Abstract
The hydrophobic C terminus of pore-forming colicins associates with and inserts into the cytoplasmic membrane and is the target of the respective immunity protein. The hydrophobic region of colicin U of Shigella boydii was mutated to identify determinants responsible for recognition of colicin U by the colicin U immunity protein. Deletion of the tip of the hydrophobic hairpin of colicin U resulted in a fully active colicin that was no longer inactivated by the colicin U immunity protein. Replacement of eight amino acids at the tip of the colicin U hairpin by the corresponding amino acids of the related colicin B resulted in colicin U(575–582ColB), which was inactivated by the colicin U immunity protein to 10% of the level of inactivation of the wild-type colicin U. The colicin B immunity protein inactivated colicin U(575–582ColB) to the same degree. These results indicate that the tip of the hydrophobic hairpin of colicin U and of colicin B mainly determines the interaction with the corresponding immunity proteins and is not required for colicin activity. Comparison of these results with published data suggests that interhelical loops and not membrane helices of pore-forming colicins mainly interact with the cognate immunity proteins and that the loops are located in different regions of the A-type and E1-type colicins. The colicin U immunity protein forms four transmembrane segments in the cytoplasmic membrane, and the N and C termini face the cytoplasm.
Pore-forming colicins form voltage-dependent ion channels in the cytoplasmic membrane of sensitive bacteria. Colicin U belongs to the family of channel-forming colicins (22), which consist of three domains responsible for translocation through the outer membrane (N-terminal domain), binding to the receptor (central domain), and channel formation (C-terminal domain). Crystal structures of the pore-forming domains of colicins A, E1, and Ia have been determined at atomic resolution (4, 16, 27). In the water-soluble state, the pore-forming domains are arranged similarly and consist of a central hydrophobic hairpin (helices 8 and 9) surrounded by eight amphipathic helices. The structure of the membrane pore is less clear. Upon contact with the cytoplasmic membrane, the colicins unfold and the hydrophobic hairpin inserts into the lipid bilayer. It is debated whether the hydrophobic hairpin is oriented parallel to the bilayer or whether it assumes a transmembrane arrangement, and how its arrangement and that of the other helices change upon voltage-dependent pore formation (1, 3, 11–13, 15). In colicin Ia, at least helices 5 and 6 are translocated across the membrane in response to a transmembrane voltage (21), whereas helices 8 and 9 are inserted voltage-independent into the membrane (9). For the purpose of this paper, the general agreement that helices 8 and 9 are embedded in the membrane is of relevance.
Sequence similarities separate the pore-forming colicins into the A-type (colicins A, B, N, and U) and the E1-type (colicins E1, 5, K, 10, Ia, and Ib) colicins (Fig. 1). The corresponding immunity proteins have been classified into the same two groups (17, 20, 23). The colicin A immunity protein (Cai) has four transmembrane segments, and its N and C termini are located in the cytoplasm (8), whereas the immunity proteins of colicin E1 (23) and colicin 5 (17) cross the cytoplasmic membrane three times, with the N terminus in the cytoplasm and the C terminus in the periplasm.
FIG. 1.
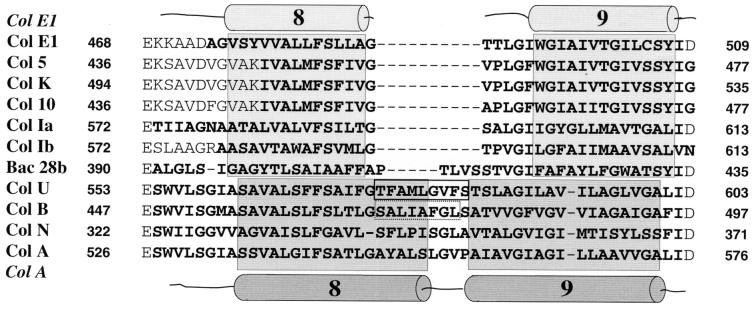
Hydrophobic hairpin sequences (helices 8 and 9) of the channel-forming domains of the E1-type (colicins E1, 5, K, 10, Ia, and Ib) and A-type (colicins B, N, U, and A) colicins and of bacteriocin 28b. The hydrophobic amino acids are indicated in boldface, and the α helices of colicin E1 (4) and colicin A (16) are indicated schematically above and below the protein sequences, respectively. The determined helical segments and the derived helices are boxed and shaded. The excised hairpin tip of colicin U (solid line) and the corresponding sequence of colicin B (interrupted line) are boxed.
