Highlights
-
•
A computer assisted approach was used for screening novel α-agarase.
-
•
AgaB was tolerant to most metal ions and retained activities at 3 M NaCl or KCl.
-
•
CBM2 could greatly decrease the Km of AgaB comparing that of CBM2 removed truncations therefore may play crucial role in substrate binding.
-
•
E468 and D333 was identified as catalytic amino acids based on conservation analysis, molecular docking result and site-directed mutagenesis.
Keywords: Agarase, CBM2, Catalytic amino acid, Biochemical characterization
Abstract
Agarooligosaccharides have great potential in food industry because of their various bio-activities, while the limited availability and diversity of α-agarases hinder agarooligosaccharides’ broader application. To overcome this limitation, a computer-assisted method was used to screen and identify novel agarases. Firstly, one novel α-agarase, AgaB, with an N-terminal CBM2 domain (the first report of this domain in agarases), was discovered. Purified agarases only exhibited activity against agarose, with optimum activity at 40℃ and pH 8.0. Analysis of hydrolysis products indicated that AgaB is an endo-type α-agarase, producing agarotetraose and agarohexaose. Secondly, AgaB truncated CBM2 showed increased Km values, suggesting that CBM2 aids in substrate binding. Thirdly, E468 and D333 are possibly catalytic amino acids, which was supported by molecular docking results and mutants. Biochemical characterization of first reported CBM2-containing agarase and catalytic mechanism study lay the foundation for the exploration and development of α-agarases in the future.
Introduction
Agar oligosaccharides have recently captured significant attention due to their potential as bioactive agents in food, pharmaceuticals, and cosmetics industries. This interest stems from several key factors (Chen, Fu, Huang, Xu, & Gao, 2021). Firstly, red algae, as the source of agar oligosaccharides, are abundant and have a large production volume (4.87 million tons/year). They do not require arable land irrigation or fertilization making them an accessible and economical non-grain biomass (Hehemann et al., 2014, Wargacki et al., 2012). Secondly, agar oligosaccharides exhibit a range of biological activities including anti-inflammatory, antioxidant, hepatoprotective, anti-diabetic, anti-tumor, anti-caries, and α-glucosidase inhibitory activities (Chen et al., 2006, Enoki et al., 2010, Enoki et al., 2012, Hong et al., 2017, Yu et al., 2019). Thirdly, due to their low molecular mass and good water solubility, agar oligosaccharides can be easily absorbed expanding their range of applications while avoiding the limitations associated with the high viscosity and poor water solubility of agar (Young et al., 1971, Yun et al., 2017).
Agarase serves as a pivotal enzyme in the enzymatic production of agar oligosaccharides, where its abundance and diversity are preconditions for its application. This enzyme is classified into α-agarase (GH96 family) and β-agarase (GH16/GH50/GH86/GH117) based on their ability to hydrolyze α-1,3 glycosidic bond and β-1,4 glycosidic bond in agar, respectively. Most reported agarases belong to β-agarases and came from marine, soil, plant and human microbiota (Allouch et al., 2003, Hosoda and Sakai, 2006, Song et al., 2015, Song et al., 2016). While, only 7 α-agarases were reported and all of them come from marine bacteria, including Alteromonas agarilytica, Gilvimarinus agarilyticus, Thalassomonas sp JAMB A33, Thalassomonas sp LD5, Catenovulum agarivorans and Catenovulum sediminis WS1-A (Cheng et al., 2015, Flament et al., 2007, Ohta et al., 2005, Xu et al., 194 (2022), Lee et al., 2019). Due to the unique bio-activities exhibited by agarooligosaccharides (with 3,6-anhydro-l-galactose at the reducing end) produced by α-agarase, researchers' interest in α-agarase has grown in recent years (Xu et al., 194 (2022), Liu et al., 2019).
Carbohydrate-binding module (CBM) domain is a non-catalytic module often found in conjunction with glycoside hydrolases and could enhance enzyme solubility and stability (Sidar et al., 2020). CBM6 and CBM13 are found in most β-agarases while only CBM6 is found in all α-agarases (Alkotaini, Han, & Kim, 2016). Most reported α-agarases face the difficulty in expression and low activities, for example, α-agarase from Alteromonas agarilytica are not solubly expressed and had very low activity (Flament et al., 2007). Meanwhile, removal of CBM6 caused insoluble expression and activity lost of AgaE from Thalassomonas sp. LD5. The activity and stability of enzymes serve as pivotal prerequisites for industrial applications, and the discovery of additional CBM families akin to the CBM6 domain represents a vital avenue for enhancing the stability and activity of α-agarases.
The current scarcity and inadequate diversity of α-agarases pose significant limitations to their further application. Meanwhile, reported α-agarases share relatively high sequence similarity with each other ranging from 44 % to 90.5 %. Considering there is no available approach to screen α-agarase specifically, in this study, we employed a computer-assisted method to find novel α-agarase from public databases. A novel α-agarase AgaB with CBM2 domain (never reported in agarase) was characterized and potential catalytic amino acids were investigated.
Materials and methods
Construction of a local α-agarase database
Firstly, we download non-redundant database containing 420 million genes from the NCBI database. Then α-agarase was identified in two approaches, one based on protein sequence similarity and the other based on protein domains.
Identification based on protein sequence similarity: In order to reduce computational time, the diamond method was used due to its faster computational speed compared to blast (Buchfink, Xie, & Huson, 2015). Experimentally characterized α-agarases with catalytically irrelevant regions such as CBM and TSP removed were employed as query sequences. The parameters were set as follows: e-value > 1e-10, match length > 50, matched protein length > 150.
Identification based on protein structural domains: Currently, α-agarases mainly came from the GH96 family, so only GH96 Hmmer model was used for searching (Finn et al., 2011, Zhang et al., 2018). The GH96 Hmmer model was downloaded from the dbCAN database and the parameters for the hmmersearch program were set as follows: e-value < 1e-10, matched length > 50, and matched protein length > 150.
Domain architecture of agarase
In order to investigate the structural domain composition of proteins in the locally constructed α-agarase database, we used Hmmer models from both pfam and dbCAN sources for analysis. The prediction results prioritized the use of dbCAN-predicted structural domains; if dbCAN could not predict them, then pfam-predicted structural domains were used. The match length was greater than 50 and the score was greater than 1e-10.
Phylogenetic tree analysis
In order to reduce the influence of non-catalytic regions on the phylogenetic analysis, regions of non-catalytic domains were removed using in-house script. Then MUSCLE was used to perform protein sequence alignment and IQtree was used for phylogenetic tree analysis. The inferred phylogenetic tree was displayed in ITOLs. Three GH16 agarases were used as outgroup.
