Abstract
An unexpectedly high proportion of TGA nonsense mutations was obtained in a collection of chemically induced mutations in the spoIIR locus of Bacillus subtilis. Of 11 different mutations obtained, TGA mutations were found in four codons, whereas only three codons yielded missense mutations. Six suppressors of the TGA mutations were isolated, and five of the suppressing mutations were mapped to the prfB gene encoding protein release factor 2. These are the first mutations shown to map to the B. subtilis prfB locus. The sequence of the prfB gene was completed, and two revisions of the published sequence were made. The five prfB mutations also resulted in suppression of the catA86-TGA mutation to between 19 and 54% of the expression of catA86+, compared to the readthrough level of 6% in the prfB+ strain. N-terminal sequencing of suppressed catA86-TGA-specified protein demonstrated that the amino acid inserted at UGA because of the prfB1 mutations was tryptophan.
The genetic code shows small but fundamental differences in various organisms, suggesting that the code has evolved to meet special requirements of the host (reference 12 and references therein). Some of the variability of the code is strikingly evident in the codon UGA (29). UGA is one of the three translation termination codons. In Escherichia coli, Salmonella typhimurium, and Bacillus subtilis, UGA is encountered less frequently than UAA and more frequently than UAG (4, 13). Although UGA is a termination codon, in E. coli and S. typhimurium UGA can be decoded at very low frequency; when this occurs, the amino acid inserted is tryptophan (29). Thus, UGA is viewed as a “leaky” termination codon (34). The extent of readthrough of UGA in wild-type E. coli appears to be on the order of 10−5 to 10−2 per termination at UGA, and the readthrough efficiency of UGA seems to depend on the context in which UGA is found (13, 29). Readthrough as Trp is distinct from the special case of UGA coding for selenocysteine, where a codon context of 40 nucleotides is involved (14).
Although UGA functions as a stop codon in B. subtilis, the efficiency of UGA readthrough is quite high. With a catA86 reporter gene with UGA inserted into two different sites, the efficiency of UGA readthrough was approximately 6% (26). Similar UGA readthrough values were obtained with Staphylococcus aureus as host (25, 26). Thus, in these very different gram-positive species, UGA is substantially leakier than is observed in the Enterobacteriaceae. N-terminal sequencing of the protein specified by catA86 containing UGA at codon 7 demonstrated that tryptophan was the inserted amino acid. Mycoplasma are “wall-less” gram-positive bacteria, and in many Mycoplasma species UGA is not a termination codon but rather directly encodes tryptophan (4, 7, 18). The very high level of readthrough of UGA in B. subtilis and S. aureus suggested that it might be difficult to obtain TGA nonsense mutations in most coding sequences within these organisms.
In the present study, we demonstrate the isolation of several mutations in spoIIR which cause a defect in sporulation and result from a change of a sense codon to UGA. The sporulation phenotype associated with the TGA mutations allowed the isolation of second-site mutations that suppress TGA nonsense mutations. We demonstrate here that five of six such UGA suppressors result from mutations in the structural gene (prfB) for release factor 2 (RF2).
MATERIALS AND METHODS
Media.
B. subtilis was grown in modified Schaeffer’s sporulation medium (2 × SG) without glucose and on Schaeffer’s sporulation agar (33). Addition of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (100 μg/ml), chloramphenicol (4 μg/ml), neomycin (3 μg/ml), erythromycin (10 μg/ml), and lincomycin (1 μg/ml) (resistances to the latter two are encoded by the erm gene and the combination is referred to as Ermr) or of spectinomycin (50 μg/ml) was done as required.
Strains and plasmids.
B. subtilis 168 strain BR151, trpC2 metB10 lys-3, was used as the parent strain in all experiments. E. coli DH5α (GIBCO/BRL) was used to maintain plasmids. pPL708C2 contains a constitutively expressed version of catA86, a gene that codes for chloramphenicol acetyltransferase (CAT). pPL708C2 UGA-7 contains TGA as replacement for cat codon 7, which is Glu (GAA) in the wild-type gene (26). Both plasmids were transformed into strain BR151 and derivatives of BR151 containing prfB1 mutations to generate strains containing either the wild-type cat gene (pPL708C2) or the mutant cat gene (pPL708C2 UGA-7).