In this study, we show that deletion of residues 575 to 583, which we propose to form the tip of the hydrophobic helical hairpin, did not alter the cytotoxic activity of colicin U. We further demonstrate that the tip sequence is a main determinant for the specific recognition of colicin U by the cognate immunity protein. In addition, we determined the transmembrane topology of the colicin U immunity protein and show that it corresponds with the immunity protein of colicin A.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown in a medium composed of 1% Bacto Tryptone–0.5% yeast extract (TY; Difco Laboratories, Detroit, Mich.) plus 0.5% NaCl (pH 7) or on TY agar plates. When required, media were supplemented with kanamycin (50 μg/ml) or chloramphenicol (50 μg/ml). The ampicillin resistance of strains carrying cui-blaM fusion genes was tested on TY agar plates supplemented with increasing concentrations of ampicillin (5, 25, 100, 200, and 400 μg/ml).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| 5K | hsdR lacZ rpsL ser thi thr | This institute |
| BL21 | F−hsdS gal | 24 |
| HP87 | 5K ompA rfa | This work |
| Plasmids | ||
| pDS2 | pBCSK+ carrying cua cui cul | 22 |
| pDS4 | pBCSK+ carrying cui cul | 22 |
| pJBS636 | blaM under phage T7 control | T. Focareta |
| pHP81 | pBCKS+ carrying cbi | This work |
| pHP140 | pDS2 cua (Δ575–583) | This work |
| pHP141 | pDS2 cua (575 SALIAFGL 582) | This work |
| pDS101 | pDS2 cua (T575S) | This work |
| pDS102 | pDS2 cua (F576A) | This work |
| pDS103 | pDS2 cua (A577L) | This work |
| pDS104 | pDS2 cua (M578I) | This work |
| pDS105 | pDS2 cua (L579A) | This work |
| pDS106 | pDS2 cua (G580F) | This work |
| pDS107 | pDS2 cua (V581G) | This work |
| pDS108 | pDS2 cua (F582L) | This work |
| pHP131 | pJBS636 cui | This work |
| pHP132 | pJBS636 cui68-blaM | This work |
| pHP133 | pJBS636 cui90-blaM | This work |
| pHP134 | pJBS636 cui139-blaM | This work |
| pHP135 | pJBS636 cui174-blaM | This work |
Recombinant DNA techniques.
Plasmid DNA was isolated with ion-exchange columns (Qiagen, Hilden, Germany). Standard methods were used for restriction endonuclease analyses, ligation, and transformation with plasmid DNA (18). DNA was sequenced by the dideoxy chain-termination method (19) with an AutoRead sequencing kit (Pharmacia Biotech, Freiburg, Germany) and an A.L.F. Automatic Sequenator (Pharmacia Biotech). Site-specific mutagenesis was performed by PCR (10). The nucleotide sequence of the primer used for the A-to-T replacement at position 1857 (colicin U8 mutant) of the cua gene (22) is given as an example of the mismatch primers used for PCR (replaced nucleotide underlined): 5′-GCTAAACTAGTCGATAAAACACCCAGC-′3. The sequences of the mutagenized fragments were verified by DNA sequencing. For the construction of cui-blaM gene fusions, restriction sites that yield blunt ends were introduced into the cui gene. After amplification by PCR, the cui gene fragments were cloned into the pJBS636 vector, which resulted in cui-blaM gene fusions. pJBS636, a β-lactamase fusion vector, was derived from vector pJBS633 (2); pJBS636 carries the T7 promoter and the multiple cloning site of plasmid pT7-7.
