Abstract
It has been previously established that Thiobacillus neapolitanus fixes CO2 by using a form I ribulose bisphosphate carboxylase/oxygenase (RuBisCO), that much of the enzyme is sequestered into carboxysomes, and that the genes for the enzyme, cbbL and cbbS, are part of a putative carboxysome operon. In the present study, cbbL and cbbS were cloned and sequenced. Analysis of RNA showed that cbbL and cbbS are cotranscribed on a message approximately 2,000 nucleotides in size. The insertion of a kanamycin resistance cartridge into cbbL resulted in a premature termination of transcription; a polar mutant was generated. The mutant is able to fix CO2, but requires a CO2 supplement for growth. Separation of cellular proteins from both the wild type and the mutant on sucrose gradients and subsequent analysis of the RuBisCO activity in the collected fractions showed that the mutant assimilates CO2 by using a form II RuBisCO. This was confirmed by immunoblot analysis using antibodies raised against form I and form II RuBisCOs. The mutant does not possess carboxysomes. Smaller, empty inclusions are present, but biochemical analysis indicates that if they are carboxysome related, they are not functional, i.e., do not contain RuBisCO. Northern analysis showed that some of the shell components of the carboxysome are produced, which may explain the presence of these inclusions in the mutant.
Ribulose bisphosphate carboxylase/oxygenase (RuBisCO), the initiating enzyme of the Calvin cycle, occurs in nature in two structural types, form I and form II (18, 33). Form I, the most common type, found in nearly all carbon dioxide-fixing organisms, including higher plants, algae, cyanobacteria, and autotrophic bacteria, is a hexadecamer consisting of eight large, highly conserved catalytic subunits (CbbL) and eight small subunits (CbbS) whose function is still not clearly understood (18, 33). The form II RuBisCO consists solely of large catalytic subunits (CbbM), the number of which varies from two to eight, depending on the organism (33). The catalytic subunits of the two forms, CbbL and CbbM, are biochemically and immunologically distinct and share only about 25% sequence identity (20). Some bacteria possess both a form I and a form II RuBisCO (7, 10, 11).
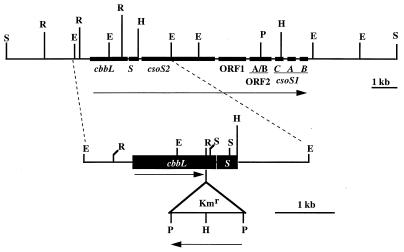
Many autotrophic bacteria and apparently all cyanobacteria sequester much of their form I RuBisCO into primitive organelles, carboxysomes, which somehow enhance carbon dioxide fixation (29). The carboxysomes of Thiobacillus neapolitanus are approximately 120 nm in diameter, are surrounded by a protein shell 3 to 4 nm thick, and consist of nine major polypeptides (3, 30). The genes encoding most of these polypeptides, including those for the large and small subunits of RuBisCO, cbbL and cbbS, appear to constitute a carboxysome operon (30) (Fig. 1).
FIG. 1.
Restriction map of T. neapolitanus putative carboxysome operon (30). The enlargement depicts the inactivation of the cbbL gene. The kanamycin resistance (Kmr) cartridge was inserted into the EcoRV site in the coding region of cbbL. The mutation was introduced into the genome by homologous recombination. E, EcoRI; H, HindIII; P, PstI; R, EcoRV; S, SalI. Arrows indicate direction of transcription.
The form II RuBisCO was first isolated from Rhodospirillum rubrum (34) and later from non-sulfur purple bacteria (10, 11, 27). For many years it was assumed that the presence of the form II enzyme was limited to these organisms. More recently, however, form II enzymes have been demonstrated in a number of other bacteria, including a symbiont of the tubeworm Riftia pachyptila (23), Hydrogenovibrio marinus (4, 38), Thiobacillus intermedius (32), and T. denitrificans (7), and in eukaryotic dinoflagellates (19, 37). Thus, the presence of the form II enzyme seems to be more widespread than originally envisioned.
T. denitrificans expresses both a form I and a form II RuBisCO when grown anaerobically with nitrate as the electron acceptor (7). The genes for both form I and form II (cbbM) RuBisCO have been demonstrated in several other thiobacilli, including T. neapolitanus, T. intermedius, T. ferrooxidans, and T. thiooxidans, via heterologous hybridization using specific DNA probes (28). The cbbM gene from one of these thiobacilli, T. intermedius, has been isolated and expressed in Escherichia coli (32). However, with the exception of the anaerobically grown T. denitrificans, none of the thiobacilli synthesize enough form II RuBisCO to be detected by common laboratory procedures, e.g., by separation of the two forms using sucrose gradients and subsequent assay of the resulting fractions (27). Immunological assays have not been accomplished. To date, we have made no attempt to alter either these thiobacilli or their growth conditions in order to increase the expression of the form II enzyme, nor have we determined if one or both forms are expressed when T. denitrificans is cultured under aerobic conditions.
Herein, we report the creation of a kanamycin insertion mutation of cbbL in T. neapolitanus. The mutant expresses a form II RuBisCO, does not possess functional carboxysomes, and requires elevated levels of CO2 for growth.
MATERIALS AND METHODS
Cultures and plasmids.
