Abstract
A 4,103-bp long DNA fragment containing the structural gene of a gentisate 1,2-dioxygenase (EC 1.13.11.4), gtdA, from Sphingomonas sp. strain RW5 was cloned and sequenced. The gtdA gene encodes a 350-amino-acid polypeptide with a predicted size of 38.85 kDa. Comparison of the gtdA gene product with protein sequences in databases, including those of intradiol or extradiol ring-cleaving dioxygenases, revealed no significant homology except for a low similarity (27%) to the 1-hydroxy-2-naphthoate dioxygenase (phdI) of the phenanthrene degradation in Nocardioides sp. strain KP7 (T. Iwabuchi and S. Harayama, J. Bacteriol. 179:6488–6494, 1997). This gentisate 1,2-dioxygenase is thus a member of a new class of ring-cleaving dioxygenases. The gene was subcloned and hyperexpressed in E. coli. The resulting product was purified to homogeneity and partially characterized. Under denaturing conditions, the polypeptide exhibited an approximate size of 38.5 kDa and migrated on gel filtration as a species with a molecular mass of 177 kDa. The enzyme thus appears to be a homotetrameric protein. The purified enzyme stoichiometrically converted gentisate to maleylpyruvate, which was identified by gas chromatography-mass spectrometry analysis as its methyl ester. Values of affinity constants (Km) and specificity constants (Kcat/Km) of the enzyme were determined to be 15 μM and 511 s−1 M−1 × 104 for gentisate and 754 μM and 20 s−1 M−1 × 104 for 3,6-dichlorogentisate. Three further open reading frames (ORFs) were found downstream of gtdA. The deduced amino acid sequence of ORF 2 showed homology to several isomerases and carboxylases, and those of ORFs 3 and 4 exhibited significant homology to enzymes of the glutathione isomerase superfamily and glutathione reductase superfamily, respectively.
Large amounts of aromatic compounds have been released into the environment during the last decades as a result of extensive production of industrial chemicals and agricultural applications of pesticides. Many of these compounds, particularly the chlorinated derivatives, are toxic, even at low concentrations, and persist in the environment (14, 39). Numerous microorganisms have been isolated which degrade xenobiotic aromatic compounds through aerobic or anaerobic degradative reactions (16, 17, 34, 46, 47). A wide variety of polycyclic and homocyclic aromatic compounds are aerobically transformed to a limited number of central dihydroxylated intermediates like catechol, protocatechuate, or gentisate. Whereas catabolic pathways for catechol and protocatechuate have been extensively characterized (22, 35), little is known about gentisate degradation.
Gentisic acid (2,5-dihydroxybenzoic acid) is a key intermediate in the aerobic degradation of such aromatic compounds as dibenzofuran (15), naphthalene (18, 48), salicylate (40, 55), anthranilate (32), and 3-hydroxybenzoate (26). Degradation of gentisate is initiated by gentisate 1,2-dioxygenase (GDO; EC 1.13.11.4, gentisate:oxygen oxidoreductase), which cleaves the aromatic ring between the carboxyl and the vicinal hydroxyl group to form maleylpyruvate (30). Maleylpyruvate can be converted to central metabolites of the Krebs cycle either by cleavage to pyruvate and maleate (5, 24) or by isomerization to fumarylpyruvate and subsequent cleavage to fumarate and pyruvate (10, 31, 51).
Of the two well-studied classes of ring cleavage dioxygenases, intradiol enzymes, such as catechol 1,2-dioxygenase or protocatechuate 3,4-dioxygenase, contain an Fe3+ atom in the catalytic center and cleave the aromatic substrate between two vicinal hydroxyl groups (7, 37, 38), whereas dioxygenases of the extradiol class, such as catechol 2,3-dioxygenase or protocatechuate 4,5-dioxygenase, contain Fe2+ and cleave the aromatic substrate adjacent to two vicinal hydroxyl groups (1, 13). Gentisate 1,2-dioxygenase cleaves aromatic rings containing hydroxyl groups situated para to one another. Although the mechanism of oxygen activation was proposed to be similar to that of enzymes of the extradiol dioxygenase class (20), and the active center contains Fe2+ (11, 21, 29, 49, 51), the Fe2+ is not bound to the enzyme by electron-donating ligands such as cysteine or tyrosine (21) as is the case for extradiol-cleaving dioxygenases (19). It is being assumed, therefore, that GDO represents a novel class of ring-cleaving dioxygenases.