Strain cultivation
Strain Agarilytica rhodophyticola (MCCC 1H00123) was purchased from the Marine Culture Collection of China (MCCC). According to instruction of MCCC, it was streaked onto Marine2216 agar medium and incubated at 20 °C for 3 days. At the same time, 16srDNA of the strain was sequenced to ensure that purchased strain is correct.
Protein expression and purification
Firstly, A. rhodophyticola from a solid plate was inoculated into liquid Marine2216 medium and incubated at 20 °C for 3 days. Subsequently, genomic DNA was extracted from using a TIANGEN DNA extraction kit, and the integrity of the extracted genomic DNA was confirmed by electrophoresis on a 0.8 % agarose gel. Secondly, primers were designed to amplify selected α-agarase DNA sequences without signal peptides or transmembrane regions (Supplementary material 1). The PCR amplification of the target gene sequence was performed using a High-Fidelity DNA Polymerase enzyme Q5 (NEB,lot number M0491). The pET-28(+) plasmids and amplified PCR products were digested by BamH I and Xhol I restriction enzymes, ligated using T4 ligase and then transformed into Escherichia coli. In this study, two expression strains E. coli BL21(DE3) and E. coli Rosetta (DE3), were investigated. The clones were sequenced to confirm the gene was successfully constructed into the vectors and used for further protein expression.
Then clone with constructed expression vector was inoculated into LB medium containing a final concentration of 100 μg/mL kanamycin and incubate at 37 °C and 180 rpm. When OD600 reached 0.60 ∼ 0.70, IPTG was added to a final concentration of 5 mM and induction was conducted at 20 °C for 14 h. Then bacteria pellet was resuspended in buffer A (20 mM Tris-HCl, 10 mM CaCl2, 100 mM NaCl) and sonicated. The supernatant was collected at 10,000 g for 5 min. Subsequently, recombinant proteins were purified using immobilized metal ion affinity chromatography. Buffer A containing different concentrations of imidazole were used for washing step. Fractions with high purity were collected, dialyzed with buffer A and used for subsequent enzymatic property analysis.
Biochemical characterization of recombinant protein
Enzyme activity assay
To determine the agarase activity, 0.2 % agarose substrate solution (20 mM Tris-HCl and 10 mM CaCl2, pH 7.0) was prepared and kept at 40 °C for later use. 20 μL of purified AgaB was added to 480 μL substrate and was kept at 40 °C for 15 min (Song et al., 2014). Then, 500 μL of DNS was added to the mixture, with the NaOH in the DNS solution acting to stop the enzyme reaction, and the mixture was subsequently boiled at 100℃ for 7 min to complete the reaction between the released reducing sugar and DNS. The absorbance was then measured at 540 nm. The unit of agarase activity is defined as: under experimental conditions, the amount of enzyme required to produce 1 μmol reducing sugar per minute is one enzyme activity unit (U). Unless otherwise specified in this study, all experiments used inactivated enzyme solution (heat inactivated for 10 min at 100℃) as the control group and had three replicates.
Substrate specificity
To determine the substrate specificity of the recombinant enzyme, we tested the polysaccharides that could be degraded by the recombinant enzyme. The substrates tested included agarose, sodium carboxymethyl cellulose, soluble starch, sodium alginate, dextran, pectin, carrageenan, xanthan gum and gum arabic. 0.2 % substrate solutions were prepared separately (20 mM Tris-HCl and 10 mM CaCl2, pH 7.0). For each 480 μL of substrate solution, 20 μL of purified AgaB was added and reacted at 40 °C for 15 min. The concentration of reducing sugar was determined by the DNS method (Miller, 1959, Song et al., 2021). If reducing sugar was released, it was considered that agarase could degrade the polysaccharide.
Effect of Ca2+ on agarase activity
Most reported α-agarases needed Ca2+ as the co-factor, therefore we firstly investigated if AgaB needed metal ions as co-factor and if Ca2+ could enhance AgaB activity. Exhaustively dialysis was used to remove Ca2+ from the purified enzyme solution. The purified α-agarase was dialyzed using a Ca2+ buffer (20 mM Tris-HCl, 100 mM NaCl, pH7.0), with a volume of 2 L each time and dialyzed 5 times. First, to confirm if AgaB was metal ion dependent, we assessed the enzyme's activity by introducing varying concentrations of a metal ion chelator (EDTA) at levels of 0 mM, 1 mM, 5 mM, 10 mM, and 20 mM. The presence of EDTA causing a decrease in AgaB's activity would signify the essentiality of metal ion as a cofactor for AgaB; conversely, its sustained activity would indicate otherwise. Then different concentrations of Ca2+ (0 mM, 1 mM, 5 mM, 10 mM, 20 mM and 50 mM) were added to the reaction system separately and the enzyme activity was measured under optimal conditions.
Optimum temperature and temperature stability
To evaluate the optimal temperature of the recombinant agarase, the enzyme activity was tested at 0 °C, 10 °C, 20 °C, 30 °C, 40 °C, 50 °C and 60 °C. The temperature with highest enzyme activity was the optimum temperature. To test the temperature tolerance of the recombinant agarase, the agarase was placed at different temperatures and its enzyme activity was measured after 2 h (Song et al., 2021).
Optimum pH and pH stability
To test the optimal reaction pH of the recombinant agarase, agarose solutions were prepared using Britton-Robinso pH buffers (pH 3–11). The agarase was added to 0.2 % agarose solution prepared using different pH buffer solutions and reacted at 40 °C for 15 min to measure the released reducing sugar. To test the stability of purified AgaB at different pHs, the agarase was placed in different pH buffers and its enzyme activity was measured after 2 h. In this experiment, the highest enzyme activity was recorded as 100 % to calculate the relative enzyme activity under other conditions (Song et al., 2021).
Effect of heavy metal ions and chemical regents on enzyme activity
To investigate the effect of metal ions and chemical reagents on the activity of recombinant agarase, 1 mM and 10 mM of Co2+, Ba2+, Ni2+, Fe2+, Mg2+, Mn2+, Cu2+, Li+, Zn2+, Cr2+, Fe3+ and SDS were added to the substrate separately. Inactivated enzyme solution was used as a blank control and the positive control was enzyme activity without the addition of metal ions. The enzyme activity was recorded as 100 % and used to calculate the relative enzyme activity at different concentrations of metal ions and chemical reagents.
Effect of salt concentration on enzyme activity
0.2 % agarose solutions with different NaCl concentrations (0.10 M, 0.20 M, 0.50 M, 1.00 M, 2.00 M and 3.00 M) were prepared and purified AgaB was added separately. The enzyme activity was measured after reacting for 15 min under optimal conditions. In this experiment, the highest enzyme activity was recorded as 100 % to calculate the relative enzyme activity of recombinant agarase at different NaCl concentrations.