Isolation of prfB mutants.
prfB1 was isolated from an ethyl methanesulfonate (EMS)-mutagenized (9) culture of MLK940. MLK940 contained spoIIR152 with a TGA mutation and a cotE-lacZ fusion (to indicate ςE activity) linked to a Camr determinant (38). The prfB1 mutant was isolated and exhibited a LacZ+ phenotype on sporulation agar plates containing X-Gal. The prfB2 suppressor strain and the prfB3, -4, and -5 suppressor strains were isolated as Spo+ colonies from UV-mutagenized derivatives of BR151 containing spoIIR151 and spoIIR74, respectively.
Cloning of the gene encoding prfB1.
A library of Camr Tn10 insertions (31) made in BR151 was transduced into MLK984 (a BR151 derivative containing prfB1) by using transduction phage PBS-1 (17), and colonies that were Camr and Spo+ at 42°C were isolated. DNA from each Camr and Spo+ clone was isolated and transformed back into MLK984, and linkage of the Tn10s was determined. One Tn10 that was 65% cotransformed to prfB1 was cloned, along with its flanking chromosomal DNA, to make pMLK252. This plasmid did not contain prfB. It was used as a probe for a lambda DNA library. A lambda clone that hybridized to pMLK252 and corrected the prfB1 mutation was subcloned to yield pMLK262, carrying the 800-bp SacI-HindIII fragment of the insert-phage junction ligated with SacI-HindIII-digested pBluescript KS (Stratagene). pMLK262 corrected the prfB1 mutation. The adjacent 900-bp EcoRI-HindIII fragment was cloned in pBluescript KS to make pMLK266.
PCR amplification and sequencing.
Both the spoIIR and prfB mutant DNAs were PCR amplified with a GeneAmp kit (Perkin-Elmer Cetus). The spoIIR gene primers were 5′CACCCTGCACGTTTATCCCAGGCTCTCC3′ and 5′GCAGTTGATAAAACATCCGTTCACCCCG3′, and the prfB primers were 5′GTGGTTGATATCGGACGAAATGCCC3′ and 5′GCAGCAGTGAAATCAAGGATATAAG3′. One primer in each reaction was phosphorylated with dATP and T4 polynucleotide kinase, and after amplification, the phosphorylated strand was degraded with lambda exonuclease (5). The remaining strand was sequenced with Sequenase 2.0 (Amersham) according to the manufacturer’s instructions.
Other methods.
Sporulation frequency was determined as heat-resistant spores per milliliter of culture 16 h after the initiation of sporulation (27). CAT assays were performed by the colorimetric procedure of Shaw (37). Protein was assayed according to the method of Bradford (8).
CAT UGA-7 protein was affinity purified with chloramphenicol caproate agarose (Sigma) and eluted with 10 mM chloramphenicol. The purified protein was subjected to automated N-terminal sequencing by Edman degradation. Typically, sequencing was allowed to proceed through 15 cycles (15 amino acid residues).
Nucleotide sequence accession number.
The complete nucleotide sequence of the prfB gene is available from GenBank under accession no. AF013188.
RESULTS AND DISCUSSION
TGA nonsense mutations in spoIIR.
The spoIIR gene is required for the coordination of transcriptional events during sporulation. Sporulation is a developmental process requiring the concerted efforts of two cells, the mother cell and the forespore (also called the prespore). These two cells express separate genetic programs that are sequentially controlled by a set of sigma factors that are coordinated by cell-cell signaling (15). The SpoIIR protein is produced by the forespore with sigma factor F and initiates transcriptional events in the mother cell by activating ςE (20, 24).
