Abstract
Prostate cancer is the leading cause of cancer-related mortality among men worldwide. In particular, castration-resistant prostate cancer presents a formidable clinical challenge and emphasizes the need to develop novel therapeutic strategies. Forkhead box M1 (FOXM1) is a multifaceted transcription factor that is implicated in the acquisition of the multiple cancer hallmark capabilities in prostate cancer cells, including sustaining proliferative signaling, resisting cell death and the activation of invasion and metastasis. Elevated FOXM1 expression is frequently observed in prostate cancer, and in particular, FOXM1 overexpression is closely associated with poor clinical outcomes in patients with prostate cancer. In the present review, recent advances in the understanding of the oncogenic role of deregulated FOXM1 expression in prostate cancer were highlighted. In addition, the molecular mechanisms by which FOXM1 regulates prostate cancer development and progression were described, thereby providing knowledge and a conceptual framework for FOXM1. The present review also provided valuable insight into the inherent challenges associated with translating biomedical knowledge into effective therapeutic strategies for prostate cancer.
Keywords: FOXM1, prostate cancer, oncogene
1. Introduction
Prostate cancer is a highly prevalent malignancy and the leading cause of cancer-related deaths among men worldwide (1,2). Localized or organ-confined prostate cancer can be effectively managed via surgical intervention or radiotherapy. Patients with de novo or recurrent metastatic prostate cancer initially exhibit favorable responses to androgen deprivation therapy, known as chemical castration. However, most patients eventually relapse and the disease progresses to an incurable or lethal castration-resistant state (3–5). The emergence of castration-resistant prostate cancer poses a substantial clinical challenge that highlights the need for the development of promising therapeutic strategies to overcome anti-cancer therapy resistance.
Forkhead box (FOX) proteins represent a superfamily of transcription factors that share an evolutionarily conserved DNA-binding domain called the ‘forkhead box’ or winged helix domain. FOX proteins are encoded by 50 genes in the human genome and may be categorized into 19 subfamilies (6) (Fig. 1). FOX proteins have crucial roles in regulating a wide spectrum of biological and developmental processes in response to environmental cues. The dysregulation of FOX alters cell fate and underlies various human diseases, particularly cancer (7–9). Furthermore, several FOX proteins, including FOXM1, have been shown to drive tumor initiation, progression, metastasis and drug resistance in various human cancers (9).
Figure 1.
Overview of the structural organization of FOXM1 subfamily members. FOX, forkhead box.
Among the FOX proteins, FOXM1 is ubiquitously expressed in various tissues during embryogenesis and its knockout leads to embryonic lethality owing to multiple developmental defects (10). FOXM1 expression is markedly decreased in adult tissues but is induced under various regenerative conditions (10). In addition, overexpression and mutation of FOXM1 are frequently observed in various human cancers, including prostate cancer (11–15). It has also been confirmed that FOXM1 has a crucial role in tumorigenesis by regulating a multitude of biological processes, such as cell cycle, proliferation, migration, differentiation, apoptosis and metabolism (12,13,16). Therefore, FOXM1 has garnered attention as a promising target for the development of anti-cancer drugs. However, to the best of our knowledge, no comprehensive review that encompasses the oncogenic role and mechanisms of action of FOXM1 in prostate cancer has so far been compiled.
In the present review, recent advances in the understanding regarding the oncogenic role of FOXM1 and its underlying mechanisms in prostate cancer were presented. The challenges associated with FOXM1 were discussed and perspectives were provided for further related research and the application of the results obtained regarding the development of therapeutic strategies for prostate cancer.
2. Dysregulation of FOXM1 expression in prostate cancer
FOXM1 expression is frequently elevated in prostate cancer tissues (17–27). It is regulated at multiple levels, including the transcription, post-transcription and protein stability (21–43) (Fig. 2). FOXM1-activatory molecules are generally upregulated in prostate cancer tissues (21,24,25,34–36), whereas FOXM1-inhibitory molecules are downregulated during prostate cancer progression (27,30,32,37). However, the molecular mechanisms associated with FOXM1 regulation in prostate cancer have remained to be fully elucidated, particularly regarding the nuclear localization and posttranslational modification of FOXM1. Therefore, further studies are required to elucidate the molecular mechanisms underlying dysregulated FOXM1 expression and accurately predict its transcriptional output in prostate cancer.
Figure 2.
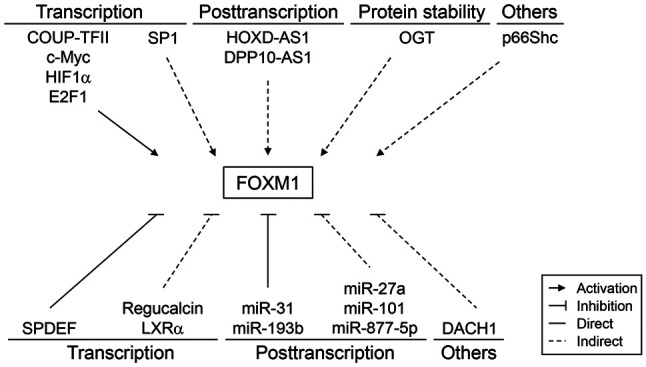
Regulation of FOXM1 expression in human prostate cancer. FOX, forkhead box. COUP-TFII, chicken ovalbumin upstream promoter transcription factor II; c-Myc, cellular myelocytomatosis; HIF1α, hypoxia-inducible factor α; E2F1, E2 promoter binding factor; SP1, specificity protein 1; HOXD-AS1, homeobox D cluster antisense RNA 1; DPP10-AS1, dipeptidyl peptidase-like 10-antisense RNA 1; OGT, O-linked β-N-acetylglucosamine transferase; SPDEF, SAM-pointed domain-containing ETS transcription factor; LXRα, liver X receptor α; DACH1, dachshund homolog 1.