Wild-type T. neapolitanus ATCC 23641 was maintained as a chemostat culture in the medium of Vishniac and Santer (36) at 30°C. The mutant, T. neapolitanus cbbL::Km, and the wild type as a control were grown as batch cultures supplied with air supplemented with 5% CO2. Growth of both the wild-type and cbbL::Km mutant strains was monitored at A600. E. coli DH5α was grown in Luria-Bertani medium at 37°C (17). Plasmids and strains used are listed in Table 1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Source or reference | |
|---|---|---|
| Strains | ||
| E. coli DH5α | F recA Δ(lacZYA-argF) | New England Biolabs |
| T. neapolitanus | ||
| ATCC 23641 | Wild type | ATCC |
| T. neapolitanus cbbL::Km | Kmr insert in cbbL | This study |
| Plasmids | ||
| pT7T3α18 | AprlacZ | Boehringer Mannheim |
| pUC4K | Apr Kmr | 15 |
| pTnI-1EE1.5 | 1.5-kb EcoRI fragment in pT7T3α18 | This study |
| pTnI-2ES0.7 | 0.7-kb EcoRI-SalI fragment in pT7T3α18 | This study |
| pTnI-2SS0.3 | 0.3-kb SalI fragment in pT7T3α18 | This study |
| pTnI-2SE1.3 | 1.3-kb SalI-EcoRI fragment in pT7T3α18 | This study |
| pcbbLT7T3 | 2.51-kb BglII fragment in the BamHI site of pT7T3α18 | This study |
| pTnI-2EE2.3 | 2.3-kb EcoRI fragment in pT7T3α18 | This study |
| pTnI-2EE2.3Km | Kmr insert in cbbL | This study |
| pcsoS1 | 300-bp PCR fragment containing csoS1A in pT7T3α18 | This study |
Isolation and sequencing of the cbbL gene.
All DNA manipulations were performed by using standard techniques (17). Screening of a λEMBL3 library of T. neapolitanus genomic DNA with the entire Synechococcus sp. strain PCC 6301 cbbL (rbcL) gene as a probe was performed as previously described (25, 32). Several clones were isolated, and one, λTnI, was selected for further study. Hybridization of the probe to Southern blots of various restriction digests of this clone allowed the construction of a restriction map of cbbL and its flanking region (Fig. 1). Automated sequencing was accomplished with an ABI PRISM Dye Terminator Cycle Sequencing Core kit, a Perkin-Elmer Cetus DNA thermal cycler, and an ABI 373a DNA sequencer. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, Iowa).
Mutant construction.
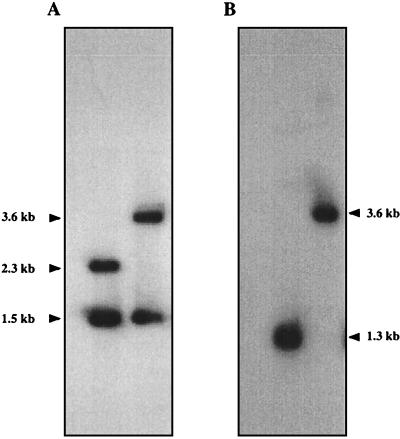
The cbbL gene was interrupted by inserting the kanamycin resistance gene (Kmr) cut from pUC4K with BamHI (35) into the EcoRV site of cbbL via blunt-end ligation (Fig. 1). The 2.3-kbp EcoRI fragment containing the cbbL interruption was subcloned into pT7T3α18 and subsequently transformed into T. neapolitanus (5). After electroporation, the cells were incubated for 24 h in culture medium sparged with air supplemented with 5% CO2. Kanamycin was added to a final concentration of 50 μg/ml, and incubation continued for 24 h. Cells (50 μl) were then plated on selective medium. After 3 days, colonies were transferred to fresh medium. Correct replacement of the wild-type gene with the mutated gene was confirmed by Southern blot analysis (Fig. 2).
FIG. 2.
Confirmation of correct replacement of the wild-type cbbL gene with the insertionally inactivated gene by hybridization with the cbbL gene (A) and the kanamycin resistance gene (B). (A) Lane 1, T. neapolitanus chromosomal DNA digested with EcoRI; lane 2, T. neapolitanus cbbL::Km chromosomal DNA digested with EcoRI. (B) Lane 1, T. neapolitanus cbbL::Km chromosomal DNA digested with PstI; lane 2, T. neapolitanus cbbL::Km chromosomal DNA digested with EcoRI. Sizes of the restriction fragments are indicated on the sides.
Isolation and analysis of RNA transcripts.
The method of Schmitt et al. (24) was used to isolate RNA from 500-ml cultures of the wild type or the mutant grown in batch culture in air supplemented with 5% CO2. Cells were harvested during early exponential phase, 16 and 22 h postinoculation for the wild type and mutant, respectively. RNA (20 μg) was resolved on a 1.2% agarose–6% formaldehyde gel and transferred to Hybond N+ nylon membrane (Amersham, Arlington Heights, Ill.), using a Bio-Rad MiniTransblot apparatus (Bio-Rad Laboratories, Hercules, Calif.) in 0.5× morpholinepropanesulfonic acid buffer (6). The resulting blot was UV cross-linked in a UV Stratalinker (Stratagene, La Jolla, Calif.) and then baked at 80°C in a vacuum oven. Blots were hybridized with either the 1.7-kb SalI (cbbL-specific) fragment from pcbbLT7T3, the 0.295-kb HindIII-NcoI (cbbS-specific) fragment from pTnI-2EE2.3, or the 0.3-kb HindIII-EcoRI (csoS1-specific) fragment from pcsoS1 and washed as described elsewhere (21).