Previously, we obtained the four mutations that defined the spoIIR locus by EMS mutagenesis of an appropriately marked strain (20). In order to obtain further mutations in spoIIR, we utilized directed mutagenesis of transforming DNA (3). We obtained two spoIIR mutants by nitrous acid mutagenesis and nine by methoxyamine mutagenesis (11). DNA sequence analysis indicated that we had obtained, in total, 10 distinct point mutations in spoIIR, of which 6 were nonsense mutations, including TGA mutations in four separate codons (Table 1). Three of the mutations obtained by the directed mutagenesis were identical to three of the mutations obtained by mutagenesis of bacteria with EMS. The SpoIIR protein is predicted to contain 224 residues, including a 23-residue leader sequence that is thought to be cleaved during protein secretion (20). In only 3 of 224 residues were missense mutations obtained, and in 2 of these residues mutations were obtained more than once (Table 1). In contrast, of the four codons that could yield TGA by a single-base transition, three did so; the fourth codon, W218, encoded the residue that is only seven residues from the C terminus, and a nonsense mutation there may have no phenotype. An additional TGA mutation resulted from a transversion of GGA which encoded G144. Of the nine codons that could yield TAA or TAG by a single-base transition, two did so. There were thus two surprises from this analysis: the disproportionate number of nonsense mutations and the occurrence of TGA mutations. The disproportionate number of isolated nonsense mutations may indicate that most amino acid substitutions in spoIIR cause no phenotypic change. We are aware of only one other report of a TGA (opal) nonsense mutation in B. subtilis (22).
TABLE 1.
Description of spoIIR mutants
| Allele | Amino acid change | Base change | Sporulation (% of parental spo+ strain BR151) | Mutagenc |
|---|---|---|---|---|
| spoIIRΔneo | Insertion at codon 111 | <2.9 × 10−6 | ||
| spoIIR11 | Deletion of TA | 1.4 × 10−1 | NA | |
| spoIIR29 | Gln→ochre | CAA→TAA | 4.0 × 10−3 | MHA |
| spoIIR40 | Gln→amber | CAG→TAG | 7.7 × 10−3 | MHA |
| spoIIR44 | Arg→Cys | CGT→TGT | 4.8 × 10−4 | MHA |
| spoIIR74 | Trp→opal | TGG→TGA | 2.9 × 10−1 | MHA |
| spoIIR144 | Gly→opal | GGA→TGA | 6.1 × 10−3 | NA |
| spoIIR151 | Trp→opal | TGG→TGA | 2.3 × 10−4 | MHA |
| spoIIR152 (spoIIR4a) | Trp→opal | TGG→TGA | 6.7 × 10−3 | EMS; MHAb |
| spoIIR153 (spoIIR3a) | Cys→Tyr | TGT→TAT | 6.1 × 10−3 | EMS; MHAb |
| spoIIR157L (spoIIR1a) | Pro→Leu | CCG→CTG | 3.5 × 10−1 | EMS |
| spoIIR157S (spoIIR2a) | Pro→Ser | CCG→TCG | 7.0 × 10−1 | EMS; MHAb |
Original allele name (20).
Same mutant allele was obtained by two different mutagenesis protocols (see text).
NA, nitrous acid; MHA, methoxyamine.
Previously, we had supposed that high readthrough levels of UGA nonsense codons in wild-type B. subtilis curtailed the ability to isolate such mutants (26). For example, there is sufficient readthrough of the catA86 UGA-7 mutation that the mutant catA86 gene confers Camr to the bacterium. However, the wild-type level of UGA readthrough in the spoIIR mutants must not be sufficient to produce enough SpoIIR to support sporulation, and thus we were able to identify several mutants with TGA nonsense mutations of this gene.
Isolation and characterization of UGA suppressor mutants.
