Abstract
Objectives
A novel morbillivirus was recently described in stray and domestic cats in Asia, the USA and Europe. Most cats infected with feline morbillivirus (FeMV) showed lower urinary tract or kidney disease. Although the association of FeMV infection and kidney diseases has been suggested, the virus pathogenicity remains unclear. The present study aimed to investigate the distribution of FeMV infection, as well as the relationship between FeMV infection and kidney diseases in cats from northwestern Italy.
Methods
A total of 153 urine samples (150 individuals and three pools) and 50 kidney samples were collected and included in the study; total RNA was extracted and a reverse transcription quantitative PCR (RT-qPCR) was performed in order to identify FeMV. Kidneys were also submitted to anatomopathological examination. Phylogenetic analysis and isolation attempts were carried out on positive samples. In FeMV-positive cats, urinalysis and blood analysis were performed.
Results
FeMV RNA was detected in 7.3% of urine samples and in 8% of kidney samples, both in healthy cats and in cats with clinical signs/post-mortem lesions compatible with kidney disease. At histopathological examination, tubulointerstitial nephritis (TIN) was shown in 3/4 positive kidney samples, but a clear relationship between FeMV and TIN was not observed. Isolation attempts were unsuccessful, although the urine sample of one castrated male cat hosted in a cattery showed a positive signal in RT-qPCR until the fourth cell passage. Phylogenetic analysis revealed that this FeMV strain belonged to genotype 1-B. In the same cattery, a second genotype 1-B variant was detected from a urine pool. Urinalysis showed proteinuria in three cats, while at blood analysis three cats presented altered creatinine levels.
Conclusions and relevance
Data reported suggest the presence of a FeMV sub-cluster distinct from the strain previously isolated in Italy, whose role in renal disorders remains uncertain.
Keywords: Clinical pathology, histopathology, kidney, morbillivirus, reverse transcription quantitative PCR, phylogenetic, urine
Introduction
The genus morbillivirus includes a consistent number of pathogens that cause some of the most devastating viral diseases of humans and animals (ie, measles virus, canine distemper virus, rinderpest virus). 1 In 2012, a novel morbillivirus was discovered in a stray cat population of Hong Kong and named feline morbillivirus (FeMV). 2 FeMV is a negative-sense, non-segmented, single-stranded RNA virus whose genome of 16,050 base pairs (bp) was recently characterised. 3 It follows the ‘rule of six’, which is typical for morbilliviruses: six genes (N, P/V/C, M, F, H, L) coding for six structural and two non-structural proteins.
A considerable number of FeMV-infected cats in Japan showed feline lower urinary tract diseases (FLUTD) with typical tubulointerstitial nephritis (TIN), 2 but possible correlations between renal diseases and FeMV infection remain unclear. 4 In recent years, different epidemiological studies have been carried out in Japan, the UK, Brazil, Turkey and Italy.3,5–8 From a diagnostic point of view, several biomolecular approaches have been developed and optimised for investigation purposes.5,6,8–10 Previous studies demonstrated that urine and the kidneys represent the best targets for biomolecular detection of FeMV by reverse transcription quantitative PCR (RT-qPCR).11,12 Concerning isolation procedures, FeMV was demonstrated to replicate into Crandell-Rees feline kidney (CRFK) cells 13 and in feline embryonic fibroblast cells, 14 causing a discrete cytopathic effect characterised by syncytia formation. However, long incubation periods for the infected CRFK cells and biological features of FeMV make isolation attempts time-consuming and often unsuccessful. 12 Indeed, Koide et al 13 demonstrated that viral titre increases exponentially between hours 18 and 30 post-inoculation in CRFK cells. Conversely, no increase in virus titre was observed 63 h post-infection. 13 However, only a few studies on FeMV have reported information on cat anamnesis or the clinicopathological features related to FeMV infection. Data regarding the correlation between kidney diseases and FeMV infection are often discordant.
The aim of this study was to investigate the presence of FeMV, as well as the relationship between FeMV infection and kidney disease, in cats from northwestern Italy by performing biomolecular analysis on kidney and urine samples, and undertaking histopathological examination of the kidneys. Phylogenetic analysis was carried out to investigate genetic correlations with other field strains from different countries.