Overexpression of FOXM1 in prostate cancer tissues
Immunohistochemical, reverse transcription-PCR and transcriptomic analyses have consistently shown elevated FOXM1 expression levels in prostate cancer tissues compared to adjacent normal tissues (17–27). A higher FOXM1 expression level has also been found to be significantly associated with tumor grade, disease severity or therapeutic resistance (18–20,22–24,28,29). In addition, FOXM1 overexpression is closely associated with poor prognosis in patients with prostate cancer (20,22,25–28,30–33) (Table I). Furthermore, several immunohistochemical studies have revealed that FOXM1 overexpression in the nucleus is strongly associated with tumor grade, disease severity and poor clinical outcomes (17,22,28). Therefore, targeting FOXM1 may be a clinically useful therapeutic approach for prostate cancer.
Table I.
Survival outcome of patients with prostate cancer based on forkhead box M1 expression level.
| First author, year | Data source | Hazard ratio; P-value | Clinical outcome | (Refs.) |
|---|---|---|---|---|
| Cheng, 2014 | GSE16560 | P=0.049 | Overall survival | (30) |
| Ketola, 2017 | GSE21032 (Taylor dataset) | P<0.001 | Disease-free survival | (20) |
| Xu, 2022 | GEPIA | 1.9; P=0.0049 | Disease-free survival | (27) |
| Tian, 2021 | Rembrandt | P=0.0107 | Overall survival | (26) |
| Sharma, 2021 | TCGA | P<0.01 | Recurrence-free survival | (32) |
| Koo, 2023 | TCGA | 3.7; P=2.8×10-5 | Overall survival | (33) |
| Kim, 2019 | Korea prostate bank | 10.524; P=0.022 | Biochemical recurrence-free survival | (22) |
| Tian, 2021 | Private data | P=0.0125 | Overall survival | (26) |
TCGA, The Cancer Genome Atlas.
FOXM1 overexpression via transcription
FOXM1 transcription is activated by various transcription factors, including chicken ovalbumin upstream promoter transcription factor II (COUP-TFII), cellular myelocytomatosis (c-Myc), hypoxia-inducible factor α (HIF1α) and E2 promoter binding factor 1 (E2F1) (21,24,34,35). These transcription factors induce FOXM1 expression by directly binding to the FOXM1 promoter. Specifically, E2F1 has been found to upregulate FOXM1 expression by recruiting su(var)3-9, enhancer-of-zeste and trithorax domain containing 1A (SETD1A), a histone H3K4 methyltransferase (25). Conversely, specificity protein 1, which is upregulated by G2 and S phase-expressed-1, also upregulates FOXM1 expression (36).
FOXM1 overexpression via derepression
Liver X receptor α (LXRα) knockdown was reported to upregulate FOXM1 expression (27). Indeed, a negative association has been observed between LXRα and FOXM1 in prostate tumor tissues (27). SAM-pointed domain-containing ETS transcription factor (SPDEF) also represses FOXM1 gene transcription by directly binding to the FOXM1 promoter (30). In addition, regucalcin and dachshund homolog 1 (DACH1), a winged helix/forkhead DNA-binding protein, suppresses FOXM1 expression (32,37). However, the expression levels of these repressive proteins are markedly reduced during prostate cancer progression and this leads to the derepression of FOXM1 expression.
FOXM1 overexpression via post-transcription
FOXM1 expression is increased by long noncoding RNAs, such as homeobox D cluster antisense RNA 1 (HOXD-AS1) and dipeptidyl peptidase-like 10-antisense RNA 1 (DPP10-AS1) (38,39). Furthermore, HOXD-AS1 mediates H3 lysine 4 (H3K4) trimethylation at the FOXM1 promoter by binding with tryptophan-aspartate repeat domain 5 (38). HOXD-AS1 also upregulates FOXM1 expression by sponging micro (mi)RNA miR-361-5p (40). By contrast, DPP10-AS1 induces cyclic AMP response element-binding protein-binding protein-mediated H3K27 acetylation at the FOXM1 promoter (39).
The post-transcriptional stability of FOXM1 mRNA is decreased by several miRNAs, including miR-31 and miR-193b (23,41). By contrast, miR-101 and miR-27a indirectly reduce FOXM1 expression by inhibiting COUP-TFII (34). MiR-877-5p also suppresses FOXM1 expression (42). These miRNAs are frequently downregulated in prostate cancer and this may partly explain the underlying mechanism of FOXM1 overexpression.
FOXM1 overexpression by protein stability
FOXM1 expression is regulated by altering O-linked β-N-acetylglucosamine transferase (OGT)-mediated protein stability (43). OGT upregulates FOXM1 expression by preventing its proteasomal degradation. However, as FOXM1 is not O-GlcNAcylated, OGT appears to indirectly regulate FOXM1 stability.
Other mechanisms
Transgenic adenocarcinoma of the mouse prostate (TRAMP) mice show upregulated FOXM1 expression, which is reversed via surgical castration (44). However, the synthetic androgen R1881 does not affect FOXM1 expression levels (45). This suggests that FOXM1 expression is upregulated in TRAMP mice through a mechanism that is independent of androgen receptor (AR) signaling. By contrast, p66Shc, an oxidative stress response protein, upregulates FOXM1 protein levels (46).
Summary of the regulation of FOXM1 expression
Several studies have contributed to the current knowledge regarding FOXM1 expression. However, the determinants of the expression levels of FOXM1 and its activity have remained to be fully elucidated. For instance, it remains unclear whether molecular signaling or nuclear transport pathways have crucial roles in the direct regulation of FOXM1 expression in prostate cancer. Therefore, efforts are required to further provide a comprehensive explanation and prediction of FOXM1-mediated biological outcomes.
3. Role of FOXM1 in prostate cancer
FOXM1 regulates various cancer hallmark-related biological processes, including cell cycle, survival, proliferation, apoptosis, autophagy, migration and invasion (Table II). In this section, the molecular mechanisms and biological roles of FOXM1 in prostate cancer were summarized.
Table II.
