Significance
Compared to the cardiac ventricle, there are significant knowledge gaps in the sarcomeric mechanisms responsible for normal atrial relaxation. The atrial myosin-binding protein, myosin-binding protein H-like, was previously implicated through genetic studies in cardiomyopathy and arrhythmias in humans and mice. We now define the precise position of myosin-binding protein H-like within the atrial sarcomere and identify a competitive binding relationship with myosin-binding protein-C. Loss of myosin-binding protein H-like also accelerates atrial myofibril relaxation from maximal calcium activation under conditions with high myofilament phosphorylation. Together, these data suggest a mechanism where myosin-binding protein H-like reduces levels of cMyBP-C in the atrial sarcomere, thereby reducing the activating effects of phosphorylated cMyBP-C.
Keywords: myosin-binding protein, atria, contractility, cardiomyopathy, sarcomere
Abstract
Mutations in atrial-enriched genes can cause a primary atrial myopathy that can contribute to overall cardiovascular dysfunction. MYBPHL encodes myosin-binding protein H-like (MyBP-HL), an atrial sarcomere protein that shares domain homology with the carboxy-terminus of cardiac myosin-binding protein-C (cMyBP-C). The function of MyBP-HL and the relationship between MyBP-HL and cMyBP-C is unknown. To decipher the roles of MyBP-HL, we used structured illumination microscopy, immuno-electron microscopy, and mass spectrometry to establish the localization and stoichiometry of MyBP-HL. We found levels of cMyBP-C, a major regulator of myosin function, were half as abundant compared to levels in the ventricle. In genetic mouse models, loss of MyBP-HL doubled cMyBP-C abundance in the atria, and loss of cMyBP-C doubled MyBP-HL abundance in the atria. Structured illumination microscopy showed that both proteins colocalize in the C-zone of the A-band, with MyBP-HL enriched closer to the M-line. Immuno-electron microscopy of mouse atria showed MyBP-HL strongly localized 161 nm from the M-line, consistent with localization to the third 43 nm repeat of myosin heads. Both cMyBP-C and MyBP-HL had less-defined sarcomere localization in the atria compared to ventricle, yet areas with the expected 43 nm repeat distance were observed for both proteins. Isometric force measurements taken from control and Mybphl null single atrial myofibrils revealed that loss of Mybphl accelerated the linear phase of relaxation. These findings support a mechanism where MyBP-HL regulates cMyBP-C abundance to alter the kinetics of sarcomere relaxation in atrial sarcomeres.
Physiological regulation of striated muscle force generation and relaxation is finely tuned for muscle subtype-specific function by differential expression of sarcomere protein isoforms (1–5). In heart muscle, mutations in genes that encode most of the components of the sarcomere have been linked to cardiomyopathies, most commonly hypertrophic and dilated cardiomyopathy (HCM and DCM, respectively) (6, 7). HCM mutations that cause hypercontractile sarcomeres are almost exclusively found in genes that encode sarcomere proteins (8, 9). In contrast, DCM mutations affect a variety of cellular processes, with sarcomere mutations generally causing a hypocontractile left ventricle (7, 8). Mutations that cause cardiomyopathy can also cause atrial dilation and atrial fibrillation (6, 10), comorbidities that are associated with a worse prognosis (11, 12).
Given the left ventricle’s critical role in generating cardiac output, the mechanisms of cardiac sarcomere function and dysfunction have focused on protein isoforms that comprise and regulate left ventricular sarcomeres. The atria express different sarcomere protein isoforms that accommodate the distinct hemodynamic requirements of the atria (4, 13). Atrial myopathy can occur in the context of atrial fibrillation and worsen many types of cardiovascular disease (14, 15). These data, and others, underscore the importance of better defining the contribution of atrial-specific dysfunction to the progression and severity of cardiomyopathies (11, 16–19).
A premature truncation in the MYBPHL gene (Arg255Stop) was previously linked with atrial dilation, DCM, and atrial and ventricular arrhythmias (20), and this allele is present in approximately 1 in 700 people. Deletion of Mybphl in mice recapitulated the DCM, atrial dilation, and conduction system dysfunction in both heterozygous and homozygous Mybphl null mice (20, 21). MYBPHL encodes myosin-binding protein-H-like (MyBP-HL) and is highly expressed in mouse atria, sharing domain homology with the carboxy-terminal domains of cardiac myosin-binding protein-C (cMyBP-C) (20, 21). Truncating mutations in MYBPC3, which encodes cMyBP-C, result in haploinsufficiency and are a major cause of HCM (22, 23). In the ventricular sarcomere, when dephosphorylated, cMyBP-C stabilizes the myosin heads in nonforce-generating conformations and acts as a “brake” on the acto-myosin interaction (24–26). When phosphorylated, cMyBP-C loses its inhibitory function and potentiates actomyosin interaction (27–30).
The homology of these two myosin-binding proteins suggests that their functions may be linked and contribute to an atrial-specific regulatory mechanism, but this relationship has never been studied. We now demonstrate a competative occupancy relationship between MyBP-HL and cMyBP-C in the atrial sarcomere, as well as the functional consequences of disrupting the stoichiometry of myosin-binding proteins in the atria. This interrelationship between MyBP-HL and cMyBP-C identifies a differentiating feature between atrial and ventricular sarcomere function.
Results
MyBP-HL Binds to the A-band in Atrial Sarcomeres.