Materials and methods
Sample collection
Urine
A total of 150 individual urine samples were collected from client-owned cats hospitalised in a veterinary clinic (n = 127) or cats living in a private cattery (n = 23). The private cattery contained a total of 55 cats at the time of the study, divided in four rooms with access to an external fenced area. Moreover, in the same cattery, three pooled urine samples were collected in order to rapidly screen the host population for FeMV, avoiding the manipulation and sedation of feral cats. Urine pools were collected by confining 10 cats per room and placing different litter boxes in each room. All cats (n = 150) included in the study were clinically examined, and sex, breed and age were recorded (Table 1). Of the 150 cats, 68% were male and 86% of the male cats were castrated. Thirty-two percent of 150 cats were female and 92% of the female cats were spayed. Urine samples were collected from animals of nine different pure breeds in addition to cross breeds (Table 1). Across the animals included in this study, 21% of cats were affected by FLUTD; acute and chronic urinary failures were detected in 3% and 25% of cats, respectively. The remaining cats (51%) were healthy or affected by non-urinary diseases. Urine samples were stored at –80°C until total RNA extraction.
Table 1.
Cat signalment, clinical diagnosis and test results of urine samples analysed in this study
| n = 150 urine samples | RT-qPCR positive | Clinical diagnosis |
||||
|---|---|---|---|---|---|---|
| FLUTD | CKD | AKD | Other* | |||
| Age (years) | ||||||
| <2 | 5 | 0 | 3 | 0 | 0 | 2 |
| 2–9 | 93 | 8 | 21 | 22 | 3 | 47 |
| >10 | 52 | 3 | 8 | 14 | 2 | 28 |
| Sex | ||||||
| Castrated male | 88 | 7 | 21 | 23 | 3 | 41 |
| Non-castrated male | 14 | 0 | 5 | 0 | 1 | 8 |
| Spayed female | 44 | 3 | 6 | 12 | 1 | 25 |
| Unspayed female | 4 | 1 | 0 | 1 | 0 | 3 |
| Breed | ||||||
| Cross | 122 | 11 | 28 | 27 | 4 | 63 |
| Pure | 28 | 0 | 4 | 9 | 1 | 14 |
| Birman | 4 | 0 | 1 | 2 | 0 | 1 |
| British Shorthair | 4 | 0 | 0 | 4 | 0 | 0 |
| Chartreux | 3 | 0 | 0 | 0 | 1 | 2 |
| Exotic Shorthair | 1 | 0 | 0 | 1 | 0 | 0 |
| Maine Coon | 2 | 0 | 0 | 0 | 0 | 2 |
| Norwegian Forest Cat | 4 | 0 | 0 | 0 | 0 | 4 |
| Ragdoll Seal Point | 3 | 0 | 2 | 0 | 0 | 1 |
| Persian | 4 | 0 | 1 | 1 | 0 | 2 |
| Siamese | 3 | 0 | 0 | 1 | 0 | 2 |
| Siberian | 0 | 0 | 0 | 0 | 0 | 0 |
Pathology not related to the urinary tract
RT-qPCR = reverse transcription quantitative PCR; FLUTD = feline lower urinary tract disease; CKD = chronic kidney disease; AKD = acute kidney disease
Kidneys
Fifty renal tissue samples (40 from client-owned cats and 10 from cattery cats) were collected from cats necropsied at the Department of Veterinary Science (University of Turin, Turin, Italy). Sex, breed, age and available anamnestic information were gathered (Table 2). Five different pure breeds were included in addition to cross breeds. Of the 50 renal tissue samples, 50% were collected from male cats, 48% of which were castrated; 50% were from female cats, of which 16% were spayed. Both gross and histopathological investigations were performed in order to describe post-mortem features of the tested kidneys. The kidney samples were kept at –80°C until total RNA extraction.
Table 2.