Radiolabeling of proteins.
cui-blaM gene fusions were under the control of the phage T7 gene 10 promoter and were transcribed by the T7 RNA polymerase, which was chromosomally encoded in Escherichia coli BL21 and was under the control of the lacI repressor. Cells (2 ml) in the exponential growth phase (optical density at 578 nm = 0.4) were collected by centrifugation and then suspended in 1 ml of a medium that contained 0.6% Na2HPO4, 0.3% KH2PO4, 0.1% NH4Cl, 0.05% NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 1 mM sodium citrate, 0.4% glucose, 20 mg of thiamine/liter, and 0.01% methionine assay medium (Difco Laboratories). T7 RNA polymerase synthesis was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside. After the culture was shaken for 60 min at 37°C, 20 μl of rifamycin solution (20 mg/ml in methanol) was added to inhibit the E. coli RNA polymerase, and the culture was shaken for an additional 30 min. Cells were then labeled by adding 185 kBq of [35S]methionine and incubating the culture for 15 min at room temperature. Cells were sedimented by centrifugation, suspended in 40 μl of sample buffer, and heated for 5 min at 100°C. Ten microliters was subjected to polyacrylamide gel electrophoresis (PAGE; 3% polyacrylamide stacking gel, 15% polyacrylamide running gel) in the presence of 0.1% sodium dodecyl sulfate (SDS). The dried gel was autoradiographed with Kodak X-Omat S100 film.
Colicin U and the mutated colicin U proteins were labeled in vitro with [35S]methionine in a bacterial cell-free transcription-translation system (Promega, Madison, Wis.) and subjected to SDS-PAGE as described above.
Colicin activity assay.
Colicin activity was tested by spotting 10-fold dilutions of colicin-containing crude cell extracts onto plates prepared with 20 ml of TY agar; the plates were then overlaid with 3 ml of low-melting-point TY agar in which 0.1 ml of an overnight culture of the indicator strain had been suspended (14).
RESULTS
Deletion of the tip of the hydrophobic hairpin of colicin U.
The colicin U determinant consists of the genes cua, which encodes the colicin; cui, which confers immunity to colicin U; and cul, which causes lysis of the colicin U-producing cells (22). To investigate the function of the hydrophobic hairpin of colicin U, amino acids 575 to 583 (henceforth designated the tip) of colicin U were deleted. E. coli 5K cells transformed with plasmid pHP140 cua [colicin U(Δ575–583)] cui cul resulted in single colonies. A second attempt to grow the transformed cells on a nutrient agar plate failed due to cell death, which indicates that the immunity protein could not fully inactivate the mutated colicin. Because of the instability of E. coli 5K(pHP140), the colicin U-resistant strain HP87, which does not take up colicin U, was transformed with plasmid pHP140. Nonimmune cells devoid of uptake are resistant to pore-forming colicins which for pore formation have to insert from the periplasmic side into the cytoplasmic membrane. Crude extracts of E. coli HP87(pHP140) killed sensitive E. coli 5K cells to the same extent as cell extracts of E. coli 5K(pDS2 cua cui cul), which synthesized wild-type colicin U. This result demonstrated that the tip of the hydrophobic hairpin is dispensable for colicin U activity. Colicin U(Δ575–583) contained in the cell extract of E. coli HP87 killed E. coli 5K(pDS4 cui) cells despite synthesis of the immunity protein, as demonstrated by immunity to wild-type colicin U (Table 2). This shows that colicin U(Δ575–583) was not recognized by the colicin U immunity protein.
TABLE 2.
