Abstract
Under anoxic conditions in the presence of an oxidizable cosubstrate such as glucose or glycerol, Escherichia coli converts citrate to acetate and succinate. Two enzymes are specifically required for the fermentation of the tricarboxylic acid, i.e., a citrate uptake system and citrate lyase. Here we report that the open reading frame (designated citT) located at 13.90 min on the E. coli chromosome between rna and the citrate lyase genes encodes a citrate carrier. E. coli transformed with a plasmid expressing citT was capable of aerobic growth on citrate, which provides convincing evidence for a function of CitT as a citrate carrier. Transport studies with cell suspensions of the transformed strain indicated that CitT catalyzes a homologous exchange of citrate or a heterologous exchange against succinate, fumarate, or tartrate. Since succinate is the end product of citrate fermentation in E. coli, it is likely that CitT functions in vivo as a citrate/succinate antiporter. Analysis of the primary sequence showed that CitT (487 amino acids, 53.1 kDa) is a highly hydrophobic protein with 12 putative transmembrane helices. Sequence comparisons revealed that CitT is related to the 2-oxoglutarate/malate translocator (SODiT1 gene product) from spinach chloroplasts and five bacterial gene products, none of which has yet been functionally characterized. It is suggested that the E. coli CitT protein is a member of a novel family of eubacterial transporters involved in the transport of di- and tricarboxylic acids.
Under oxic growth conditions, most Escherichia coli strains are not able to utilize citrate due to the lack of a functional transport system. This is a key characteristic of E. coli among enterobacteria (15). Some E. coli strains capable of aerobic growth on citrate possess plasmid-encoded citrate uptake systems. The citrate carrier genes from two of these plasmids were cloned and sequenced (13, 29). The deduced proteins, which consisted of 431 amino acids and differed in six positions only, exhibited 93% sequence identity to CitA from Salmonella typhimurium (30) and 66% to CitH from Klebsiella pneumoniae (35), both of which are chromosomally encoded. Citrate transport by CitH was shown to occur in symport with protons (36), and a similar mechanism is likely to apply for the plasmid-encoded CitA carriers from E. coli.
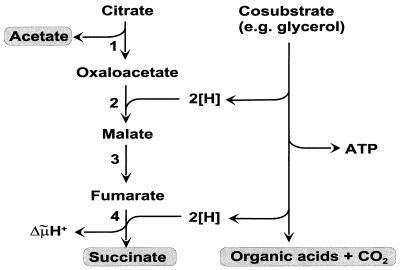
Under anoxic conditions, E. coli can utilize citrate if an oxidizable cosubstrate is present. The corresponding fermentation pathway is shown in Fig. 1. After uptake into the cell, citrate is split by citrate lyase to acetate and oxaloacetate. The latter is subsequently converted via malate and fumarate to succinate by malate dehydrogenase, fumarase, and fumarate reductase. The reducing equivalents required for this conversion must be provided by the oxidation of the cosubstrate, e.g., glucose or glycerol (17). Two enzymes must be specifically induced for anaerobic citrate dissimilation, i.e., a citrate uptake system and citrate lyase. The latter enzyme has been purified from E. coli and was shown to consist of three subunits (α [55.5 kDa], β [35 kDa], and γ [12.5 kDa]), similar to citrate lyase from other bacterial species (25).
FIG. 1.
Cosubstrate-dependent citrate fermentation by E. coli. The enzymes involved in the conversion of citrate to succinate are (1) citrate lyase, (2) malate dehydrogenase, (3) fumarase, and (4) fumarate reductase.
