Abstract
Tet(M) protein interacts with the protein biosynthesis machinery to render this process resistant to tetracycline by a mechanism which involves release of the antibiotic from the ribosome in a reaction dependent on GTP hydrolysis. To clarify this resistance mechanism further, the interaction of Tet(M) with the ribosome has been examined by using a gel filtration assay with radioactively labelled Tet(M) protein. The presence of GTP and 5′-guanylyl imido diphosphate, but not GDP, promoted Tet(M)-ribosome complex formation. Furthermore, thiostrepton, which inhibits the activities of elongation factor G (EF-G) and EF-Tu by binding to the ribosome, blocks stable Tet(M)-ribosome complex formation. Direct competition experiments show that Tet(M) and EF-G bind to overlapping sites on the ribosome.
Tetracycline inhibits protein synthesis by interfering with the binding of aminoacyl tRNA to ribosomes (20). It has been shown that the ribosomal 30S subunit binds tetracycline (9, 10, 22), and experiments in which single ribosomal proteins have been omitted during reconstitution have established that proteins S3, S7, S8, and S14 (3) are involved in this binding.
Tet(M)-mediated tetracycline resistance reverses the inhibitory effects of the antibiotic at the level of protein synthesis (4, 5) both in the original streptococcal host (4) and in Escherichia coli (5). Previous studies in our laboratory have shown that Tet(M) catalyzes release of tetracycline from the ribosome in a reaction dependent on GTP (6); however, release of the antibiotic does not take place in the presence of a nonhydrolyzable GTP analog, 5′-guanylyl imido diphosphate (GMPPNP) (6). This result could be explained either by the failure of Tet(M) to bind to the ribosome or by the necessity for hydrolysis of GTP for Tet(M)-promoted release of tetracycline from ribosomes. We show here that Tet(M) binds to the ribosome in the presence of GTP or GMPPNP and that Tet(M) and elongation factor G (EF-G) compete for binding.
MATERIALS AND METHODS
Construction of pET16b-Tet(M).
A version of the tet(M) gene containing a 5′ extension encoding 10 contiguous histidine codons was constructed with the vector pET16b (Novagen) by using standard procedures (1). A 2.8-kbp fragment obtained by digestion of plasmid pSH52 (5) with BamHI and partial digestion with NdeI was inserted into pET16b (Novagen) similarly digested with NdeI and BamHI to yield pET16b-Tet(M). The resulting fusion protein expressed from this construct, His10-Tet(M), includes the amino-terminal extension MGHHHHHHHHHHSSHIEGRH on the full-length Tet(M) polypeptide. The junction between tet(M) and the vector was sequenced to ensure that the construction was as expected.
Preparation of protein.
E. coli BL21(DE3) (19) transformed with pET16b-Tet(M) was grown at 37°C to an optical density at 590 nm of 1.0 in Luria-Bertani (LB) medium. Expression of His10-Tet(M) was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by an additional 3 h of incubation. Cells from 250 ml of the culture were harvested by centrifugation and resuspended in buffer A (25 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) containing 0.2-mg ml−1 lysozyme. Following incubation for 20 min at 20°C, Triton X-100 was added to 0.1% and the cells were disrupted by three cycles of freezing at −80°C and thawing at room temperature. The crude extract (10 ml) recovered following centrifugation at 30,000 × g for 30 min was mixed with 1 ml of packed Talon resin (Clontech). After 1 h of mixing at 4°C, the resin was recovered by centrifugation at 3,000 × g for 2 min, transferred to a column, and washed to remove unbound protein. The column was eluted by using four sequential steps (3 ml each) of 10, 25, 50, and 100 mM imidazole in buffer A. Near-homogeneous His10-Tet(M) protein elutes from this resin at 50 mM imidazole. Recovered His10-Tet(M) protein was dialyzed and stored at −20°C in 20 mM potassium phosphate (pH 7.2)–0.1 mM EDTA–0.15 M NaCl–0.5 mM dithiothreitol–50% glycerol (5).
Preparation of radioactive proteins.
Cultures [BL21(DE3)/pET16b-Tet(M) or BL21(DE3)pLysS/pRSET-EF-G (15)] were grown in M9 glucose minimal medium (12) containing all amino acids (40 μg ml−1) except leucine to an optical density at 590 nm of 1.0. IPTG was added to a 0.1 mM final concentration. Incubation was continued for a further 15 min before [3H]leucine (10 μCi ml−1; NEN, Boston, Mass.) was added to the culture. After incubation for a further 3 h, cells were harvested by centrifugation. [3H]His10-Tet(M) was purified as outlined above. [3H]His6–EF-G was purified under native conditions to >95% purity by affinity chromatography on Talon resin (Clontech) as described above; EF-G was stored at −20°C in 10 mM Tris-HCl (pH 7.5)–75 mM KCl–1 mM dithiothreitol–50% glycerol.