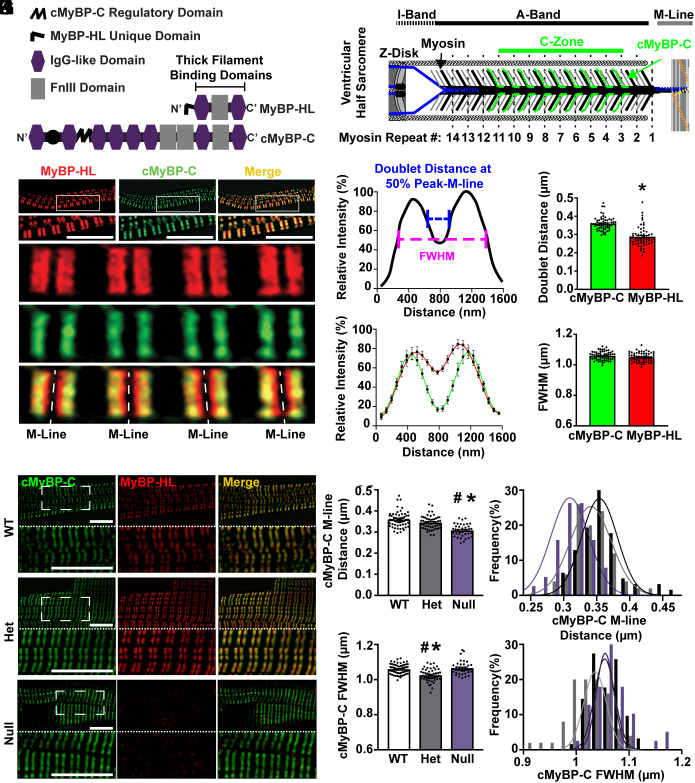
MyBP-HL shares homology with the thick filament binding carboxy-terminal domains of cMyBP-C (Fig. 1A) (20). To assess MyBP-HL’s subcellular sarcomere binding properties, we generated and validated a rabbit polyclonal antibody that recognizes a unique set of hydrophilic amino acids at the amino-terminus of mouse MyBP-HL (21). We isolated atrial cardiomyocytes from adult wild-type mice and used immunostaining and super-resolution structured illumination microscopy to study sarcomere localization of MyBP-HL. Immunostaining with α-actinin and MyBP-HL antibodies shows an A-band doublet pattern of MyBP-HL, interdigitated between α-actinin stained Z-disks (SI Appendix, Fig. S1). The C-zone is defined as the region of the A-band where three cMyBP-C molecules bind to the myosin thick filament at nine specific repeats per half sarcomere that are spaced 43 nm apart (Fig. 1B). We found that MyBP-HL and cMyBP-C colocalized in the C-zone (Fig. 1C) and identified that MyBP-HL also occupies additional space closer to the sarcomere’s central M-line (cMyBP-C doublet distance across M-line: 359 ± 5 nm; MyBP-HL doublet distance: 288 ± 7 nm) (Fig. 1 D and E). The MyBP distribution suggests that in wild-type atrial sarcomeres MyBP-HL might occupy an additional 43 nm repeat of myosin heads closer to the M-line compared to cMyBP-C. Both MyBPs appear to have a similar lateral boundary, approximately 1,050 nm at full width of half-maximal fluorescence intensity (Fig. 1G). This value agrees with immuno-labeled electron micrographs showing that the lateral edges of cMyBP-C C-zone doublets are 1,100 ± 2 nm wide (31).
Fig. 1.
MyBP-HL localizes to the C-zone of atrial sarcomeres. (A) Protein domain schematic showing the similarities between the thick filament binding regions at the carboxyl terminus of MyBP-HL and cMyBP-C. The MyBP-HL antibody recognizes the MyBP-HL unique domain. (B) Schematic of a ventricular half-sarcomere illustrating the known cMyBP-C binding localization to nine repeats, spaced 43 nm, at every third level of myosin heads. (C) Structured illumination microscopy on isolated atrial cardiomyocytes stained with MyBP-HL (red) and cMyBP-C (green) shows that MyBP-HL co-localizes with the C-zone pattern of cMyBP-C but also occupies space closer to the M-line (Scale bar, 10 µm). (D) Schematic of a fluorescence intensity trace along one sarcomere showing A-band doublet patterns. The distance across the M-line was calculated at the points between the 50% vales of the left and right peaks and the nadir of the fluorescence trace at the M-line (blue line) and the full-width of the fluorescence trace was calculated at the half-maximum fluorescence value (FWHM, pink line). (E) Averaged intensity plots of MyBP-HL and cMyBP-C localization over six sarcomeres. (F) Quantification of the doublet distance across the M-line shows MyBP-HL doublets have less distance between them compared to cMyBP-C doublets. (G) Full-width at half max fluorescence intensity shows no difference in the lateral boundary of the MyBP-HL and cMyBP-C doublets. N = 62 sarcomeres. * = P < 0.05 by Student’s t test. (H) Structured illumination microscopy images of permeabilized isolated atrial cardiomyocytes from WT, heterozygous, and homozygous Mybphl null mice stained for cMyBP-C (green) and MyBP-HL (red). Magnified region indicated with dashed line box (Scale bar, 10 µm). (I and J) Quantification of the doublet distance of cMyBP-C in each genotype showing reduced M-line distance of cMyBP-C in Mybphl null sarcomeres. (K and L) The full-width at half maximal intensity showing a smaller cMyBP-C C-zone width in heterozygous sarcomeres. * = P < 0.05 by One-Way ANOVA vs. WT; # = P < 0.05 vs. Null.
Loss of MyBP-HL Allows cMyBP-C to Occupy Additional Binding Sites Closer to the M-line.
To assess whether cMyBP-C can occupy the same sarcomere binding sites as MyBP-HL, we evaluated atrial cardiomyocytes from wild-type and Mybphl heterozygous and homozygous null mice using structured illumination microscopy. Costaining for MyBP-HL and cMyBP-C in wild-type cardiomyocytes showed data consistent with the experiment in Fig. 1 C–G. Predictably, no MyBP-HL was detected in the Mybphl-null atrial cardiomyocytes (Fig. 1H). However, cMyBP-C was still present and showed A-band doublets that were significantly closer together across the M-line compared to wild-type cMyBP-C doublets (Fig. 1 I and J). No significant changes in distance across the M-line between cMyBP-C doublets were observed in Mybphl heterozygous cardiomyocytes (Fig. 1 I and J). In Mybphl null sarcomeres, the cMyBP-C doublet distance was ~308 nm, closer to the 288 nm MyBP-HL doublet distance than the 359 nm cMyBP-C doublet distance observed in wild-type atrial cardiomyocytes (Fig. 1E), suggesting that cMyBP-C occupies the medial binding sites normally occupied by MyBP-HL in atrial sarcomeres. Curiously, the Mybphl heterozygous images showed a small but significant 34 nm reduction in cMyBP-C full-width half-max (Fig. 1 K and L), although the biological basis for this alteration is unclear.
MyBP-HL and cMyBP-C Demonstrate a Competitive Stoichiometry.