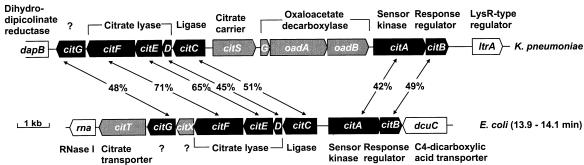
After the completion of the E. coli genome sequence (2), a gene cluster which showed a high degree of similarity to the citCDEFG operon of K. pneumoniae was identified between 13.9 and 14.2 min (Fig. 2). These genes encode the three subunits of citrate lyase (CitD, CitE, and CitF), a ligase required for the acetylation of the 2-(5"-phosphoribosyl)-3′-dephosphocoenzyme-A prosthetic group (CitC), and a protein (CitG) which presumably is involved in the biosynthesis or the covalent attachment of the prosthetic group (4). The proteins deduced from the E. coli genes citC, citD, citE, citF, and citG exhibited 50.6, 44.9, 65.1, 70.7, and 48.3% sequence identity to the corresponding K. pneumoniae proteins, respectively. A noticeable difference between the two gene clusters was the presence of an additional open reading frame (designated citX in Fig. 2) between the E. coli citF and citG genes. The citAB genes located upstream and divergent to E. coli citC encode proteins which are most closely related to the K. pneumoniae CitA-CitB two-component signal transduction system (42.0 and 48.7% identity, respectively). The K. pneumoniae CitA and CitB proteins are essential for expression of the genes specifically involved in citrate fermentation, including citCDEFG (5). The sensor kinase CitA was proposed to function as a citrate sensor (5), and the response regulator CitB was shown to bind to two sites extending from −50 to −96 upstream of the citC transcription start site and from −55 to −89 upstream of the citS transcription start site (20). Phosphorylation led to 10- to 100-fold increase of the apparent binding affinity (20). The E. coli citB gene has also been designated criR, because the deduced amino acid sequence is identical to the sequence of the CriR protein from Shigella flexneri, which has been implicated in the regulation of the ipa genes (24). Since E. coli does not possess an invasion plasmid carrying ipa genes, the primary function of the citAB gene products is presumably the regulation of the citrate lyase genes, as in K. pneumoniae. This supposition is supported by the similarity of the DNA-binding helix-turn-helix motifs of the two CitB response regulators and by the similarity of the citC upstream regions. With respect to the gene designations, one should be aware that in E. coli citA denotes both a plasmid-linked gene encoding a citrate carrier and a chromosomally located gene encoding a histidine sensor kinase. Similarly, citB denotes a gene associated with the plasmid-linked citA and also a chromosomally located gene encoding the cognate response regulator of the sensor kinase CitA.
FIG. 2.
E. coli genes required for citrate fermentation and comparison with the corresponding K. pneumoniae genes. Genes shaded in dark gray represent those K. pneumoniae genes involved in citrate fermentation which are also present in E. coli; genes shaded in light gray are those present only in the E. coli or only in the K. pneumoniae cluster. All other indicated genes are presumably not directly involved in citrate fermentation.
An important difference between the K. pneumoniae and E. coli cit gene clusters is the lack of genes encoding a Na+-dependent citrate carrier (citS) and oxaloacetate decarboxylase (oadGAB) in the latter species (Fig. 2). In fact, these genes are not present on the whole E. coli chromosome. The absence of oxaloacetate decarboxylase provides an explanation for the different fermentation pathways in these organisms (3), and the absence of a CitS-type protein necessitates the use of a different citrate uptake system. Inspection of the E. coli DNA region downstream of citG revealed a gene (ybdS) encoding a highly hydrophobic protein with 34% sequence identity to the 2-oxoglutarate/malate translocator from spinach chloroplasts. The ybdS start codon is located only 50 bp downstream of the stop codon of citG, indicating that ybdS is cotranscribed with citCDEFXG. In this report, we present evidence that the protein encoded by ybdS functions as a citrate carrier; we therefore renamed the gene citT, for citrate transporter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli DH5α (Bethesda Research Laboratories) was routinely used as the host for the cloning procedures. E. coli JM83 (38) was used for the preparation of chromosomal DNA by the method of Marmur (18). E. coli BL21(DE3), which contains the phage T7 RNA polymerase gene under the control of the lacUV5 promoter (33), served as the host for the expression of citT and citS from pET-derived plasmids (Novagen). The strains were routinely grown at 37°C in Luria-Bertani (LB) medium (21) with shaking at 180 rpm. For testing the ability to grow with citrate as the sole carbon and energy source, we used either Simmons’ citrate agar (31) supplemented with 12 μM thiamine or a liquid medium (adjusted to pH 6.8 with Na+-poor KOH) that contained 7 mM citric acid, 20 mM KH2PO4, 11 mM (NH4)2SO4, 1 mM MgSO4, 12 μM thiamine, 0.4% (vol/vol) trace elements solution (7), and either 25 mM Na2SO4 or 25 mM K2SO4. The latter medium contained less than 50 μM Na+ as determined by atomic absorption spectroscopy. The antibiotics ampicillin (100 to 200 μg/ml) and kanamycin (50 μg/ml) were used as appropriate. Cells used for growth studies with citrate minimal medium were pregrown in LB medium and washed with Na+-poor minimal medium before use as inoculum.
Recombinant DNA work.