Summary of the biological roles of FOXM1 in human prostate cancer.
| First author, year | Phenomenon | Effect | Possible molecular mechanism | Cell model | Animal model | (Refs.) |
|---|---|---|---|---|---|---|
| Tian, 2021 | Apoptosis | Inhibition | Increase in Bcl-2 and | 22Rv1, C4-2, | siFOXM1-expressing | (26) |
| RRM2 expression | DU145, DU145-DR, | PC3-DR- | ||||
| Xu, 2022 | LNCaP, PC3, PC3- | xenografted mouse | (27) | |||
| Yu, 2020 | DR, VCaP, VCaP- | (47) | ||||
| Yu, 2020 | DR | (48) | ||||
| Lin, 2020 | (49) | |||||
| Mazzu, 2019 | (50) | |||||
| Wu, 2018 | (54) | |||||
| Lin, 2020 | Autophagy | Activation | Increase in AMPK | PC3, PC3-DR, | / | (49) |
| activity; Decrease in | VCaP, VCaP-DR | |||||
| mTOR activity | ||||||
| Kalin, 2006 | Cell | Activation | Increase in AR, | 22Rv1, C4-2, | LNCaP-xenografted | (17) |
| Pan, 2018 | proliferation | EXO1,CDC6, | DU145, DU145-DR, | mouse; LNCaP-AI- | ||
| Mazzu, 2019 | and tumor | 11β-HSD2, | LNCaP, Myc-CaP, | xenografted mouse; | (21) | |
| Tian, 2021 | growth | KIF20A, RRM2, | PC3, TRAMP C2, | DU145-DR-xenografted | ||
| Xu, 2022 | cyclin A2, cyclin | VCaP, VCaP-DR | mouse; FOXM1- | (23) | ||
| Aytes, 2014 | B1, cyclin B2, | overexpressing PC3- | (26) | |||
| Cheng, 2014 | cyclin E1, cyclin D1, | xenografted mouse; | (27) | |||
| Lai, 2021 | Cdc25b and CDK1 | FOXM1-overexpressing | (28) | |||
| Yu, 2020 | expression | LADY mouse; FOXM1- | (30) | |||
| Yu, 2020 | overexpressing; TRAMP | |||||
| Mazzu, 2019 | mouse; FOXM1-deleted | (36) | ||||
| Kim, 2021 | TRAMP mouse; | (47) | ||||
| Wu, 2018 | shFOXM1-expressing | (48) | ||||
| Li, 2011 | DU145-xenografted | (50) | ||||
| Zhou, 2017 | mouse | (52) | ||||
| Cai, 2013 | (54) | |||||
| Liu, 2014 | (55) | |||||
| (56) | ||||||
| (58) | ||||||
| (59) | ||||||
| Wang, 2014 | Invasion and | Activation | Increase in EXO1, | 22Rv1, C4-2, | 22Rv1-xenografted | (19) |
| Pan, 2018 | metastasis | KIF20A, RRM2, | DU145, DU145-DR, | mouse; FOXM1- | (21) | |
| Tang, 2019 | vimentin, SLUG, | LNCaP, PC3, PC3- | overexpressing | (24) | ||
| LOX, VCAN, ZEB2 | ML, TRAMP C2, | TRAMP mouse; TRAMP | ||||
| and VEGF expression; | VCaP, VCaP-DR | FOXM1-deleted mouse | ||||
| Tian, 2021 | Decrease in | (26) | ||||
| Cheng, 2014 | E-cadherin expression | (30) | ||||
| Lin, 2016 | (34) | |||||
| Lynch, 2012 | (43) | |||||
| Yu, 2020 | (48) | |||||
| Mazzu, 2019 | (50) | |||||
| Kim, 2021 | (52) | |||||
| Li, 2011 | (55) | |||||
| Zhou, 2017 | (56) | |||||
| Cai, 2013 | (58) | |||||
| Qu, 2018 | (60) | |||||
| Yuan, 2018 | Drug | Activation | Increase in UHRF1 | DU145-DR, LNCaP- | siFOXM1-expressing | (29) |
| Lin, 2016 | resistance | and AR expression | ER, PC3-DR, VCaP- | PC3-DR-xenografted | (34) | |
| Gu, 2017 | DR | mouse | (38) | |||
| Liu, 2017 | (45) | |||||
| Yu, 2020 | (48) | |||||
| Lin, 2020 | (49) | |||||
| Yuan, 2018 | Cancer | Activation | Increase in ALDH1, | DU145, DU145-DR, | / | (29) |
| Koo, 2023 | stemness and | NANOG, SOX2 | PC3 | (30) | ||
| energy | and SHH expression; | |||||
| metabolism | Increase in HK2, PKM2, | |||||
| and LDHA expression |
DR, docetaxel-resistant; ER, enzalutamide resistance; AI, androgen-independent; FOX, forkhead box; RRM2, ribonucleotide reductase small subunit M2; EZH2, enhancer of zeste homolog 2; AMPK, adenosine monophosphate-activated protein kinase; mTOR, mammalian target of rapamycin; AR, androgen receptor; EXO1, exonuclease 1; CDC6, cell division cycle 6; 11β-HSD2, 11β-hydroxysteroid dehydrogenase 2; KIF20A, kinesin family member 20A; cdc25b, cell division cycle 25b; CDK1, cyclin dependent kinase 1; LOX, lysyl oxidase; VCAN, versican; ZEB2, zinc finger E-box binding homeobox 2; VEGF, vascular endothelial growth factor; ALDH1, aldehyde dehydrogenase 1; SOX2, sex-determining region of Y-related high mobility group-box; SHH, sonic hedgehog; HK2, hexokinase 2; PKM2, pyruvate kinase M2; LDHA, lactate dehydrogenase A.
Apoptosis and autophagy
In cell culture models, FOXM1 was observed to enable prostate cancer cells to acquire a cancer hallmark capability of resistance or evasion of apoptosis. Furthermore, FOMX1 overexpression suppresses apoptosis (47–49) by inducing the ribonucleotide reductase small subunit M2 (RRM2) or enhancer of zeste homolog 2 (EZH2) (26,50). The expression levels of RRM2 and EZH2 are frequently elevated in prostate cancer and their upregulation is closely associated with poor clinical outcomes in patients with prostate cancer (50,51). By contrast, FOXM1 knockdown induces prostate cancer cell apoptosis (26,48). In addition, FOXM1 inhibition by miRNAs (e.g., miR-193b) or chemical compounds [e.g., forkhead domain inhibitory compound-6 (FDI-6), niclosamide, siomycin A, SR9009, morusin, cinnamaldehyde, cinnamic acid and eugenol] was reported to induce apoptosis (23,26,27,33,50,52,53).