GDOs have been purified and characterized from gram-positive bacteria of the genera Bacillus and Rhodococcus (29, 50) and gram-negative bacteria of the genera Klebsiella, Comamonas, and Moraxella (11, 21, 49), and amino-terminal sequences of GDOs from Comamonas testosteroni and Comamonas acidovorans have been determined (21), but until now, no complete sequence of any GDO or of a gene encoding GDO has been reported. Here we describe the cloning and sequencing of the gene encoding the GDO from Sphingomonas sp. strain RW5 and its partial characterization. This GDO represents a novel class of dioxygenases with very low similarity to any other known ring-cleaving dioxygenases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Sphingomonas sp. strain RW5 was isolated from aerobic sediment samples obtained from the Elbe River in Hamburg, Germany, on the basis of its ability to grow with Dicamba (3,6-dichloro-2-methoxybenzoate) as the sole source of carbon and energy. Other bacterial strains used in this study were Escherichia coli BL21(DE3)[pLys] (host for the T7 expression system, obtained from Stratagene, Heidelberg, Germany) F− dcm ompT hsdS(rB− mB−) galλ(DE3) and E. coli DH5α endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argFV169) φ80δlacZΔM15 F− λ− (obtained from Clontech, Palo Alto, Calif.). Plasmids used were pBluescript II SK(+) (Apr) and pCR-Script SK(+) (Apr), both from Stratagene, and pCR2.1 (Apr Kmr) from Invitrogen. Plasmid pT7-7 (Apr Cmr) was kindly supplied by Stan Tabor, Harvard Medical School, Boston, Mass.
Culture conditions.
Strain RW5 was routinely grown in mineral salts medium, pH 7.3, containing (per liter): 14.0 g of Na2HPO4 · 12H2O, 2.0 g of KH2PO4, 50 mg of Ca(NO3)2 · 4H2O, 0.5 g of (NH4)2SO4, 0.1 g of MgCl2 · 6H2O, 20 mg of Fe2NH4-citrate, and 0.1 ml of a trace-element solution (44). Agar plates contained 1.5% (wt/vol) purified agar (Oxoid, Hampshire, United Kingdom). Liquid cultures were incubated at 30°C on a rotary shaker at 160 rpm. E. coli strains were grown on Luria broth (LB) plates containing appropriate antibiotics. Liquid cultures were grown in baffled Erlenmeyer flasks.
Enzyme assays, kinetic measurements, and protein determination.
Gentisate 1,2-dioxygenase activity was assayed spectrophotometrically at 25°C by measuring the formation of maleylpyruvate at 330 nm (30). Cleavage of 3,6-dichlorogentisate by gentisate 1,2-dioxygenase was assayed by measuring product formation at 361 nm under the same conditions. The molar extinction coefficient used for maleylpyruvate was 10,200 M−1 cm−1 (54), and that for the chlorinated derivative was estimated to be 6,620 M−1 cm−1 (this work). Km values were determined from initial reaction rates by fitting initial velocities to the Michaelis-Menten equation by nonlinear regression. One unit of activity was defined as the turnover of 1 μmol of (chloro)gentisate per min. Protein concentrations were determined by the method of Bradford (6), with bovine serum albumin as the standard.
Enzyme purification.
All purification steps were performed at 4°C with a Fast Protein Liquid Chromatography system from Pharmacia, Uppsala, Sweden.
(i) Preparation of cell extracts.
Cultures (3 liters each) of strain RW5 were grown to mid-exponential phase with Dicamba as the sole source of carbon and energy and harvested by centrifugation (13,000 × g). Cell pellets were washed three times with 20 mM Tris-HCl buffer (pH 7.0) and resuspended in 3 ml of the same buffer. The suspension was passed twice through a precooled French pressure cell (Aminco, Silver Spring, Md.) at a pressure of 76 MPa. Cell debris was removed by centrifugation at 35,000 × g for 30 min, and the resulting supernatant fluid was immediately used. Cell extracts of E. coli BL21(DE3) containing plasmid pJW48 were induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and disrupted by the freeze-thaw method previously described (2).
(ii) Ion-exchange chromatography I.