RuBisCO determination.
Cells from both the mutant and the wild type were harvested by centrifugation, washed with TEMB buffer (10 mM Tris-HCl, 1 mM EDTA, 15 mM MgCl2, 20 mM NaHCO3 [pH 8.0]), and broken by sonication. The cell debris was removed by centrifugation, and the resulting supernatant was loaded onto the surface of linear sucrose gradients (10 to 60% [wt/vol] in TEMB) in 40-ml quick seal tubes (Beckman) and centrifuged in a VTi50 rotor as previously described (27). The tubes were fractionated from the bottom in 1-ml aliquots, and the fractions were assayed for RuBisCO activity (26). Cell extracts of Rhodobacter capsulatus and T. denitrificans grown under conditions where both form I and form II RuBisCO are synthesized were used as controls (7, 27).
To analyze the amount of RuBisCO assembled inside carboxysomes relative to the amount of free enzyme, the ratio of soluble to particulate RuBisCO activities (S/P ratio) was determined as previously described (2).
Electron microscopy.
Cells were fixed in 2.5% cacodylate-buffered glutaraldehyde (pH 7.2) and postfixed in 1% OsO4, both for 30 min at room temperature. Following dehydration in an ethanol series, the samples were embedded in Spurr low-viscosity resin. Samples were stained overnight at room temperature with 1% uranyl acetate in 75% ethanol during dehydration.
Blocks were sectioned on a LKB Nova microtome equipped with a diamond knife and poststained for 5 min on a Formvar-coated grid with lead citrate. All preparations were observed and photographed on a Zeiss EM-10CA transmission electron microscope.
Immunoblot analysis.
Cell extracts and sucrose gradient fractions containing either the form I or form II enzyme were used for the immunoblot analysis of RuBisCO. Since we observed no form II peak of activity in the wild type and no form I peak in the mutant, fractions that could potentially contain the form I RuBisCO (fractions 24 to 28; see Fig. 5) or form II RuBisCO (fractions 19 to 23) were analyzed. Samples were resolved on either a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel or a 6% nondenaturing gel. After electrophoresis, the proteins were transferred to nitrocellulose, using a Bio-Rad MiniTransblot apparatus (Bio-Rad). The primary antibody was specific for either Synechococcus sp. strain PCC 6301 form I RuBisCO or Rhodobacter sphaeroides form II RuBisCO. The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma, St. Louis, Mo.). The desired proteins were detected by using an enhanced chemiluminescence detection kit (Amersham).
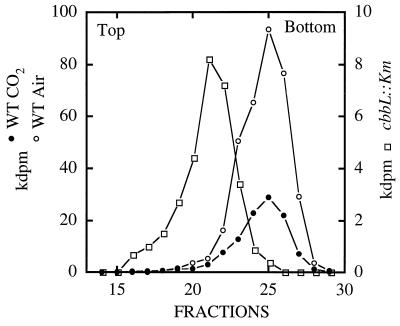
FIG. 5.
RuBisCO activity profiles of sucrose gradients from wild-type (WT) T. neapolitanus grown in air and wild-type and cbbL::Km T. neapolitanus grown in air supplemented with 5% CO2. The left peak represents the form II enzyme; the right peaks represent the form I enzyme.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this article are available from GenBank under accession no. AF038430.
RESULTS
Nucleotide sequences of cbbL and cbbS.
The 1,422-bp cbbL gene of T. neapolitanus encodes a 473-amino-acid peptide with a deduced molecular mass of 52,694 Da. The 333-bp cbbS gene, which encodes a 110-amino-acid peptide with a deduced molecular mass of 12,958 Da, is separated from cbbL by a 36-bp spacer region. The transcriptional start site was determined by primer extension (1) to be located 54 bp upstream of the ATG start codon of cbbL (data not shown).
Transcriptional analysis.
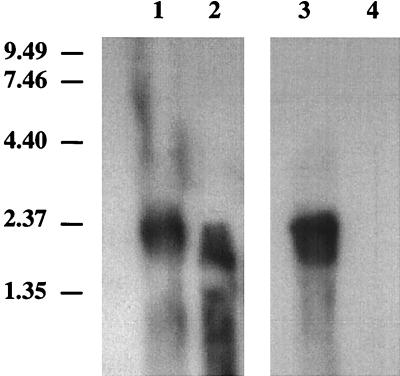
Total RNA isolated from wild-type T. neapolitanus hybridized with either the cbbL gene or cbbS gene revealed a 2.0-kb message (Fig. 3). With the mutant, a smeared signal slightly smaller than that observed for the wild type was present when the blot was probed with cbbL, whereas no signal was observed with cbbS as the probe.
FIG. 3.
Northern blot analysis of cbbL and cbbS. RNA (20 μg) was resolved on a denaturing 1.5% agarose gel and transferred to a nylon membrane. Lane 1, wild-type RNA probed with the cbbL gene; lane 2, T. neapolitanus cbbL::Km RNA probed with the cbbL gene; lane 3, wild-type RNA probed with the cbbS gene; lane 4, T. neapolitanus cbbL::Km RNA probed with the cbbS gene. Sizes (in kilobases) of RNA markers (Life Technologies, Grand Island, N.Y.) are indicated to the left.