It has also been observed that FOXM1 attenuates cell death by inducing protective autophagy (49). Furthermore, FOXM1 overexpression activates adenosine monophosphate-activated protein kinase (AMPK) and inhibits mammalian target of rapamycin (mTOR) activity, leading to autophagy. However, AMPK inhibitor compound C and mTOR activator MHY1485 were observed to abolish FOXM1-mediated autophagy and trigger apoptosis (49).
Cell proliferation and tumor growth
FOXM1 exerts oncogenic effects by sustaining proliferative signaling and evading growth suppressors in various experimental models, including cell cultures, xenografts and genetically engineered mouse models. Furthermore, FOXM1 knockdown suppresses cell cycle progression, cell viability, proliferation, colony formation or tumor growth (17,23,26,27,36,48–50,52,54). In addition, FOXM1 inhibition by miRNAs (e.g., miR-877-5p) or chemical compounds [e.g., natura-α, tetramethylpyrazine (TMP), thiostrepton, monensin, FDI-6, thiostrepton, SR9009, morusin and baicalin] also reproduces knockdown phenotypes (20,27,33,36,42,47,50,55,56). By contrast, upregulation of FOXM1 by upstream molecules, such as c-Myc, DPP10-AS1, HOXD-AS1 and SETD1A, or dibutyl phthalate, was observed to promote cell proliferation, colony formation and tumor growth (21,25,38,39,57). Furthermore, FOXM1 upregulation following SPDEF inhibition also stimulated cell proliferation and tumor growth (30).
The possible mechanisms of cell proliferation and tumor growth mediated by FOXM1 include the induction of 11β-hydroxysteroid dehydrogenase 2, cell division cycle 6 (CDC6) and exonuclease 1 by FOXM1 (52,58,59), which also cooperates with other oncogenes, including AR and centromere protein F (CENPF) (28,31,34,59). The FOXM1-AR interaction has a crucial role in CDC6 upregulation (59), whereas the FOXM1-CENPF interaction activates various signaling pathways associated with prostate cancer malignancy, including the cell cycle and the PI3K and MAPK pathways (28). The combined inhibition of FOXM1 and AR or FOXM1 and CENPF using small interfering RNAs or chemical inhibitors significantly inhibit cell proliferation, colony formation and tumor growth (28,31,59).
Invasion and metastasis
FOXM1 has been shown to accelerate tumor malignancy by inducing angiogenesis and activating invasion and metastasis in cell cultures, xenografts and genetically engineered mouse models, while its knockdown inhibits cell migration and invasion (19,24,26,48,50). Furthermore, FOXM1 inhibition by miRNAs (miR-193b and miR-877-5p) or chemical compounds (FDI-6, TMP, thiostrepton, SR9009, natura-α and docetaxel plus anestat) also suppresses cell migration and invasion (23,27,42,46,55,56,60). In addition, FOXM1 inactivation via OGT depletion, regucalcin overexpression or FOXM1 gene ablation was reported to reduce angiogenesis, cell invasion and tumor metastasis (32,43,58). By contrast, ectopic FOXM1 expression or FOXM1 upregulation by COUP-TFII, c-Myc, HIF1α and exosomal HOXD-AS1 was observed to stimulate cell migration and invasion (21,24,34,40,48). Furthermore, FOXM1 upregulation following SPDEF inhibition also promoted cell migration and invasion (30).
The possible mechanisms of FOXM1-activated angiogenesis, invasion and metastasis include the upregulation of vascular endothelial growth factor, lysyl oxidase, versican and RRM2, as well as the stimulation of TGFβ-mediated epithelial-mesenchymal transition by FOXM1 (19,24,43,50,58). Of note, FOXM1 inhibition suppressed the expression of E-cadherin and upregulates the expression of vimentin, Slug and zinc finger E-box binding homeobox 2 (19,24).
Drug resistance
FOXM1 has been shown to confer resistance to chemical castration and nonselective chemotherapy in prostate cancer cells. FOXM1 knockdown enhances the efficacy of enzalutamide, an anti-androgen drug, as well as that of docetaxel (34,49). Furthermore, FOXM1 inhibition by miRNAs (e.g., miR-101 and miR-27a) or chemicals (e.g., thiostrepton) also increases sensitivity to docetaxel (34,48). HOXD-AS1 knockdown also represses resistance to bicalutamide and paclitaxel, possibly by suppressing FOXM1 expression (38). Furthermore, inhibition of FOXM1-induced autophagy via the knockdown of autophagy-related (ATG) protein 7 or beclin-1 or using chloroquine, compound C or MHY1485 restored sensitivity to docetaxel in FOXM1-overexpressing cells (49). Conversely, FOXM1 overexpression leads to enzalutamide and docetaxel resistance (34,48,49).
The upregulation of the plant homeodomain and an interesting new gene, finger domain-containing 1 is involved in FOXM1-mediated therapeutic resistance (29). In addition, FOXM1-mediated AR upregulation provides a possible explanation for resistance to chemical castration in prostate cancer (45).
Other biological processes
FOXM1 regulates cancer stemness and metabolic programs in cell culture and xenograft models. Specifically, inhibition of FOXM1 by thiostrepton or monensin suppresses cancer stemness (20), while its overexpression increases the expression levels of cancer stem cell-associated molecules, such as aldehyde dehydrogenase 1 (ALDH1), NANOG homeobox, sex-determining region of Y-related high mobility group-box (SOX) and sonic hedgehog (SHH) (29). Furthermore, FOXM1 inhibition using morusin suppresses glycolysis by reducing the expression of hexokinase 2 (HK2), pyruvate kinase M2 (PKM2) and lactate dehydrogenase A (LDHA) (33), implicating FOXM1 in deregulating cellular energetics, a cancer hallmark.