Kinetic parameter measurement
AgaB was added to a mixture of different concentrations of agarose solution (0.05 %, 0.08 %, 0.10 %, 0.15 %, 0.20 %, 0.25 % and 0.30 %) containing purified AgaB (without Ca2+), purified AgaB (with Ca2+). The enzyme activity was measured after incubating for 15 min under optimal conditions using the Lineweaver-Burk double reciprocal plot method in origin software (Jumper et al., 2021). A plot of 1/V against 1/[S] was used to determine Vmax and Km.
Characterization of hydrolyzed product
5 U AgaB was added to a 10 mL 0.2 % agarose solution (20 mM Tris-HCl and 10 mM CaCl2, pH 7.0). Samples were taken at different time points: 0, 3, 5, 10, 15, 30, 60 and 120, 240 min. The supernatants were collected by centrifugation and applied to thin layer chromatography using n-butanol/acetic acid/distilled water (volume ratio = 2:1:1) as developing agent and aniline-diphenylamine solution as coloring agent. Freeze-dried products were dissolved in D2O and then analyzed using JNM-ECZ400S (400 M) spectrometer. Meanwhile, the freeze-dried products were also analyzed using electrospray-ionization mass spectrometry (ESI-MS) in negative ion mode.
Structure prediction and molecular docking of AgaB
Homology modeling was not used to predict AgaB structure because no 3D structure of GH96 family protein was available currently. Therefore, AgaB model was produced using AlphaFold Colab notebook using the default settings with Amber relaxation (msa_method = mmseqs2, homooligomer = 1, pair_mode = unpaired, max_msa = 512:1024, num_relax = 5, use_ptm = True, rank_by = pLDDT, num_models = 5, max_recycles = 3, use_templates = False) (Mirdita et al., 2022) (Jumper et al., 2021). Consurf online service was used to analyze conservation of AgaB, Prankweb3 was used to predict the substrate binding pocket and Autodock Vina was used to conduct the molecular docking (Ashkenazy et al., 2016, Jakubec et al., 2022, Trott et al., 2009).
Construction of truncated α-agarase
Three truncations were constructed, including Truncation-1 (30–108), Truncation-2 (109–789), and Truncation-3 (160–786), each containing CBM2, linker + GH96, and GH96, respectively. The plasmid construction, protein expression, protein purification and enzyme kinetic assay were similar to those for AgaB. Corresponding primers were showed in Supplementary material 1.
Site directed mutagenesis
Overlapping PCR was used for construct site directed mutagenesis and the mutants were sequenced to confirm. The plasmid construction, protein expression, protein purification and enzyme kinetic assay were similar to those for AgaB. Corresponding primers were showed in Supplementary material 1.
Results and discussion
Screen of novel α-agarase with novel CBM domain
Phyla distribution of α-agarases
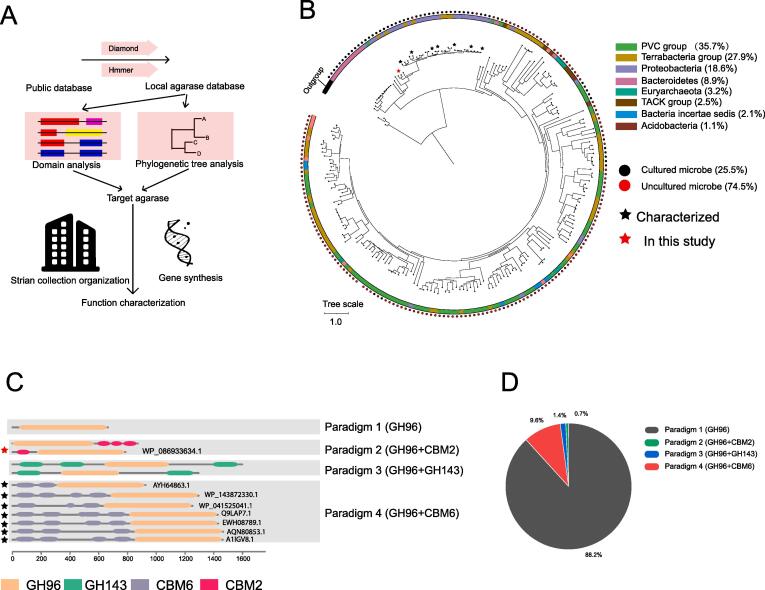
The enzymatic degradation of agar by α-agarase produces oligosaccharides with unique biological activities and significant economic potential, making α-agarase a vital enzyme. Therefore, developing accurate and efficient methods to explore α-agarase is crucial for enhancing its diversity and quantity, and is an indispensable prerequisite for its industrial application. In this study, we utilized bioinformatic ways to find 298 α-agarases, (Fig. 1A). The phylogenetic tree analysis revealed that the 7 previously studied α-agarases and AgaB were grouped within the same clade, indicating that they shared a common ancestor. Meanwhile, the 7 agarases formed a smaller distinct subclade, and the short branch lengths suggested they were closely related. This was consistent with their high sequence similarity with each other (44 % to 90.5 %). Based on phylogenetic topology, AgaB was distinct with the reported agarases and selected for subsequent study (Fig. 1B). Meanwhile, the short branch lengths in this clade might because they have high sequence similarities. The taxonomic distribution of these explored α-agarases revealed that the majority came from bacteria, with the PVC group (35.7 %), Terrabacteria group (27.9 %), and Proteobacteria (18.6 %) being the most common. In contrast, only a small proportion of α-agarases (3.2 %) were from the Euryarchaeota. Remarkably, although α-agarases were found across various bacterial and archaeal phyla, all reported α-agarases to date are from Proteobacteria. Furthermore, a significant proportion of the microbes carrying α-agarases (74.5 %) were from metagenomes, while only 25.5 % can be cultivated in the laboratory. The prevalence of Proteobacteria among cultivable microorganisms corresponds to the observed pattern of α-agarase exclusivity to Proteobacteria. Thus, the reliance on screening coupled with sequencing may introduce a bias towards bacterial species that are easier to cultivate in laboratory settings when exploring α-agarase, potentially skewing α-agarase discovery.
Fig. 1.
A: Scheme of novel α-agarase screening pipeline. B: Phylogenetic tree analysis of collected 298 α-agarases. IQ-tree was used to infer the phylogenetic tree and ITOL was used for visualization. The color of the outer ring of the evolutionary tree represents the species of the corresponding protein origin, the outermost circle represents whether the protein origin species can be cultured, and the different coloured pentagrams represent the studied proteins and AgaB. Three GH16 agarases were used as outgroup. C: Paradigms of α-agarases domain architecture. D: Distribution of α-agarases domain architecture paradigms.
