in the N-terminal domain. CBM48 contains several carbohydrate binding sites that interacts with substrates, and is considered to play a critical role in enzyme-substrate binding (Palomo et al., 2009, Ruiz-Gayosso et al., 2018).
Fig. 2.
Structure diagram of the wild-type Gt-GBE and mutants L25R, L25A, and L25K. A: Structure visualization of the wild-type Gt-GBE. The red balls represent Pro5, Glu10, Ser22, Glu24, and Leu25; Glu10 and Leu25 are indicated in silver; hydrophobic residues are indicated in purple. B: Variation in interactions caused by mutations at 25 sites. The dashed pink lines indicate hydrogen bonds, while the dashed blue lines indicate salt bridges. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
From the structural model of the wild-type Gt-GBE (Fig. 2B), Leu25 could form a hydrogen bond with Ser22. After the mutation of Leu25 to Arg25, the mutant L25R introduced two salt bridges, Arg25-Glu10 and Arg25-Glu24, which increased the local electrostatic interactions. Replacement of Leu with Lys added a hydrogen bond at this position, Pro5-Lys25. Comparison of the homology models of the wild-type Gt-GBE and L25A revealed that interactions of L25A at position 25 were unchanged.
Discussion
Enzymes with high catalytic activity have significant advantages in industrial applications, which prompts researchers to explore GBEs with enhanced catalytic activity. However, most GBEs have low catalytic activity and cannot be applied in industrial productions, such as GBE from Oryza sativa L, Bifidobacterium longum (see Table 4 for details). GBE from G. thermoglucosidans STB02 is derived from thermophilic bacteria. It shows optimal enzyme activity at 60°C, which is higher than that of many GBEs, and has a wide range of pH tolerance (Ban et al., 2016). However, compared with GBEs from Geobacillus mahadia Geo-05 and Rhodothermus obamensis STB05, the catalytic activity of Gt-GBE is lower (Mohtar et al., 2016, Wang et al., 2019). Thus, it is essential to further improve the catalytic activity of Gt-GBE by molecular modification to obtain a broad range of applications. Currently, most researches have focused on the catalytic center of enzymes, and few have focused on the non-catalytic domains such as the N-terminal domain (Binderup and Preiss, 1998, Liu et al., 2017). Based on the results obtained from previous studies, we aimed to construct salt bridges in the N-terminal domain to enhance the catalytic activity. A previous study reported that the N-terminal domain of Gt-GBE plays the key role in enzyme activity of Gt-GBE (Xin et al., 2019). In the present study, we aimed to creat a series of point mutants in the N-terminal region of Gt-GBE to increase catalytic activity. The results showed that L25R and L25A exhibited an increase in specific activity by 28.46% and 23.46%, respectively. However, L25K displayed a reduction in activity.
Table 4.
Enzymatic properties of GBEs from different sources.
| Sources | Enzyme activity | Optimal temperature | Reference |
|---|---|---|---|
| Oryza sativa L | 20.80 U/mg | 25°C | (Vu et al., 2008) |
| Bifidobacterium longum | 19.60 U/mg | 25°C | (Li et al., 2020) |
| Geobacillus stearothermophilus TC-91 | 36.00 U/mg | 50°C | (Aga et al., 2010) |
| M. tuberculosis H37Rv | 81.66 U/mg | 30°C | (Garg, Alam, Kishan, & Agrawal, 2007) |
| T. thermophilus HB8 | 0.29 U/mg | 65°C | (Palomo et al., 2011) |
| Rhizomucor miehei | 10.80 U/mg | 25°C | (Wu, Liu, Yan, & Jiang, 2014) |
| Geobacillus thermoglucosidans STB02 | 1790.16 U/mg | 60°C | (Ban et al., 2016) |
Gt-GBE contains CBM48 in the N-terminal domain, which plays an important role in the substrate-binding capacity and catalytic capacity of GBEs (Jiang et al., 2021). Homology modeling analysis showed that Glu10 and Leu25 are located on CBM48, which are located far away from the substrate binding pocket and do not form interactions with the key amino acid residues near the catalytic pocket. We considered that the changes in enzyme activity were related to conformational changes of CBM48. As shown in Fig. 2 and 3, it is evident from ball-and-stick models and electrostatic potential maps of the wild-type Gt-GBE and mutants that Glu10 and Leu25 are located on two opposite α-helices, and these regions are distributed with several hydrophobic amino acids with strong hydrophobic interaction. After mutation of Leu25 to Arg25, the distance of loop17-21 which connected the two α-helices became shorter, altering the local conformation of CBM48. Moreover, Arg is a positively charged amino acid, and the Leu-to-Arg mutation also affected the surface charge distribution and local electrostatic interactions (introduced two salt bridges). These changes contributed to a more compact spatial structure of CBM48, which enhanced the catalytic activity of L25R. In addition, the formation of salt bridges increased the local structural rigidity of Gt-GBE and compensated for the negative impact on structural stability caused by the weakened hydrophobic interaction in the region where Glu10 and Leu25 were located, making the heat resistance of L25R not significantly different from that of the wild-type Gt-GBE.