Summary of the role of FOXM1 in prostate cancer
FOXM1 is a crucial determinant of tumor cell physiology in established prostate cancer cells, as evidenced by loss-of-function experiments, which showed reduced cell survival, proliferation, migration and invasion. However, the role of FOXM1 in driving prostate cancer remains inconclusive, as FOXM1 transgenic mice do not develop prostate tumors or hyperplasia (17,58). Furthermore, FOXM1 overexpression significantly accelerates tumor development and growth in TRAMP or LADY prostate cancer mouse models (17). These findings suggest that the precise role of FOXM1 and its underlying molecular mechanisms in prostate cancer development and progression are yet to be fully elucidated.
4. Therapeutic agents targeting FOXM1 in prostate cancer
Numerous synthetic and naturally occurring compounds have been shown to inhibit FOXM1 expression in prostate cancer cells. These compounds include siomycin A (61), thiostrepton (48,61), natura-α (55), metformin (19), TMP (39,56), monensin (20), FDI-6 (46), mocetinostat (23), baicalin (47), niclosamide (52), dilazep (62), MYCi975 (35), SR9009 (27) and morusin (33). In addition, combination therapies with rapamycin and PD0325901 (31) or docetaxel and aneustat (60) have also been shown to inhibit FOXM1 activity or expression.
These compounds exert anti-cancer effects by modulating various biological processes, including cell proliferation, migration, invasion or apoptosis (Table III). Among them, siomycin A has been found to potentiate the anti-cancer effects of bicalutamide (45,59), whereas thiostrepton has been shown to increase sensitivity to docetaxel (48). However, only a small number of compounds have been validated via FOXM1 rescue experiments, which demonstrated their ability to reverse phenotypic alterations. For instance, natura-α inhibits cell proliferation and invasion (55); tetramethylpyrazine suppresses cell proliferation, colony formation, migration and invasion (56), and combined treatment with docetaxel and aneustat reduces cell migration (60). It has also been noted that these alterations can be reversed via forced FOXM1 expression.
Table III.
Anti-tumor activity of the compounds inhibiting FOXM1 expression.
| Effective concentration or dose for inhibiting FOXM1 expression | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| In vitro experiment | In vivo experiment | ||||||
|
|
|
||||||
| First author, year | Compound name | Model | Concentration | Model | Dose, mg/kg | Anti-tumor activity | (Refs.) |
| Pandit, 2010 | Siomycin A | DU145, | 5 µM | / | / | Apoptosis↑ | (61) |
| LNCaP, PC3 | Proliferation↓ | ||||||
| Pandit, 2010 | Thiostrepton | DU145, | 5 µM | / | / | Apoptosis↑ | (61) |
| LNCaP, PC3 | Proliferation↓ | ||||||
| Yu, 2020 | DU145-DR, | 3 µM | DU145-DR- | 30 | Proliferation↓ | (48) | |
| VCaP-DR | xenografted mouse | ||||||
| Li, 2011 | Natura-α | LNCaP, | 4-10 mmol/l | LNCaP and | 100 | Proliferation↓ | (55) |
| LNCaP-AI | LNCaP-AI- | Invasion↓ | |||||
| xenografted mouse | |||||||
| Bu, 2023 | C4-2 | 5.067 µM | / | / | Apoptosis↑ | (57) | |
| PC3 | 4.523 µM | / | / | Proliferation↓ | |||
| Wang, 2014 | Metformin | DU145 | 20 mM | / | / | Proliferation↓ | (19) |
| Zhou, 2017 | Tetramethylpyrazine | PC3 | 100-1,000 | PC3-xenografted | 10-100 | Proliferation↓ | (56) |
| g/l | mouse | Invasion↓ | |||||
| Migration↓ | |||||||
| Zhou, 2020 | / | / | DPP10-AS1- | 50 | Proliferation↓ | (39) | |
| overexpressing | |||||||
| PC3-xenografted | |||||||
| mouse | |||||||
| Ketola, 2017 | Monensin | 42D-ER | 10-100 nM | 42D-ER- | 10 | Proliferation↓ | (20) |
| xenografted mouse | |||||||
| Ingersoll, 2018 | FDI-6 | LNCaP | 5 µM | / | / | Proliferation↓ | (46) |
| Migration↓ | |||||||
| Mazzu, 2019 | Mocetinostat | LNCaP, 22Rv1 | 1 µM | / | / | Proliferation↓ | (23) |
| Yu, 2020 | Baicalin | LNCaP, PC-3 | 10 µg/ml | LNCaP- | 10-40 | Proliferation↓ | (47) |
| xenografted mouse | |||||||
| Kim, 2021 | Niclosamide | 22Rv1, PC3 | 0.25–10 µM | 22Rv1- | 20-50 | Apoptosis↑ | (52) |
| xenografted mouse | Proliferation↓ | ||||||
| Kaochar, 2021 | Dilazep | LNCaP, | 50 µM | MDA PCa337A- | 50 | Proliferation↓ | (62) |
| LNCaP-Abl, | xenografted mouse | ||||||
| LNCaP-ER | |||||||
| Holmes, 2022 | MYCi975 | 22Rv1 | 10 µM | 22Rv1- | 100 | Proliferation↓ | (35) |
| xenografted mouse | |||||||
| Xu, 2022 | SR9009 | 22Rv1, PC3 | 20 µM | 22Rv1- | 100 | Apoptosis↑ | (27) |
| xenografted mouse | Proliferation↓ | ||||||
| Migration↓ | |||||||
| Koo, 2023 | Morusin | DU145, PC3 | 5-10 µM | / | / | Apoptosis↑ | (33) |
| Proliferation↓ | |||||||
| Mitrofanova, | Rapamycin + | 22Rv1, | 3 µM + | / | / | Proliferation↓ | (31) |
| 2015 | PD0325901 | DU145, PC3 | 1 µM | ||||
| Qu, 2018 | Aneustat + | C4-2 | 100 µg/ml + | / | / | Invasion↓ | (60) |
| Docetaxel | 5 nM | Migration↓ | |||||
DR, docetaxel-resistant; AI, androgen-independent; ER, enzalutamide-resistant; FDI-6, forkhead domain inhibitory compound-6; Abl, ablation of androgen; FOX, foxhead box.