GTP hydrolysis.
Ribosome-dependent GTP hydrolytic activity was monitored as previously described (6). Briefly, reaction mixtures contained 50 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM magnesium acetate, 0.3 mM [γ-32P]GTP, and 0.5 μM ribosomes. Tet(M), His10-Tet(M), and inhibitors were added as indicated. Hydrolysis was initiated by the addition of GTP, and samples were withdrawn at timed intervals and pipetted into a slurry of activated charcoal in 1 M HCl–0.1 M sodium pyrophosphate to terminate the reaction. The charcoal was pelleted by centrifugation, and the radioactivity present in the supernatant was quantitated by liquid scintillation counting. Hydrolysis in the presence of factor (<1%) or ribosomes (<5%) alone was subtracted from identical samples so that only the ribosome-dependent activity is reported. Ribosomes were prepared from E. coli MRE600 (18).
Polymerization assays.
The ability of Tet(M) or His10-Tet(M) to relieve tetracycline inhibition of polyphenylalanine synthesis was tested as described previously (5). One unit of activity was that amount of Tet(M) permitting incorporation of 1 pmol of phenylalanine into polyphenylalanine in the presence of 100 μM tetracycline.
Analysis of [3H]Tet(M) and [3H]EF-G binding to ribosomes.
The ability of [3H]Tet(M) and [3H]EF-G to associate with ribosomes was monitored in 50-μl reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 75 mM KCl, 75 mM NH4Cl, 10 mM magnesium acetate, 5 mM dithiothreitol, and ribosomes, protein, guanine nucleotide, and inhibitors as indicated. Samples were applied to a Sephacryl S300 column (14 by 0.7 cm) equilibrated with the buffer described above that also included nucleotide and inhibitors if they were present in the binding reaction mixture. The column was eluted at a flow rate of 5 ml/h, and fractions were collected and analyzed for radioactivity to localize Tet(M), while ribosomes were localized by measuring A260 as appropriate.
Antibiotic resistance levels.
Cultures were grown in 100 μl of LB broth in a sterile microtiter plate for 6 to 7 h prior to the transfer of a droplet of the culture with a 48-prong inoculator onto the surfaces of LB agar plates containing twofold serially increasing antibiotic concentrations (23). The resistance level was taken as the highest drug concentration showing growth comparable to that observed in the absence of an antibiotic. This antibiotic level is also referred to as the subinhibitory concentration.
RESULTS
Construction and expression of His10-Tet(M).
Tet(M) protein catalyzes the release of tetracycline from ribosomes in a reaction dependent on GTP (6), but drug release does not occur in the presence of the nonhydrolyzable GTP analog GMPPNP. The failure of this GTP analog to promote drug release could be due either to a requirement for GTP hydrolysis or to the failure of Tet(M) to bind to ribosomes. We have therefore prepared radioactive Tet(M) to directly study the nucleotide dependence of Tet(M)-ribosome interaction.
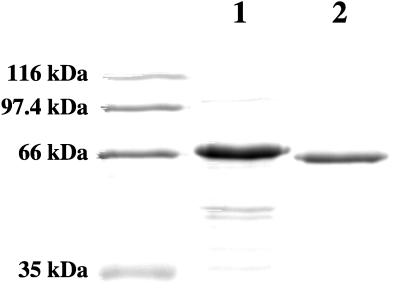
To facilitate the preparation of radioactively labelled Tet(M), a recombinant His10-Tet(M) protein was constructed which contained an amino-terminal polyhistidine tag (see Materials and Methods). Synthesis of the recombinant protein is under transcriptional control of the T7 gene 10 promoter present in the vector. This construct permitted isolation of near-homogeneous His10-Tet(M) in a single step by immobilized metal affinity chromatography (Fig. 1) and has facilitated the purification of radioactively labelled near-homogeneous preparations of the activity.
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of protein samples. Lanes: 1, molecular size standards (Bio-Rad); 2, His10-Tet(M) purified by metal ion affinity chromatography (see Materials and Methods); 3, native Tet(M) purified by conventional chromatography (4).