In order to determine how SpoIIR acts, we attempted to identify second-site suppressor mutants of spoIIR. To do so, we screened EMS-mutagenized (9) colonies of our original four spoIIR mutants (20) for the activation of ςE by using a lacZ fusion to a ςE-directed gene, cotE, that was present in each of the strains. After mutagenesis, the bacteria were plated on Schaeffer’s sporulation agar containing X-Gal, and colonies exhibiting β-galactosidase activity were isolated. Only one of the spoIIR mutants, spoIIR4 (20), produced a β-galactosidase-positive colony that was neither a revertant nor an up mutant of the endogenous β-galactosidase gene. The spoIIR4 mutant had a TGA mutation at codon 152 (Table 1) (this mutation is hereafter referred to as spoIIR152). As shown below, the suppressor mutation was found to be a UGA suppressor and to map in prfB. For clarity, the name prfB is used here and throughout, even though the evidence placing the mutation in prfB is presented later. This suppressor mutation has been named prfB1. ςE transcriptional activity during sporulation was increased in the presence of prfB1 over that of the spoIIR152 strain, although it remained approximately one-third that of the spoIIR+ strain (Fig. 1). The presence of the suppressor mutation also increased the sporulation frequency of the spoIIR152 mutant approximately 450-fold (Table 2). We obtained no suppressors of the spoIIR152 mutation that did not need spoIIR for ςE activation.
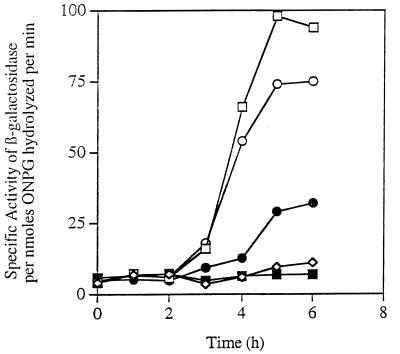
FIG. 1.
Restoration of ςE activity to spoIIR bacteria by prfB1. β-Galactosidase activity from the ςE-controlled p1 promoter of cotE fused to lacZ in the following BR151 derivatives: □, spoIIR+; ■, spoIIR152; •, spoIIR152 prfB1; ○, spoIIR+ prfB1; and ◊, endogenous β-galactosidase activity of the parent strain BR151 that does not contain the lacZ fusion. ONPG, o-nitrophenyl-β-d-galactopyranoside.
TABLE 2.
Sporulation of suppressed spoIIR mutants at different temperatures
| Relevant genotypea | Spores per mlb
|
Relative increase in spores per ml
|
||
|---|---|---|---|---|
| 37°C | 42°C | 37°C | 42°C | |
| spoIIR152 | 6.2 × 103 | 1.5 × 103 | ||
| spoIIR152 prfB1 | 2.7 × 106 | 1.4 × 102 | 428 | −11 |
| spoIIR152 prfB2 | 2.8 × 106 | 1.2 × 106 | 444 | 800 |
| spoIIR152 prfB3 | 9.8 × 105 | 1.1 × 105 | 156 | 73 |
| spoIIR152 prfB4 | 1.8 × 106 | 4.0 × 105 | 290 | 267 |
| spoIIR152 prfB5 | 4.3 × 105 | 1.8 × 105 | 69 | 120 |
| spoIIR151 | 4.2 × 104 | 1.2 × 104 | ||
| spoIIR151 prfB2 | 1.3 × 107 | 2.5 × 106 | 309 | 208 |
| spoIIR74 | 4.0 × 105 | 1.3 × 105 | ||
| spoIIR74 prfB3 | 5.9 ×107 | 1.3 × 107 | 148 | 100 |
| spoIIR74 prfB4 | 1.4 × 108 | 2.6 × 107 | 350 | 200 |
| spoIIR74 prfB5 | 3.0 × 107 | 1.7 × 107 | 75 | 131 |
Strains are isogenic sets derived from B. subtilis 168 strain BR151.
Spores per milliliter were determined as heat-resistant spores per milliliter of culture. A strain with an insertion mutation in spoIIR produced less than 10 spores per ml.