Growth of the cbbL mutant.
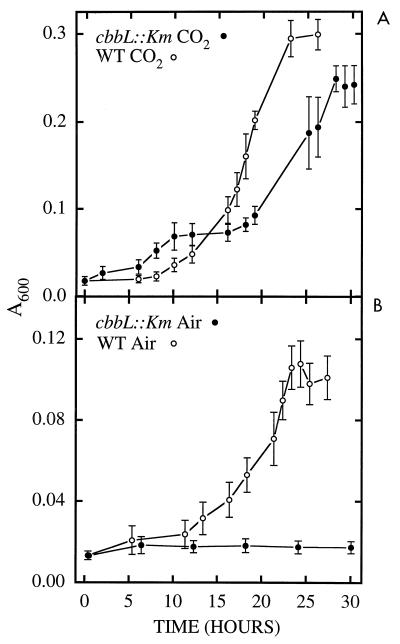
Wild-type cultures reached maximum density at approximately the same time whether they were grown in air or air supplemented with 5% CO2; however, the maximum density of the culture grown in air was about half of that of the culture grown in air supplemented in 5% CO2 (Fig. 4). The mutant was unable to grow in air and grew more slowly than the wild type in air supplemented with 5% CO2.
FIG. 4.
Growth curves of wild-type (WT) T. neapolitanus and T. neapolitanus cbbL::Km grown in air supplemented with 5% CO2 (A) and in air (B). Growth was monitored at A600.
RuBisCO activity in mutant.
Since the interruption of cbbL did not result in a lethal phenotype, it was concluded that an alternate CO2-fixing enzyme was expressed. In view of the earlier observation that T. neapolitanus possessed a form II RuBisCO gene (28), it was hypothesized that the organism was responding to the cbbL mutation by expressing a form II RuBisCO. To determine if a form II enzyme was expressed, wild-type and mutant cells were broken by sonication, the cell-free supernatants were subjected to sucrose gradient centrifugation, and the resulting fractions assayed for RuBisCO activity (Fig. 5). The wild type exhibited only a form I peak of activity when grown in air or in air supplemented with 5% CO2; no peak of activity corresponding to a form II enzyme could be detected. In contrast, the mutant exhibited only a form II peak of activity.
The S/P ratio, a biochemical approach developed to determine the presence or absence of carboxysomes (2), was determined for the T. neapolitanus wild type and mutant and for the non-carboxysome-forming Paracoccus versutus (16) (formerly Thiobacillus versutus). The S/P ratio of the mutant was significantly higher than that of the wild type (29 and 1.67, respectively). The value for the mutant was essentially the same as for the non-carboxysome former (S/P = 30), thus indicating that RuBisCO is not packaged inside carboxysomes. Although the mutant’s RuBisCO specific activity was approximately one-third of the wild type’s specific activity, the S/P ratio determined by using specific activities was still greater than 12.
Immunoblot analysis.
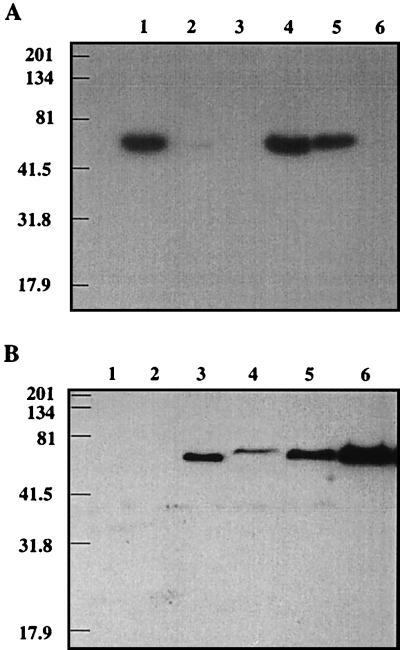
Immunoblot analysis of all of the sucrose gradient fractions that could potentially contain form I enzyme (fractions 24 to 28 [Fig. 5]) or form II enzyme (fractions 19 to 24) confirmed the RuBisCO activity results but in addition demonstrated that although the form I enzyme predominated in the wild type grown in either air or air supplemented with 5% CO2, a small amount of form II RuBisCO was also present (Fig. 6; blots containing only the peak fractions [either fraction 26 for form I RuBisCO or fraction 21 for form II RuBisCO] are shown). In the wild type grown in air supplemented with 5% CO2, form I RuBisCO expression was decreased whereas the expression of form II RuBisCO was increased. Only the form II enzyme was detected in the mutant. The absence of the CbbL protein from the mutant was additionally confirmed by probing immunoblots containing cell extracts from both the wild type and the mutant (data not shown). Again, no CbbL protein, not even a truncated form, was detected in the mutant.
FIG. 6.