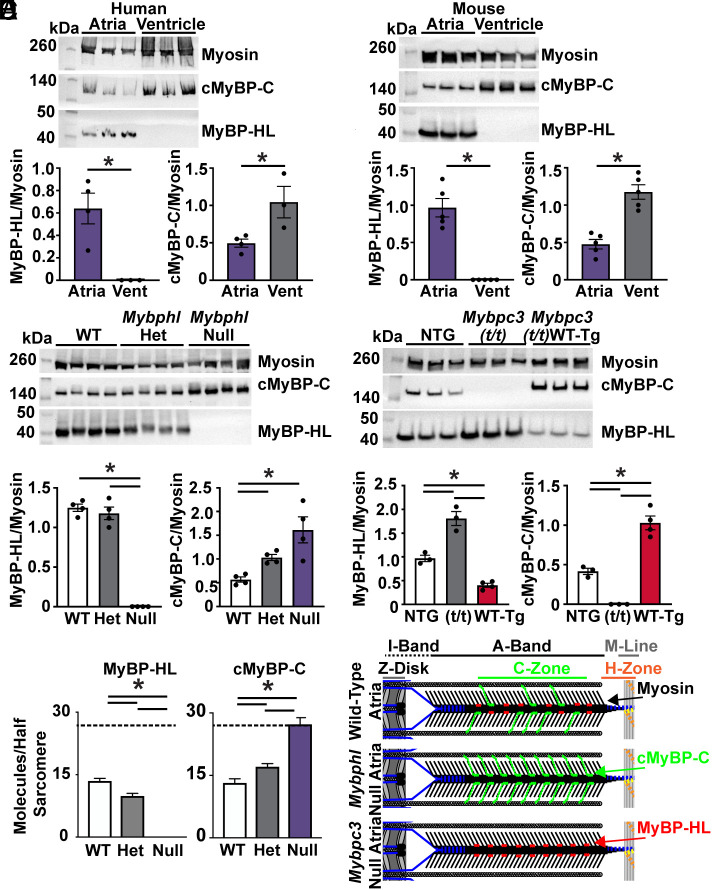
The observation that the loss of MyBP-HL allowed cMyBP-C to bind to the medial portion of the C-zone raised the question of whether these two proteins compete for binding sites throughout the C-zone. We used immunoblotting on sarcomere protein fractions from human and mouse atria and ventricles to determine whether atria have less cMyBP-C normalized to total myosin than the ventricle. If MyBP-HL binds sites in the atria beyond the nine 43 nm cMyBP-C sites previously established in ventricular sarcomeres, we would expect no alteration in cMyBP-C levels between the chambers (32). In both species, the level of cMyBP-C was significantly less abundant in the atria (41% mouse, 48% human) compared to the ventricles (100%) (Fig. 2 A and B).
Fig. 2.
MyBP-HL and cMyBP-C have an inverse stoichiometry. (A) Western blot of atrial and ventricular total protein from nonfailing human hearts shows MyBP-HL enriched and cMyBP-C reduced in the atria compared to ventricle; N = 4 atria, 3 ventricles. (B) Western blot of wild-type mouse atrial and ventricular total protein lysate to detect total myosin heavy chain, cMyBP-C, and MyBP-HL shows the atrial enrichment of MyBP-HL, as well a reduction in cMyBP-C levels in the atria; N = 5. (C) Western blot using total atrial protein lysates from WT, heterozygous, and homozygous Mybphl null mice shows an increase in cMyBP-C levels as MyBP-HL is reduced; N = 4. (D) Western blot of total atrial protein lysates from wild-type nontransgenic controls, mice lacking cMyBP-C (Mybpc3 t/t), and mice with a transgenic overexpression of wild-type cMyBP-C (Mybpc3 t/t/WT-Tg) shows that MyBP-HL levels increase or are reduced in an inverse relation to cMyBP-C levels; N = 3 WT, 3 t/t, 4 WT(t/t). (E) Mass spectrometry analysis of total atrial protein from WT, heterozygous, and homozygous Mybphl null mice shows an inverse stoichiometric relationship between MyBP-HL and cMyBP-C, with both protein signals normalized to total myosin content; N = 3. Dashed line at 27 molecules (i.e., full MyBP complement). (F) Schematic of potential inverse stoichiometric binding of cMyBP-C and MyBP-HL in a half sarcomere.
Immunoblotting of total atrial protein from Mybphl wild-type, heterozygous, and homozygous null mice showed that as MyBP-HL levels decreased, cMyBP-C levels increased normalized to total myosin content (Fig. 2C), such that complete loss of MyBP-HL resulted in approximate doubling of cMyBP-C levels. To assess the inverse of this relationship, we performed immunoblotting on total atrial protein from a mouse model with a Mybpc3 truncating mutation that results in no cMyBP-C protein (Mybpc3(t/t)) (33, 34). We also used a mouse line with a wild-type Mybpc3 transgene expressed on the somatic truncating Mybpc3(t/t) allele background (WT-Mybpc3-Tg(t/t)). This model (WT-Mybpc3-Tg(t/t)) expresses approximately 15-fold more Mybpc3 mRNA than nontransgenic wild-type mice, but this does not result in increased levels of cMyBP-C in the ventricle of these mice due to the defined stoichiometry of cMyBP-C in the sarcomere (35). We used strain-matched wild-type nontransgenic mice (NTG) as controls. Compared to NTG controls, we found an approximate doubling of MyBP-HL levels in the absence of cMyBP-C in the atria of Mybpc3(t/t) mice and a reduction of MyBP-HL in atria of mice with transgenic overexpression of wild-type cMyBP-C (Fig. 2D).
To assess the stoichiometry of these myosin-binding proteins more quantitatively, we used mass spectrometry to measure the molecular abundance of MyBP-HL and MyBP-C relative to myosin heavy chain in the atria of Mybphl wild type, heterozygous, and homozygous null mice. In ventricular sarcomeres, each half-thick filament is composed of 306 myosin heavy chain monomers and 27 molecules of cMyBP-C for an 11:1 ratio of myosin:cMyBP-C. Three cMyBP-C molecules are found per nine myosin dimers that compose one 43-nm repeat of myosin heads in the C-zone (Fig. 1B) (26). The mass spectrometry data demonstrated that wild type mice have 45% as much cMyBP-C in the atria compared to the ventricle. We confirmed that as the abundance of MyBP-HL decreases in the Mybphl heterozygous and homozygous null mice, the abundance of cMyBP-C increases while the stoichiometry of total myosin-binding proteins relative to myosin is maintained (Total MyBP molecules/half-sarcomere: WT 27.1 ± 3.1, Het 26.7 ± 0.5, and Null 27.5 ± 2.8. Mean ± SD) (Fig. 2E). These data demonstrate that MyBP-HL and cMyBP-C can bind the same sites per half-thick filament in the atria (Fig. 2F).