The prfB1 mutation on its own impaired sporulation, and interestingly, this effect was much greater at higher temperatures. prfB1 bacteria produced 8.2 × 106 heat-resistant spores per ml of culture at 37°C and only 180 spores per ml at 42°C, whereas the wild-type strain produced about 2 × 108 spores per ml at both temperatures (Table 3). The sporulation of prfB1 bacteria appears to be blocked at an early stage of sporulation. There was no detectable formation of the asymmetric septum, one of the earliest sporulation-specific morphological markers (32). In contrast to the sporulation phenotypes, there was only a slight effect on the growth rate of prfB1 at 42°C, with a growth rate (mass doublings/hour) of 1.34 for the prfB1 strain compared to 1.52 for the prfB+ parent. At 37°C, the growth rates were similar for the two strains, 1.32 for the prfB1 strain and 1.31 for the prfB+ parent.
TABLE 3.
Sporulation of prfB mutants at different temperatures
| Relevant genotypea | Spores per mlb
|
% of parental strain BR151
|
||
|---|---|---|---|---|
| 37°C | 42°C | 37°C | 42°C | |
| prfB+ | 2.0 × 108 | 2.4 × 108 | ||
| prfB1 | 8.2 × 106 | 1.8 × 102 | 5.5 | 0.00014 |
| prfB2 | 5.6 × 107 | 5.7 × 107 | 24 | 33 |
| prfB3 | 1.2 × 108 | 8.0 × 107 | 60 | 33 |
| prfB4 | 1.9 × 108 | 5.7 × 107 | 95 | 24 |
| prfB5 | 1.6 × 108 | 1.2 × 108 | 80 | 50 |
Strains are an isogenic set derived from B. subtilis 168 strain BR151.
Spores per milliliter were determined as heat-resistant spores per milliliter of culture.
The spoIIR152 mutant and strains containing TGA mutations affecting codons 74 and 151 were used in a second type of screen in which suppressors that exhibited a Spo+ phenotype after UV mutagenesis were isolated. In this screen, the spoIIR152 mutant did not yield any Spo+ colonies that contained extragenic suppressors, although five intragenic revertants were isolated from the 5 × 107 colonies that were screened. With spoIIR151, we isolated one Spo+ extragenic suppressor, and with spoIIR74 we isolated four. The spoIIR151 suppressor and three of the four spoIIR74 suppressors were found to map in prfB (data not shown). The location of the fourth spoIIR74 suppressor was not determined. We have named the spoIIR151 suppressor mutation prfB2 and the three spoIIR74 mutations prfB3, -4, and -5. In the presence of these suppressors, sporulation of the spoIIR mutants was increased 70- to 450-fold (Table 2). Thus, spoIIR152 strains with the new prfB mutations sporulated at a frequency similar to or less than that of the spoIIR152 strain with prfB1. This indicates that the Spo+ phenotype used to isolate these suppressors did not closely reflect the activity of the suppressors and also indicates the fact that spoIIR74 and spoIIR151 were leakier than spoIIR152. None of these Spo+ suppressors exhibited the strong Ts− sporulation phenotype exhibited by prfB1 (Table 2 and 3).
Mapping and cloning of the gene affected by prfB1.
By utilizing the Ts− sporulation phenotype of prfB1, a Tn10 insert was identified that was 65% linked to prfB1 by transformation. The Tn10 and its flanking chromosomal DNA were cloned and used to probe the ordered YAC library of B. subtilis (6). The DNA hybridized to one YAC clone, 10-119, which carries DNA of the 305° region of the chromosome.