Sensitivity of E. coli strains to colicin U and colicin U mutants
| Colicin | Amino acid sequencea | Colicin activityb
|
||
|---|---|---|---|---|
| 5K | 5K(pDS4 cui) | 5K(pHP81 cbi) | ||
| Colicin U-WT | 575 T F A M L G V F S 583 | 2 | i | 2 |
| Colicin U(Δ575–583) | 575 - - - - - - - - - 583 | 2 | 2 | 2 |
| Colicin U(575–582ColB) | 575 S A L I A F G L S 583 | 2 | 1 | 1 |
| Colicin U1 | 575 S F A M L G V F S 583 | 2 | i | 2 |
| Colicin U2 | 575 T A A M L G V F S 583 | 2 | (1) | 1 |
| Colicin U3 | 575 T F L M L G V F S 583 | 2 | i | 2 |
| Colicin U4 | 575 T F A I L G V F S 583 | 2 | i | 2 |
| Colicin U5 | 575 T F A M A G V F S 583 | 2 | i | 2 |
| Colicin U6 | 575 T F A M L F V F S 583 | 2 | 0 | 2 |
| Colicin U7 | 575 T F A M L G G F S 583 | 2 | i | 2 |
| Colicin U8 | 575 T F A M L G V L S 583 | 2 | i | 2 |
Identities are indicated in boldface.
A 102-fold-diluted colicin sample yielded a clear zone of growth inhibition. 0, undiluted colicin solution; i, immune. (1), both an undiluted and a 10-fold-diluted colicin sample yielded a turbid zone of growth inhibition. The experiments were repeated at least three times with the same results.
Mutational analysis of the hydrophobic colicin U hairpin.
Although colicins U and B display 73% sequence identity in the pore-forming domains, they show no cross-immunity (22). Since the hydrophobic hairpins of the two colicins are only 35% identical, they may determine the specificity for the immunity proteins (Fig. 1). To investigate whether the tip of the colicin B hydrophobic hairpin specifies interaction with the colicin B immunity protein, the tip of the colicin U hairpin was replaced by the tip of the colicin B hairpin (SALIAFGL), which resulted in colicin U(575–582ColB). Transformation of E. coli 5K with plasmid pHP141 Col U(575–582ColB) yielded unstable cells, similar to transformants carrying pHP140. Therefore, strain HP87 was transformed with pHP141, and the crude colicin extract was tested on sensitive E. coli 5K cells; colicin U(575–582ColB) was as active as wild-type colicin U (Table 2). The colicin B immunity protein reduced colicin U(575–582ColB) activity 10-fold (Table 2). The SALIAFGL sequence was also recognized by the colicin U immunity protein, which reduced the activity of colicin U(575–582ColB) 10-fold (Table 2). Since the colicin B immunity protein did not fully inactivate colicin U(575–582ColB) and the colicin U immunity protein did not inactivate colicin B, either the SALIAFGL sequence is not the only recognition site for the colicin B immunity protein or SALIAFGL assumes somewhat different conformations in the two colicins.
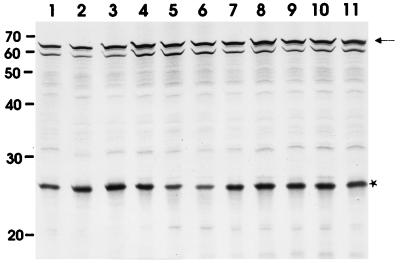
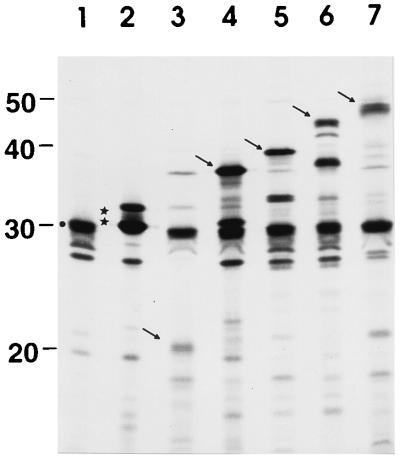
To investigate which of the eight inserted amino acids of colicin U(575–582ColB) are responsible for the interaction with the immunity proteins, single point mutations were introduced in the tip of the hydrophobic hairpin of colicin U (Table 1). The eight mutants isolated were fully active on E. coli 5K cells (Table 2). Colicins U1, U3, U4, U5, and U7 were inactivated by the colicin U immunity protein to the same extent as wild-type colicin U (Table 2). Colicin U6 was less inactivated by the colicin U immunity protein (Table 2) and rendered E. coli 5K[pDS106 cua (G580F) cui cul] unstable. Colicin U2 cross-reacted with the colicin B immunity protein, which indicates that the F576A replacement altered the immunity specificity. Although E. coli 5K(pDS4 cui cul) was also not fully immune to colicin U2 (Table 2), no instability was observed after transformation with DS102 cua (F576A). All mutant colicin U proteins were synthesized in similar amounts and showed the expected size, as determined by SDS-PAGE (Fig. 2). The unknown band below the colicin U proteins was formed in the in vitro protein synthesis system used and was also observed previously (22).