Screen of novel α-agarases
Due to the impracticality of studying all 298 individual α-agarases, representative α-agarases with specific domain architectures were selected for investigation (Fig. 1C). The domain architecture of α-agarase had four different paradigms, which were Paradigm-1 (GH96, 88.2 %), Paradigm-2 (GH96 + CBM2, 0.7 %), Paradigm-3 (GH96 + GH143, 1.4 %), and Paradigm-4 (GH96 + CBM6, 9.6 %) (Fig. 1D). The most abundant paradigm was Paradigm-1, which mostly consisted of members with one or two GH96 domains. Paradigm-4 had the second-largest number of members and included all currently reported α-agarases, with 3–4 consecutive CBM6 domains. Paradigms 2 and 3 had the fewest members, with only 2 and 4 members, respectively, but they also contained domains that have never been found in agarase, namely CBM2 and GH143 domains.
Paradigm 2 (GH96 + CBM2) was selected for this study based on two considerations: firstly, CBM2 was not found in any reported agarases, and secondly, the presence of CBM could enhance both the solubility and stability of agarase, thereby increasing success probability of experiments. Consequently, Paradigm 1 and Paradigm 3 were not considered due to the absence of the CBM domain, and Paradigm 4 was not considered because CBM6-containing agarases were well studied in previous studies. There were 2 proteins from Paradigm 2, including PCK04292.1 from a metagenomic assemble genome Alteromonadaceae bacterium and WP_086933634.1 from A. rhodophyticola. Considering the A. rhodophyticola was deposited in MCCC, we chose A. rhodophyticola for later analysis of α-agarase’s role in agar degradation process. Protein AgaB (WP_086933634.1) had a N-terminal CBM2 domain, shared 49–65 % sequence identities and 41–65 % coverage with known α-agarases, indicating that it is a novel α-agarase.
Since the first discovery of α-agarase in the 1993, only a few α-agarases have been reported. Compared to the traditional method of screening agarases by culturing bacteria and sequencing, constructing α-agarase database for screening has several advantages. First, this approach utilizes current sequencing data and enables researchers to target the screening of α-agarases with novel domain architecture or low sequence similarity. If the bacterium is cultivable, researchers may obtain the strain from a bacteria culture collection center and investigate the role of α-agarase in corresponding strain’s agar utilization. Second, the computer-assisted approach is not limited by the bacterial screening method or sampling, making it possible to explore the “dark matter” of metagenomes and enabling researchers to quickly identify novel proteins of interest. In particular, for proteins such as α-agarases, which are sparsely reported and studied, the use of computer-assisted approaches is crucial to augment the diversity of α-agarases.
Biochemical characterization of AgaB
Purification and expression of AgaB
After CBM2-containing AgaB is selected, we purchased and cultured the corresponding bacterial strain A. rhodophyticola from MCCC, not only to clone AgaB gene, but also to study its role in agar degradation. Despite our attempts to alter the culture medium, adjust the culture temperature, and re-inoculate the strain, A. rhodophyticola failed to propagate after one passage. We were able to extract ample genomic DNA from a colony on a plate, enabling us to proceed with gene amplification and vector construction. Moreover, a lot of effort was made to obtain soluble and active AgaB. Firstly, we attempted to express AgaB in different bacterial strains, and recombinant AgaB was only expressed actively in E. coli Rosetta (DE3) strain while not in E coli BL21 (DE3). This was probably because AgaB had rare codons, especially continuous occurrence of rare codon (Supplementary material 2). Secondly, the expression conditions for AgaB were strict, as it only expressed at OD600nm of 0.4–0.6, induction temperature of 16-20℃, and IPTG concentration of 1–5 mM. Activity sharply decreased or was absent when these conditions were not met. Following metal affinity chromatography purification, AgaB exhibited a single band with over 90 % purity on SDS-PAGE with 90 kDa molecular mass, which was close to predicted 89.8 kDa (Fig. 2A). Furthermore, AgaB was found to be active only against agarose, but not pectin, starch, beta-glucan, cellulose, or carrageenan, indicating that it is an agarase (Fig. 2B).
Fig. 2.
A: SDS-PAGE analysis of purified AgaB. B: Relative activity of AgaB against different polysaccharide substrates. C: Effects of EDTA on AgaB activity. D: Effects of Ca2+ on AgaB activity. E: Optimum temperature of AgaB activity. F: Thermostability of AgaB. G: Optimum pH of AgaB activity. H: pH stability of AgaB. I: Effects of different concentration of KCl on AgaB activity. J: Effects of different concentration of NaCl on AgaB activity. Data are mean ± SD of three independent experiments.
Dependent of Ca2+ for enzyme activity
In previous reports, Ca2+ was characterized as critical co-factor for α-agarase (Lee et al., 2019, Liu et al., 2019, Zhang et al., 2018). Firstly, our results showed that the relative enzymatic activity of AgaB was 52 %, 48 %, and 47 % at EDTA concentrations of 1 mM, 5 mM, and 10 mM, respectively (Fig. 2C). Thus, although EDTA can reduce AgaB activity, it does not completely abolish it. Secondly, the addition of Ca2+ resulted in AgaB activity increment, with the highest activity observed at 10 mM, which was 1.75 times higher than the activity without Ca2+ (Fig. 2D). However, when the calcium ion concentration exceeded 10 mM, the activity slightly decreased. Thirdly, the presence of Ca2+ significantly enhanced the substrate affinity of AgaB enzyme, as evidenced by a lower Km value (0.34 mg/mL) compared to that in the absence of Ca2+ (3.18 mg/mL) (Table 1). In conclusion, our results suggested that Ca2+ may enhance the binding between AgaB and its substrate.
Table 1.
Km and Vmax of AgaB, AgaB mutants and truncations.
| Km (mg/mL) | Vmax (U/mg) | |
|---|---|---|
| AgaBa | 3.18 ± 0.23 | 0.24 ± 0.13 |
| AgaBb | 0.35 ± 0.01 | 0.63 ± 0.11 |
| Truncation-2 | 4.30 ± 0.37 | 0.79 ± 0.04 |
| Truncation-3 | 5.47 ± 0.28 | 3.96 ± 0.30 |
| E468Q | ND | ND |
| D259N | 0.11 ± 0.01 | 0.55 ± 0.03 |
| D333N | ND | ND |
| E452Q | 0.27 ± 0.02 | 1.21 ± 0.05 |
AgaB activity was detected in 20 mM Tri-HCl buffer without Ca2+.