Cat signalment, histopathological findings and test results of kidney samples analysed in this study
| n = 50 kidney samples | RT-qPCR positive | Histopathological
findings |
|||||
|---|---|---|---|---|---|---|---|
| Lymphoma | TIN | GN | Other* | ASL | |||
| Age (years) | |||||||
| <2 | 10 | 0 | 0 | 2 | 3 | 3 | 2 |
| 2–9 | 25 | 3 | 2 | 10 | 4 | 3 | 6 |
| >10 | 15 | 1 | 0 | 13 | 0 | 2 | 0 |
| Sex | |||||||
| Castrated male | 12 | 1 | 0 | 8 | 2 | 0 | 2 |
| Non-castrated male | 13 | 0 | 0 | 7 | 3 | 1 | 2 |
| Spayed female | 4 | 2 | 0 | 2 | 0 | 1 | 1 |
| Unspayed female | 21 | 1 | 2 | 8 | 2 | 6 | 3 |
| Breed | |||||||
| Cross | 42 | 3 | 2 | 22 | 5 | 8 | 5 |
| Pure | 8 | 1 | 0 | 3 | 2 | 0 | 3 |
| Birman | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| British Shorthair | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chartreux | 3 | 0 | 0 | 2 | 0 | 0 | 1 |
| Exotic Shorthair | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Maine Coon | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| Norwegian Forest Cat | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| Ragdoll Seal Point | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Persian | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Siamese | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Siberian | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Tubular calcification, hyperaemia, tubular steatosis and metastasis
RT-qPCR = reverse transcription quantitative PCR; TIN = tubulointerstitial nephritis; GN = granulomatous nephritis; ASL = absence of significant lesions
Biomolecular assays
Total RNA was extracted from 350 µl of urine or 1 mg of renal tissue using Trizol RNA isolation reagents (Life Technologies) following the manufacturer’s instructions, with a final elution in 30 µl of DNase/RNase-free water. The extracted RNA was tested by One Step RT-qPCR according to De Luca et al, 12 with minor modifications. The diagnostic assay target was represented by a fragment of the P/V/C gene (74 bp) and the original protocol was optimised for use with Superscript III Platinum One Step rt-qPCR System (Invitrogen). The reaction volume of 25 µl contained 5 µl of purified total RNA, 12.5 µl of Superscript III One-Step RT-qPCR Invitrogen Reaction Mix, a final concentration of 0.6 µM for each primer (FeMVrt forward 5′-GGGATCCAGAGGGTAACCT-3′ and FeMVrt Reverse 5′-CCGGCCATTAATCTCTGAA-3′), 0.225 µM of FeMVrt TaqMan Probe (5′-TATTCGAAAGCGATGATGATGAAAACCATTA-3′), 4 µM of MgSO4, 0.5 µl of Taq mix and nuclease-free water up to the final volume. The assay was carried out on a CFX96 Touch RT-PCR Detection System (Bio-Rad Laboratories), setting the following thermal conditions: one cycle of reverse transcription at 50°C for 30 mins, one cycle of PCR (initial activation step) at 95°C for 15 mins, followed by 45 cycles at 95°C for 30 s, 55°C for 40 s and 60°C for 30 s. In the present study, samples with a Cq value <40 were considered positive, while samples with a Cq value >40 were classified as negative.
FeMV isolation attempts
From each RT-qPCR-positive sample, FeMV isolation was carried out according to Woo et al. 2 Urine samples were diluted 1 to 10 into Minimum Essential Medium Earle (MEM-Earle; Biowest) supplemented with a 5% solution of penicillin (100,000 U/l) and streptomycin (100 mg/l) (Sigma Aldrich). The mixture was filtered through 450 nm disc filters (Millipore), further diluted 1 to 2 in MEM-Earle supplemented with 1 µg/ml of Polybrene Infection Reagent (Sigma Aldrich) and inoculated into a flask of 25 cm2 CRFK cells. After 1 h of incubation with gentle agitation at 37°C, the mixture was decanted and the CRFK cells were washed twice with PBS ×1. Seven millilitres of MEM-Earle supplemented with 0.1 µg/ml of L-1-tosylamide-2-phenylethylchloromethyl ketone-treated trypsin and with 2% heat-inactivated fetal bovine serum (Gibco; Thermofisher Scientific) and antibiotics were added directly on the monolayer. The CRFK cells were incubated at 37°C in a humidified atmosphere with 5% CO2 and observed daily for cytopathic effect by microscopy. Each passage consisted of 12 days of incubation and then the cell lysate was tested by the FeMV RT-qPCR protocol, 12 and further cell passages were performed until the cell lysate remained positive during biomolecular investigations. The mean Cq values of cell passages were compared in order to semi-quantitatively detect a potential increase in virus titre, indicative of efficient viral replication.