The wild-type gene was isolated from a B. subtilis lambda library by using the Tn10-flanking region as a probe. A subclone (pMLK262, from a lambda clone) rescued the prfB1 phenotype, and the insert in pMLK262 was sequenced. Comparison of the resulting sequence with those in GenBank (1, 16) indicated that pMLK262 carried a region internal to the prfB gene encoding RF2. This region was originally cloned and sequenced by Sadaie et al. (35) as part of the operon carrying the B. subtilis secA gene (GenBank accession no. D90218). Pel et al. (30) later recognized that the gene encoded prfB and that it contained a region similar to the frameshift site found in most prfB genes, thus extending the predicted open reading frame 5′ to include codons for an additional 27 amino acids. The published sequence did not include the 3′ end of the prfB gene. We sequenced the complete prfB gene and found that it is followed by a sequence similar to that of Rho-independent terminators, indicating this is most likely the end of the operon. Two parts of the sequence that differed from the published sequence were found. One is an extra G after base pair 3137 (following the numbering of Sadaie et al. [35]) that extends the open reading frame 5′ to include 12 more codons, extending the encoded protein to a length similar to that of E. coli, S. typhimurium, Streptomyces coelicolor, and Haemophilus influenzae (Fig. 2). The new suggested AUG initiation codon is preceded by a good ribosome-binding site with a ΔG for binding equal to −17.2 kcal/mol. The other difference is a loss of a G (residue 4176 [35]), changing the reading frame such that the downstream encoded protein is now also similar to other RF2 sequences (Fig. 2).
FIG. 2.
Alignment of RF2 amino acid sequences. The amino acid sequences of RF2 from B. subtilis (Bs) (this work and reference 35), S. coelicolor (Sc) (28), E. coli (Ec) (10), S. typhimurium (St) (21), and H. influenzae (Hi) (GenBank accession no. P43918) are shown. Amino acids that are identical in all five sequences are shown in bold type; the overall identity is 35%. The asterisks with numbers located above the B. subtilis sequences identify the amino acids changed in prfB1, -3, and -5. The frameshift region found in all of the sequences except that of S. coelicolor is marked with a dashed line above the B. subtilis amino acid sequence.
Sequencing of the prfB mutants.
To determine the nature of the prfB mutations, the mutant prfB genes were amplified by PCR and the products were sequenced. The prfB1 mutant contained a transversion of T to A, changing Tyr (TAT) 325 to Asn (AAT). Both prfB2 and prfB4 affected the proposed ribosome-binding site, with prfB2 containing a T-to-C transition at −13 (with +1 as the A of the ATG translation initiation codon) and prfB4 containing a G-to-A transition at −12. The prfB3 mutant contained a G-to-A transition that altered the second amino acid residue of the protein, changing it from Glu (GAA) to Lys (AAA). The prfB5 mutant contained a G-to-C transversion, changing residue 21 from Arg (AGG) to Thr (ACG). Thus, three of the mutations likely impair the formation of RF2, prfB2, and prfB4 by weakening the ribosome-binding site and prfB5 by affecting frame shifting. Such impaired RF2 formation would be expected to increase UGA suppression (14).
Nonsense suppression during vegetative growth of the prfB mutants.
To determine if these suppressors could act under conditions of vegetative growth as well as to quantify misreading by the prfB mutants, we used a TGA nonsense mutation that had been constructed in codon 7 of the CAT gene catA86. This mutant, named UGA-7, is carried on pPL708C2 UGA-7 (26) and was introduced into the prfB1 strain MLK1013 and its parent strain BR151. In the presence of prfB1, the level of CAT activity from UGA-7-containing bacteria grown at 37°C was increased 6.6-fold over that from the isogenic prfB+ strain (Table 4). Analysis of the other prfB suppressor mutants indicated that they too increased the level of UGA readthrough from 3.8- to 7.1-fold. This analysis was complicated by an unexpected effect of some of the suppressor mutations on the constitutive expression of wild-type catA86 from pPL708C2. When this is taken into consideration, the prfB mutants increased CAT activity to 19 to 54% of that of the wild type from the readthrough level of 6% of that of the wild type. These results indicate that the suppressors were not acting specifically during sporulation and could also suppress TGA mutations during vegetative growth. The high level of UGA readthrough did not affect the growth rate, even though UGA is used as a stop codon for about 20% of B. subtilis genes (36). We also tested the effects of the prfB1 mutation on readthrough of a UAA codon at position 7 and found no suppression (data not shown), indicating that suppression is limited to UGA codons. To explore the nature of this suppression, we sequenced purified intact protein (UGA-7) from the prfB1 pPL708C2 UGA-7 strain. The sole residue at position 7 was identified as tryptophan, indicating that tRNATrp was decoding the UGA codon in the suppressor mutant. The three spoIIR UGA mutants that were analyzed (Table 2) were codon changes from UGG, encoding Trp. Thus, if suppression at these UGA codons occurs also by insertions of tryptophan, it would lead not only to the elongation of the protein to its full length but also to the synthesis of a wild-type SpoIIR protein.