AgaB activity was detected in 20 mM Tri-HCl buffer without 10 mM Ca2+.
We hypothesized that Ca2+ improved AgaB activity by enhancing the binding affinity between substrate and enzyme, while AgaB could exhibit activity without Ca2+. This was similar to AgaE from Thalassomonas sp. LD5 and AgaWS5 from Thalassomonas agarivorans (Lee et al., 2019, Zhang et al., 2018). However, in some reports, EDTA could completely inhibit α-agarase activity (Liu et al., 2019, Zhang et al., 2018). This is likely due to the low activity of the enzyme itself making it undetectable when Ca2+ was removed and activity decreased, or the agarase was not stable after removal of Ca2+. Therefore, the mechanism by which Ca2+ affect AgaB activity requires further investigation.
Effects of metal ions and chemical regeants on enzyme activities
AgaB was tolerant to most metal ions and exhibited 26.82 %-87.35 % enzymatic activity in the presence different metal ions (1 mM or 10 mM) (Table 2). For example, AgaB maintained activity in the presence of metal ions at a concentration of 10 mM, with Cu2+ (58.85 % activity), Mg2+ (86.91 %), Co2+ (57.67 %), Ni+ (52.54 %), Cr3+ (32.71 %), Fe3+ (26.82 %), Fe2+ (33.02 %), and Zn2+ (37.30 %). Only the presence of 10 mM Mn2+ led to complete inactivation of the enzyme, which was also observed in AgaD from Thalassomonas sp. LD5 (Zhang et al., 2018). Notably, 1 mM and 10 mM Ba2+ significantly enhanced the activity of AgaB, by 21.27 % and 22.43 %, respectively. Meanwhile, the enzymatic activity of AgaB was significantly reduced in the presence of 1 mM or 10 mM SDS, with respective decreases of 71.36 % and 19.70 %. This suggested that the enzyme was sensitive to the presence of detergents, which may limit its application.
Table 2.
Effects of different metal ions and reagents on AgaB activity.
| 1 mM | 10 mM | |
|---|---|---|
| Control | 100.01 ± 3.13 | 100.00 ± 7.93 |
| Cu2+ | 62.47 ± 3.53 | 58.85 ± 2.97 |
| Mg2+ | 80.26 ± 3.68 | 86.91 ± 1.93 |
| Mn2+ | 58.55 ± 5.36 | 0.00 ± 0.00 |
| Co2+ | 87.35 ± 5.50 | 57.67 ± 3.11 |
| Ba2+ | 121.27 ± 6.75 | 122.43 ± 5.97 |
| Ni+ | 78.90 ± 4.05 | 52.54 ± 4.65 |
| Cr3+ | 60.51 ± 1.71 | 32.71 ± 2.66 |
| Li+ | 61.71 ± 1.59 | 106.97 ± 1.87 |
| Fe3+ | 60.66 ± 1.63 | 26.82 ± 1.63 |
| Fe2+ | 82.22 ± 6.02 | 33.02 ± 9.88 |
| Zn2+ | 72.27 ± 1.71 | 37.30 ± 3.13 |
| SDS | 71.36 ± 4.53 | 19.70 ± 0.53 |
Effects of temperature and pH on enzyme activities
The optimum temperature of α-agarase was 40 °C, with over 80 % enzymatic activity retained at 30–40 °C (Fig. 2E). The optimal temperature for AgaB was similar to the reported optimal temperature for most α-agarases. This was possible that the predominant α-agarases came from marine environments, where temperatures typically range from 0 to 35℃. α-agarases had evolved to adapt its natural habitat. Meanwhile, like other α-agarases, AgaB was relative stable at 0–20 °C and its activity declined sharply when exposed to temperatures of 50 °C or higher (Fig. 2F).
The optimum pH for AgaB was 7, which was also the optimum pH for other reported α-agarases. AgaB showed over 50 % enzymatic activity between pH 4–8, indicating its relative wide pH range (Fig. 2G). AgaB retained over 80 % activity after being incubated for 2 h at pH 7–11, indicating AgaB was relative stable in alkaline environment (Fig. 2H). This was similar to CaLJ96 from C. agarivorans and better than AgaE from Thalassomonas sp. LD5 (Xu et al., 194 (2022), Lee et al., 2019). The fact that AgaB is stable within an alkaline environment could be advantageous for its industrial applications.
Effects of salt concentration on enzyme activities
Most agarases came from marine environments and therefore the salt concentration might affect the enzyme activities. Both NaCl and KCl could enhance AgaB enzymatic activity (Fig. 2I and Fig. 2J). For instance, 0.1 M NaCl increased the enzyme activity by 23.44 %, while 0.2 M KCl increased it by 17.54 %. As the salt concentration increased, the activity of AgaB gradually decreased. However, even at a salt concentration of 3 M, AgaB retained activity. For example, AgaB showed 29.86 % and 43.38 % activities in the presence of 3 M NaCl and 3 M KCl. Tolerance to high salt concentration was not reported in any reported α-agarases and is also advantageous in industrial application.
Characterization of degradation products
TLC was used to determine whether AgaB is an endo- or exo-type enzyme. It was observed that large oligosaccharides appeared after 10 min hydrolysis and low molecular mass oligosaccharides increased with time on TLC plate (Fig. 3A). At 240 min, reducing sugar amounts were not increased and only two bands were observed. Therefore, like other reported α-agarases, AgaB was an endo-type α-agarase rather than exo-type α-agarase. ESI-MS analysis of hydrolysis product showed that two products might be A4 and A6, because the appearance of 664.97 mz ([M + Cl]-) and 971.05 mz ([M + Cl]-) (Fig. 3B). Similarly, hydrolysis products of AgaE from Thalassomonas sp. LD5 were also A4 and A6, while other α-Agarases mainly produced A4 as the major hydrolysis product.
Fig. 3.
A: TLC analysis of agarose degradation products by AgaB. B: ESI-MS analysis of agarose degradation products by AgaB. C: C-NMR analysis of agarose degradation products by AgaB. G β-d-galactopyranose, A 3,6-anhydro-a-L- aglactopyranose, r reducing end, nr non-reducing end, α α anomer, β β anomer.
In addition, NMR analysis of the products showed appearance of a characteristic peak of α-agarase at 90.72 ppm (corresponding to hydrated form of the aldehyde), indicating the AgaB was an α-agarase (Rochas, Potin, & Kloareg, 1994) (Fig. 3C). The signals (97.11 ppm and 93.12 ppm) for C-1 position of galactose residue confirmed that 3,6-anhydropyranose was the reducing end of purified oligosaccharides (Li et al., 2007). Therefore, combining the MS results and CNMR results, we confirmed that AgaB was an endo-type α-agarase which produced A4 and A6 as main products.