Morphological and histopathological investigations
Systemic post-mortem examination was performed on the cats, and kidneys were grossly evaluated. All the kidneys also underwent histopathological investigation in order to confirm macroscopic diagnosis. Cross-sections of the renal cortex, medulla and pelvis were collected, fixed in 10% buffered formalin solution, paraffin embedded, sectioned using a microtome, and stained with haematoxylin and eosin. The samples were observed by means of light microscopy. The histopathological evaluation was mainly focused on the presence of acute or chronic TIN with associated glomerulopathies, sclerosis, calcifications, granulomas or tumour lesions.
Urine and blood analysis
Urinalysis was performed in RT-qPCR-positive samples in order to try to correlate FeMV with urinary tract infections and subsequent altered urine composition, as reported by Yilmaz et al. 8 Samples were obtained by cystocentesis or spontaneous urination. Each sample was placed in a sterile universal tube and processed within 6 h of sampling. Urinalysis was performed using dipstick tests (Multistix-10-SG; Siemens Healthcare Diagnostics) read by a Clinitek Status analyser (Siemens Healthcare Diagnostics). Urine specific gravity (USG) was measured by a refractometer. Microscopic examination of the urine sediment on 10 field at ×100 magnification was also performed in order to identify casts, crystals or epithelial cells. Moreover, two blood samples were collected in positive cats: one at the time of urine sampling and the other 30 days later. Blood samples were placed in sterile tubes to evaluate blood creatinine (mg/dl) as a marker of renal function according to the International Renal Interest Society (IRIS) guidelines. 15
Phylogenetic analysis
The total RNA of PCR-positive samples was reverse-transcribed by a First Strand cDNA Synthesis Kit (Roche). The cDNA was subsequently used for the amplification of a fragment (401 bp) of the L gene using a double-step nested PCR, as described by Furuya et al. 5
An agarose gel electrophoresis allowed detection of nested PCR products with GelGreen Nucleic Acid Gel Stain (Biotium) using Uvitec technology for image capture. The amplified 401 bp fragment was sequenced after DNA purification from agarose gel, performed with a High Pure PCR Product Purification Kit (Roche). The Sanger sequencing was conducted using the BigDye Terminator v3.1 Cycle Sequencing Kit on a 3130 Genetic Analyzer (Applied Biosystems). Two software packages were used for sequence analysis of the detected basis (Sequencing Analysis) and for the manual correction (FinchTV, Geospiza), respectively. From the obtained sequences of the amplified products, the primer sequences were removed and the Clustal Omega software 16 allowed the alignment with 30 sequences of FeMV genotypes available in the public National Center for Biotechnology Information (NCBI) database and three sequences from the region of Lombardy, 17 kindly provided by Dr Stefania Lauzi (Università degli Studi di Milano Statale, Milan, Italy). For phylogenetic inference, the best nucleotide substitution model was estimated by jModelTest2. 18 The phylogenetic tree was constructed by MrBayes version 3.2.6, 19 with the following settings: two parallel runs of four chains each for 5,000,000 generations and TPM1uf+Ias nucleotide substitution model, as selected by jModelTest2, and a 10% burn-in to summarise parameter and tree log files. Convergence and effective sample size were controlled with Tracer v1.7.1. The tree was then visualised with Figtree v1.4.4.
Results
Biomolecular assays
Eleven of the 150 individual urine samples (from five client-owned cats and six cats from the cattery) and two urine pools were positive for FeMV RNA according to the cut-off level established by Lorusso et al (Table 1). 10 As for the kidney samples, 4/50 (from three client-owned cats and one cat from the cattery) were positive for FeMV RNA (Table 2).
FeMV isolation attempts
The entire panel of RT-PCR-positive samples were used for isolation attempts (Table 3); nevertheless, only the urine sample of ‘cat Totò’ showed a promising biomolecular outcome for isolation purposes. The first two cell passages, tested by RT-qPCR, had lower mean Cq values (23.48 and 24.22, respectively) than the starting urine sample (29.78). However, no FeMV-associated cytopathic effect was observed during 13 days of daily observations. Isolation attempts from the remaining positive samples (four kidney, 10 individual urine samples and two pooled urine samples with higher Cq values) were unsuccessful from the first cell passage.
Table 3.