Immunoblot analysis of RuBisCO form I- and form II-enriched fractions from sucrose gradients. (A) Immunoblot of proteins transferred from an SDS–10% polyacrylamide gel and probed with form I antibodies. Lane 1, purified Synechococcus sp. strain 6301 RuBisCO; lane 2, purified R. sphaeroides form I RuBisCO; lane 3, purified R. sphaeroides form II RuBisCO; lane 4, form I peak fraction (fraction 26 in Fig. 5) from wild-type T. neapolitanus grown in air; lane 5, form I peak fraction (fraction 26) from wild-type T. neapolitanus grown in air supplemented with 5% CO2; lane 6, corresponding fraction (fraction 26) from T. neapolitanus cbbL::Km. (B) Immunoblot of proteins from an SDS–10% polyacrylamide gel and probed with form II antibodies. Lane 1, purified Synechococcus sp. strain 6301 RuBisCO; lane 2, purified R. sphaeroides form I RuBisCO; lane 3, purified R. sphaeroides form II RuBisCO; lane 4, corresponding fraction (fraction 21 in Fig. 5) from wild-type T. neapolitanus grown in air; lane 5, corresponding fraction (fraction 21) from wild-type T. neapolitanus grown in air supplemented with 5% CO2; lane 6, form II peak fraction (fraction 21) from T. neapolitanus cbbL::Km. Sizes (in kilodaltons) of Kaleidoscope prestained standards (Bio-Rad) are indicated to the left.
Electron microscopy.
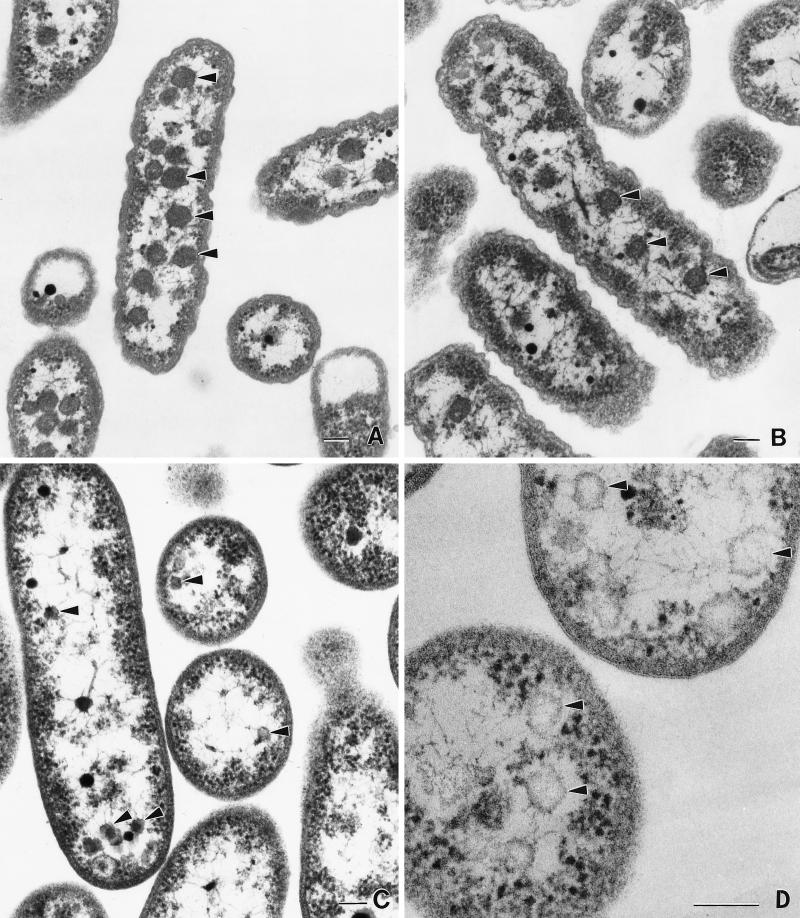
The wild type produces carboxysomes approximately 120 nm in diameter when grown in air or air supplemented with 5% CO2 (Fig. 7A and B). As demonstrated previously (2), CO2 supplementation results in a reduction in the number of carboxysomes. This was also noted for the S/P ratio (S/P of air-grown T. neapolitanus = 1.1; S/P of 5% CO2-grown T. neapolitanus = 1.67). Carboxysomes were not observed in the mutant, but smaller (60 to 70 nm in diameter), empty inclusions were visible (Fig. 7C and D).
FIG. 7.
Electron micrographs of wild-type and cbbL::Km T. neapolitanus. (A) Wild-type T. neapolitanus grown in batch culture in air. (B) Wild-type T. neapolitanus grown in batch culture supplemented with 5% CO2. (C) T. neapolitanus cbbL::Km grown in batch culture supplemented with 5% CO2. Arrowheads mark empty polyhedral inclusions. (D) Enlargement of the empty inclusions (arrowheads) observed in T. neapolitanus cbbL::Km. The scale bar represents 100 nm.
Analysis of downstream gene products.