Fig. 3.
Electrostatic surface potential of the wild-type Gt-GBE and its mutants.
Lys is also a positively charged amino acid, but after substituting Leu for Lys at position 25, Lys25 did not form intramolecular salt bridges with the surrounding negatively charged amino acid residues. And L25K, in contrast to L25R, did not display greater catalytic activity. This may be caused by the fact that Lys is a hydrophilic amino acid, and replacing Leu with Lys disrupted the local hydrophobic interaction, which adversely affected the structural stability of CBM48. Although, L25k formed an additional hydrogen bond, it did not offset the negative effect of the weakened hydrophobic interaction on the structural stability. This also led to a decrease in the ability of L25K to resist thermal inactivation. Whereas, the increase in the catalytic activity of Gt-GBE after the mutation of Leu25 to Ala25 may be attributed to the fact that the mutation changed the amino acid side-chain length and expanded the cavity structure of this region, which in turn affected the structure of CBM48. And, leucine and alanine are hydrophobic amino acids, the Leu25-Ala mutation had no significant effect on the hydrophobic interactions in this region, and thus did not significantly affect the thermostability of Gt-GBE. These results suggested that enzyme activity of Gt-GBE can be enhanced by rational structural modification of the N-terminal domain.
Conclusion
GBEs that can be used for industrial applications should have excellent enzymatic properties, especially high catalytic activity. The stabilized GBE mutants with a mutation at position 25, L25R and L25A, showed 1.28- and 1.23-fold increase in specific activity, respectively, compared with that of the wild-type Gt-GBE. The α-1,6 glycosidic linkage ratios of maltodextrin samples modified by L25R and L25A were increased to 5.71%, which were significantly increased by 19.96% compared with that of the wild-type Gt-GBE. The mutants, L25R and L25A, did not change their optimum temperature and transfer pattern. Therefore, these two mutants are more suitable for future industrial applications. Homologous structural analysis showed that appropriate structural modification of the N-terminal region of GBEs could increase the catalytic activity of GBEs. The results of the present study are helpful to further determine the role of the N-terminal domain, develop an effective protein engineering strategy based on the N-terminal domain, and increase the industrial applications of GBEs.