Reverse transcription quantitative PCR (RT-qPCR) Cq values from the positive urine and kidney samples across isolation attempts
| Samples | Cq value RT-qPCR | Cq value of the first cell passage | Cq value of the second cell passage | Cq value of the third cell passage | Cq value of the fourth cell passage | Cq value of the fifth cell passage |
|---|---|---|---|---|---|---|
| Us1 (cat Totò) | 29.78 | 23.48 | 24.22 | 32.42 | 39.02 | >40 |
| Us2 | 34.20 | >40 | NP | NP | NP | NP |
| Us3 | 35.01 | >40 | NP | NP | NP | NP |
| Us4 | 35.72 | >40 | NP | NP | NP | NP |
| Us5 | 39.53 | >40 | NP | NP | NP | NP |
| Us6 | 39.98 | >40 | NP | NP | NP | NP |
| Us7 | 33.66 | >40 | NP | NP | NP | NP |
| Us8 | 35.21 | >40 | NP | NP | NP | NP |
| Us9 | 36.00 | >40 | NP | NP | NP | NP |
| Us10 | 39.73 | >40 | NP | NP | NP | NP |
| Us11 | 37.78 | >40 | NP | NP | NP | NP |
| Pooled Us1 | 30.83 | >40 | NP | NP | NP | NP |
| Pooled Us2 | 29.66 | >40 | NP | NP | NP | NP |
| Ks1 | 33.86 | >40 | NP | NP | NP | NP |
| Ks2 | 37.53 | >40 | NP | NP | NP | NP |
| Ks3 | 38.21 | >40 | NP | NP | NP | NP |
| Ks4 | 32.80 | >40 | NP | NP | NP | NP |
Us = individual urine samples; pooled Us = pooled urine samples; Ks = kidney samples; NP = not performed
Morphological and histopathological investigations
At gross examination, 20 cats showed no macroscopic lesions (40%), while 30 cats had different renal injuries (60%). In particular, pale, firm hypertrophic/hypotrophic kidneys, multifocal cortical granulomas and renal infarcts were detected.
At histological examination, TIN was detected in 50% of samples (Figure 1a), while granulomatous nephritis (Figure 1b) and tumour lesions were detected in 14% and 4% of examined cats, respectively. Finally, 16% of the kidney samples showed other lesions (tubular calcification, hyperaemia, tubular steatosis, metastasis) and 16% did not present any histopathological lesions (Table 2).
Figure 1.
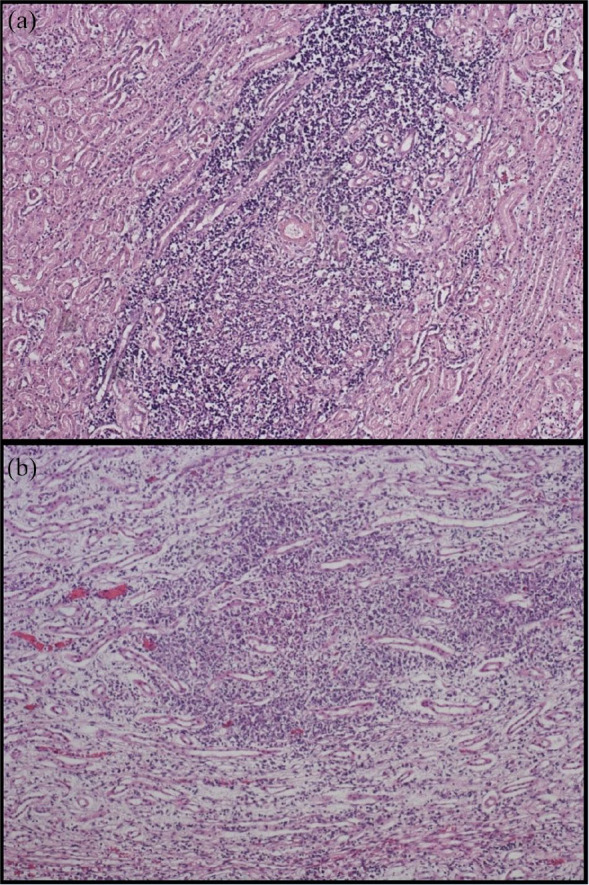
Cat kidney samples. (a) Multifocal moderate interstitial nephritis. Haematoxylin and eosin ×100. (b) Multifocal moderate granulomatous nephritis. Haematoxylin and eosin ×100
Of the four cats with RT-qPCR-positive kidneys, one cat showed no macroscopic lesions and one had pale firm hypotrophic kidneys, while two cats presented bilateral hypertrophy and multifocal cortical granulomas. Histopathological findings revealed the presence of TIN in 3/4 animals, while the last cat was affected by a systemic lymphoma with renal metastatic lesions.