Numerous compounds that suppress FOXM1 expression exhibit anti-tumor activity against prostate cancer, suggesting that therapeutic strategies targeting FOXM1 may be useful in the treatment of prostate cancer. However, further studies are necessary to establish whether these anti-tumor effects are solely attributable to FOXM1 inhibition. Moreover, the specific mechanisms by which these compounds inhibit FOXM1 expression need to be elucidated.
5. Summary and future direction
Altered transcriptional programs improve the biological fitness of prostate cancer cells under various stress conditions, providing survival benefits to these cells in a given microenvironment. FOXM1, a representative transcription factor, enhances prostate cancer cell survival by regulating transcription. Increased FOXM1 expression, which is frequently observed in prostate cancer cells, is associated with disease severity and a poor prognosis in patients. FOXM1 also mediates cancer hallmarks, including sustaining proliferative signaling, resisting cell death and activating invasion and metastasis. Furthermore, FOXM1 enhances sensitivity to anti-androgen therapy or nonselective chemotherapy. Therefore, these results suggest that FOXM1 holds promising potential as a therapeutic target in prostate cancer. In addition, FOXM1 has been indicated to have clinical utility as both a prognostic and predictive marker in prostate cancer.
However, several challenges still exist with respect to understanding the role of FOXM1 in prostate cancer. First, current research has mainly focused on identifying the molecular mechanisms that regulate FOXM1 expression. Therefore, the molecular signaling mechanisms that control FOXM1 activity in prostate cancer, such as post-translational modifications and subcellular localization, require further elucidation. Second, FOXM1 has four different splicing variants (11), and it remains unclear whether these different variants have specific oncogenic functions. Third, FOX proteins may act as monomers or dimers with other interacting partners (8). Therefore, additional studies are necessary to determine which transcription and chromatic remodeling factors cooperate with FOXM1 during prostate cancer progression. Furthermore, it is necessary to investigate changes in FOXM1-mediated transcription programs and outputs that drive tumor development and progression. Fourth, it is important to clarify the molecular mechanisms by which FOXM1 contributes to therapeutic resistance and androgen independence in prostate cancer. Finally, the tumor microenvironment (TME) has an important role in cancer evolution, with hypoxia leading to the selection of more malignant prostate cancer cells. However, the role of FOXM1 in the TME remains poorly understood. Therefore, further research should focus on answering these questions to improve our understanding of the role of FOXM1 in prostate cancer biology and treatment.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by grants from the National Research Foundation of Korea funded by the Korean government (MSIT; grant nos. 2018R1A4A1023822 and 2020R1A2C1102574) and the Education and Research Encouragement Fund of Seoul National University Hospital.
Availability of data and materials
Not applicable.
Authors' contributions
DYL, JNC and JHJ conceived and designed the article. DYL, JNC and JHJ reviewed the literature and wrote the manuscript. DYL, JNC and IS surveyed the literature and provided suggestions. All authors have read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Berenguer CV, Pereira F, Camara JS, Pereira JAM. Underlying features of prostate cancer-statistics, risk factors, and emerging methods for its diagnosis. Curr Oncol. 2023;30:2300–2321. doi: 10.3390/curroncol30020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Jiang X, Liang X, Jiang G. Molecular and cellular mechanisms of castration resistant prostate cancer. Oncol Lett. 2018;15:6063–6076. doi: 10.3892/ol.2018.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanagisawa T, Kawada T, Rajwa P, Kimura T, Shariat SF. Emerging systemic treatment for metastatic castration-resistant prostate cancer: A review of recent randomized controlled trials. Curr Opin Urol. 2023;33:219–229. doi: 10.1097/MOU.0000000000001080. [DOI] [PubMed] [Google Scholar]
- 5.Cai M, Song XL, Li XA, Chen M, Guo J, Yang DH, Chen Z, Zhao SC. Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist Updat. 2023;68:100962. doi: 10.1016/j.drup.2023.100962. [DOI] [PubMed] [Google Scholar]
- 6.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 8.Herman L, Todeschini AL, Veitia RA. Forkhead transcription factors in health and disease. Trends Genet. 2021;37:460–475. doi: 10.1016/j.tig.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Castaneda M, Hollander PD, Mani SA. Forkhead box transcription factors: Double-Edged swords in cancer. Cancer Res. 2022;82:2057–2065. doi: 10.1158/0008-5472.CAN-21-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: Lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gartel AL. FOXM1 in Cancer: Interactions and vulnerabilities. Cancer Res. 2017;77:3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, Hu CJ, Bai JY. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16:57. doi: 10.1186/s12964-018-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalathil D, John S, Nair AS. FOXM1 and cancer: Faulty cellular signaling derails homeostasis. Front Oncol. 2021;10:626836. doi: 10.3389/fonc.2020.626836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Barger CJ, Karpf AR. FOXM1: A multifunctional oncoprotein and emerging therapeutic target in ovarian cancer. Cancers (Basel) 2021;13:3065. doi: 10.3390/cancers13123065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzenellenbogen BS, Guillen VS, Katzenellenbogen JA. Targeting the oncogenic transcription factor FOXM1 to improve outcomes in all subtypes of breast cancer. Breast Cancer Res. 2023;25:76. doi: 10.1186/s13058-023-01675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan MA, Khan P, Ahmad A, Fatima M, Nasser MW. FOXM1: A small fox that makes more tracks for cancer progression and metastasis. Semin Cancer Biol. 2023;92:1–15. doi: 10.1016/j.semcancer.2023.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, Lyubimov A, Costa RH. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Yao B, Wang Y, Zhang M, Fu S, Gao H, Peng R, Zhang L, Tang J. Increased FoxM1 expression is a target for metformin in the suppression of EMT in prostate cancer. Int J Mol Med. 2014;33:1514–1522. doi: 10.3892/ijmm.2014.1707. [DOI] [PubMed] [Google Scholar]