To determine whether the polyhistidine tag influenced the properties of Tet(M), various activities of the protein were examined in vivo and in vitro. As summarized in Table 1, the polyhistidine-tagged Tet(M) construct is able to confer tetracycline resistance on E. coli in vivo. It is important to point out that the resistance levels observed were not due to highly overproduced amounts of Tet(M). Even though expression of the Tet(M) fusion protein is regulated by the T7 φ10 promoter, tetracycline resistance was observed in cells in which the T7 RNA polymerase gene, in turn controlled by the lacUV5 promoter, is present but not induced, and resistance was significantly lower in a strain which also expressed T7 lysozyme. The latter activity is known to decrease the levels of available T7 polymerase (16). Immunoblot experiments (data not shown) demonstrated that fusion protein is present in extracts of uninduced BL21(DE3) cells at levels comparable to those observed in extracts from strains expressing similar levels of tetracycline resistance from a wild-type tet(M) gene.
TABLE 1.
Tetracycline resistance of Tet(M) fusion constructs in expression hosts
| Host strain | Tetracycline resistancea
|
||
|---|---|---|---|
| No plasmid | pET16b vector | pET16b-Tet(M) | |
| BL21 | 0.4 | 0.4 | 0.4 |
| BL21(DE3) | 0.4 | 0.4 | 6.25 |
| BL21(DE3)pLysS | 0.4 | 0.4 | 0.8 |
Tetracycline resistance was measured as described in Materials and Methods. Resistance levels are given as the drug amounts at which growth was equivalent to that in the absence of the drug following overnight incubation.
Biochemical activities of the purified His-tagged Tet(M) protein were also similar to those of native Tet(M). The ribosome-dependent GTPase activities of the two proteins were nearly identical (specific activities of 950 and 1,050, respectively), as were their abilities to protect protein synthesis from tetracycline inhibition (specific activities of 500 and 700). Taken together, these results indicate that the presence of an amino-terminal His10 tag on Tet(M) does not significantly alter the in vivo or in vitro activity of Tet(M). It is worth noting that amino- and carboxy-terminal His-tagged EF-G and EF-Tu are fully functional and have been used in a number of recent biochemical studies (2, 15, 17, 25, 27, 28).
Ribosome binding.
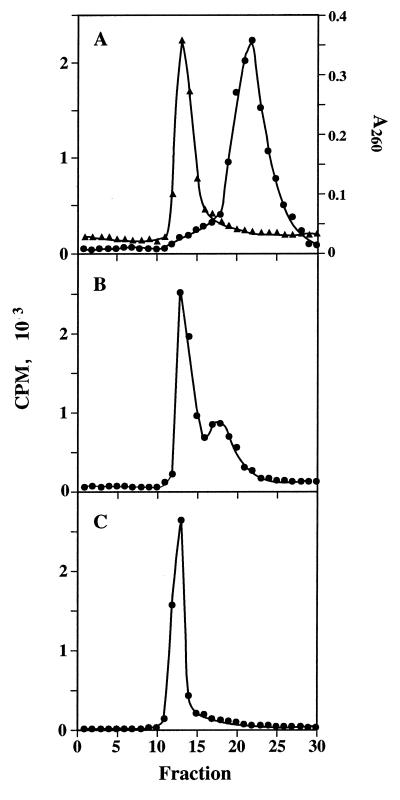
We have previously demonstrated that Tet(M) catalyzes the release of tetracycline from ribosomes in a reaction dependent on GTP. To clarify the interaction of the protein with the ribosome, radioactively labelled His10-Tet(M) protein was used to monitor its interaction with ribosomes by a rapid gel filtration assay. When run separately in the presence or absence of nucleotide or together in the absence of nucleotide, ribosomes eluted in the column void volume while Tet(M) eluted within the included volume of the column (Fig. 2A). In some samples, a small amount (<5%) of Tet(M) protein chromatographed in the same position as ribosomes even in the absence of ribosomes or nucleotide. When chromatographed together in the presence of GDP, elution profiles of Tet(M) and ribosomes are identical to those in the absence of nucleotide. Thus, Tet(M) does not associate with ribosomes in the presence of GDP.
FIG. 2.
Gel filtration chromatography of His10-Tet(M) and ribosomes. (A) [3H]Tet(M) (•, 5 pmol) and ribosomes (▴, 26 pmol) were chromatographed together in the absence of nucleotide as described in Materials and Methods. (B) [3H]Tet(M) (•, 5 pmol) and ribosomes (26 pmol) were chromatographed together in the presence of 1 mM GTP. (C) Ribosomes (26 pmol) and [3H]Tet(M) (•, 5 pmol) were chromatographed in the presence of 250 μM GMPPNP. Ribosomes and ribosome-bound Tet(M) chromatograph about fraction 13; free Tet(M) peaks in fraction 23.