For routine work with recombinant DNA, established protocols were used (27). For the construction of a citT expression plasmid, the citT gene was amplified from chromosomal E. coli DNA by using the oligonucleotides ec-citT-for (5′-GATTCGAAGCTTCATATGTCTTTAGCAAAAGATAATATATGG-3′) and ec-citT-rev (5′-CCGCGAATTCTTAGTTCCACATGGCGAGAATCGGCCAG-3′). In ec-citT-for, the ATG start codon of citT is part of an NdeI restriction site, which is preceded by a HindIII site and five additional nucleotides, allowing increased restriction efficiency. In ec-citT-rev, a BamHI restriction site is introduced after the citT stop codon. The PCR mixture contained 500 ng of genomic DNA of E. coli JM83, 0.5 μM each primer, 0.2 mM deoxynucleoside triphosphates, 1× buffer for cloned Pfu DNA polymerase, and 2.5 U of Pfu DNA polymerase (Stratagene). After an initial denaturation step (2 min at 95°C), 30 cycles consisting of 15 s at 95°C, 15 s at 62°C, and 4 min at 72°C were performed, followed by a terminal elongation step (4 min at 72°C). The complete PCR mix was subsequently separated on a 1% agarose gel, and the expected 1.49-kb fragment was isolated with Qiaex (Qiagen). After restriction with HindIII and BamHI, the PCR product was purified with a QIA-Quick spin column and ligated with HindIII/BamHI-restricted pUC19 (38), resulting in pUC19-citT. Since the citT gene in pUC19-citT was not preceded by a well-conserved ribosome binding site, a 1.46-kb NdeI/BamHI fragment from pUC19-citT was cloned in pET24b (Novagen) restricted with the same enzymes, resulting in pET24-citT. Plasmid pCitSHis-3 is a derivative of pET16b (Novagen) and is used for synthesis of the K. pneumoniae CitS citrate carrier modified with an N-terminal His10 tag (23).
DNA sequence analysis.
The sequence of the citT gene present in plasmid pET24-citT was determined by the dideoxynucleotide chain termination method (28), using the protocols and equipment for automated DNA sequencing (Sequencer 310 and PRISM Ready Reaction Dye-Deoxy terminator cycle sequencing kit from Applied Biosystems). For this purpose, pET24-citT was purified with a Qiagen Tip-500 column. A primer-walking strategy involving eight primers derived from citT and two primers derived from pET24b was applied. Computer-assisted DNA and protein sequence analysis was performed with the software package of the University of Wisconsin Genetics Computer Group. Prediction of the transmembrane helices indicated in Fig. 3 was performed with the TopPred II software (6) and the TMpred software (12).
FIG. 3.
Amino acid sequence alignment of the E. coli (Ec) CitT protein with the 2-oxoglutarate/malate translocator from spinach (Spinacia oleraceae [So]) chloroplasts and five eubacterial gene products. Identical residues present in at least four of the sequences are framed in black; conservative exchanges are framed in gray. Asterisks indicate residues conserved in all sequences. Putative transmembrane helices within the CitT sequence are overscored. Details on the aligned sequences can be found in the text and in Table 1. Hi, Haemophilus influenzae; Bs, Bacillus subtilis; Hp, Helicobacter pylori.
Transport experiments.
For transport experiments, E. coli BL21(DE3) transformed with either pET24b, pET24-citT, or pCitSHis-3 was grown in LB medium with appropriate antibiotics to an optical density at 600 nm (OD600) of ca. 0.7 to 1.0. Subsequently, cells were washed once with 50 mM morpholineethanesulfonic acid-Tris buffer (pH 7.0) and concentrated 10-fold in the same buffer. The protein concentration of the resulting cell suspension was calculated by assuming that an OD600 of 1.4 corresponds to 109 cells/ml and those 109 cells contain 150 μg of protein (21). At time zero, 98 μl of the concentrated cell suspension preincubated at 25°C was added to 2 μl of 1.2 mM [1,5-14C]citrate (145 cpm/pmol). After various times at 25°C, transport was terminated by the addition of 0.9 ml of ice-cold 0.1 M LiCl followed by rapid filtration through 0.45-μm-pore-size cellulose nitrate filters (diameter, 25 mm; Sartorius). The filters were washed once with 1 ml of ice-cold 0.1 M LiCl and then placed into scintillation vials. Immediately afterwards, 4 ml of scintillation fluid (Irga-Safe Plus; Packard) was added, and the entrapped [1,5-14C]citrate was determined by liquid scintillation counting. The values obtained in this way were corrected for a time zero value obtained as follows: 98 μl of cell suspension was first mixed with 0.9 ml of ice-cold 0.1 M LiCl and then applied to 2 μl of 1.2 mM [1,5-14C]citrate. This mixture was rapidly filtered as described above.