Catalytic mechanism study of AgaB
Prediction of AgaB three-dimension structure
The investigation of the catalytic mechanism of α-agarase has been limited in previous reports due to the lack of a 3D structure. The scarcity of α-agarases and difficulties in their expression have greatly impeded progress in characterizing their crystal structures. Thanks to the recent development of AI-based algorithms for 3D structure prediction, it is now possible to simulate the 3D structure of α-agarase and investigate its catalytic mechanism even in the absence of experimentally verified structures. In this study, the three-dimensional structure of AgaB was predicted using the widely used AlphaFold2 algorithm. The prediction results revealed that the AgaB 3D structure comprised a N-terminal CBM2 domain and a C-terminal GH96 domain, connected by a long linker (amino acids 108–160) (Fig. 4A and B). The CBM2 structure consisted mainly of β-sheets and was attached to the surface of GH96 structure, possibly through hydrogen bonds and salt bridges. The GH96 structure was comprised of multiple β-sheets and α-helices. Both structures (CBM2 and GH96) had a high confidence score, and only the confidence score of the linker random coil was low (Supplementary material 3). Since the low confidence region was not located within the functional domains, this predicted structure can be used for subsequent analysis.
Fig. 4.
A: Domain architecture of AgaB and different truncations. B: SDS-PAGE analysis of purified Truncation-2 and Truncation-3. C: Predicted three dimension structure of AgaB.
Enzyme kinetic analysis of truncations
We sought to examine the impact of the CBM2 domain on AgaB activity by constructing three truncations, Truncation-1 (30–108), Truncation-2 (109–789), and Truncation-3 (160–786), comprising CBM2, linker + GH96, and GH96 domain, respectively (Fig. 4C). However, only Truncation-2 and Truncation-3 were successfully expressed, while Truncation-1 failed to be expressed even though the expression conditions were exclusively optimized. Interestingly, we found that purified Truncation-2 and Truncation-3 had agarase activity, indicating that α-agarase could be expressed solubly without the CBM2 domain and retained activity (Fig. 4D). This finding contrasts with previous studies that showed CBM was essential for α-agarase stability, such as AgaE from Thalassomonas sp. LD5, which did not have activity after CBM was removed. Therefore, we hypothesized that CBM2 domain might not be essential for the soluble expression and activity of AgaB.
Moreover, our findings also suggested that the CBM2 domain could enhance the affinity between AgaB and substrate. The quantitative measurement of binding affinity between CBM2 and agarose was unknown because CBM2 was not successfully expressed. Km values of CBM2-removed truncations (Truncation-2 and Truncation-3) increased 12.32- and 16.44-fold compared to that of AgaB (with Ca2+), respectively, and 1.35 and 1.8-fold increases for AgaB (without Ca2+) (Table 1). Therefore, our results indicated that CBM2 could increase substrate binding affinity evidenced by increased Km value of CBM2-removed truncations, which was similar to previous report (Alkotaini, Han, & Kim, 2016). Moreover, our findings extended the substrate spectrum of CBM2 family, as known CBM2 could only bind to chitin (Hanazono et al., 2016), cellulose (Courtade, Forsberg, Heggset, Eijsink, & Aachmann, 2018), xylan (Simpson, Xie, Bolam, Gilbert, & Williamson, 2000), and β-glucan (Courtade et al., 2018).
In our study, we found that Truncation-2 and Truncation-3 had higher Vmax values compared to full-length AgaB (with Ca2+), with 1.25-fold and 6.26-fold increases, respectively (Table 1). These results suggested that the linker region may impede enzyme catalytic efficiency. The removal of the flexible linker region may have removed an obstacle to enzyme-substrate interactions or conformational changes required for catalytic activity. Additionally, we found that this discovery provided a strategy for the industrial application of AgaB, which usually requires high concentrations of agarose for biomass conversion and enzymes that are usually substrate saturated. Therefore, Truncation-3, which had higher catalytic efficiency and lower molecular mass, may be a more optimal choice.
Key catalytic amino acid of AgaB
Studies on the catalytic mechanism of alpha-agarase have been limited, but using the protein structure predicted by AlphaFold, we attempted to elucidate its catalytic mechanism. Firstly, we used consurf analysis to identify the conserved amino acid residues. Most conserved amino acid residues located in the cleft formed by alpha helices and beta folds in the GH96 domain, which was adjacent to the CBM2 structure (Fig. 5A). Meanwhile, 5 substrate binding pockets were predicted and all of them were located on one side of CBM2 and GH96 (Fig. 5B and Supplementary material 4). Pocket 1, Pocket 2 and Pocket 4 were in the conserved region predicted by consurf, making them likely to be involved in agarose hydrolysis. However, the predicted binding pockets involved too many amino acids, making it difficult to identify the key catalytic amino acid. Therefore, we used Truncation-3 which had catalytic activity and a simpler structure for further substrate binding pocket prediction. Truncation-3 only had 1 predicted binding pocket, which matched the conserved region predicted by consurf, greatly reducing the number of amino acid candidates (Fig. 5C).
Fig. 5.
A: Conservation analysis of AgaB using Consurf. B: Predicted substrate binding pockets of AgaB using Prankweb. C: Predicted substrate binding pockets of Truncation-3 using Prankweb. D: Results of Autodock-vina results using Truncation-3 as macromolecule and NA6 as ligand. E: Protein-ligand Interactions between NA6 and amino acids within the substrate binding pocket. F: 2D interactions between ligand NA6 and interacted amino acids. G: SDS-PAGE analysis of 4 site-directed mutants.
The conservation of catalytic amino acids in glycoside hydrolases (GH) is essential during protein evolution process. In this study, we aimed to identify conserved aspartic acids and glutamic acids in the substrate-binding pocket of AgaB. We employed Prank and Consurf analyses to identify the most conserved amino acids in the substrate-binding pocket. In predicted substrate-binding pocket, 2 aspartic acids (D333 and D259) and 2 glutamic acids (E452 and E468) were found. Subsequently, the Consurf analysis revealed that D333 and E468 were the most conserved residues (100 %), while D259 and E452 were relatively conserved (Supplementary material 5). To confirm the potential catalytic amino acids, we used molecular docking and selected NA6 as the substrate because AgaB could only degrade oligosaccharides equal to or longer than NA6. Autodock-vina results showed that the best match had an affinity score of −8.5 kcal/mol, indicating high affinity between the ligand and AgaB. Among the amino acids that interacted with NA6, D333, E468, D259, E452, and D349 were identified (Fig. 5D, 5E and 5F). Finally, based on the results of the substrate-binding pocket analysis, protein conservation analysis, and molecular docking, we selected D333, E468, D259, and E452 as candidate sites for subsequent mutagenesis studies.