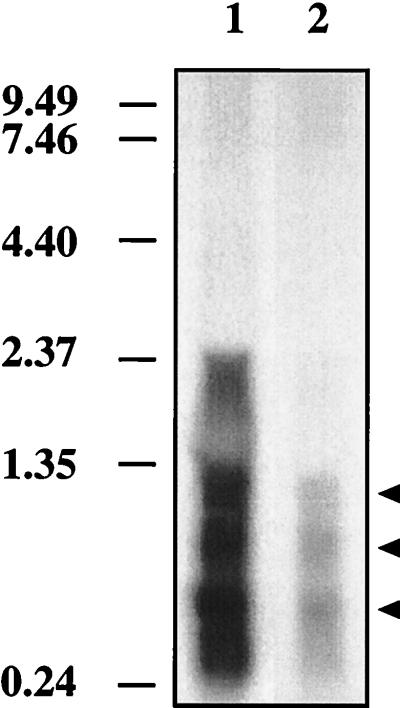
Northern analysis was performed to determine if the insertion into cbbL affected the transcription of downstream genes in the carboxysome operon. English and coworkers (6) showed that the three csoS1 shell genes are initially cotranscribed and then processed to the individual messages. To determine if the shell genes are transcribed in the mutant, csoS1A was used to probe a Northern blot. Signals approximately 400, 650, and 1,100 bases in size were detected in both the wild type and the mutant (Fig. 8). Approximately equal amounts of RNA were loaded on the gel, but the wild type gave a much stronger signal with the probe than the mutant (Fig. 8).
FIG. 8.
Detection of downstream gene products of the T. neapolitanus carboxysome operon by Northern blotting with the carboxysome gene csoS1A as a probe. Lane 1, wild-type T. neapolitanus RNA (20 μg); lane 2, T. neapolitanus cbbL::Km RNA (20 μg). Sizes (in kilobases) of RNA markers are indicated to the left.
DISCUSSION
T. neapolitanus is a chemoautotrophic bacterium which grows aerobically with thiosulfate as an energy source and CO2 as its sole carbon source. Snead and Shively (31) demonstrated that T. neapolitanus fixes CO2 via the Calvin cycle, using the hexadecameric form I RuBisCO enzyme. The genes encoding the form I RuBisCO (cbbL and cbbS) were cloned and sequenced. As in other prokaryotic autotrophs (12), cbbL and cbbS are transcribed as a single polycistronic message. The size of the transcript, approximately 2.0 kb, is in agreement with the predicted size.
The cbbL gene was insertionally inactivated, resulting in the mutant T. neapolitanus cbbL::Km. Northern analysis of the RNA isolated from the mutant revealed that a slightly smaller cbbL RNA transcript was produced, suggesting that the insertion of the antibiotic cartridge resulted in a premature termination of transcription, i.e., a polar mutation. The smeared pattern suggests that the message is unstable, resulting in rapid turnover. Instability of the message could explain why no CbbL protein was observed in the mutant as determined by Western blot analysis. No transcript was detected with the cbbS probe, confirming that a polar mutation was created.
To compensate for the loss of the form I enzyme, T. neapolitanus cbbL::Km synthesizes an increased quantity of form II RuBisCO. It has been shown previously that R. capsulatus and T. denitrificans express both forms of RuBisCO, resulting in two peaks of activity in sucrose gradients (7, 27). Comparison of T. denitrificans and R. capsulatus gradients to those of wild-type T. neapolitanus showed a single peak of activity in T. neapolitanus corresponding to form I RuBisCO, while a similar comparison of T. neapolitanus cbbL::Km showed one peak corresponding to form II RuBisCO. The results were confirmed by using antibodies specific for CbbL and CbbM.
RuBisCO is a bifunctional enzyme capable of fixing either CO2 or O2 (18). The specificity factor is a measure of the ability of RuBisCO to discriminate between the two substrates at a given CO2/O2 ratio; a high specificity factor indicates a preference for CO2 (15). As a general rule, the form II enzyme has a lower specificity factor and thus requires a higher CO2/O2 ratio to function as an efficient carboxylase (15). Utilizing its form I RuBisCO, wild-type T. neapolitanus grows well in atmospheric levels of CO2. The cbbL::Km mutant, however, requires higher levels of CO2 for growth, suggesting that like the T. denitrificans form II enzyme, the form II RuBisCO of T. neapolitanus has a low specificity factor (13).
Since T. neapolitanus is an obligate aerobe and form II RuBisCO has a low specificity factor, why has T. neapolitanus retained a functional cbbM? As demonstrated by immunoblot analysis, T. neapolitanus synthesizes both form I and form II RuBisCO when cultured in air supplemented with 5% CO2. Even under these conditions, the level of the form II enzyme is negligible in comparison to that of form I. Perhaps, if the O2 level were lowered to the absolute minimum necessary to support growth, the majority of the RuBisCO synthesized would be of the form II type. When grown in air, the expression of form II is reduced and that of form I is markedly increased. In this case, the form I enzyme, being better equipped to discriminate between CO2 and O2, becomes, for all practical purposes, solely responsible for the assimilation of CO2. The further enhancement of CO2 fixation by T. neapolitanus is accomplished by packaging a large percentage of its form I RuBisCO inside carboxysomes. Considerable evidence has been gathered to support the conclusion that the carboxysome enhances CO2 fixation: as the level of CO2 supplied for growth decreases, the amount of RuBisCO sequestered inside the carboxysome increases (2), and an insertion mutation of the shell gene, csoS1A, results in cells that have a markedly reduced number of carboxysomes and a requirement of supplemental CO2 for growth (5). With regard to the latter, carboxysomeless mutants of Synechococcus sp. strain PCC 7942 also require elevated levels of CO2 for growth (8).