To determine whether certain di- and tricarboxylic acids are able to trigger the efflux of [1,5-14C]citrate previously taken up by the cells, 98 μl of cell suspension was incubated with 2 μl of 1.2 mM [1,5-14C]citrate for 30 s. Subsequently, these preloaded cells (100 μl) were applied to 1 μl of a 1 M solution of either citric acid, succinic acid, fumaric acid, or fumarate (Na+-salt). After various times, 0.9 ml of ice-cold 0.1 M LiCl was added, and the mixtures were filtered and treated as described above.
The transport experiments with 0.91 mM dl-[1,4-14C]tartrate (163 cpm/pmol; custom synthesized by Anawa) were performed in the same way as described above for [1,5-14C]citrate.
RESULTS AND DISCUSSION
Aerobic growth of E. coli harboring a citT expression plasmid on citrate as the sole carbon and energy source.
The open reading frame located at 13.9 min on the E. coli chromosome (designated citT), starting 50 bp downstream of the citG stop codon, encoded a protein of 487 amino acids (53.1 kDa) that was very hydrophobic and contained 12 putative transmembrane helices (Fig. 3). The physical proximity of citT to the citrate lyase genes and the fact that CitT showed 34% amino acid sequence identity to the 2-oxoglutarate/malate translocator from spinach chloroplasts (37) suggested to us that CitT might function as a citrate carrier. To test this assumption, the citT gene was amplified by PCR from chromosomal DNA of E. coli and ligated as a 1.5-kb HindIII/BamHI fragment in pUC19, which allows transcription of citT from the vector-encoded lac promoter. The ligation mixture was transformed into E. coli DH5α and plated on Simmons’ citrate agar. After 48 h at 37°C, several citrate-positive colonies were identified by the color change of the agar from green to blue. This color change of the pH indicator bromthymol blue is observed only with cells able to utilize citrate as a carbon and energy source, which results in alkalinization of the medium. Cells unable to utilize citrate, such as E. coli DH5α containing only the vector pUC19, form only very small colonies (diameter, <1 mm), presumably by using residual carbon sources present in the agar, and bromthymol blue remains green. Restriction analysis of plasmid DNA isolated from several Cit+ clones showed that all contained pUC19 with the 1.5-kb HindIII/BamHI insert carrying citT. One of the pUC19-citT plasmids was transformed again into E. coli DH5α and plated on Simmons’ citrate agar. In this case, all transformants were able to utilize citrate, confirming that citT is responsible for the Cit+ phenotype. Besides citrate, isocitrate could also be used as the sole carbon and energy source by E. coli DH5α harboring pUC19-citT.
To provide a good ribosome binding site (5′-AAGGAG-3′) upstream of the citT start codon, a 1.46-kb NdeI/BamHI fragment obtained from pUC19-citT was cloned into pET24b. Plasmid pET24-citT isolated from one of the resulting Cit+ clones was used for DNA sequence analysis of the region encompassing citT. The sequence was 100% identical to the one present in the database, and thus this plasmid was suitable for further studies. E. coli BL21(DE3) harboring pET24-citT was able to grow aerobically in citrate minimal medium. After a lag phase of about 40 h, the cells grew within 24 h from an OD600 of 0.05 to an OD600 of about 0.5, whereas the control cells containing the vector pET24b were unable to multiply in this medium (data not shown). To find out whether mutations had occurred within citT during the long lag phase, pET24-citT was isolated from citrate-grown cells, and the region encompassing citT was sequenced again. No mutations were detected, showing that other adaptation processes must be responsible for the 40-h lag phase. Growth of E. coli/pET24-citT was independent of sodium ions in the concentration range tested (50 μM to 50 mM).
Transport studies with cell suspensions.