In the presence of GTP, >80% of the [3H]Tet(M) eluted with ribosomes (Fig. 2B). Some protein not associated with ribosomes chromatographed between the positions of the ribosome-bound protein and the free protein. This broad peak of material may represent Tet(M) which has dissociated from the ribosome following GTP hydrolysis. Since we know from other experiments that Tet(M) is a ribosome-dependent GTPase (5), it is expected that some hydrolysis of GTP takes place during sample preparation and chromatography. Tet(M) also associates with the ribosome in the presence of GMPPNP, a nonhydrolyzable GTP analog (Fig. 2C). This association is stoichiometric; all of the Tet(M) protein binds to ribosomes under these conditions [Tet(M)/ribosome ratio of 1:5].
Several antibiotics have been tested for their effects on the stability of the ribosome-Tet(M) complexes formed in the presence of GTP or GMPPNP. Tetracycline, which is released from ribosomes by Tet(M) in the presence of GTP but not GMPPNP, was tested for its effect on Tet(M)-ribosome interaction. As expected, tetracycline is without effect on the formation of Tet(M)-ribosome-GMPPNP complexes (data not shown). Similarly, fusidic acid and kirromycin have no effect on the ability of Tet(M) to associate with ribosomes in the presence of GTP or its nonhydrolyzable analog. Fusidic acid is known to stabilize EF-G on the ribosome as a complex with GDP (24); in like manner, the binding of EF-Tu–aminoacyl-tRNA–GTP complexes is stabilized by kirromycin (26). We know from previous experiments that the ribosome-dependent GTPase activity associated with Tet(M) is fusidic acid resistant while that of EF-G is fusidic acid sensitive (6).
Binding of elongation factors to the ribosome is inhibited by thiostrepton, which binds to the 1060 region of the 23S rRNA (8) which is part of the ribosomal GTPase center. The effect of thiostrepton on Tet(M) binding to ribosomes has been tested and has been found to completely block the formation of stable Tet(M)-ribosome complexes in the presence of GMPPNP and GTP (data not shown). This result may be expected since the activities of Tet(M) are dependent on GTP hydrolysis following binding to the ribosome and suggests that stable complexes cannot be formed under these conditions.
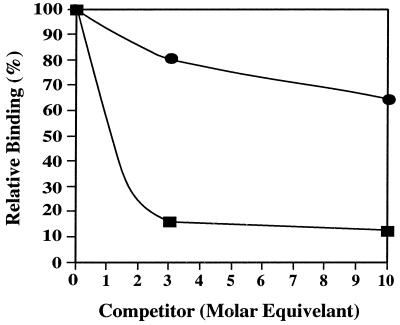
Several lines of evidence suggest that Tet(M) and EF-G occupy overlapping sites on the ribosome. There is considerable homology throughout the length of the two proteins (5), both are ribosome-dependent GTPases (5), and high levels of Tet(M) inhibit protein synthesis, even in the absence of tetracycline, although no inhibition of EF-Tu-dependent aminoacyl-tRNA binding to ribosomes (6) is seen. To directly assess whether Tet(M) and EF-G share a binding site on the ribosome, mixtures containing [3H]Tet(M) and unlabelled EF-G or [3H]EF-G and unlabelled Tet(M) were added to ribosomes in the presence of GTP and fusidic acid, and the extent of ribosome binding by the labelled protein was monitored by gel filtration chromatography (Fig. 3). The results clearly demonstrate that Tet(M) and EF-G compete for binding to the ribosome and, furthermore, that Tet(M) appears to form more stable complexes.
FIG. 3.
Competition between Tet(M) and EF-G. [3H]Tet(M) (•, 20 pmol) was mixed with unlabelled EF-G (0, 60, or 200 pmol) in the presence of 1 mM GTP and 2 mM fusidic acid. In the alternate competition, [3H]EF-G (■, 20 pmol) was mixed with unlabelled Tet(M) (0, 60, or 200 pmol) in the same manner. These solutions were supplemented with ribosomes (20 pmol) in 1 mM GTP and 2 mM fusidic acid for 5 min at 37°C and chromatographed as described in Materials and Methods. In the absence of a competitor, 70% of the Tet(M) was ribosome bound while 57% of the EF-G was bound.