Urine and blood analysis
Of the 11 cats with RT-qPCR-positive urine, nine samples were available for urinalysis (six cats from the cattery and three client-owned cats). In particular, eight cats had a normal urine pH (6–7), while one showed acidic urine (pH 5); six showed normal USG (⩾1035), while three presented lower USG (1014–1027) with proteinuria, and six showed urine sediment with crystals, epithelial cells and lipid droplets. Regarding blood creatinine levels, six cats were classified as IRIS stage 1 (<1.6 mg/dl), one cat was IRIS stage 3 and two cats were IRIS stage 4 (see Table S1 in the supplementary material).
Phylogenetic analysis
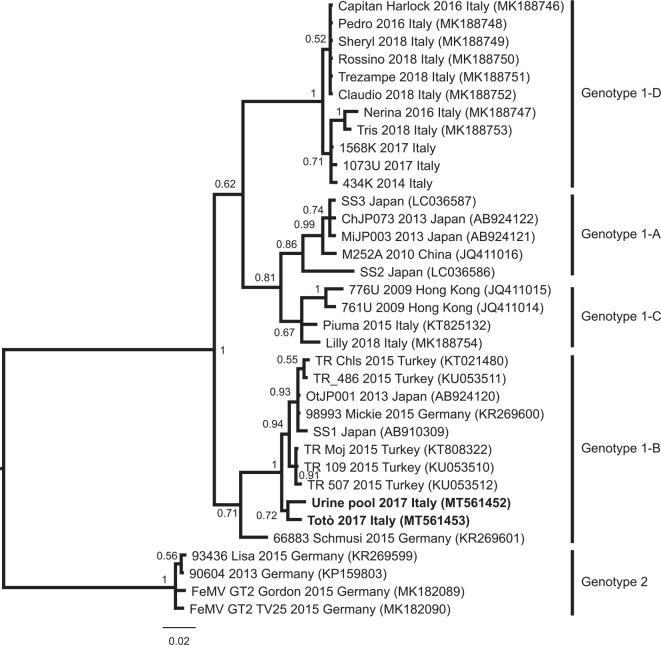
Across the entire set of RT-qPCR-positive samples, sequencing of a portion of the FeMV L gene (401 bp) was possible only for two. The first sequenced sample was obtained from the urine of the cat, Totò, collected by cystocentesis (GenBank accession number: MT561452); the second sequenced sample was obtained from a pool of urine collected from cats cohabiting with Totò (GenBank accession number MT561453). The percentage of sequence similarity between the two samples was calculated to be 97.9%. The obtained phylogenetic tree topology is similar to trees previously published,17,20 and the same FeMV genotypes 1 A–D and 2 were observed (Figure 2). The two sequences of this study belong to the clade of the isolates from the cats Schmusi (Germany) and Moj (Turkey), but they are phylogenetically distant from the first Italian isolate from the cat, Piuma, and all the other Italian strains.3,17 Focusing on the Italian samples, they are present in 3/4 sub-clusters (B, C and D). Sub-cluster D is composed of Italian samples only. Ours are the only Italian samples in sub-cluster B.
Figure 2.
Bayesian phylogenetic tree based on a 339 base pair-long fragment of the L gene of the feline morbillivirus. Node support is reported as posterior probability for the node. The strains characterised in this study are marked as bold. Genotype and sub-cluster of the clades is reported as described by Sieg et al 20 and Stranieri et al. 17 The novel clade described by Stranieri et al 17 has been named here as genotype 1-D, in order to continue the previous nomenclature
Discussion
FeMV represents a novel negative single-stranded RNA virus with little available information on the dynamics of infection, host susceptibility in the target population, and time and methods of excretion by infected animals. After its first discovery in Hong Kong, 2 FeMV was reported in the USA 9 and Japan, 4 with a urine prevalence varying from 3% to 23%. In Germany, urine samples collected from two cats with chronic kidney disease (CKD) (15 and 19 years old, respectively) scored positive for FeMV RNA. 21 A recent study carried out in Italy, in the Abruzzo region, detected FeMV RNA both in urine (15.1%) and blood samples (0.37%) collected from 264 cats. In the same study, 35/385 tissue samples (9.1%) collected from dead cats were positive for FeMV by RT-qPCR. 12
In 2015, FeMV was identified in Italy in the urine of a 15-year-old stray cat with suspected CKD, 10 and, recently, Stranieri et al 17 investigated the presence of FeMV in northern Italy, reporting a lower prevalence (1.2%) than previously published studies.