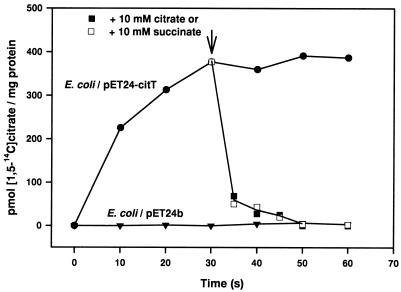
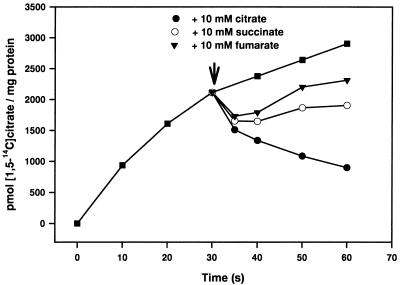
The growth experiments described above confirmed our suggestion that CitT catalyzes the uptake of citrate. Further evidence was obtained by transport studies with cell suspensions. As shown in Fig. 4, cell suspensions of E. coli BL21(DE3)/pET24-citT catalyzed [1,5-14C]citrate uptake with an initial rate of 1.4 nmol min−1 (mg of protein)−1, whereas cells containing only the vector pET24b did not show any [1,5-14C]citrate accumulation. Citrate uptake by cells containing CitT was independent of Na+ ions in the concentration range tested (10 μM to 50 mM; data not shown). The similarity of CitT to the 2-oxoglutarate/malate translocator from spinach chloroplasts and the fact that the final product of citrate fermentation in E. coli is succinate led us to assume that CitT may function as a citrate/succinate antiporter. Indeed, the addition of 10 mM succinate to cells that previously had taken up [1,5-14C]citrate led to a complete efflux of the 14C label with an initial rate of 3.7 nmol min−1 (mg of protein)−1 (Fig. 4). The same effect was obtained by the addition of 10 mM citrate. Fumarate (10 mM) caused a comparable effect, but the rate was somewhat lower and efflux was not complete (data not shown). In a control experiment, uptake and efflux of [1,5-14C]citrate were analyzed with E. coli BL21(DE3)/pCitSHis-3. These cells contain the citrate carrier CitS from K. pneumoniae modified by an N-terminal His10 tag (22, 23). Since the CitS carrier was shown to be highly specific for citrate as a substrate (1), we expected that only citrate, not succinate or fumarate, would be able to trigger an efflux of [1,5-14C]citrate previously taken up. As shown in Fig. 5, cells containing CitS catalyzed the uptake of citrate with an initial rate of 5.6 nmol min−1 (mg of protein)−1. Addition of 10 mM citrate led to a partial efflux of the 14C label from [1,5-14C]citrate-loaded cells [initial rate, 7.3 nmol min−1 (mg of protein)−1], whereas succinate or fumarate did not elicit such a response. This result confirms that the succinate- and fumarate-induced efflux observed with the E. coli/pET24-citT cells (Fig. 4) is catalyzed by the CitT protein rather than by other carriers present in the cytoplasmic membrane. The fact that only a partial efflux of the 14C label was observed in the experiment shown in Fig. 5 can be explained by the fact that part of [1,5-14C]citrate had been converted to other intermediates of the tricarboxylic acid cycle within the 30 s before addition of the unlabeled citrate. These other intermediates are transported not by CitS but apparently by CitT, as indicated by the complete efflux shown in Fig. 4.
FIG. 4.
Uptake and efflux of [1,5-14C]citrate by cell suspensions of E. coli BL21(DE3) containing either pET24b or pET24-citT. The transport experiments were performed as described in Materials and Methods.
FIG. 5.
Uptake and efflux of [1,5-14C]citrate by cell suspensions of E. coli BL21(DE3) containing pCitSHis-3. The transport experiments were performed as described in Materials and Methods.
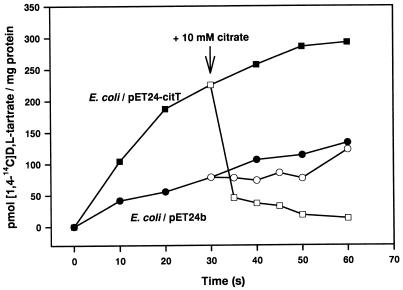
As outlined below, the protein most similar to CitT is the product of the E. coli ygjE gene (Fig. 3), which we predict to function as a tartrate carrier. We therefore tested whether CitT also catalyzes the uptake of dl-[1,4-14C]tartrate. In contrast to citrate, E. coli can utilize tartrate as a carbon and energy source under oxic conditions and contains an appropriate transport system (14). This was confirmed by the fact that dl-[1,4-14C]tartrate uptake was observed with E. coli BL21(DE3) cells harboring the control plasmid pET24b at a rate of 0.25 nmol min−1 (mg of protein)−1 (Fig. 6). Addition of 10 mM citrate did not lead to an efflux of dl-[1,4-14C]tartrate from these cells. With cells harboring pET24-citT, a significantly higher rate of dl-[1,4-14C]tartrate uptake [0.62 nmol min−1 (mg of protein)−1] was found, and addition of 10 mM citrate led to a complete efflux of the 14C label (Fig. 6). As in the case with [1,5-14C]citrate, efflux was significantly faster [2.5 nmol min−1 (mg of protein)−1] than uptake.
FIG. 6.