FIG. 2.
SDS-PAGE of radiolabeled colicin U proteins. The genes of colicin U (lane 1), colicin U(Δ575–583) (lane 2), colicin U(575–582 ColB) (lane 3), and the colicin U mutant proteins U1 to U8 (lanes 4 to 11) were synthesized in vitro. The 25-kDa bands (marked by a star) represent the chloramphenicol transacetylase encoded on pBCSK+. The arrow indicates the position of the colicin proteins. Numbers on the right indicate positions of molecular mass standards in kilodaltons.
Topology of the colicin U immunity protein (Cui) in the cytoplasmic membrane.
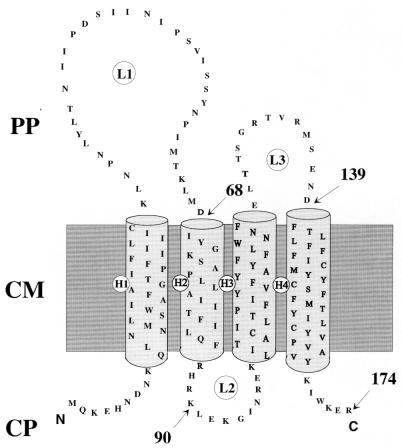
The hydropathy profile of the Cui immunity protein shows four hydrophobic segments that are predicted to form four α helices across the cytoplasmic membrane (Fig. 3). To support this model, hybrid proteins between the colicin U immunity protein and β-lactamase (BlaM) were constructed. Only hybrid proteins with fusion sites located in the periplasm should allow cells to grow as single colonies on nutrient agar plates supplemented with at least 5 μg of ampicillin/ml (2). The Cui68-BlaM hybrid in loop 1 conferred resistance to 200 μg of ampicillin/ml, whereas cells expressing the Cui-BlaM hybrids with fusion sites at positions 90 and 174 formed no colonies on plates containing 5 μg of ampicillin/ml. Cells that synthesized the Cui139-BlaM hybrid were resistant to 100 μg of ampicillin/ml, indicating a localization of residue 139 of Cui in the periplasm. The transmembrane topology of Cui corresponds to that of the colicin A immunity protein (Cai) (8), with which it shares 45% sequence identity (22). When examined by SDS-PAGE (Fig. 4), the hybrid proteins each displayed the molecular mass as calculated considering the fusion sites in the Cui immunity protein and the mature form of the BlaM β-lactamase. There were also proteolytic degradation products of the hybrid proteins (bands below the hybrid proteins) and bands corresponding to the resistance proteins.
FIG. 3.
Predicted arrangement of the colicin U immunity protein in the cytoplasmic membrane of E. coli. H1, H2, H3, and H4 denote the transmembrane α helices; L1, L2, and L3 denote loops. PP, periplasmic space; CM, cytoplasmic membrane; CP, cytoplasm. Arrows and numbers indicate the locations of the fusion sites of the constructed Cui-BlaM hybrid proteins.