The cell extract was applied to a DEAE-Sepharose column prepared as described by the manufacturer (Pharmacia). The column was equilibrated in 20 mM Tris-HCl buffer, pH 7.0 (buffer A), and developed with a linear gradient of 0 to 1 M NaCl in buffer A. Fractions were assayed for GDO activity, and active fractions were pooled and concentrated to less than 1 ml with the Centriprep-30 system (Amicon Inc., Beverly, Mass.).
(iii) Ion-exchange chromatography II.
Pooled fractions from the DEAE Sepharose column were applied to a prepacked MonoQ HR10/10 column (Pharmacia) equilibrated with buffer A. Proteins were eluted with NaCl in buffer A, first with a 40-ml linear 0 to 0.25 M gradient, then with a 160-ml linear 0.25 to 0.35 M gradient, and finally with 40 ml of 1 M NaCl. Active fractions were pooled and concentrated to less than 1 ml with the Centriprep-30 system.
(iv) Gel filtration chromatography.
The pooled fractions obtained from the Mono Q column were loaded onto a prepacked Superdex 200 column (Pharmacia) equilibrated with 50 mM Tris-HCl buffer (pH 7.0) containing 100 mM NaCl and eluted with the same buffer and analyzed for activity. Active fractions were pooled and used for further studies.
Amino acid sequencing.
For N-terminal sequencing, the purified protein was transferred onto a ProBlot polyvinylidene difluoride membrane (Applied Biosystems, Weiterstadt, Germany) for 70 min at a constant current of 120 mA as described by the manufacturer. The polyvinylidene difluoride-blotted protein was sequenced by Edman degradation in the blot cartridge of an Applied Biosystems 494A Procise sequencer with the standard gas-phase cycles recommended by the manufacturer (Applied Biosystems). Peptide fragments were generated by carboxyamidomethylation of the protein and digestion in situ with endoproteinase Lys-C (sequencing grade; Promega, Heidelberg, Germany).
Determination of molecular mass. (i) Denatured polypeptide.
The subunit molecular mass of the GDO was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturating conditions with 12% (wt/vol) acrylamide gels according to the method of Laemmli (33). Gels were stained with Coomassie brilliant blue R-250. Broad-range molecular standards from Pharmacia were used to determine the sizes of the subunits.
(ii) Native polypeptide.
The native molecular mass of the enzyme was estimated by gel filtration chromatography on a prepacked Superdex-200 column (Pharmacia) with protein standards of the gel filtration calibration kit from Pharmacia.
Genetic methods and sequence analysis.
Standard procedures such as Southern blot experiments, DNA subcloning, and DNA manipulations were performed by the method of Sambrook et al. (45). Hybridization was performed at 68°C with an RPN2510 oven (Amersham, Buckinghamshire, United Kingdom). Total DNA from strain RW5 was extracted according to the CTAB protocol (4). [α-32P]dCTP and the Megaprime DNA labeling system used for the labeling of gene probes were purchased from Amersham. Biodyne B membrane (Pall, Glen Cove, N.Y.) and uncharged Qiabrane membrane (Qiagen, Hilden, Germany) were used for DNA blotting experiments. Plasmid DNA for sequencing was extracted with the Plasmid Maxi kit (Qiagen). Sequencing reactions on both strands were performed with the Applied Biosystems 373A DNA sequencer according to the protocols of the manufacturer for Taq cycle sequencing with fluorescent dye-labeled dideoxynucleotides, as described previously (28). Sequences were compiled and analyzed with the GeneWorks software package V2.45 (IntelliGenetics, Mountain View, Calif.). The computational resources of the European Molecular Biology Laboratory (Heidelberg, Germany) and the National Center for Biotechnology Information (Bethesda, Md.) were used for initial similarity searches. Sequence comparisons, calculations of evolutionary distances, and drawing of unrooted phylogenetic trees were done with GeneWorks software and the Phylips programs package.
Cloning of the gentisate 1,2-dioxygenase gene.