The presence of a putative carboxysome operon in T. neapolitanus which begins with cbbL and continues through csoS1B has been hypothesized (30). However, it should be noted that although our insertion mutation of cbbL also completely eliminated the expression of cbbS, Northern analysis showed that the csoS1 genes were transcribed, albeit at a lower level than in the wild type grown with 5% CO2. This finding suggests that an additional promoter(s) downstream of cbbS allows for the independent but reduced transcription of the other carboxysome genes. Evidence from studies of the cbbXYZ operon involved in CO2 fixation in R. sphaeroides (9) support this reasoning.
Pierce et al. (22) found that when the wild-type form I RuBisCO locus of the cyanobacterium Synechocystis sp. strain 6803 was replaced with the Rhodospirillum rubrum form II RuBisCO gene, the mutant “cyanorubrum” required higher levels of CO2 in order to grow. Neither the cyanorubrum mutant nor our T. neapolitanus cbbL::Km mutant produces functional carboxysomes. The T. neapolitanus cbbL::Km mutant possesses small inclusions with polyhedral profiles, many of which appear to be empty carboxysomes. This finding suggests that some of the necessary components of the carboxysome are synthesized and assembly is attempted. As noted above, although expression of the genes downstream of cbbS is definitely perturbed, some expression was detected.
It seems plausible that assembly of the carboxysome requires the specific participation of form I RuBisCO; form II RuBisCO will not substitute. This conclusion is supported by our data, by the data on cyanorubrum (22), and by the data of Holthuijzen et al. (14), who demonstrated the very tight association between the CbbS of RuBisCO and carboxysome shell polypeptides. However, whether the lack of functional carboxysomes in T. neapolitanus cbbL::Km is due solely to the lack of form I RuBisCO is unknown. The reduced level of the expression of the carboxysome shell genes downstream of cbbS may also interfere with proper carboxysome production.
In conclusion, there is still a great deal to be learned about how the thiobacilli regulate carbon dioxide fixation. The growth conditions which favor the synthesis of form II RuBisCO must be thoroughly investigated in order to fully understand why T. neapolitanus and other thiobacilli have retained the genes encoding both forms of RuBisCO; the mechanism whereby the carboxysome enhances CO2 fixation must be addressed; and to understand the regulation of the synthesis and assembly of the carboxysome, the putative carboxysome operon must be unequivocally defined.
ACKNOWLEDGMENTS
We thank F. R. Tabita for kindly providing the anti-Synechococcus strain 6301 form I antibodies and anti-R. sphaeroides form II antibodies and the purified Synechococcus sp. RuBisCO, R. sphaeroides form I, and R. sphaeroides form II proteins and Sally Brock, Department of Biological Sciences, Clemson University, Clemson, S.C., for assistance with figure preparation.
This work was supported by the Cooperative State Research Service, U.S. Department of Agriculture, under agreement 92-37306-7663 and NSF grant MCB-9513481.
REFERENCES
- 1.Alam J, Whitaker R A, Krogmann D W, Curtis S E. Isolation and sequence of the gene for ferredoxin I from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1986;168:1265–1271. doi: 10.1128/jb.168.3.1265-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beudeker R F, Codd G A, Kuenen J G. Quantification and intracellular distribution of ribulose-1,5-bisphosphate carboxylase in Thiobacillus neapolitanus, as related to possible functions of carboxysomes. Arch Microbiol. 1981;129:361–367. [Google Scholar]
- 3.Cannon G C, Shively J M. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. [Google Scholar]
- 4.Chung S Y, Yaguchi T, Nishihara H, Igarashi Y, Kodama T. Purification of form L2 RubisCO from a marine obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenovibrio marinus strain MH-110. FEMS Microbiol Lett. 1993;109:49–53. doi: 10.1111/j.1574-6968.1993.tb06142.x. [DOI] [PubMed] [Google Scholar]
- 5.English R S, Jin S, Shively J M. Use of electroporation to generate a Thiobacillus neapolitanus carboxysome mutant. Appl Environ Microbiol. 1995;61:3256–3260. doi: 10.1128/aem.61.9.3256-3260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English R S, Lorbach S C, Qin X, Shively J M. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 7.English R S, Williams C A, Lorbach S C, Shively J M. Two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase from Thiobacillus denitrificans. FEMS Microbiol Lett. 1992;94:111–119. doi: 10.1016/0378-1097(92)90593-d. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg D, Jager K M, Kessel M, Silman N J, Bergman B. Rubisco but not Rubisco activase is clustered in the carboxysomes of the cyanobacterium Synechococcus sp. PCC 7942: Mud-induced carboxysomeless mutants. Mol Microbiol. 1993;9:1193–1201. doi: 10.1111/j.1365-2958.1993.tb01248.x. [DOI] [PubMed] [Google Scholar]
- 9.Gibson J L, Tabita F R. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J Bacteriol. 1997;179:663–669. doi: 10.1128/jb.179.3.663-669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson J L, Tabita F R. Different molecular forms of d-ribulose-1,5-bisphosphate carboxylase from Rhodopseudomonas sphaeroides. J Biol Chem. 1977;252:943–949. [PubMed] [Google Scholar]