According to an in vitro study, the ability of the virus to cross species barriers and to infect humans seems to be remote. 21 However, new epidemiological, histopathological and phylogenetic investigations could improve field strain characterisation by increasing current knowledge about this new member of the genus morbillivirus. Our data show that at least three different FeMV sub-clusters are circulating in Italy, and the samples collected in our study do not share the same clade with the other Italian strains. Hence, multiple introductions of the virus may have occurred in Italy, but further analyses, especially at the genomic level, are needed.
In this scenario, this study focused on different diagnostic aspects of FeMV infection: biomolecular detection in biological samples, isolation attempts from RT-qPCR-positive matrices (both urine and kidney samples) and sequencing of target genes in order to reveal phylogenetic relationships.
Considering the group of cats involved in the present study, the rate of infection appears to be relatively low (7.3%), but quite similar to the prevalence data reported in the literature, particularly in stray cats, which are believed to be more easily infected than client-owned cats. 2 The case of ‘Totò’ and of the cohabitant cats from the private cattery (mostly negative for FeMV detection) leads to speculation that just a few animals can develop an effective infection and/or an effective excretion of FeMV. Moreover, according to the detected sequence data, a second FeMV variant, different from Totò’s sequence, was present in the cattery. Even though the other five cats cohabiting with Totò were positive on the biomolecular assays, it can be hypothesised that cat-to-cat transmission is not a common occurrence, or that FeMV infection could have a long incubation period before viraemia and viral shedding. 8
A serological investigation could clarify how many cohabiting cats have developed at least a humoral response against FeMV, but, unfortunately, no commercial serological ELISA is currently available for this purpose. Moreover, the development of an ‘in-house ELISA’ would require a considerable number of antibody-positive FeMV serum samples, which are not available. Concerning the clinical features of FeMV infection, Totò showed no signs of renal disease or FLUTD and was healthy for the entire study. Regarding positive urine samples, three cats were affected by CKD, while the remaining FeMV-positive cats were negative to renal disease and three cats presented proteinuria. These findings are in agreement with a recent study reporting an unclear correlation between chronic urinary disease and FeMV infection. 8
Regarding the possible co-presence of FeMV infection and TIN, TIN was detected in 3/4 RT-qPCR-positive kidney samples, and this feature could support the hypothesis of a co-existence of FeMV infection with renal disease.
However, the presence of FeMV viral RNA in one kidney affected by lymphoma could suggest that FeMV is possibly able to cause lesions other than TIN. Clinically, only one positive cat was affected by CKD. Moreover, in the present study, TIN was detected in 50% of all processed kidneys, and granulomatous nephritis was observed in 14%. Therefore, a clear relationship between the presence of FeMV infection and kidney diseases in cats was not observed.5,7,8,17,20
Concerning host susceptibility in cats, little information is currently available about the mechanism involved in early infection or feline viraemia. Therefore, further studies should clarify the real efficiency of FeMV transmission and its persistence in different environmental contexts.
Conclusions
This is the first study to report the FeMV genotype 1-B in northwestern Italy, suggesting that at least three different FeMV sub-clusters are circulating in Italy (sub-clusters B, C and D). Further studies are required to better investigate the potential role of FeMV in the pathogenesis of kidney disease.
Supplemental Material
Blood creatinine level of cats with RT-qPCR positive urine samples
Acknowledgments
We thank Mr Walter Fiore, owner of the cattery, for his support in the sample collection. We thank Dr Stefania Lauzi (Department of Veterinary Medicine, University of Milan, Milan, Italy) for kindly providing the Italian feline morbillivirus sequences identified by Stranieri et al.17
Footnotes
Accepted: 5 October 2020
Supplementary material: The following file is available online:
Table S1: Blood creatinine level of cats with RT-qPCR positive urine samples.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. The authors have performed this study on the basis of their freedom of research.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article
Ethical approval: This work involved the use of non-experimental animals only (including owned or unowned animals and data from prospective or retrospective studies). Established internationally recognised high standards (‘best practice’) of individual veterinary clinical patient care were followed. Ethical approval from a committee was therefore not necessarily required.