Point-spread Function Modeling Suggests that MyBP-HL Occupies One Additional 43-nm Repeat Toward the Center of the Atrial Sarcomere Compared to cMyBP-C.
We wanted to determine whether MyBP-HL localizes closer to the M-line than cMyBP-C due to MyBP-HL binding preferentially to myosin repeat # 3 (the medial-most repeat where cMyBP-C binds in ventricular sarcomeres) or whether MyBP-HL can bind to an additional medial 43-nm myosin repeat (i.e., repeat # 2). We generated a model in which the cMyBP-C and MyBP-HL molecules were spaced along each half of the thick filament with three MyBP molecules per 43 nm repeat. The position and number of molecules within each repeat were iteratively adjusted to best-match reference confocal images. The molar abundance of MyBP-HL and cMyBP-C was taken from the mass spectrometry data (Fig. 2E) and used to define relative abundance of the proteins across the thick filament. We first modeled ventricular cardiomyocytes with 27 molecules of cMyBP-C across nine C-zone repeats (43 nm myosin repeat # 3–11) per half sarcomere (Fig. 1B and SI Appendix, Fig. S2A) (32, 36). A point-spread function was used to model the fluorescence intensity trace of this distribution (SI Appendix, Fig. S2A, green line). The result was in good agreement with measured fluorescence intensity traces from confocal images of ventricular cardiomyocytes immunostained for cMyBP-C. We extended this analysis to wild-type atrial cardiomyocytes (SI Appendix, Fig. S2B). We modeled MyBP-HL occupying one additional 43-nm myosin repeat (repeat # 2). We further used this model to simulate reduced and abolished distribution of MyBP-HL in the heterozygous and homozygous Mybphl null mice (SI Appendix, Fig. S2 C and D). In this model, MyBP-HL occupies an additional 43 nm repeat closer to the M-line than cMyBP-C in WT atrial sarcomeres, but this additional repeat is occupied by cMyBP-C in the absence of MyBP-HL. However, cMyBP-C does not bind to 43 nm repeat # 2 in the ventricle, making it unlikely to be able to replace MyBP-HL at this location in the atria (32).
Electron Microscopy of MyBP-HL Immunolabeled Sarcomeres Reveals a Unique Sarcomere Binding Pattern.
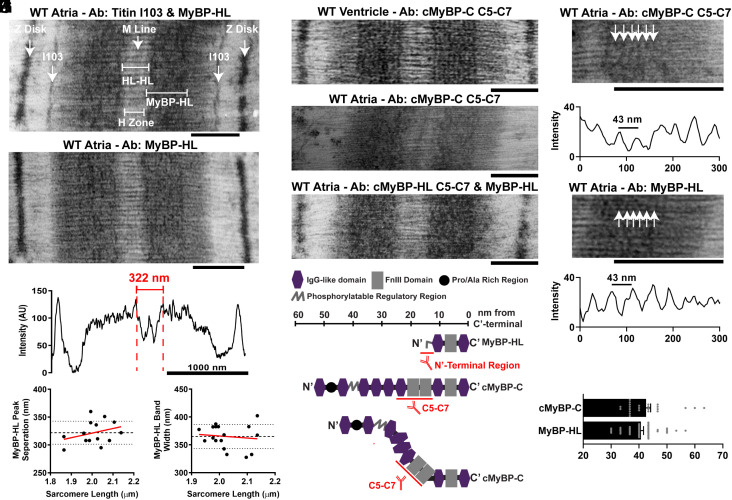
To resolve the sarcomere localization of MyBP-HL, we used immunolabeled electron micrographs to identify the myosin repeats where MyBP-HL is localized. The localization of myosin-binding proteins within the sarcomere have been established using immunolabeled electron micrographs in skeletal muscle and cardiac ventricles (32, 36). Immunolabeling with the MyBP-HL antibody on wild-type mouse atrial tissue shows enrichment in the C-zone of the A band (Fig. 3 A and B). Costaining with the titin I103 antibody delineated the A/I band junction (Fig. 3A). The distance between I103 bands is 1,650 nm, and this distance was used to calibrate the measurements and evaluate the sample shrinkage over a range of sarcomere lengths (SI Appendix, Fig. S3 A and B) (37). Intensity traces of the MyBP-HL staining pattern showed a prominent peak in the medial portion of the A-band with a 322-nm distance between these peaks or 161 nm from peak to M-line (Fig. 3 C and D). This is the known distance from the center of the M-line to 43 nm myosin repeat # 3 (31, 36). The MyBP-HL stained region in each half of the A-band was 365 ± 5 nm wide (Fig. 3E).
Fig. 3.
Myosin-binding proteins are distributed throughout the C-zone in the atria. (A) Immuno electron microscopy from wild-type mouse atria costained with MyBP-HL antibody and the titin A/I band junction I103 antibody and (B) staining with MyBP-HL antibody alone (Scale bar, 500 nm). (C) Intensity trace across one sarcomere stained with MyBP-HL antibody. (D) The prominent medial MyBP-HL peaks were separated by 322 nm across the M-line, or 161 nm from peak to M-line. (E) The MyBP-HL stained region on each side of the A-band measured 365 ± 5 nm wide. (F) Representative IEM of mouse ventricular tissue stained with cMyBP-C (C5-C7) antibody shows clear 43 nm repeating pattern. (G) Micrograph of wild-type atrial tissue stained with cMyBP-C (C5-C7) does not delineate clear 43 nm repeating pattern. (H) Micrograph of wild-type atrial tissues costained with cMyBP-C (C5-C7) and MyBP-HL antibodies produces dense staining in the A-band but does not delineate clear axial repeats. (I) Schematic of the protein domains of cMyBP-C and MyBP-HL with the location of the epitope recognized by the respective antibodies. Scale is approximate. Immuno electron micrographs of wild-type mouse atrial tissue stained with the cMyBP-C C5-C7 antibody (G) or MyBP-HL antibody (B) shows C-zone localization but without the characteristic strong axial repeats observed in the ventricle (F). (J–L) Despite no strong 43-nm axial repeats throughout the C-zone, some areas of the filaments in the C-zone had periodic staining regions with an average distance between peaks of 42.5 ± 1.6 nm with cMyBP-C staining and 40.7 ± 1 nm with MyBP-HL staining, which were not significantly different. N = distances between 24 cMyBP-C adjacent repeats, 45 MyBP-HL adjacent repeats. Student’s t test.