- 11.Gibson J L, Tabita F R. Isolation and preliminary characterization of two forms of ribulose-1,5-bisphosphate carboxylase from Rhodopseudomonas capsulata. J Bacteriol. 1977;132:818–823. doi: 10.1128/jb.132.3.818-823.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson J L, Tabita F R. The molecular regulation of the reductive pentose phosphate pathway in Proteobacteria and Cyanobacteria. Arch Microbiol. 1996;166:141–150. doi: 10.1007/s002030050369. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez J M, Baker S H, Lorbach S C, Shively J M, Tabita F R. Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J Bacteriol. 1996;178:347–356. doi: 10.1128/jb.178.2.347-356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holthuijzen Y A, Van Breeman J F L, Kuenen J G, Konings W N. Protein composition of the carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- 15.Jordan D B, Ogren W L. Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature. 1981;291:513–515. [Google Scholar]
- 16.Katayama Y, Hiraishi A, Kuraishi H. Paracoccus thiocyanatus sp. nov., a new species of thiocyanate-utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus. Microbiology. 1995;141:1469–1477. doi: 10.1099/13500872-141-6-1469. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.Miziorko H H, Lorimer G H. Ribulose-1,5-bisphosphate carboxylase-oxygenase. Annu Rev Biochem. 1983;52:507–535. doi: 10.1146/annurev.bi.52.070183.002451. [DOI] [PubMed] [Google Scholar]
- 19.Morse D, Salois P, Markovic P, Hastings J W. A nuclear-encoded form II RuBisCO in dinoflagellates. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 20.Nargang F, McIntosh L, Somerville C. Nucleotide sequence of the ribulosebisphosphate carboxylase gene from Rhodospirillum rubrum. Mol Gen Genet. 1984;193:220–224. [Google Scholar]
- 21.O’Brien S N, Welter B H, Mantzke K A, Price T M. Identification of progesterone receptor in human subcutaneous adipose tissue. J Clin Endocrinol Metab. 1998;83:509–513. doi: 10.1210/jcem.83.2.4561. [DOI] [PubMed] [Google Scholar]
- 22.Pierce J, Carlson T J, Williams J G K. A cyanobacterial mutant requiring the expression of ribulose bisphosphate carboxylase from a photosynthetic anaerobe. Proc Natl Acad Sci USA. 1989;86:5753–5757. doi: 10.1073/pnas.86.15.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson J J, Stein J L, Cavanaugh C M. Cloning and sequencing of a form II ribulose-1,5-bisphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tube worm Riftia pachyptila. J Bacteriol. 1998;180:1596–1599. doi: 10.1128/jb.180.6.1596-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt M E, Brown T A, Trumpower B L. Rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinozaki K, Yamada C, Takahata N, Sugiura M. Molecular cloning and sequence analysis of the cyanobacterial gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1983;80:4050–4054. doi: 10.1073/pnas.80.13.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shively J M, Ball F, Brown D H, Saunders R E. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) in Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 27.Shively J M, Davidson E, Marrs B L. Depression of the synthesis of the intermediate and large forms of ribulose-1,5-bisphosphate carboxylase/oxygenase in Rhodopseudomonas capsulata. Arch Microbiol. 1984;138:233–236. doi: 10.1007/BF00402127. [DOI] [PubMed] [Google Scholar]
- 28.Shively J M, Devore W, Stratford L, Porter L, Medlin L, Stevens S E. Molecular evolution of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) FEMS Microbiol Lett. 1986;37:251–257. [Google Scholar]
- 29.Shively J M, English R S. The carboxysome, a prokaryotic organelle: a mini review. Can J Bot. 1991;69:957–962. [Google Scholar]
- 30.Shively J M, Lorbach S C, Jin S, Baker S H. Carboxysomes: the genes of Thiobacillus neapolitanus. In: Lidstrom M, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer; 1996. pp. 56–63. [Google Scholar]
- 31.Snead R M, Shively J M. d-Ribulose-1,5-bisphosphate carboxylase from Thiobacillus neapolitanus. Curr Microbiol. 1978;1:309–314. [Google Scholar]
- 32.Stoner M T, Shively J M. Cloning and expression of the d-ribulose-1,5-bisphosphate carboxylase/oxygenase form II gene from Thiobacillus intermedius in Escherichia coli. FEMS Microbiol Lett. 1993;107:287–292. doi: 10.1111/j.1574-6968.1993.tb06044.x. [DOI] [PubMed] [Google Scholar]
- 33.Tabita F R. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol Rev. 1988;52:155–189. doi: 10.1128/mr.52.2.155-189.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabita F R, McFadden B A. d-Ribulose-1,5-disphosphate carboxylase from Rhodospirillum rubrum. J Biol Chem. 1974;249:3459–3464. [PubMed] [Google Scholar]
- 35.Viera J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 36.Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitney S M, Shaw D C, Yellowless D. Evidence that some dinoflagellates contain a ribulose-1,5-bisphosphate carboxylase/oxygenase related to that of the alpha-proteobacteria. Proc R Soc Lond Ser B. 1995;259:271–275. doi: 10.1098/rspb.1995.0040. [DOI] [PubMed] [Google Scholar]
- 38.Yaguchi T, Chung S Y, Igarashi Y, Kodama T. Cloning and sequence of the L2 form of RubisCO from a marine obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenovibrio marinus strain MH-110. Biosci Biotechnol Biochem. 1994;58:1733–1737. doi: 10.1271/bbb.58.1733. [DOI] [PubMed] [Google Scholar]