FIG. 4.
SDS-PAGE of [35S]methionine-labeled wild-type Cui (lane 3), Cui68-BlaM (lane 4), Cui90-BlaM (lane 5), Cui139-BlaM (lane 6), and Cui174-BlaM (lane 7) (indicated by arrows) expressed in E. coli BL21 transformed with the corresponding plasmids. Lane 1 shows the neomycin phosphotransferase (indicated by a dot) expressed by the vector plasmid pJBS636; lane 2 shows the precursor and the processed form of β-lactamase (BlaM) (indicated by stars). Numbers on the right indicate positions of molecular mass standards in kilodaltons.
The colicin U immunity protein confers cross-immunity to colicin A but not to colicin B (22). The immunity of Cui should therefore be specified by amino acids that are common to Cai but not to Cbi. There are only 30 such residues, and they are distributed along the entire Cui sequence (data not shown), indicating that different parts of the Cui protein may participate in the specific recognition of the colicins.
DISCUSSION
In this study, we showed that residues 575 to 583 (designated the tip) of the hydrophobic hairpin are not required for colicin U activity. Deletion of nine residues which included five residues of the predicted helix 8 and the loop between helix 8 and helix 9 (Fig. 1) resulted in a fully active colicin. Full activity of colicin U(Δ575–583) demonstrates that deletion of the tip does not alter the relative arrangement of helices 8 and 9 and of the other membrane helices in a way that prevents pore formation, which includes binding to the cytoplasmic membrane, unfolding, insertion into the membrane, and assembly of the helices to a pore. The length of the truncated helix 8 is similar to the length of helix 8 of the E1-type colicins (Fig. 1). Helix 9 could also be shortened when helix 9 residues are included in the connecting loop of the deletion derivative. Either full-length helices of 17 and 18 residues required to span a lipid bilayer are not necessary for pore formation of the A-type colicins and of the E1-type colicins or the helices lengthen upon binding to the membrane (3, 4). Replacement of the eight colicin U residues by eight colicin B residues did not alter colicin U activity, which supports the conclusion that there is no strict requirement regarding the length and the sequence of the tip.
Deletion of residues 575 to 583 abolished inactivation of colicin U by the colicin U immunity protein. Lack of immunity to colicin U(Δ575–583) probably indicates recognition of the tip sequence, or a portion of it, by the immunity protein. Replacement of the 575 to 582 region by the corresponding region of colicin B expanded immunity specificity in that the resulting colicin U(575–582ColB) was inhibited by the colicin B immunity protein and by the colicin U immunity protein. This result indicates that the tip represents an important immunity-conferring region of colicin U, and correspondingly of colicin B, but it is not the only specificity-determining region because of the cross-immunity. Analysis of single point mutations in the tip of the hairpin of colicin U revealed that phenylalanine 576 contributes to immunity specificity since its replacement by alanine in colicin U2 caused cross-immunity with the colicin B immunity protein and a decrease in inhibition by the colicin U immunity protein. The G580F substitution in colicin U6 somewhat lowers immunity conferred by the colicin U immunity protein but does not alter the immunity specificity. Reduction of specificity and reactivity may be caused by a distortion of the colicin B tip conformation in colicin U, and/or colicin regions outside the tip contribute to the interaction with the immunity protein. Furthermore, deletion of the tip may affect the conformation of a binding region of the immunity protein outside the tip which results in a reduced reactivity and specificity with the immunity protein. Apart from the unsolved molecular details, our data lead us to propose that the tip of the hydrophobic hairpin in colicin U and in colicin B represents a major determinant for the interaction with the corresponding immunity proteins. We consider these results as representative for the interaction of the A-type colicins with their immunity proteins.