Immunolabeled electron micrographs of mouse ventricle tissue stained with the cMyBP-C C5-C7 antibody produces the 43 nm repeating pattern (Fig. 3F). Staining atrial tissue with either the MyBP-HL antibody (Fig. 3B) or the cMyBP-C C5-C7 antibody alone (Fig. 3G) did not produce clear regular repeats like those observed in the ventricle (Fig. 3F). Because neither myosin-binding protein alone showed clear repeating patters, we investigated whether costaining with MyBP-HL and cMyBP-C would produce clear 43-nm repeats in atrial tissue. The costained micrograph showed dense staining in the C-zone, but no clear pattern emerged (Fig. 3H). The lack of clear 43-nm repeats with this costaining may be due to the location of the epitopes for these antibodies. The cMyBP-C C5-C7 antibody recognizes three central domains of cMyBP-C, each roughly four nm in diameter that can enter the interfilament space, and the MyBP-HL antibody recognizes a 50 amino acid amino terminal domain of MyBP-HL (Fig. 3I). If MyBP-HL and cMyBP-C bind at the nine myosin repeat sites along the sarcomere randomly, the spatial offset of antibody binding to the two proteins may result in loss of the coherent 43 nm repeat pattern between adjacent thick filaments. However, MyBP-HL and cMyBP-C randomly occupying the myosin-binding protein sites on the nine myosin repeats in the C-zone should, by chance, result in some thick filaments having the same myosin-binding protein occupying several consecutive binding sites. Examination of the atrial sarcomeres stained only for cMyBP-C or MyBP-HL revealed occasional instances of labeling that had a 43-nm repeating periodicity (Fig. 3 J and K). Quantification of intensity plots taken along these dense regions revealed regularly spaced peaks that were separated by 40.7 ± 1 nm for MyBP-HL and 42.5 ± 1.6 nm for cMyBP-C, which was not significantly different between the two myosin-binding proteins (Fig. 3L).
Loss of MyBP-HL and Increased cMyBP-C Causes Faster Relaxation in Atrial Myofibrils.
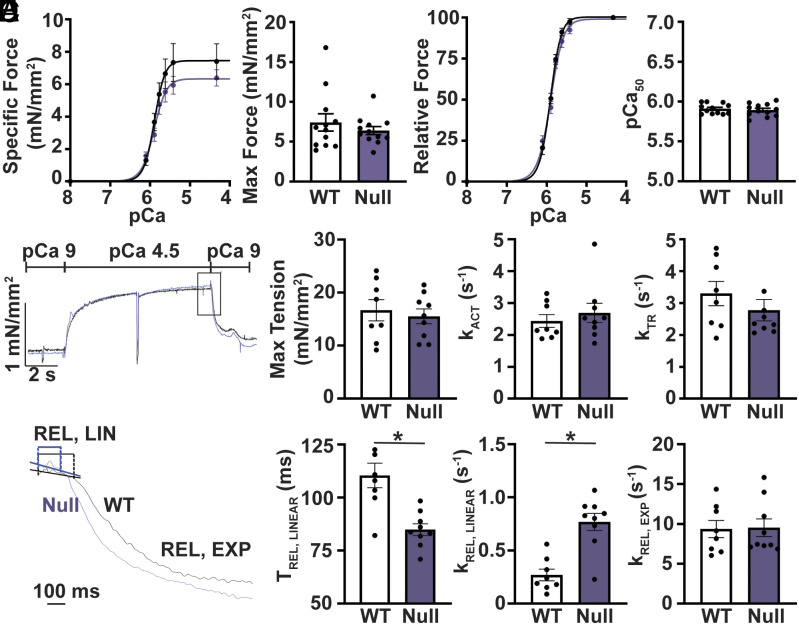
We performed isometric force–calcium relationship measurements in permeabilized atrial and ventricular cardiomyocytes to assess the functional consequences of the altered stoichiometry of myosin-binding proteins. Peak isometric specific force was not significantly different between atrial and ventricular cardiomyocytes from wild-type mice (SI Appendix, Fig. S4A). Wild-type atrial cardiomyocytes showed a significant increase in the calcium sensitivity of force development compared to ventricle (SI Appendix, Fig. S4B). Comparisons of wild-type and Mybphl homozygous null atrial cardiomyocytes showed no significant change in peak specific force or calcium sensitivity of force development (Fig. 4 A and B). In ventricular cardiomyocytes, cMyBP-C has been shown to affect the rate of myofibril contraction and relaxation (27, 38). We assessed the kinetics of contractile function in single myofibrils from WT and Mybphl homozygous null mouse atria. Myofibrils underwent activation, tension redevelopment after rapid stretch and slack, and relaxation (Fig. 4C). Myofibril relaxation is illustrated with the slow linear phase of relaxation (REL, LIN) and the fast exponential phase (REL, EXP) marked (Fig. 4D). The myofibrils showed no significant changes in maximal tension or the rate of force development and redevelopment (Fig. 4E). However, Mybphl homozygous null myofibrils did show significantly faster rates of the linear relaxation phase with concomitantly shortened durations of the linear relaxation phase compared to wild-type myofibrils.
Fig. 4.
Loss of Mybphl increases relaxation kinetics. (A) Calcium-isometric force relationship of permeabilized isolated atrial cardiomyocytes from wild-type and Mybphl homozygous null hearts. N = WT: 5 mice, 16 cells; null: 4 mice, 13 cells. (B) Relative force traces from permeabilized isolated atrial cardiomyocytes from wild-type and Mybphl homozygous null hearts. N = WT: 5 mice, 16 cells; null: 4 mice, 13 cells. (C) Representative trace of single myofibril force measurements from WT and homozygous Mybphl null atria. The trace shows kinetics of force activation (kACT), redevelopment (kTR), and relaxation of WT and Mybphl myofibrils. (D) Expanded traces of myofibril relaxation showing the time and rate of the short linear phase (tREL, LINEAR, kREL, LINEAR) and the exponential phase (kREL, EXP) of relaxation. (E) Quantification of the single myofibril data. N = WT: 7 mice per group, averaged from 3 to 6 fibers per mouse. Null: 9 mice, averaged from 3 to 6 fibers per mouse. * = P < 0.05 by Welch’s t test.
The faster rate of relaxation in Mybphl null myofibrils suggests that additional cMyBP-C within atrial myofibrils favors the activating effects of cMyBP-C resulting in faster cross-bridge cycling. Because phosphorylated cMyBP-C mediates these activating effects, we assessed cMyBP-C phosphorylation at serine 273, 282, and 302 with immunoblotting. Importantly, we used the same samples prepared for the myofibril studies. While the ratio of phosphorylated cMyBP-C to total cMyBP-C was not changed between groups, because cMyBP-C levels were significantly increased in Mybphl null atria, absolute levels of phosphorylated cMyBP-C are also increased (SI Appendix, Fig. S5).