Uptake and efflux of dl-[1,4-14C]tartrate by cell suspensions of E. coli BL21(DE3) containing either pET24b or pET24-citT. The transport experiments were performed as described in Materials and Methods.
The transport experiments described above show that the E. coli CitT protein functions as a Na+-independent antiporter with relatively broad substrate specificity for C4-dicarboxylates and tricarboxylates. Whether the initial uptake is in fact unidirectional uptake or exchange against the internal pool of dicarboxylates and tricarboxylates is not known. Nevertheless, the significantly higher rates of efflux compared to uptake clearly favor exchange as the usual transport mode of CitT. In order to characterize the catalytic properties of CitT in more detail, a proteoliposomal system with purified CitT seems more suitable than studies with whole cells, as shown before for the CitS protein from K. pneumoniae (22, 23). Since the citT gene is physically linked to the citrate lyase genes and probably coregulated with them, the in vivo function of CitT is likely to be citrate-succinate exchange. The use of an antiport system for the excretion of succinate formed by fumarate respiration seems to be the general rule in E. coli. Three secondary carriers for anaerobic C4-dicarboxylate transport (DcuA, DcuB, and DcuC), all of which preferentially catalyze exchange but also catalyze unidirectional uptake of C4-dicarboxylates, have been identified (8, 9, 32, 39). None of these carriers shows significant sequence similarity to CitT (identity of <20%). The dcuC gene is located immediately downstream of citA in inverse orientation (Fig. 2), but there is no evidence at present for involvement of DcuC in citrate fermentation.
Proteins related to CitT.
A search for proteins related to CitT from E. coli led to the identification of five bacterial polypeptides, none of which has been functionally characterized hitherto, and the 2-oxoglutarate/malate translocator (SOT1) from Spinacia oleraceae (37). The SOT1 protein is located in the inner envelope membrane of spinach chloroplasts, where it catalyzes the import of 2-oxoglutarate in exchange for stromal malate. The protein has recently been purified and functionally reconstituted into liposomes (19). Besides malate, succinate, fumarate, and 2-oxoglutarate can be used as counterions (37). From the sequence alignment shown in Fig. 3, it is obvious that the chloroplast protein is related to CitT and the other bacterial proteins described below. Moreover, preliminary characterization of the catalytic properties of CitT indicates that the transport mechanism of this carrier is similar to that of the SOT1 protein.
The E. coli YgjE protein shows 44% sequence identity to CitT (Fig. 3). It is located at 69.08 min on the E. coli chromosome immediately downstream of the ttdAB genes encoding an oxygen-labile l-tartrate dehydratase (26). This enzyme converts l-tartrate to oxaloacetate and is induced by l- and meso-tartrate during anaerobic growth with glycerol as cosubstrate. Oxaloacetate is subsequently converted to succinate, using the reducing equivalents provided by the oxidation of the cosubstrate. Thus, the tartrate fermentation pathway is very similar to the citrate fermentation pathway depicted in Fig. 1 and leads to the same end product. In view of these facts, it seems plausible that YgjE could function as a tartrate/succinate antiporter.
Besides YgjE, another protein (designated YbhI) with 35% sequence identity to CitT is encoded by the E. coli chromosome at 17.27 min. Interestingly, a spontaneous E. coli K-12 mutant (strain D2004) that is able to utilize citrate, cis-aconitate, trans-aconitate, and isocitrate aerobically has been described (11). Genetic analysis of strain D2004 indicated that the mutations responsible for the Cit+ phenotype are located in cit genes that are linked to the gal operon. Since the galETKM genes are located between 16.9 and 17.1 min, ybhI is likely to be one of the genes expressed in mutant D2004. Consequently, a function of YbhI as tricarboxylic acid transporter is very likely. Additional support for this assumption is provided by the fact that the open reading frame downstream of ybhI encodes a protein of 761 amino acids that exhibits similarity to aconitase from several organisms.
The HI0020 protein of Haemophilus influenzae exhibits 38% sequence identity to CitT. It was tentatively identified as a 2-oxoglutarate/malate translocator due to its similarity to the SOT1 protein from spinach chloroplasts (10). However, since the HI0020 gene is located immediately downstream of the citrate lyase genes, as is the citT gene of E. coli, it seems more likely that the HI0020 protein is functionally related to citrate metabolism.
The YflS protein from Bacillus subtilis (16) exhibits 35% sequence identity to CitT. The corresponding gene is located at 828.9 kb on the chromosome downstream of the pel gene encoding pectate lyase. Remarkably, the two genes downstream of yflS (citS and citT) encode a two-component regulatory system with significant similarity to the CitA/CitB system from K. pneumoniae (5). The proteins derived from citS and citT exhibit 28 and 33% amino acid sequence identity to the sensor kinase CitA and the response regulator CitB, respectively.