CRediT authorship contribution statement
Wenjuan Fan: Conceptualization, Methodology, Data curation, Writing – original draft. Zhaofeng Li: Conceptualization, Validation, Supervision, Writing – review & editing. Caiming Li: Software, Investigation, Writing – review & editing. Zhengbiao Gu: Validation, Resources, Project administration. Yan Hong: Resources, Supervision. Li Cheng: Resources, Visualization, Supervision. Xiaofeng Ban: Data curation, Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by National Key R&D Program of China (2021YFD2101001-2), the National Natural Science Foundation of China (No. 22378162), the Fundamental Research Funds for the Central Universities (JUSRP121004), Science and Technology Support Program (Modern Agriculture) of Jiangsu Province (BE2022323) and Project of Jiangsu Provincial Center of Technology Innovation for Future Food (BM2020023).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.100888.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- Abad M.C., Binderup K., Rios-Steiner J., Arni R.K., Preiss J., Geiger J.H. The X-ray crystallographic structure of Escherichia coli branching enzyme. Journal of Biological Chemistry. 2002;277(44):42164–42170. doi: 10.1074/jbc.M205746200. [DOI] [PubMed] [Google Scholar]
- Aga H., Okamoto I., Taniguchi M., Kawashima A., Abe H., Chaen H., Fukuda S. Improved yields of cyclic nigerosylnigerose from starch by pretreatment with a thermostable branching enzyme. Journal of Bioscience and Bioengineering. 2010;109(4):381–387. doi: 10.1016/j.jbiosc.2009.09.047. [DOI] [PubMed] [Google Scholar]
- Ban X., Li C., Gu Z., Bao C., Qiu Y., Hong Y.…Li Z. Expression and biochemical characterization of a thermostable branching enzyme from Geobacillus thermoglucosidans. Journal of Molecular Microbiology and Biotechnology. 2016;26(5):303–311. doi: 10.1159/000446582. [DOI] [PubMed] [Google Scholar]
- Ban X., Liu Y., Zhang Y., Gu Z., Li C., Cheng L.…Li Z. Thermostabilization of a thermophilic 1,4-α-glucan branching enzyme through C-terminal truncation. International Journal of Biological Macromolecules. 2018;107:1510–1518. doi: 10.1016/j.ijbiomac.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Ban X., Wu J., Kaustubh B., Lahiri P., Dhoble A.S., Gu Z.…Li Z. Additional salt bridges improve the thermostability of 1,4-α-glucan branching enzyme. Food Chemistry. 2020;316 doi: 10.1016/j.foodchem.2020.126348. [DOI] [PubMed] [Google Scholar]
- Binderup K., Preiss J. Glutamate-459 is important for Escherichia coli branching enzyme activity. Biochemistry. 1998;37(25):9033–9037. doi: 10.1021/bi980199g. [DOI] [PubMed] [Google Scholar]
- Feng L., Fawaz R., Hovde S., Sheng F., Nosrati M., Geiger J.H. Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins. Acta Crystallographica Section D-Structural Biology. 2016;72:641–647. doi: 10.1107/S2059798316003272. [DOI] [PubMed] [Google Scholar]
- Feng Y., Liu S., Pang C., Gao H., Wang M., Du G. Improvement of catalytic efficiency and thermal stability of L-asparaginase from Bacillus subtilis 168 through reducing the flexibility of the highly flexible loop at N-terminus. Process Biochemistry. 2019;78:42–49. doi: 10.1016/j.procbio.2019.01.001. [DOI] [Google Scholar]
- Garg S.K., Alam M.S., Kishan K.V.R., Agrawal P. Expression and characterization of α-(1,4)-glucan branching enzyme Rv1326c of Mycobacterium tuberculosis H37Rv. Protein Expression and Purification. 2007;51(2):198–208. doi: 10.1016/j.pep.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Suzuki R., Colleoni C., Ball S.G., Fujita N., Suzuki E. Bound substrate in the structure of cyanobacterial branching enzyme supports a new mechanistic model. Journal of Biological Chemistry. 2017;292(13):5465–5475. doi: 10.1074/jbc.M116.755629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Xie X., Ban X., Gu Z., Cheng L., Hong Y.…Li Z. Flexible loop in carbohydrate-binding module 48 allosterically modulates substrate binding of the 1,4-α-glucan branching enzyme. Journal of Agricultural and Food Chemistry. 2021;69(20):5755–5763. doi: 10.1021/acs.jafc.1c00293. [DOI] [PubMed] [Google Scholar]