Discussion
Atrial dysfunction is a comorbidity in HCM, DCM, and many other conditions (6, 10–12, 14, 15, 17). The sarcomeric mediators of atrial dysfunction have not been well understood. Here, we define an interrelationship between two myosin-binding proteins, MyBP-HL and cMyBP-C, within the atrial sarcomere, demonstrating that these two proteins compete for the same binding sites within the C-zone and maintain a stoichiometry of 27 myosin-binding proteins per half-thick filament. This interrelationship indicates that myosin regulation in the atrial sarcomere is mediated by cMyBP-C and MyBP-HL. The finding that loss of MyBP-HL hastens the linear phase of myofibril relaxation suggests that MyBP-HL alters the off-rate of myosin (39, 40). The shift in atrial relaxation seen in the absence of MyBP-HL could reflect a primary atrial defect from loss of MyBP-HL and/or the increased cMyBP-C content in the atria. Currently, primary atrial myopathies are still a nascent concept, and our understanding of the genes that are highly enriched in atrial cardiomyocytes and their atrial-specific function is incomplete (6, 14, 16). The human atrial sarcomere differs from the ventricular sarcomere in both myosin heavy-chain and light-chain content, which likely impacts how MyBP-HL regulates myofilament function.
Myosin-binding Protein Localization.
Our modeling of cMyBP-C and MyBP-HL distribution supports the hypothesis that both myosin-binding proteins are distributed equally throughout the nine 43-nm myosin repeats that comprise the C-zone, except for MyBP-HL that exclusively binds the medial myosin repeat # 3. In the absence of MyBP-HL, cMyBP-C can occupy medial repeat # 3. The ability of cMyBP-C to bind to this medial site in the absence of MyBP-HL suggests that MyBP-HL is not binding to a novel repeat (i.e., myosin repeat # 2), as cMyBP-C does not bind repeat # 2 in the ventricle (32). This was confirmed by the ultrastructural data, as MyBP-HL staining showed its strongest and medial-most peak 161 nm from the M-line where 43 nm myosin repeat # 3 is located (37). The highly homologous skeletal muscle protein MyBP-H was shown to localize to 43 nm myosin repeat # 3 in rabbit psoas muscle (36). Our current finding of MyBP-HL binding preferentially to repeat # 3 in mouse atria suggests that MyBP-H and MyBP-HL are skeletal and cardiac isoforms of H protein.
Functional Implications of MyBP-HL Regulating cMyBP-C Abundance.
We show that MyBP-HL can regulate cMyBP-C abundance and, to a lesser extent, localization in atrial sarcomeres. The foreshortened MyBP-HL lacks the elongated amino terminus found in cMyBP-C that regulates actomyosin binding and contractility (41, 42). Therefore, MyBP-HL can exert a functional effect by statically attenuating the ability of cMyBP-C to regulate the sarcomere. Functions for cMyBP-C include facilitating homogenous contraction across the sarcomere (43) and promoting sensitization of thin filaments to calcium (29, 30). Dephosphorylated cMyBP-C slows crossbridge attachment rates and promotes the super-relaxed state of myosin, reducing rates of tension development (24). Whether MyBP-HL has any other intrinsic regulatory function remains to be determined.
We found that atrial myofibrils without MyBP-HL have faster linear relaxation rates and shorter linear relaxation times. The linear phase of force decay is mediated by the off-rate of myosin heads from actin (i.e., crossbridge detachment rates) (40, 44). Phosphorylated cMyBP-C accelerates crossbridge detachment rates and overall accelerates cross bridge cycling (27, 28). Loss of cMyBP-C or transgenic overexpression of constitutively phosphorylated cMyBP-C in ventricular cardiomyocytes also showed accelerated rates of myosin detachment compared to wild-type cMyBP-C (28). Levels of phosphorylated to total cMyBP-C were not different between wild-type and Mybphl null myofibrils, but the increased abundance of cMyBP-C and therefore increased abundance of phosphorylated cMyBP-C in the Mybphl null myofibrils may be sufficient to account for the faster linear relaxation rate. Importantly, myofibril relaxation was measured by changing from maximal activating calcium to diastolic calcium levels. Phosphorylated cMyBP-C has a more profound effect on accelerating relaxation at lower, more physiological calcium concentrations (45). Therefore, the difference between wild-type and Mybphl null atrial myofibrils is likely larger at lower activating calcium concentrations.
Phosphorylated cMyBP-C not only accelerates linear relaxation but also accelerates overall crossbridge cycling rates (27, 28). In the myofibrils from Mybphl null atria, we do not observe a corresponding acceleration in the rate of tension redevelopment that would be expected. While a trend toward accelerated rate of tension redevelopment exists in our data, additional experiments specifically controlling for cMyBP-C phosphorylation, potentially with genetic models of cMyBP-C phosphorylation mutants, may be necessary to explain this observation.
Overall, these studies identify that the atrial sarcomere contains two different myosin-binding proteins that compete for the same binding sites within the C-zone. Loss of MyBP-HL increases levels of cMyBP-C and accelerates the rate of linear relaxation in atrial myofibrils. This may be due to the ability of phosphorylated cMyBP-C to accelerate crossbridge cycling, and MyBP-HL is expected to reduce this effect, resulting in prolonged detachment. The contribution of atrial relaxation to normal cardiac physiology and in cardiomyopathy is relatively unexplored. Therefore, there is minimal context to speculate on the consequence of MyBP-HL statically slowing the linear rate of relaxation at the whole organ level. The physiology of early atrial relaxation is fascinating, as the atria do not experience isovolumic relaxation periods like the ventricles. Because of this, some atrial sarcomeres may remain in a force-generating state when the tissue begins to lengthen. Regulation of this early relaxation period may provide a physiological benefit for atrial function. Future work measuring hemodynamic function in control and Mybphl null atria would shed light on the relevance of atrial relaxation to whole organ function. These data also provide a strong rationale for evaluating the effect of cardiomyopathy causing mutations in MYBPC3 on atrial function, as the presence of MyBP-HL may mitigate or exacerbate these effects.
Materials and Methods
Animal Use.