The killing activity of a series of colicin A-colicin B chimeric proteins on immune indicator strains has revealed the hydrophobic region between amino acids 530 and 577 as the main immunity-specifying determinant of the colicin A sequence (7). Our data confine the region that specifies interaction with the immunity protein to the tip of the hairpin of colicins U and B. A nontoxic derivative of colicin A bound to the cytoplasmic membrane does not form an open channel but reacts with the immunity protein, which shows that the immunity protein can inactivate the colicins without opening the channel (5, 6). In colicin E1, which represents another type of pore-forming colicins, amino acid substitutions of residues 440, 443, 444, 474, and 477, located in the segments between helices 5 and 6 and helices 7 and 8, respectively (4), reduced protection by the immunity protein (28). For colicin 5 (E1-type colicin), we have shown that residues 405 to 424 of helix 6, corresponding to residues 437 to 456 of colicin E1, are important for the inactivation by the immunity protein (17). Since the site of interaction on the immunity protein is located in the cytoplasmic loop and the inner leaflet of the cytoplasmic membrane (17), residues 405 to 424 of colicin 5 have to be deeply inserted in the cytoplasmic membrane. Apparently, E1-type and A-type colicins differ with regard to the membrane locations of the interaction sites with the cognate immunity proteins, and the reacting interhelical loops are located in different regions of the E1-type colicins and the A-type colicins.
The amino acid sequence of bacteriocin 28b (probably identical to colicin L) of Serratia marcescens displays the highest sequence similarity to colicin A (26). However, it lacks five residues in the tip of the hairpin (Fig. 1) which, according to the results presented in this paper, it would not need because producer cells are not protected by an immunity protein (25).
With the identification of the hydrophobic hairpin tip as a site of interaction with the immunity protein, the location of the tip within the membrane gains interest. During the multistep membrane insertion process, the location of the tip most likely differs after the voltage-independent primary insertion of the hydrophobic hairpin and after the subsequent response to the transmembrane voltage. The known transmembrane arrangement of the immunity protein and the identification of the site of interaction on the immunity protein should allow determination of whether A-type colicins are inactivated prior to or after pore formation. β-Lactamase hybrid proteins revealed four transmembrane segments of the colicin U immunity protein and that the N and C termini of the immunity protein face the cytoplasm. This arrangement agrees with the transmembrane topology of the colicin A immunity protein (8) and suggests that all immunity proteins of A-type colicins display similar structures.
ACKNOWLEDGMENTS
We thank K. Hantke for fruitful discussions and K. A. Brune for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 323, project B1, foreign guest grant to D.S.; Graduiertenkolleg “Mikrobiologie,” fellowship to H.P.).
REFERENCES
- 1.Benedetti H, Geli V. Colicin transport, channel formation and inhibition. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 666–691. [Google Scholar]
- 2.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 3.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Structure-function of the channel-forming colicins. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 4.Elkins P, Bunker A, Cramer W A, Stauffacher C V. A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domain of colicin E1. Structure. 1997;15:443–458. doi: 10.1016/s0969-2126(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 5.Espesset D, Piet P, Lazdunski C, Geli V. Immunity proteins to pore-forming colicins: structure-function relationships. Mol Microbiol. 1994;13:1111–1120. doi: 10.1111/j.1365-2958.1994.tb00502.x. [DOI] [PubMed] [Google Scholar]
- 6.Espesset D, Duche D, Baty D, Geli V. The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane. EMBO J. 1996;15:2356–2364. [PMC free article] [PubMed] [Google Scholar]