Through expression and purification of 4 mutants (D333, E468, D259, E452), we found that E468Q and D333N, exhibited no enzymatic activity. Considering that these two amino acids were highly conserved and located within the predicted pocket, and the molecular docking results predicedt their interaction with NA6 through hydrogen bonding, we speculated that they play a role as a proton donor or a nucleophile/base. Interestingly, previous reported AgaE from Thalassomonas sp. LD5 inferred D779 was critical for catalysis, which was different with our finds. Therefore, further experimental studies are needed, particularly on experimentally confirmed crystal structures, to clarify the catalytic mechanism of α-agarase.
Meanwhile, the Km and Vmax values of D259N and E452Q mutants were changed compared to the wild type. For example, Km values of D259N (0.11 mg/mL) and E452Q (0.27 mg/mL) were lower than that of AgaB (0.35 mg/mL). Meanwhile, Vmax of D259N (0.55 U/mg) was slightly decreased and the Vmax of E452Q (1.21 U/mg) was doubled comparing with Vmax of AgaB (0.63 U/mg) (Table 1). These results indicated that mutation of key amino acids in the substrate binding pocket may be a feasible strategy for improving the catalytic properties of AgaB.
Conclusion and perspective
The absence of specific screening methods is the primary reason for the scarcity and inadequate diversity of α-agarases. We employed a computer-assisted screen method to find a novel α-agarase with CBM2 domain, which was never found in agarases. The following enzyme characterization indicates its potential in industrial application due to its small molecular weight and tolerance to most heavy metal ions. The identification of crucial catalytic amino acids and substrate binding pocket also lay a foundation for improving its catalytic efficiency and enzyme stability.
CRediT authorship contribution statement
Dezhi Yuan: Conceptualization, Writing – review & editing. Hua Lv: Visualization, Methodology. Tiantian Wang: Formal analysis, Investigation. Yulu Rao: Data curation, Methodology. Yibo Tang: Project administration, Funding acquisition. Yiwen Chu: Funding acquisition. Xinrong Wang: Funding acquisition. Jiafu Lin: Funding acquisition. Peng Gao: Conceptualization, Writing – review & editing, Supervision. Tao Song: Conceptualization, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was founded by the Open Project Program of Irradiation Preservation Key Laboratory of Sichuan Province, Sichuan Institute of Atomic Energy (NO. FZBC2022003), the general Project of Sichuan Education Department(18ZB0150), Natural Science Foundation of Sichuan Province(No.2022NSFSC0591), Funding from Department of Ecology and Environment of Sichuan Province (2022HB05), Sichuan key research and development program (2023YFS0384), National Natural Science Foundation of Sichuan Province (2023NSFSC1237), Zunyi Technology and Big data Bureau, Moutai institute Joint Science and Technology Research and Development Project (ZSKHHZ[2021] No.320), Research Foundation for Scientific Scholars of Moutai Institute (mygccrc[2022] No.066), Guizhou Provincial Basic Research Program(Natural Science).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.100915.
Contributor Information
Peng Gao, Email: ppenggao@163.com.
Tao Song, Email: songtao@cdu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Chen X., Fu X., Huang L., Xu J., Gao X. Agar oligosaccharides: A review of preparation, structures, bioactivities and application. Carbohydrate Polymers. 2021;265 doi: 10.1016/j.carbpol.2021.118076. [DOI] [PubMed] [Google Scholar]
- A.J. Wargacki, E. Leonard, M.N. Win, D.D. Regitsky, C.N.S. Santos, P.B. Kim, S.R. Cooper, R.M. Raisner, A. Herman, A.B. Sivitz, A. Lakshmanaswamy, Y. Kashiyama, D. Baker, Y. Yoshikuni, An engineered microbial platform for direct biofuel production from brown macroalgae, Science (80-.). 335 (2012) 308–313. https://doi.org/10.1126/science.1214547. [DOI] [PubMed]
- Hehemann J.H., Boraston A.B., Czjzek M. A sweet new wave: Structures and mechanisms of enzymes that digest polysaccharides from marine algae. Current Opinion in Structural Biology. 2014;28:77–86. doi: 10.1016/j.sbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Enoki T., Okuda S., Kudo Y., Takashima F., Sagawa H., Kato I. Oligosaccharides from agar inhibit pro-inflammatory mediator release by inducing heme oxygenase 1. Bioscience, Biotechnology, and Biochemistry. 2010;74:766–770. doi: 10.1271/bbb.90803. [DOI] [PubMed] [Google Scholar]
- Chen H., Yan X., Zhu P., Lin J. Antioxidant activity and hepatoprotective potential of agaro-oligosaccharides in vitro and in vivo. Nutrition Journal. 2006;5:31. doi: 10.1186/1475-2891-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.J., Lee J.-H., Kim E.J., Yang H.J., Park J.-S., Hong S.-K. Anti-Obesity and anti-diabetic effect of neoagarooligosaccharides on high-fat diet-induced obesity in mice. Marine Drugs. 2017;15:90. doi: 10.3390/md15040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki T., Tominaga T., Takashima F., Ohnogi H., Sagawa H., Kato I. Anti-tumor-promoting activities of agaro-oligosaccharides on two-stage mouse skin carcinogenesis. Biological & Pharmaceutical Bulletin. 2012;35:1145–1149. doi: 10.1248/bpb.b12-00188. [DOI] [PubMed] [Google Scholar]
- Yu S., Yun E.J., Kim D.H., Park S.Y., Kim K.H. Anticariogenic activity of agarobiose and agarooligosaccharides derived from red macroalgae. Journal of Agricultural and Food Chemistry. 2019;67:7297–7303. doi: 10.1021/acs.jafc.9b01245. [DOI] [PubMed] [Google Scholar]
- Yun E.J., Yu S., Kim K.H. Current knowledge on agarolytic enzymes and the industrial potential of agar-derived sugars. Applied Microbiology and Biotechnology. 2017;101:5581–5589. doi: 10.1007/s00253-017-8383-5. [DOI] [PubMed] [Google Scholar]
- Young K., Duckworth M., Yaphe W. The structure of agar. Carbohydrate Research. 1971;16:446–448. doi: 10.1016/S0008-6215(00)81179-9. [DOI] [Google Scholar]
- Allouch J., Jam M., Helbert W., Barbeyron T., Kloareg B., Henrissat B., Czjzek M. The Three-dimensional Structures of Two β-Agarases. The Journal of Biological Chemistry. 2003;278:47171–47180. doi: 10.1074/jbc.M308313200. [DOI] [PubMed] [Google Scholar]