All experiments using mice were performed on male and female animals between 10 and 14 wk of age. The Mybphl null mouse model was obtained from the Knock-Out Mouse Project and is maintained on a C57/Bl6-J background and have been previously described (20). All procedures were approved by the Institutional Animal Care and Use Committee at Northwestern University.
Human Tissue Sample.
Deidentified samples from post-mortem nonfailing human donor hearts that were not used for transplant were used for this study.
Superresolution Microscopy.
Atrial cardiomyocytes were isolated and immunostained as previously described and covered in the data supplement (21). Superresolution microscopy was performed using a Nikon Structured Illumination Microscopy imaging platform in the Nikon Imaging Center in Northwestern University’s Center for Advanced Microscopy. The microscope was fitted with a 100× oil objective (NA 1.49) and 488-nm and 561-nm wavelength lasers were used for imaging. All images were acquired and processed using the Nikon Elements software. Fluorescence intensity traces were measured and annotated in Microsoft Excel to measure distances between fluorescent peaks.
Quantitative Mass Spectrometry.
The number of MyBP-HL and MyBP-C molecules per half of a thick filament in the atria of Mybphl wild-type, heterozygous, and homozygous mice was estimated by quantitative mass spectrometry. Methods for sample preparation is provided in supplementary methods. The LC peak area of the top MyBP-HL peptide (Q5FW53; TSHQQEAGSPSLQLLPSIEEHPK) and average LC peak area of the top three cMyBP-C peptides (Q3UIK0; VAGASLLKPPVVK, IAFQHGVTDLR, VIDVPDAPAAPK; SD = 6.5%) were divided by the average LC peak area of the top 3 myosin heavy-chain peptide (EAEASLEHEEGK; SD = 8.5%) to estimate the relative molar abundance of each protein. Only the LC peak area for the most abundant MyBP-HL peptide was used due to the high degree of variance between the top three peptides (SD = 49%), as could be expected from a lower molecular weight protein that produces a limited number of tryptic peptides. The number of MyBP-HL and MyBP-C molecules per half of a thick filament was estimated from these ratios assuming that there are 153 myosin molecules or 306 monomers of myosin heavy chain per half of a thick filament. This assumes nine myosin molecules exist in 17 helical repeats, spaced every 43 nm along each half of the thick filament. The mean number of MyBP-HL and MyBP-C molecules and SDs were determined from the three biological and two instrumental replicates from the tissue from the Mybphl wild-type, heterozygous, and homozygous mice.
Single Myofibril Force Measurements.
Buffer composition is described in the data supplement. Myofibril mechanical function was assessed using the fast solution switching method as described (39, 46). For each measurement, a small bundle of myofibrils (average diameter of 6.72 µm and average length of 43.2 µm) were mounted between a calibrated cantilever force probe (10.31 μm/μN) and a microtool attached to a motor that produces rapid length changes at 15 °C in relaxing solution (pCa 9.0). Myofibril length was set at ~2.0 µm. Mounted myofibrils were activated and relaxed by rapidly translating the interface between two flowing streams of pCa 4.5 full activating calcium concentration solution and pCa 9.0 relaxation solution. Data were collected and analyzed using customized LabView software and defined as follows: resting tension (mN/mm2), myofibril basal tension in fully relaxing condition; maximal tension (mN/mm2), maximal tension generated at full activating calcium activation (pCa 4.5); the rate constant of tension development following maximal calcium activation (kACT); the rate constant of tension redevelopment following a release–restretch applied to the activated myofibril (kTR) (47); duration of early slow force decline measured from onset of solution change to the beginning of the exponential force decay (tREL, LINEAR); the rate constant of the first linear phase of relaxation (kREL, LINEAR); and the rate constant of the final exponential phase of force decline (kREL, LINEAR).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by NIH National Heart Lung and Blood institute grants F32HL131304, K99HL141698, and R00151698 (D.Y.B.); R35HL144998 (H.L.G.); HL128075, HL124041, and R01HL157487 (E.M.M.); American Heart Association grants ASCD SFRN (E.M.M.); Leducq Foundation Transatlantic Network on Editing the Failing Heart (E.M.M.). Electron microscopy was performed in the University of Arizona Imaging Cores—Electron RRID: SCR_023279. Structured illumination microscopy was performed at Northwestern University’s Center for Advanced Microscopy (NIH 1S10OD016342-01). We thank Dr. Carol Gregorio for the titin I103 antibody, Dr. Samantha Harris for the cMyBP-C (C5-C7) antibody, and Dr. Sakthivel Sadayappan for the phosphorylation specific cMyBP-C S273, S282, and S302 antibodies.
Author contributions
D.Y.B., M.J.P., H.L.G., and E.M.M. designed research; D.Y.B., P.T., K.C.W., S.R., T.S.O., and H.V.B. performed research; D.Y.B., P.T., K.C.W., J.A.W., J.A.K., M.J.P., H.L.G., and E.M.M. contributed new reagents/analytic tools; D.Y.B., K.C.W., S.R., T.S.O., H.V.B., M.J.P., and E.M.M. analyzed data; and D.Y.B., P.T., K.C.W., S.R., and E.M.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
David Y. Barefield, Email: dbarefield@luc.edu.
Elizabeth M. McNally, Email: elizabeth.mcnally@northwestern.edu.
Data, Materials, and Software Availability
Complete mass spectrometry data set is available publicly on Barefield Lab Database (48).