TABLE 4.
Suppression of catA86 UGA-7 by prfB mutants
| prfBa mutation | CAT activityb
|
% of suppression | |
|---|---|---|---|
| C2 (WT)c | UGA-7d | ||
| prfB+ | 10.8 | 0.7 | 6 |
| prfB1 | 23.9 | 4.6 | 19 |
| prfB2 | 9.5 | 5.0 | 53 |
| prfB3 | 14.7 | 2.9 | 20 |
| prfB4 | 14.5 | 2.9 | 20 |
| prfB5 | 5.1 | 2.7 | 54 |
prfB+ is in strain BR151; prfB mutations are in derivatives of B. subtilis 168 strain BR151.
Specific activity of CAT is expressed as micromoles per minute per milligram of protein. Data are the average of four experiments, with specific activities varying ±10 to 15% between experiments. In each experiment, the relative level of expression between the different samples remained the same.
Strain containing pPL708C2 (see Materials and Methods).
Strain containing pPL708C2 UGA-7.
The nucleotide 3′ to the UGA triplet affects the extent of readthrough in E. coli in the order A>G>C>U (23) and may explain the leakiness of catA86-UGA7 where the sequence is UGAA (2). The corresponding sequences for spoIIR74, spoIIR151, and spoIIR152 are UGAG, UGAU, and UGAU, respectively; the position 3′ to UGA might also explain, at least partly, why the spoIIR74 mutant exhibits a leakier sporulation phenotype than the spoIIR151 and spoIIR152 mutants as well as the relatively tight phenotype the spoIIR mutants display compared to catA86-UGA7.
To our knowledge, these are the first examples of mutations mapping in a release factor gene in gram-positive bacteria. Identification of UGA-suppressor mutations mapping in prfB provides the first experimental support for the supposition that prfB codes for RF2. The finding that the majority of the UGA-suppressor mutations obtained (five of six) mapped in prfB and not in a trn gene was surprising. It contrasts with results for Enterobacteriaceae when nearly all nonsense suppressors are altered tRNAs (13, 19). It suggests subtle differences in translational machinery between B. subtilis and Enterobacteriaceae.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI09111 (to M.L.K.), GM42925 (to P.S.L.), and GM43577 (to P.J.P.) from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ambulos N P, Mongkolsuk S, Lovett P S. A transcription termination signal immediately precedes the coding sequence for the chloramphenicol-inducible plasmid gene cat-86. Mol Gen Genet. 1985;199:70–75. doi: 10.1007/BF00327512. [DOI] [PubMed] [Google Scholar]
- 3.Anagnostopoulos C, Crawford I P. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc Natl Acad Sci USA. 1961;47:378–390. doi: 10.1073/pnas.47.3.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson S G E, Kurland C G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 6.Azevado V, Alvarez E, Zumstein E, Damiani G, Sgaramella V, Ehrlich S D, Serror P. An ordered collection of Bacillus subtilis DNA segments cloned in yeast artificial chromosomes. Proc Natl Acad Sci USA. 1993;90:6047–6051. doi: 10.1073/pnas.90.13.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard A. Ureaplasma urealyticum urease genes; use of a UGA tryptophan codon. Mol Microbiol. 1990;4:669–670. doi: 10.1111/j.1365-2958.1990.tb00636.x. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 9.Corran J. The induction of supersuppressor mutants of Bacillus subtilis by ethyl methanesulphonate and the posttreatment modification of mutation yield. Mol Gen Genet. 1968;103:42–57. doi: 10.1007/BF00271156. [DOI] [PubMed] [Google Scholar]
- 10.Craigen W J, Cook R G, Tate W P, Caskey C T. Bacterial peptide chain release factors: conserved primary structure and possible regulation of release factor 2. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley and Sons; 1990. pp. 27–74. [Google Scholar]
- 12.Di Giulio M. The origin of the genetic code. Trends Biochem Sci. 1997;22:49–50. doi: 10.1016/s0968-0004(97)84911-0. [DOI] [PubMed] [Google Scholar]