- Song T., Xu H., Wei C., Jiang T., Qin S., Zhang W.…Cao Y.Y. Horizontal transfer of a novel soil agarase gene from marine bacteria to soil bacteria via human microbiota. Scientific Reports. 2016;6:34103. doi: 10.1038/srep34103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda A., Sakai M. Isolation of Asticcacaulis sp SA7, a novel agar-degrading alphaproteobacterium. Bioscience, Biotechnology, and Biochemistry. 2006;70:722–725. doi: 10.1271/bbb.70.722. [DOI] [PubMed] [Google Scholar]
- Song T., Zhang W., Wei C., Jiang T., Xu H., Cao Y.…Qiao D. Isolation and characterization of agar-degrading endophytic bacteria from plants. Current Microbiology. 2015;70:275–281. doi: 10.1007/s00284-014-0713-6. [DOI] [PubMed] [Google Scholar]
- D. Flament, T. Barbeyron, M. Jam, P. Potin, M. Czjzek, B. Kloareg, G. Michel, α-agarases define a new family of glycoside hydrolases, distinct from β-agarase families, Appl. Environ. Microbiol. 73 (2007) 4691–4694. https://doi.org/AEM.00496-07 [pii] 10.1128/AEM.00496-07. [DOI] [PMC free article] [PubMed]
- Cheng H., Zhang S., Huo Y.-Y.-Y., Jiang X.-W.-W., Zhang X.-Q.-Q., Pan J.…Wu M. Gilvimarinus polysaccharolyticus sp. nov., an agar-digesting bacterium isolated from seaweed, and emended description of the genus Gilvimarinus. International Journal of Systematic and Evolutionary Microbiology. 2015;65:562. doi: 10.1099/ijs.0.065078-0. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Hatada Y., Miyazaki M., Nogi Y., Ito S., Horikoshi K. Purification and characterization of a novel α-agarase from a Thalassomonas sp. Current Microbiology. 2005;50:212–216. doi: 10.1007/s00284-004-4435-z. [DOI] [PubMed] [Google Scholar]
- Xu J., Cui Z., Zhang W., Lu J., Lu X., Yu W. Characterizing of a new α-agarase AgaE from Thalassomonas sp. LD5 and probing its catalytically essential residues. International Journal of Biological Macromolecules. 2022;194:50–57. doi: 10.1016/j.ijbiomac.2021.11.194. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Lee C.R., Hong S.K. Biochemical characterization of a novel cold-adapted agarotetraose-producing α-agarase, AgaWS5, from Catenovulum sediminis WS1-A. Applied Microbiology and Biotechnology. 2019;103:8403–8411. doi: 10.1007/s00253-019-10056-1. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu Z., Jiang C., Mao X. Biochemical characterization and substrate degradation mode of a novel α-agarase from Catenovulum agarivorans. Journal of Agricultural and Food Chemistry. 2019;67:10373–10379. doi: 10.1021/acs.jafc.9b03073. [DOI] [PubMed] [Google Scholar]
- Sidar A., Albuquerque E.D., Voshol G.P., Ram A.F.J., Vijgenboom E., Punt P.J. Carbohydrate binding modules: diversity of domain architecture in amylases and cellulases from filamentous microorganisms. Frontiers in Bioengineering and Biotechnology. 2020;8 doi: 10.3389/fbioe.2020.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkotaini B., Han N.S., Kim B.S. Enhanced catalytic efficiency of endo-β-agarase I by fusion of carbohydrate-binding modules for agar prehydrolysis. Enzyme and Microbial Technology. 2016;93:142–149. doi: 10.1016/j.enzmictec.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Research. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z.…Yin Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Cao Y., Xu H., Zhang W., Fei B., Qiao D., Cao Y. Purification and characterization of a novel β-agarase of Paenibacillus sp. SSG-1 isolated from soil. Journal of Bioscience and Bioengineering. 2014;118:125–129. doi: 10.1016/j.jbiosc.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Song T., Wang X., Wu M., Zhao K., Wang X., Chu Y., Lin J. Agarase cocktail from agar polysaccharide utilization loci converts homogenized Gelidium amansii into neoagarooligosaccharides. Food Chemistry. 2021;352 doi: 10.1016/j.foodchem.2020.128685. [DOI] [PubMed] [Google Scholar]
- Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: Making protein folding accessible to all. Nature Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O.…Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research. 2016;44(2016):W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J., Vina A. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2009:NA-NA. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubec D., Skoda P., Krivak R., Novotny M., Hoksza D. PrankWeb 3: Accelerated ligand-binding site predictions for experimental and modelled protein structures. Nucleic Acids Research. 2022;50:W593–W597. doi: 10.1093/nar/gkac389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Xu J., Liu D., Liu H., Lu X., Yu W. Characterization of an α-agarase from Thalassomonas sp. LD5 and its hydrolysate. Applied Microbiology and Biotechnology. 2018;102:2203–2212. doi: 10.1007/s00253-018-8762-6. [DOI] [PubMed] [Google Scholar]
- Rochas C., Potin P., Kloareg B. NMR spectroscopic investigation of agarose oligomers produced by an α-agarase. Carbohydrate Research. 1994 doi: 10.1016/0008-6215(94)80056-1. [DOI] [PubMed] [Google Scholar]
- Li J., Han F., Lu X., Fu X., Ma C., Chu Y., Yu W. A simple method of preparing diverse neoagaro-oligosaccharides with beta-agarase. Carbohydrate Research. 2007;342:1030–1033. doi: 10.1016/j.carres.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Alkotaini B., Han N.S., Kim B.S. Enhanced catalytic efficiency of endo-β-agarase I by fusion of carbohydrate-binding modules for agar prehydrolysis. Enzyme and Microbial Technology. 2016;93–94:142–149. doi: 10.1016/j.enzmictec.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Hanazono Y., Takeda K., Niwa S., Hibi M., Takahashi N., Kanai T.…Miki K. Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1. FEBS Letters. 2016;590:298–304. doi: 10.1002/1873-3468.12055. [DOI] [PubMed] [Google Scholar]
- Courtade G., Forsberg Z., Heggset E.B., Eijsink V.G.H., Aachmann F.L. The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. The Journal of Biological Chemistry. 2018;293:13006–13015. doi: 10.1074/jbc.RA118.004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P.J., Xie H., Bolam D.N., Gilbert H.J., Williamson M.P. The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. The Journal of Biological Chemistry. 2000;275:41137–41142. doi: 10.1074/jbc.M006948200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.