The predicted HP0143 protein from Helicobacter pylori possesses 42% sequence identity to CitT, if the predicted frameshift within the coding sequence is ignored (34). In the vicinity of the HP0143 gene (located at 156 kb on the chromosome), no genes involved in citrate or tartrate metabolism which might give a clue to the function of the HP0143 protein are present.
In Table 1, some properties of the proteins described above are summarized. It is evident that they all consist of 477 to 487 amino acids, 70 to 75% of which are apolar. Moreover, all of these proteins are basic, with calculated pIs of between 8.9 and 10.3. Together with the overall sequence similarity, the data support the assumption that these proteins form a new family of secondary transporters.
TABLE 1.
Bacterial transporters related to the 2-oxoglutarate/malate translocator from spinach chloroplasts
| Protein | SwissProt accession no. | Organism | Amino acids | Mol wt (103) | % Apolar amino acids | pI | % Identity with:
|
Demonstrated (anticipated) function | |
|---|---|---|---|---|---|---|---|---|---|
| CitT | SOT1 | ||||||||
| CitT (YbdS) | P77405 | Escherichia coli | 487 | 53.09 | 72 | 8.9 | 100 | 34 | Citrate/succinate antiporter |
| YgjE | P39414 | E. coli | 487 | 52.91 | 75 | 9.5 | 44 | 34 | (Tartrate/succinate antiporter) |
| YbhI | P75763 | E. coli | 477 | 51.35 | 73 | 10.3 | 35 | 36 | (Tricarboxylate transporter) |
| YbhI (HI0020) | Q57048 | Haemophilus influenzae | 479 | 51.35 | 71 | 10.2 | 37 | 35 | (Citrate/succinate antiporter) |
| YflS | NAa | Bacillus subtilis | 478 | 51.43 | 70 | 10.2 | 35 | 53 | (Di- and tricarboxylate transporter) |
| HP0143b | NA | Helicobacter pylori | 482 | 52.35 | 73 | 9.8 | 42 | 33 | |
| SOT1 | Q41364 | Spinacia oleraceae | 477 | 50.69 | 73 | 9.9 | 34 | 100 | 2-Oxoglutarate/malate antiporter |
NA, not available.
The gene encoding HP0143 contains a frameshift.
REFERENCES
- 1.Bandell M, Ansanay V, Rachidi N, Dequin S, Lolkema J S. Membrane potential-generating malate (MleP) and citrate (CitP) transporters of lactic acid bacteria are homologous proteins. J Biol Chem. 1997;272:18140–18146. doi: 10.1074/jbc.272.29.18140. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 4.Bott M, Dimroth P. Klebsiella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequencing, and expression. Mol Microbiol. 1994;14:347–356. doi: 10.1111/j.1365-2958.1994.tb01295.x. [DOI] [PubMed] [Google Scholar]
- 5.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 6.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 7.Dimroth P. Preparation, characterization, and reconstitution of oxaloacetate decarboxylase from Klebsiella aerogenes, a sodium pump. Methods Enzymol. 1986;125:530–540. doi: 10.1016/s0076-6879(86)25042-9. [DOI] [PubMed] [Google Scholar]
- 8.Engel P, Krämer R, Unden G. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from the aerobic dicarboxylate uptake system. J Bacteriol. 1992;174:5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel P, Krämer R, Unden G. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli: energetics and mechanism of exchange, uptake and efflux. Eur J Biochem. 1994;222:605–614. doi: 10.1111/j.1432-1033.1994.tb18903.x. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae RD. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Hall B G. Chromosomal mutation for citrate utilization by Escherichia coli K-12. J Bacteriol. 1982;151:269–273. doi: 10.1128/jb.151.1.269-273.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 13.Ishiguro N, Sato G. Nucleotide sequence of the gene determining plasmid-mediated citrate utilization. J Bacteriol. 1985;164:977–982. doi: 10.1128/jb.164.3.977-982.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kay W W, Kornberg H L. The uptake of C4-dicarboxylic acids by Escherichia coli. Eur J Biochem. 1971;18:274–281. doi: 10.1111/j.1432-1033.1971.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 15.Koser S A. Correlation of citrate-utilization by members of the colon-aerogenes group with other differential characteristics and with habitat. J Bacteriol. 1924;9:59–77. doi: 10.1128/jb.9.1.59-77.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S-K, Codani J-J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Düsterhöft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Hénaut A, Hilbert H, Holsappel S, Hosono S, Hullo M-F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S-M, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S-H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A-M, Presecan E, Pujic P, Purnelle B, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Lütgens M, Gottschalk G. Why a co-substrate is required for anaerobic growth of Escherichia coli on citrate. J Gen Microbiol. 1980;119:63–70. doi: 10.1099/00221287-119-1-63. [DOI] [PubMed] [Google Scholar]