- 13.Eggertsson G, Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988;52:354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka H, Schoulaker-Schwarz R. Suppression of termination codons. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Cellular and molecular biology. 2nd ed. I. Washington, D.C: American Society for Microbiology; 1996. pp. 909–921. [Google Scholar]
- 15.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gish W, States D J. Identification of protein coding regions by database search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 17.Hoch J A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 18.Inamine J M, Ho K-C, Loechel S, Hu P-C. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J Bacteriol. 1990;172:504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston H M, Roth J R. UGA suppressor that maps within a cluster of ribosomal protein genes. J Bacteriol. 1980;144:300–305. doi: 10.1128/jb.144.1.300-305.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karow M L, Glaser P, Piggot P J. Identification of a gene, spoIIR, that links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawakami K, Nakamura Y. Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor 2. Proc Natl Acad Sci USA. 1990;87:8432–8436. doi: 10.1073/pnas.87.21.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L, Siranosian K J, Grossman A D, Dubnau D. Sequence and properties of mecA, a negative regulator of genetic competence in Bacillus subtilis. Mol Microbiol. 1993;9:365–373. doi: 10.1111/j.1365-2958.1993.tb01697.x. [DOI] [PubMed] [Google Scholar]
- 23.Kopelowitz J, Hampe C, Goldman R, Reches M, Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992;225:261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- 24.Londono-Vallejo A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 25.Lovett, P. S. Unpublished results.
- 26.Lovett P S, Ambulos N P, Jr, Mulbry W, Noguchi N, Rogers E J. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J Bacteriol. 1991;173:1810–1812. doi: 10.1128/jb.173.5.1810-1812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biology methods for Bacillus. Chichester, England: John Wiley and Sons; 1990. pp. 391–450. [Google Scholar]
- 28.Ogawara H, Urabe H, Ohtaki R, Nakamura Y. Properties of peptide chain release factor 2 from Streptomyces coelicolor A3(2): conserved primary structure but no frameshift regulation. J Bacteriol. 1995;177:5342–5345. doi: 10.1128/jb.177.18.5342-5345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker J. Errors and alternative in reading the universal genetic code. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pel H J, Rep M, Grivell L A. Sequence comparison of new prokaryotic and mitochondrial members of the polypeptide chain release factor family predicts a five-domain model for release factor structure. Nucleic Acids Res. 1992;20:4423–4428. doi: 10.1093/nar/20.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petit M-A, Bruand C, Janniere L, Ehrlich S D. Tn10-derived transposons active in Bacillus subtilis. J Bacteriol. 1990;172:6736–6740. doi: 10.1128/jb.172.12.6736-6740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piggot P J, Curtis C A M. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth J R. UGA nonsense mutations in Salmonella typhimurium. J Bacteriol. 1970;102:467–475. doi: 10.1128/jb.102.2.467-475.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadaie Y, Takamatsu H, Nakamura K, Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secA gene. Gene. 1991;98:101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- 36.Sharp P M, Higgins D G, Shields D C, Devine K M, Hoch J A. Bacillus subtilis gene sequences. In: Zukowski M M, Ganesan A T, Hoch J A, editors. Genetics and biotechnology of bacilli. Vol. 3. San Diego, Calif: Academic Press; 1990. pp. 89–98. [Google Scholar]
- 37.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Higgins M L, Piggot P J, Karow M L. Analysis of the role of prespore gene expression in the compartmentalization of mother cell-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1996;178:2813–2817. doi: 10.1128/jb.178.10.2813-2817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]