- 20.Ketola K, Munuganti RSN, Davies A, Nip KM, Bishop JL, Zoubeidi A. Targeting prostate cancer subtype 1 by forkhead box M1 pathway inhibition. Clin Cancer Res. 2017;23:6923–6933. doi: 10.1158/1078-0432.CCR-17-0901. [DOI] [PubMed] [Google Scholar]
- 21.Pan H, Zhu Y, Wei W, Shao S, Rui X. Transcription factor FoxM1 is the downstream target of c-Myc and contributes to the development of prostate cancer. World J Surg Oncol. 2018;16:59. doi: 10.1186/s12957-018-1352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MY, Jung AR, Kim GE, Yang J, Ha US, Hong SH, Choi YJ, Moon MH, Kim SW, Lee JY, Park YH. High FOXM1 expression is a prognostic marker for poor clinical outcomes in prostate cancer. J Cancer. 2019;10:749–756. doi: 10.7150/jca.28099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzu YZ, Yoshikawa Y, Nandakumar S, Chakraborty G, Armenia J, Jehane LE, Lee GM, Kantoff PW. Methylation-associated miR-193b silencing activates master drivers of aggressive prostate cancer. Mol Oncol. 2019;13:1944–1958. doi: 10.1002/1878-0261.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang C, Liu T, Wang K, Wang X, Xu S, He D, Zeng J. Transcriptional regulation of FoxM1 by HIF-1α mediates hypoxia-induced EMT in prostate cancer. Oncol Rep. 2019;42:1307–1318. doi: 10.3892/or.2019.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Jin M, Park SJ, Seo SY, Jeong KW. SETD1A promotes proliferation of castration-resistant prostate cancer cells via FOXM1 transcription. Cancers (Basel) 2020;12:1736. doi: 10.3390/cancers12071736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian JH, Mu LJ, Wang MY, Zeng J, Long QZ, Bin-Guan Wang W, Jiang YM, Bai XJ, Du YF. FOXM1-Dependent transcriptional regulation of EZH2 induces proliferation and progression in prostate cancer. Anticancer Agents Med Chem. 2021;21:1835–1841. doi: 10.2174/1871520620666200731161810. [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Zhang J, Zheng X, Tan P, Xiong X, Yi X, Yang Y, Wang Y, Liao D, Li H, et al. SR9009 inhibits lethal prostate cancer subtype 1 by regulating the LXRα/FOXM1 pathway independently of REV-ERBs. Cell Death Dis. 2022;13:949. doi: 10.1038/s41419-022-05392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan B, Liu Y, Yu X, Yin L, Peng Y, Gao Y, Zhu Q, Cao T, Yang Y, Fan X, Li X. FOXM1 contributes to taxane resistance by regulating UHRF1-controlled cancer cell stemness. Cell Death Dis. 2018;9:562. doi: 10.1038/s41419-018-0631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng XH, Black M, Ustiyan V, Le T, Fulford L, Sridharan A, Medvedovic M, Kalinichenko VV, Whitsett JA, Kalin TV. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 2014;10:e1004656. doi: 10.1371/journal.pgen.1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitrofanova A, Aytes A, Zou M, Shen MM, Abate-Shen C, Califano A. Predicting drug response in human prostate cancer from preclinical analysis of in vivo mouse models. Cell Rep. 2015;12:2060–2071. doi: 10.1016/j.celrep.2015.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S, Pei X, Xing F, Wu SY, Wu K, Tyagi A, Zhao D, Deshpande R, Ruiz MG, Singh R, et al. Regucalcin promotes dormancy of prostate cancer. Oncogene. 2021;40:1012–1026. doi: 10.1038/s41388-020-01565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koo JI, Sim DY, Lee HJ, Ahn CH, Park J, Park SY, Lee D, Shim BS, Kim B, Kim SH. Apoptotic and anti-Warburg effect of Morusin via ROS mediated inhibition of FOXM1/c-Myc signaling in prostate cancer cells. Phytother Res. 2023;37:4473–4487. doi: 10.1002/ptr.7913. [DOI] [PubMed] [Google Scholar]
- 34.Lin SC, Kao CY, Lee HJ, Creighton CJ, Ittmann MM, Tsai SJ, Tsai SY, Tsai MJ. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat Commun. 2016;7:11418. doi: 10.1038/ncomms11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes AG, Parker JB, Sagar V, Truica MI, Soni PN, Han H, Schiltz GE, Abdulkadir SA, Chakravarti D. A MYC inhibitor selectively alters the MYC and MAX cistromes and modulates the epigenomic landscape to regulate target gene expression. Sci Adv. 2022;8:eabh3635. doi: 10.1126/sciadv.abh3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai W, Zhu W, Li X, Han Y, Wang Y, Leng Q, Li M, Wen X. GTSE1 promotes prostate cancer cell proliferation via the SP1/FOXM1 signaling pathway. Lab Invest. 2021;101:554–563. doi: 10.1038/s41374-020-00510-4. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Jiao X, Robertson AG, Di Sante G, Ashton AW, DiRocco A, Wang M, Zhao J, Addya S, Wang C, et al. The DACH1 gene is frequently deleted in prostate cancer, restrains prostatic intraepithelial neoplasia, decreases DNA damage repair, and predicts therapy responses. Oncogene. 2023;42:1857–1873. doi: 10.1038/s41388-023-02668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu P, Chen X, Xie R, Han J, Xie W, Wang B, Dong W, Chen C, Yang M, Jiang J, et al. lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol Ther. 2017;25:1959–1973. doi: 10.1016/j.ymthe.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Zhou Z, Ji Z, Yan W, Li H, Yu X. Tetramethylpyrazine reduces prostate cancer malignancy through inactivation of the DPP10-AS1/CBP/FOXM1 signaling pathway. Int J Oncol. 2020;57:314–324. doi: 10.3892/ijo.2020.5036. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Zhao H, Chen Y, Li K, Li T, Chen J, Zhang B, Guo C, Qing L, Shen J, et al. Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis. 2021;12:1129. doi: 10.1038/s41419-021-04421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–1244. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B, Diao H, Wang P, Guan F, Liu H. microRNA-877-5p exerts tumor-suppressive functions in prostate cancer through repressing transcription of forkhead box M1. Bioengineered. 2021;12:9094–9102. doi: 10.1080/21655979.2021.1989969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–11081. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan L, Tan HL, Thomas-Ahner JM, Pearl DK, Erdman JW, Jr, Moran NE, Clinton SK. Dietary tomato and lycopene impact androgen signaling- and carcinogenesis-related gene expression during early TRAMP prostate carcinogenesis. Cancer Prev Res (Phila) 2014;7:1228–1239. doi: 10.1158/1940-6207.CAPR-14-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Liu Y, Yuan B, Yin L, Peng Y, Yu X, Zhou W, Gong Z, Liu J, He L, Li X. FOXM1 promotes the progression of prostate cancer by regulating PSA gene transcription. Oncotarget. 2017;8:17027–17037. doi: 10.18632/oncotarget.15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingersoll MA, Chou YW, Lin JS, Yuan TC, Miller DR, Xie Y, Tu Y, Oberley-Deegan RE, Batra SK, Lin MF. p66Shc regulates migration of castration-resistant prostate cancer cells. Cell Signal. 2018;46:1–14. doi: 10.1016/j.cellsig.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Z, Zhan C, Du H, Zhang L, Liang C, Zhang L. Baicalin suppresses the cell cycle progression and proliferation of prostate cancer cells through the CDK6/FOXM1 axis. Mol Cell Biochem. 2020;469:169–178. doi: 10.1007/s11010-020-03739-1. [DOI] [PubMed] [Google Scholar]
- 48.Yu H, Xu Z, Guo M, Wang W, Zhang W, Liang S, Xu Z, Ye J, Zhu G, Zhang C, Lin J. FOXM1 modulates docetaxel resistance in prostate cancer by regulating KIF20A. Cancer Cell Int. 2020;20:545. doi: 10.1186/s12935-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin JZ, Wang WW, Hu TT, Zhu GY, Li LN, Zhang CY, Xu Z, Yu HB, Wu HF, Zhu JG. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020;469:481–489. doi: 10.1016/j.canlet.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 50.Mazzu YZ, Armenia J, Chakraborty G, Yoshikawa Y, Coggins SA, Nandakumar S, Gerke TA, Pomerantz MM, Qiu X, Zhao H, et al. A novel mechanism driving poor-prognosis prostate cancer: Overexpression of the DNA repair gene, ribonucleotide reductase small subunit M2 (RRM2) Clin Cancer Res. 2019;25:4480–4492. doi: 10.1158/1078-0432.CCR-18-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 52.Kim MY, Jung AR, Shin D, Kwon H, Cho HJ, Ha US, Hong SH, Lee JY, Kim SW, Park YH. Niclosamide exerts anticancer effects through inhibition of the FOXM1-mediated DNA damage response in prostate cancer. Am J Cancer Res. 2021;11:2944–2959. [PMC free article] [PubMed] [Google Scholar]
- 53.Gopalakrishnan S, Ismail A. Aromatic monophenols from cinnamon bark act as proteasome inhibitors by upregulating ER stress, suppressing FoxM1 expression, and inducing apoptosis in prostate cancer cells. Phytother Res. 2021;35:5781–5794. doi: 10.1002/ptr.7236. [DOI] [PubMed] [Google Scholar]
- 54.Wu M, Zhao H, Guo L, Wang Y, Song J, Zhao X, Li C, Hao L, Wang D, Tang J. Ultrasound-mediated nanobubble destruction (UMND) facilitates the delivery of A10-3.2 aptamer targeted and siRNA-loaded cationic nanobubbles for therapy of prostate cancer. Drug Deliv. 2018;25:226–240. doi: 10.1080/10717544.2017.1422300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Ligr M, McCarron JP, Daniels G, Zhang D, Zhao X, Ye F, Wang J, Liu X, Osman I, et al. Natura-alpha targets forkhead box m1 and inhibits androgen-dependent and -independent prostate cancer growth and invasion. Clin Cancer Res. 2011;17:4414–4424. doi: 10.1158/1078-0432.CCR-11-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Ji Z, Yan W, Zhou Z, Li H, Xiao Y. Tetramethylpyrazine inhibits prostate cancer progression by downregulation of forkhead box M1. Oncol Rep. 2017;38:837–842. doi: 10.3892/or.2017.5768. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Bu H, Tang S, Liu G, Miao C, Zhou X, Yang H, Liu B. In silico, in vitro and in vivo studies: Dibutyl phthalate promotes prostate cancer cell proliferation by activating Forkhead Box M1 and remission after Natura-alpha pretreatment. Toxicology. 2023;488:153465. doi: 10.1016/j.tox.2023.153465. [DOI] [PubMed] [Google Scholar]
- 58.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, Zhang Y, Paluch AM, Waltz SE, Kasper S, Kalin TV. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem. 2013;288:22527–22541. doi: 10.1074/jbc.M113.455089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Gong Z, Sun L, Li X. FOXM1 and androgen receptor co-regulate CDC6 gene transcription and DNA replication in prostate cancer cells. Biochim Biophys Acta. 2014;1839:297–305. doi: 10.1016/j.bbagrm.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 60.Qu S, Ci X, Xue H, Dong X, Hao J, Lin D, Clermont PL, Wu R, Collins CC, Gout PW, Wang Y. Treatment with docetaxel in combination with Aneustat leads to potent inhibition of metastasis in a patient-derived xenograft model of advanced prostate cancer. Br J Cancer. 2018;118:802–812. doi: 10.1038/bjc.2017.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandit B, Gartel AL. New potential anti-cancer agents synergize with bortezomib and ABT-737 against prostate cancer. Prostate. 2010;70:825–833. doi: 10.1002/pros.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaochar S, Rusin A, Foley C, Rajapakshe K, Robertson M, Skapura D, Mason C, Berman De Ruiz K, Tyryshkin AM, Deng J, et al. Inhibition of GATA2 in prostate cancer by a clinically available small molecule. Endocr Relat Cancer. 2021;29:15–31. doi: 10.1530/ERC-21-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.