Informed consent: Informed consent (either verbal or written) was obtained from the owner or legal custodian of all animal(s) described in this work (either experimental or non-experimental animals) for the procedure(s) undertaken (either prospective or retrospective studies). No animals or humans are identifiable within this publication, and therefore additional informed consent for publication was not required.
ORCID iD: Elena Colombino  https://orcid.org/0000-0002-6371-2000
https://orcid.org/0000-0002-6371-2000
References
- 1. Tatsuo H, Yanagi Y. The morbillivirus receptor SLAM (CD150). Microbiol Immunol 2002; 46: 135–142. [DOI] [PubMed] [Google Scholar]
- 2. Woo PCY, Lau SKP, Wong BHL, et al. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc Natl Acad Sci U S A 2012; 109: 5435–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marcacci M, De Luca E, Zaccaria G, et al. Genome characterization of feline morbillivirus from Italy. J Virol Methods 2016; 234: 160–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sakaguchi S, Nakagawa S, Yoshikawa R, et al. Genetic diversity of feline morbilliviruses isolated in Japan. J Gen Virol 2014; 95: 1464–1468. [DOI] [PubMed] [Google Scholar]
- 5. Furuya T, Sassa Y, Omatsu T, et al. Existence of feline morbillivirus infection in Japanese cat populations. Arch Virol 2014; 159: 371–373. [DOI] [PubMed] [Google Scholar]
- 6. Darold GM, Alfieri AA, Muraro LS, et al. First report of feline morbillivirus in South America. Arch Virol 2017; 162: 469–475. [DOI] [PubMed] [Google Scholar]
- 7. McCallum KE, Stubbs S, Hope N, et al. Detection and seroprevalence of morbillivirus and other paramyxoviruses in geriatric cats with and without evidence of azotemic chronic kidney disease. J Vet Intern Med 2018; 32: 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yilmaz H, Tekelioglu BK, Gurel A, et al. Frequency, clinicopathological features and phylogenetic analysis of feline morbillivirus in cats in Istanbul, Turkey. J Feline Med Surg 2017; 19: 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharp CR, Nambulli S, Acciardo AS, et al. Chronic infection of domestic cats with feline morbillivirus, united states. Emerg Infect Dis 2016; 22: 760–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lorusso A, Di Tommaso M, Di Felice E, et al. Prima segnalazione di morbillivirus felino in Europa. Vet Ital 2015; 51: 235–237. [DOI] [PubMed] [Google Scholar]
- 11. Park ES, Suzuki M, Kimura M, et al. Epidemiological and pathological study of feline morbillivirus infection in domestic cats in Japan. BMC Vet Res 2016; 12: 1–11. DOI: 10.1186/s12917-016-0853-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Luca E, Crisi PE, Di Domenico M, et al. A real-time RT-PCR assay for molecular identification and quantitation of feline morbillivirus RNA from biological specimens. J Virol Methods 2018; 258: 24–28. [DOI] [PubMed] [Google Scholar]
- 13. Koide R, Sakaguchi S, Miyazawa T. Basic biological characterization of feline morbillivirus. J Vet Med Sci 2015; 77: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donato G, De Luca E, Crisi PE, et al. Isolation and genome sequences of two feline morbillivirus genotype 1 strains from Italy. Vet Ital 2019; 55: 179–182. [DOI] [PubMed] [Google Scholar]
- 15. IRIS. Staging of chronic kidney disease. http://www.iris-kidney.com/guidelines/staging.html (2019, accessed October 15, 2020).
- 16. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539. DOI: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stranieri A, Lauzi S, Dallari A, et al. Feline morbillivirus in Northern Italy: prevalence in urine and kidneys with and without renal disease. Vet Microbiol 2019; 233: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darriba D, Taboada GL, Doallo R, et al. jModelTest 2: more models, new heuristics and high-performance computing Europe PMC Funders Group. Nat Methods 2012; 9: 772. DOI: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ronquist F, Teslenko M, Van Der Mark P, et al. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012; 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sieg M, Heenemann K, Rückner A, et al. Discovery of new feline paramyxoviruses in domestic cats with chronic kidney disease. Virus Genes 2015; 51: 294–297. [DOI] [PubMed] [Google Scholar]
- 21. Sakaguchi S, Koide R, Miyazawa T. In vitro host range of feline morbillivirus. J Vet Med Sci 2015; 77: 1485–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blood creatinine level of cats with RT-qPCR positive urine samples