- 7.Geli V, Lazdunski C. An α-helical hydrophobic hairpin as a specific determinant in protein-protein interaction occurring in Escherichia coli colicin A and B immunity systems. J Bacteriol. 1992;174:6432–6437. doi: 10.1128/jb.174.20.6432-6437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geli V, Baty D, Pattus F, Lazdunski C. Topology and function of the integral membrane protein conferring immunity to colicin A. Mol Microbiol. 1989;3:679–687. doi: 10.1111/j.1365-2958.1989.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 9.Kienker P K, Qiu X-Q, Slatin S L, Finkelstein A, Jakes K. Transmembrane insertion of the colicin Ia hydrophobic hairpin. J Membr Biol. 1997;157:27–37. doi: 10.1007/s002329900213. [DOI] [PubMed] [Google Scholar]
- 10.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakey J H, Duche D, Gonzales-Manas J-M, Baty D, Pattus F. Fluorescence energy transfer distance measurements: the hydrophobic helical hairpin of colicin A in the membrane bound state. J Mol Biol. 1993;230:1055–1067. doi: 10.1006/jmbi.1993.1218. [DOI] [PubMed] [Google Scholar]
- 12.Lakey J H, van der Goot F G, Pattus F. All in the family: the toxic activity of pore-forming colicins. Toxicology. 1994;87:85–108. doi: 10.1016/0300-483x(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 13.Lambotte S, Jasperse P, Bechinger B. Orientational distribution of α-helices in the colicin B and E1 channel domains: a one and two dimensional 15N solid-state NMR investigation in uniaxially aligned phospholipid bilayers. Biochemistry. 1998;37:16–22. doi: 10.1021/bi9724671. [DOI] [PubMed] [Google Scholar]
- 14.Mende J, Braun V. Import-defective colicin B derivatives mutated in the TonB box. Mol Microbiol. 1990;4:1523–1533. doi: 10.1111/j.1365-2958.1990.tb02063.x. [DOI] [PubMed] [Google Scholar]
- 15.Palmer L R, Merrill A R. Mapping the membrane topology of the closed state of the colicin E1 channel. J Biol Chem. 1994;269:4187–4193. [PubMed] [Google Scholar]
- 16.Parker M W, Postma J P M, Pattus F, Tucker A D, Tsernoglou D. Refined structure of the pore-forming domain of colicin A at 2.4Å resolution. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 17.Pilsl H, Braun V. Evidence that the immunity protein inactivates colicin 5 immediately prior to the formation of the transmembrane channel. J Bacteriol. 1995;177:6966–6972. doi: 10.1128/jb.177.23.6966-6972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Sanger F S, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramm E, Ölschläger T, Tröger W, Braun V. Sequence, expression and localization of the immunity protein of colicin B. Mol Gen Genet. 1988;211:176–182. doi: 10.1007/BF00338410. [DOI] [PubMed] [Google Scholar]
- 21.Slatin S L, Qiu X Q, Jakes K, Finkelstein A. Identification of a translocated protein segment in a voltage-dependent channel. Nature. 1994;371:158–161. doi: 10.1038/371158a0. [DOI] [PubMed] [Google Scholar]
- 22.Šmajs D, Pilsl H, Braun V. Colicin U, a novel colicin produced by Shigella boydii. J Bacteriol. 1997;179:4919–4928. doi: 10.1128/jb.179.15.4919-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier F W, Moffat B A. Use of bacteriophage T7-RNA-polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 25.Viejo M B, Enfedaque J, Guasch J F, Ferrer S, Regue M. Protection against bacteriocin 28b in Serratia marcescens is apparently not related to the expression of an immunity gene. Can J Microbiol. 1995;41:217–226. doi: 10.1139/m95-030. [DOI] [PubMed] [Google Scholar]
- 26.Viejo M B, Gargallo D, Ferrer S, Enfedaque J, Regue M. Cloning and DNA sequence analysis of a bacteriocin gene of Serratia marcescens. J Gen Microbiol. 1992;138:1737–1743. doi: 10.1099/00221287-138-8-1737. [DOI] [PubMed] [Google Scholar]
- 27.Wiener M, Freymann D, Ghosh P, Stroud R M. Crystal structure of colicin Ia. Nature. 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y L, Cramer W A. Intramembrane helix-helix interactions as the basis of the inhibition of the colicin E1 ion channel by its immunity protein. J Biol Chem. 1993;268:10176–10184. [PubMed] [Google Scholar]