Supporting Information
References
- 1.Miyata S., Minobe W., Bristow M. R., Leinwand L. A., Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ. Res. 86, 386–390 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Metzger J. M., Michele D. E., Rust E. M., Borton A. R., Westfall M. V., Sarcomere thin filament regulatory isoforms. Evidence of a dominant effect of slow skeletal troponin I on cardiac contraction. J. Biol. Chem. 278, 13118–13123 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Bang M. L., et al. , The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 89, 1065–1072 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Piroddi N., et al. , Tension generation and relaxation in single myofibrils from human atrial and ventricular myocardium. Pflugers Arch. 454, 63–73 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Li A., et al. , Skeletal MyBP-C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc. Natl. Acad. Sci. U.S.A. 116, 21882–21892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pensa A. V., Baman J. R., Puckelwartz M. J., Wilcox J. E., Genetically based atrial fibrillation: Current considerations for diagnosis and management. J. Cardiovasc. Electrophysiol. 33, 1944–1953 (2022). [DOI] [PubMed] [Google Scholar]
- 7.McNally E. M., Barefield D. Y., Puckelwartz M. J., The genetic landscape of cardiomyopathy and its role in heart failure. Cell Metab. 21, 174–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spudich J. A., Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106, 1236–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helms A. S., et al. , Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 7, 434–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho C. Y., et al. , Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 138, 1387–1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patten M., Pecha S., Aydin A., Atrial fibrillation in hypertrophic cardiomyopathy: Diagnosis and considerations for management. J. Atr. Fibrillation 10, 1556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aleksova A., et al. , Impact of atrial fibrillation on outcome of patients with idiopathic dilated cardiomyopathy: Data from the Heart Muscle Disease Registry of Trieste. Clin. Med. Res. 8, 142–149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dernellis J. M., Stefanadis C. I., Zacharoulis A. A., Toutouzas P. K., Left atrial mechanical adaptation to long-standing hemodynamic loads based on pressure–volume relations. Am. J. Cardiol. 81, 1138–1143 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Shen M. J., Arora R., Jalife J., Atrial myopathy. JACC Basic Transl. Sci. 4, 640–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peigh G., Shah S. J., Patel R. B., Left atrial myopathy in atrial fibrillation and heart failure: Clinical implications, mechanisms, and therapeutic targets. Curr. Heart Fail Rep. 18, 85–98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin D., Mansour M. C., Ruskin J. N., Heist E. K., Atrial fibrillation-mediated cardiomyopathy. Circ. Arrhythm. Electrophysiol. 12, e007809 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Buckley B. J. R., et al. , Atrial fibrillation in patients with cardiomyopathy: Prevalence and clinical outcomes from real-world data. J. Am. Heart Assoc. 10, e021970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huurman R., et al. , Prognostic significance of left atrial strain in sarcomere gene variant carriers without hypertrophic cardiomyopathy. Echocardiography 39, 1209–1218 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou D., et al. , Left atrial dysfunction may precede left atrial enlargement and abnormal left ventricular longitudinal function: A cardiac MR feature tracking study. BMC Cardiovasc. Disord. 22, 99 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barefield D. Y., et al. , Experimental modeling supports a role for MyBP-HL as a novel myofilament component in arrhythmia and dilated cardiomyopathy. Circulation 136, 1477–1491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barefield D. Y., et al. , Partial and complete loss of myosin binding protein H-like cause cardiac conduction defects. J. Mol. Cell Cardiol. 169, 28–40 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barefield D., et al. , Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice. J. Mol. Cell Cardiol. 79, 234–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helms A. S., et al. , Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy. JCI Insight 5, e133782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamara J. W., Singh R. R., Sadayappan S., Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl. Acad. Sci. U.S.A. 116, 11731–11736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNamara J. W., et al. , Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J. Mol. Cell Cardiol. 94, 65–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Previs M. J., Beck Previs S., Gulick J., Robbins J., Warshaw D. M., Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337, 1215–1218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moss R. L., Fitzsimons D. P., Ralphe J. C., Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 116, 183–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner B. C. W., Previs M. J., Wang Y., Robbins J., Palmer B. M., Cardiac myosin binding protein-C phosphorylation accelerates beta-cardiac myosin detachment rate in mouse myocardium. Am. J. Physiol. Heart Circ. Physiol. 320, H1822–H1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mun J. Y., et al. , Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 2170–2175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sequeira V., et al. , ADP-stimulated contraction: A predictor of thin-filament activation in cardiac disease. Proc. Natl. Acad. Sci. U.S.A. 112, E7003–7012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonino P., Kiss B., Gohlke J., Smith J. E. III, Granzier H., Fine mapping titin’s C-zone: Matching cardiac myosin-binding protein C stripes with titin’s super-repeats. J. Mol. Cell Cardiol. 133, 47–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luther P. K., et al. , Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. U.S.A. 108, 11423–11428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McConnell B. K., et al. , Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 104, 1235–1244 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barefield D., Kumar M., de Tombe P. P., Sadayappan S., Contractile dysfunction in a mouse model expressing a heterozygous MYBPC3 mutation associated with hypertrophic cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 306, H807–815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barefield D. Y., et al. , Ablation of the calpain-targeted site in cardiac myosin binding protein-C is cardioprotective during ischemia-reperfusion injury. J. Mol. Cell Cardiol. 129, 236–246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett P., Craig R., Starr R., Offer G., The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J. Muscle Res. Cell Motil. 7, 550–567 (1986). [DOI] [PubMed] [Google Scholar]
- 37.Tonino P., et al. , The giant protein titin regulates the length of the striated muscle thick filament. Nat. Commun. 8, 1041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korte F. S., McDonald K. S., Harris S. P., Moss R. L., Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ. Res. 93, 752–758 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Tesi C., Colomo F., Nencini S., Piroddi N., Poggesi C., The effect of inorganic phosphate on force generation in single myofibrils from rabbit skeletal muscle. Biophys. J. 78, 3081–3092 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poggesi C., Tesi C., Stehle R., Sarcomeric determinants of striated muscle relaxation kinetics. Pflugers Arch. 449, 505–517 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Lynch T. L. t., et al. , Amino terminus of cardiac myosin binding protein-C regulates cardiac contractility. J. Mol. Cell Cardiol. 156, 33–44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touma A. M., et al. , Nanosurfer assay dissects beta-cardiac myosin and cardiac myosin-binding protein C interactions. Biophys. J. 121, 2449–2460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Previs M. J., et al. , Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation–contraction coupling. Sci. Adv. 1, e1400205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huxley A. F., Simmons R. M., Rapid “give” and the tension “shoulder” in the relaxation of frog muscle fibres. J. Physiol. 210, 32P–33P (1970). [PubMed] [Google Scholar]
- 45.Previs M. J., et al. , Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc. Natl. Acad. Sci. U.S.A. 113, 3239–3244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colomo F., Nencini S., Piroddi N., Poggesi C., Tesi C., Calcium dependence of the apparent rate of force generation in single striated muscle myofibrils activated by rapid solution changes. Adv. Exp. Med. Biol. 453, 373–381 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Brenner B., Effect of Ca2+ on cross-bridge turnover kinetics in skinned single rabbit psoas fibers: Implications for regulation of muscle contraction Proc. Natl. Acad. Sci. U.S.A. 85, 3265–3269 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barefield D. Y., et al. , Mybphl null mouse atrial mass spectrometry data. Barefield Lab Database. https://www.barefieldlab.org/_files/ugd/f24ee7_afcf45448b544244b1d45b04d41adb86.xlsx. Deposited 28 November 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Complete mass spectrometry data set is available publicly on Barefield Lab Database (48).