DISCUSSION
Tet(M)-mediated tetracycline resistance occurs at the level of the ribosome, where Tet(M) catalyzes the release of bound tetracycline in a reaction dependent on GTP (6). Since the release of tetracycline by Tet(M) does not occur in the presence of nonhydrolyzable GTP analogs, we wished to determine whether Tet(M) fails to bind to ribosomes under these conditions or whether the protein is able to bind to the ribosome but GTP hydrolysis is necessary for antibiotic release. A second motivation for these studies has been to determine conditions which promote stable Tet(M) binding to ribosomes in order to characterize the ribosome binding site used by Tet(M).
The experiments presented here demonstrate that Tet(M) binds ribosomes in the presence of either GTP or GMPPNP but that GDP cannot promote this interaction. Under the conditions used for binding, association of Tet(M) with ribosomes is stoichiometric in the presence of GMPPNP. Although the affinity of free Tet(M) for GTP and GDP has not been determined, it is clear from this study that GTP and not GDP is necessary for Tet(M) binding to ribosomes; thus, GTP hydrolysis is required for tetracycline release from the ribosome. This type of binding and catalysis is similar to that observed for EF-G and EF-Tu, which bind to the ribosome in association with GTP while GTP catalysis accompanies activity. During the elongation phase of translation, it is the GTP-bound form of EF-G that binds to ribosomes; GTP hydrolysis probably provides the energy for translocation after which EF-G–GDP dissociates from the ribosome (11, 14, 15). Similarly, when complexed with GTP, EF-Tu binds an aminoacyl-tRNA which is then delivered to the ribosome. Shortly after binding, GTP is cleaved and EF-Tu–GDP is released from the ribosome. It is well established that the overlapping binding site for EF-G and EF-Tu is at the base of the L7/L12 stalk (see reference 13).
Insight into factor function has also been obtained from studies of antibiotics which inhibit EF-G and EF-Tu function. Thiostrepton binds to the 1060 loop in the 50S ribosomal subunit (21). In the presence of thiostrepton, EF-G–GTP forms only a transient complex with the ribosome; thus, GTP hydrolysis and translocation are inhibited (15). EF-Tu–tRNA–GTP ternary complex binding is similarly blocked (see reference 7). Thus, the thiostrepton binding site on the ribosome is part of the machinery which enables ribosomes to translocate and is important for factor binding. We have shown here that the ability of Tet(M) to form a stable complex with ribosomes in the presence of GMPPNP or GTP is similarly blocked by thiostrepton. We have also found (6a) that GTP-dependent tetracycline release from ribosomes is inhibited by thiostrepton. Thus, Tet(M) must be able to form a stable complex with the ribosome so that GTP hydrolysis can occur, catalyzing tetracycline release.
We have shown previously that Tet(M) has a number of similarities to elongation factors, especially EF-G. For example, Tet(M) and EFs require GTP for their respective activities, and these activities are inhibited by thiostrepton, which binds to the 1060 loop of 23S rRNA. In fact, we have observed that Tet(M) inhibits in vitro protein synthesis in the absence of tetracycline (6). Tet(M) does not inhibit EF-Tu-dependent tRNA binding in the absence of tetracycline, while Tet(M) does protect this reaction from antibiotic inhibition (6). Following ternary complex binding to ribosomes, codon recognition by cognate aminoacyl-tRNA triggers hydrolysis of GTP with subsequent release of EF-Tu–GDP from the ribosome. These observations, taken together, strongly suggest that Tet(M) and elongation factors bind to the same site (or overlapping sites) on the ribosome. We have directly tested this possibility and found that the binding site on the ribosome can be occupied by either Tet(M) or EF-G but not by both proteins simultaneously. Our ability to prepare stable Tet(M)-ribosome complexes will enable us to further characterize this ribosomal site by using physical methods, as well as to examine the interaction of Tet(M) with elongation factors on the ribosome.
ACKNOWLEDGMENTS
We thank Paul Modrich for useful discussions and Kristin Garrett for technical assistance.
This work was supported by Public Health Service grant AI 15619 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Boon K, Vijgenboom E, Madsen L V, Talens A, Kraal B, Bosch L. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur J Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- 3.Buck M A, Cooperman B S. Single protein omission reconstitution studies of tetracycline binding to the 30S subunit of Escherichia coli ribosomes. Biochemistry. 1990;29:5374–5379. doi: 10.1021/bi00474a024. [DOI] [PubMed] [Google Scholar]
- 4.Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986;165:564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991;266:2872–2877. [PubMed] [Google Scholar]
- 6.Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol. 1996;178:3246–3251. doi: 10.1128/jb.178.11.3246-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Burdett, V. Unpublished data.