- Jo A.R., Kim H.R., Choi S.J., Lee J.S., Chung M.N., Han S.K.…Moon T.W. Preparation of slowly digestible sweet potato Daeyumi starch by dual enzyme modification. Carbohydrate Polymers. 2016;143:164–171. doi: 10.1016/j.carbpol.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Kong H., Zhou Y., Gu Z., Li Z., Jiang Z., Cheng L.…Li C. Liquefaction concentration impacts the fine structure of maltodextrin. Industrial Crops & Products. 2018;123:687–697. doi: 10.1016/j.indcrop.2018.07.042. [DOI] [Google Scholar]
- Li D., Fei T., Wang Y., Zhao Y., Dai L., Fu X., Li X. A cold-active 1,4-α-glucan branching enzyme from Bifidobacterium longum reduces the retrogradation and enhances the slow digestibility of wheat starch. Food Chemistry. 2020;324 doi: 10.1016/j.foodchem.2020.126855. [DOI] [PubMed] [Google Scholar]
- Li W., Li C., Gu Z., Qiu Y., Cheng L., Hong Y., Li Z. Relationship between structure and retrogradation properties of corn starch treated with 1,4-α-glucan branching enzyme. Food Hydrocolloids. 2016;52:868–875. doi: 10.1016/j.foodhyd.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ban X., Li C., Gu Z., Cheng L., Hong Y., Li Z. Met349 mutations enhance the activity of 1,4-α-glucan branching enzyme from Geobacillus thermoglucosidans STB02. Journal of Agricultural and Food Chemistry. 2017;65(28):5674–5680. doi: 10.1021/acs.jafc.7b01227. [DOI] [PubMed] [Google Scholar]
- Lonhienne T., Gerday C., Feller G. Psychrophilic enzymes: Revisiting the thermodynamic parameters of activation may explain local flexibility. Biochimica et Biophysica Acta-Protein Structure and Molecular Enzymology. 2000;1543(1):1–10. doi: 10.1016/S0167-4838(00)00210-7. [DOI] [PubMed] [Google Scholar]
- Ming-Mao S., Abdula S.E., Hye-Jung L., Young-Chan C., Long-Zhi H., Hee-Jong K., Yong-Gu C. Molecular aspect of good eating quality formation in japonica rice. PLoS One. 2011;6(4):e18385. doi: 10.1371/journal.pone.0018385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtar N.S., Rahman M.B.A., Abd Rahman R.N.Z.R., Leow T.C., Salleh A.B., Isa M.N.M. Expression and characterization of thermostable glycogen branching enzyme from Geobacillus mahadia Geo-05. PeerJ. 2016;4:e2714. doi: 10.7717/peerj.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni D., Kirtel O., Yin D., Xu W., Chen Q., Oner E.T., Mu W. Improving the catalytic behaviors of Lactobacillus-derived fructansucrases by truncation strategies. Enzyme and Microbial Technology. 2021;149 doi: 10.1016/j.enzmictec.2021.109857. [DOI] [PubMed] [Google Scholar]
- Palomo M., Kralj S., van der Maarel M.J.E.C., Dijkhuizen L. The unique branching patterns of Deinococcus glycogen branching enzymes are determined by their N-terminal domains. Applied and Environmental Microbiology. 2009;75(5):1355–1362. doi: 10.1128/AEM.02141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo M., Pijning T., Booiman T., Dobruchowska J.M., van der Vlist J., Kralj S.…Leemhuis H. Thermus thermophilus glycoside hydrolase family 57 branching enzyme: Crystal structure, mechanism of action, and products formed. Journal of Biological Chemistry. 2011;286(5):3520–3530. doi: 10.1074/jbc.M110.179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Na Y., Kim J., Kang S.D., Park K.-H. Properties and applications of starch modifying enzymes for use in the baking industry. Food Science and Biotechnology. 2018;27(2):299–312. doi: 10.1007/s10068-017-0261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Chen S., Li C., Gu Z., Cheng L., Hong Y., Li Z. A two-stage modification method using 1,4-α-glucan branching enzyme lowers the in vitro digestibility of corn starch. Food Chemistry. 2020;305 doi: 10.1016/j.foodchem.2019.125441. [DOI] [PubMed] [Google Scholar]