- 18.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 19.Menzlaff E, Flügge U-I. Purification and functional reconstitution of the 2-oxoglutarate/malate translocator from spinach chloroplasts. Biochim Biophys Acta. 1993;1147:13–18. doi: 10.1016/0005-2736(93)90310-v. [DOI] [PubMed] [Google Scholar]
- 20.Meyer M, Dimroth P, Bott M. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J Mol Biol. 1997;269:719–731. doi: 10.1006/jmbi.1997.1076. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Pos K M, Bott M, Dimroth P. Purification of two active fusion proteins of the Na+-dependent citrate carrier of Klebsiella pneumoniae. FEBS Lett. 1994;347:37–41. doi: 10.1016/0014-5793(94)00502-8. [DOI] [PubMed] [Google Scholar]
- 23.Pos K M, Dimroth P. Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry. 1996;35:1018–1026. doi: 10.1021/bi951609t. [DOI] [PubMed] [Google Scholar]
- 24.Qi M S, Yoshikura H, Watanabe H. Identification of a Shigella flexneri criR gene increasing ipa gene expression: a novel member of response regulators of the two-component signal transduction family. Jpn J Med Sci Biol. 1996;49:219–239. doi: 10.7883/yoken1952.49.219. [DOI] [PubMed] [Google Scholar]
- 25.Quentmeier A, Holzenburg A, Mayer F, Antranikian G. Reevaluation of citrate lyase from Escherichia coli. Biochim Biophys Acta. 1987;913:60–65. doi: 10.1016/0167-4838(87)90232-9. [DOI] [PubMed] [Google Scholar]
- 26.Reaney S K, Begg C, Bungard S J, Guest J R. Identification of the l-tartrate dehydratase genes (ttdA and ttdB) of Escherichia coli and evolutionary relationship with the class I fumarase genes. Microbiology. 1993;139:1523–1530. doi: 10.1099/00221287-139-7-1523. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasatsu M, Misra T K, Chu L, Laddaga R, Silver S. Cloning and DNA sequence of a plasmid-determined citrate utilization system in Escherichia coli. J Bacteriol. 1985;164:983–993. doi: 10.1128/jb.164.3.983-993.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimamoto T, Izawa H, Daimon H, Ishiguro N, Shinagawa M, Sakano Y, Tsuda M, Tsuchiya T. Cloning and nucleotide sequence of the gene (citA) encoding a citrate carrier from Salmonella typhimurium. J Biochem. 1991;110:22–28. doi: 10.1093/oxfordjournals.jbchem.a123537. [DOI] [PubMed] [Google Scholar]
- 31.Simmons J S. A culture medium for differentiating organisms of the typhoid-colon aerogenes groups and for isolation of certain fungi. J Infect Dis. 1926;39:209–214. [Google Scholar]
- 32.Six S, Andrews S C, Unden G, Guest J R. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct) J Bacteriol. 1994;176:6470–6478. doi: 10.1128/jb.176.21.6470-6478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 35.van der Rest M E, Schwarz E, Oesterhelt D, Konings W N. DNA sequence of a citrate carrier of Klebsiella pneumoniae. Eur J Biochem. 1990;189:401–407. doi: 10.1111/j.1432-1033.1990.tb15502.x. [DOI] [PubMed] [Google Scholar]
- 36.van der Rest M E, Abee T, Molenaar D, Konings W N. Mechanism and energetics of a citrate-transport system of Klebsiella pneumoniae. Eur J Biochem. 1991;195:71–77. doi: 10.1111/j.1432-1033.1991.tb15677.x. [DOI] [PubMed] [Google Scholar]
- 37.Weber A, Menzlaff E, Arbinger B, Gutensohn M, Eckerskorn C, Flügge U-I. The 2-oxoglutarate/malate translocator of chloroplast envelope membranes: molecular cloning of a transporter containing a 12-helix motif and expression of the functional protein in yeast cells. Biochemistry. 1995;34:2621–2627. doi: 10.1021/bi00008a028. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Zientz E, Six S, Unden G. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J Bacteriol. 1996;178:7241–7247. doi: 10.1128/jb.178.24.7241-7247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]