- 7.Cundliffe E. Involvement of specific portions of ribosomal RNA in defining ribosomal functions: a study utilizing antibiotics. In: Hardesty B, Kramer G, editors. Structure, function, and genetics of ribosomes. New York, N.Y: Springer Verlag; 1986. pp. 586–604. [Google Scholar]
- 8.Czworkowski J, Moore P B. The elongation phase of protein synthesis. Prog Nucleic Acid Res Mol Biol. 1996;54:293–332. doi: 10.1016/s0079-6603(08)60366-9. [DOI] [PubMed] [Google Scholar]
- 9.Epe B, Woolley P, Hornig H. Competition between tetracycline and tRNA at both P and A sites of the ribosome of Escherichia coli. FEBS Lett. 1987;213:443–447. doi: 10.1016/0014-5793(87)81539-9. [DOI] [PubMed] [Google Scholar]
- 10.Goldman R A, Hasan T, Hall C C, Strycharz W A, Cooperman B. Photoincorporation of tetracycline into Escherichia coli ribosomes: identification of the major proteins photolabeled by native tetracycline and tetracycline photoproducts and implications for the inhibitory action of tetracycline on protein synthesis. Biochemistry. 1983;22:359–368. doi: 10.1021/bi00271a020. [DOI] [PubMed] [Google Scholar]
- 11.Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- 12.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 13.Möller W, Maassen J A. On the structure, function, and dynamics of L7/L12 from Escherichia coli ribosomes. In: Hardesty B, Kramer G, editors. Structure, function, and genetics of ribosomes. New York, N.Y: Springer Verlag; 1986. pp. 309–325. [Google Scholar]
- 14.Oswald G B, Rohrbach M S, Bodley J W. Equilibrium measurements of the interactions of guanine nucleotides with Escherichia coli elongation factor G and the ribosome. Biochemistry. 1976;15:4570–4574. doi: 10.1021/bi00666a004. [DOI] [PubMed] [Google Scholar]
- 15.Rodnina M V, Savelsbergh A, Katunin V I, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg A H, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 17.Semenkov Y P, Rodnina M V, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staehelin T, Maglott D T. The preparation of Escherichia coli ribosomal subunits active in polypeptide synthesis. Methods Enzymol. 1971;20:449–456. [Google Scholar]
- 19.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 20.Suarez G, Nathans D. Inhibition of aminoacyl-sRNA binding to ribosomes by tetracycline. Biochem Biophys Res Commun. 1965;18:743–750. [Google Scholar]
- 21.Thompson J, Cundliffe E, Stark M. Binding of thiostrepton to a complex of 23S rRNA with ribosomal protein L11. Eur J Biochem. 1979;98:261–265. doi: 10.1111/j.1432-1033.1979.tb13184.x. [DOI] [PubMed] [Google Scholar]
- 22.Tritton T R. Ribosome-tetracycline interactions. Biochemistry. 1977;16:4133–4138. doi: 10.1021/bi00637a029. [DOI] [PubMed] [Google Scholar]
- 23.Washington J A, II, Barry L A. Dilution test procedures. In: Lennette E H, Spaulding E H, Traunt J P, editors. Manual of clinical microbiology. Washington, D.C: American Society for Microbiology; 1974. pp. 410–417. [Google Scholar]
- 24.Willie G R, Richman N, Godtfredsen W O, Bodley J W. Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome · elongation factor G complexes. Biochemistry. 1975;14:1713–1718. doi: 10.1021/bi00679a025. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K S, Noller H F. Mapping the position of translation elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 26.Wolf H, Chinali G, Parmeggiani A. Kirromycin, an inhibitor of protein synthesis that acts on elongation factor Tu. Proc Natl Acad Sci USA. 1974;71:4910–4914. doi: 10.1073/pnas.71.12.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Li X, Spremulli L. Role of the conserved aspartate and phenylalanine residues in prokaryotic and mitochondrial elongation factor Ts in guanine nucleotide exchange. FEBS Lett. 1996;391:330–332. doi: 10.1016/0014-5793(96)00789-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Sun V, Spremulli L L. Role of domains in Escherichia coli and mammalian mitochondrial elongation factor Ts in the interaction with elongation factor Tu. J Biol Chem. 1997;272:21956–21963. doi: 10.1074/jbc.272.35.21956. [DOI] [PubMed] [Google Scholar]