- Ren J., Li Y., Li C., Gu Z., Cheng L., Hong Y., Li Z. Pasting and thermal properties of waxy corn starch modified by 1,4-α-glucan branching enzyme. International Journal of Biological Macromolecules. 2017;97:679–687. doi: 10.1016/j.ijbiomac.2017.01.087. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gayosso A., Rodriguez-Sotres R., Martinez-Barajas E., Coello P. A role for the carbohydrate-binding module (CBM) in regulatory SnRK1 subunits: The effect of maltose on SnRK1 activity. Plant Journal. 2018;96(1):163–175. doi: 10.1111/tpj.14026. [DOI] [PubMed] [Google Scholar]
- Suzuki E., Suzuki R. Distribution of glucan-branching enzymes among prokaryotes. Cellular and Molecular Life Sciences. 2016;73(14):2643–2660. doi: 10.1007/s00018-016-2243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I.J., Emes M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life. 2014;66(8):546–558. doi: 10.1002/iub.1297. [DOI] [PubMed] [Google Scholar]
- Tian Y., Chen H., Zhang X., Zhan J., Jin Z., Wang J. Highly branched dextrin prepared from high-amylose maize starch using waxy rice branching enzyme (WRBE) Food Chemistry. 2016;203:530–535. doi: 10.1016/j.foodchem.2016.02.061. [DOI] [PubMed] [Google Scholar]
- Vu N.T., Shimada H., Kakuta Y., Nakashima T., Ida H., Omori T.…Kimura M. Biochemical and crystallographic characterization of the starch branching enzyme I (BEI) from Oryza sativa L. Bioscience Biotechnology and Biochemistry. 2008;72(11):2858–2866. doi: 10.1271/bbb.80325. [DOI] [PubMed] [Google Scholar]
- Wang Z., Xin C., Li C., Gu Z., Cheng L., Hong Y.…Li Z. Expression and characterization of an extremely thermophilic 1,4-α-glucan branching enzyme from Rhodothermus obamensis STB05. Protein Expression and Purification. 2019;164 doi: 10.1016/j.pep.2019.105478. [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Yan Q., Jiang Q. Gene cloning, functional expression and characterisation of a novel glycogen branching enzyme from Rhizomucor miehei and its application in wheat breadmaking. Food Chemistry. 2014;159:85–94. doi: 10.1016/j.foodchem.2014.02.161. [DOI] [PubMed] [Google Scholar]
- Xin C., Ban X., Gu Z., Li C., Cheng L., Hong Y., Li Z. Non-classical secretion of 1,4-α-glucan branching enzymes without signal peptides in Escherichia coli. International Journal of Biological Macromolecules. 2019;132:759–765. doi: 10.1016/j.ijbiomac.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Xiong K., Hou J., Jiang Y., Li X., Teng C., Li Q.…Zhang C. Mutagenesis of N-terminal residues confer thermostability on a Penicillium janthinellum MA21601 xylanase. BMC Biotechnology. 2019;19(1):51. doi: 10.1186/s12896-019-0541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Kong H., Gu Z., Li C., Ban X., Cheng L.…Li Z. Two 1,4-α-glucan branching enzymes successively rearrange glycosidic bonds: A novel synergistic approach for reducing starch digestibility. Carbohydrate Polymers. 2021;262(2) doi: 10.1016/j.carbpol.2021.117968. [DOI] [PubMed] [Google Scholar]
- Zheng F., Huang J., Liu X., Hu H., Long L., Chen K., Ding S. N- and C-terminal truncations of a GH10 xylanase significantly increase its activity and thermostability but decrease its SDS resistance. Applied Microbiology and Biotechnology. 2016;100(8):3555–3565. doi: 10.1007/s00253-015-7176-y. [DOI] [PubMed] [Google Scholar]
- Zhong C., Song S., Fang N., Liang X., Zhu H., Tang X., Tang B. Improvement of low-temperature caseinolytic activity of a thermophilic subtilase by directed evolution and site-directed mutagenesis. Biotechnology and Bioengineering. 2009;104(5):862–870. doi: 10.1002/bit.22473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.