Two degenerate oligonucleotide primers, 5′-AARCCIYTITGGGARGT-3′ and 5′-GTICCIACCATICCRTC-3′, were designed that corresponded to the amino acid sequences representing the lowest-possible nucleotide redundancy of the N-terminal fragment and several Lys-C-generated fragments thought to be derived from the C-terminal region of the GDO. The primers were used for PCR under low-stringency conditions, with total DNA from RW5 as a template. A resulting 1.2-kb PCR product containing the determined N-terminal sequence of the GDO was cloned into the pCR-Script SK(+) vector and used as a gene probe for colony blots. Southern blots of SfuI-digested DNA of RW5 were hybridized under stringent conditions with the probe, and signals were analyzed on Biomax MR X-ray films (Kodak, Rochester, N.Y.).
For the construction of a partial gene library, SfuI-restricted DNA from strain RW5 was electrophoretically separated and a slice containing DNA fragments with sizes of 3.5 to 5.0 kb was excised from the gel. The DNA was electroeluted, dephosphorylated with calf intestine phosphatase, and cloned into pBluescript II KS(+) cleaved with AccI. The hybrid plasmids thereby generated were transformed into electro-competent E. coli DH5α cells, which were subsequently plated on LB plates containing 1.0 mM IPTG, 0.004% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and 0.1 mg of ampicillin per ml. Colony lifts were hybridized with the PCR-generated gene probe, and clones showing the strongest signals were selected for further studies.
Construction of overexpressing plasmid pJW48.
The synthetic oligonucleotide primers 5′-GGACATATGCAGCCAGTATTGGCCAATGATCAGC-3′, which contains an engineered NdeI site (underlined), and 5′-AAAGGATCCTCATAGATTTTCCTCTCGGAACAGC-3′, which contains an engineered BamHI site (underlined), were used for PCR, with pJW39 as a template. In order to minimize nucleotide misincorporation errors by DNA polymerase, the 1,052-bp fragment was amplified with VENT polymerase from New England Biolabs (Beverly, Mass.). Plasmid pJW45 was constructed by cloning the PCR product into the pCR-Script SK(+) vector. The plasmid insert was resequenced to control possible polymerase errors. The NdeI-BamHI fragment containing the complete sequence of the GDO gene was prepared by digesting pJW45 with NdeI and BamHI. Expression vector pJW48 was constructed by ligating this fragment into plasmid pT7-7, which had been previously digested with NdeI and BamHI. Hyperexpression of the recombinant GDO gene of pJW48 in E. coli BL21(DE3) was induced with 0.4 mM IPTG.
Production and identification of maleylpyruvate.
Purified GDO was used for the production of maleylpyruvate from gentisate in an enzyme assay containing 1 mM gentisate. The formation of a single product was monitored by reversed-phase high-pressure liquid chromatography (LC-10AD liquid chromatograph, DGU-3A degasser, SPD-M10A diode array detector, FCV-10AL solvent mixer, all from Shimadzu, Kyoto, Japan) on a reversed-phase type SC 125 RP8 column (125 by 4.6 cm; Bischoff, Leonberg, Germany) (Lichrospher 100 beads with a diameter of 5.0 μm). The aqueous solvent system (flow rate, 1.0 ml/min) contained 0.1% (vol/vol) ortho-phosphoric acid and 20% (vol/vol) methanol. The UV spectrum from 200 to 400 nm was monitored. Direct derivatization of the reaction mix (100 μl) for gas chromatography-mass spectrometry (GC-MS) analysis was performed with trimethylsulfonium hydroxide (Machery-Nagel, Düren, Germany). GC-MS analysis was performed with a GC-17A gas chromatograph (Shimadzu, Kyoto, Japan) equipped with an XTI-5 column from Resteck (Bellefonte, Pa.) and coupled with the QP-5000 mass spectrometer (Shimadzu) operating in the electron impact mode at 70 eV. The ion source temperature was 320°C. Helium was used as carrier gas, with a flow rate of 1.0 ml/min. The oven temperature was maintained at 60°C for 2 min and then increased to 150°C at a rate of 20°C/min, followed by an increase to 320°C at a rate of 6°C/min. Samples (1.0 μl each) were injected into the GC, operating in the splitless mode with an injector temperature of 270°C.
Chemicals, enzymes, and antibiotics.
3,6-Dichlorogentisate was synthesized as follows: 3,6-dichloro-2-methoxybenzoic acid (obtained from Promochem, Wesel, Germany) was nitrated in position 5 with nitric acid in concentrated sulfuric acid. The resulting nitro group was reduced to the amino group with Raney nickel in ethanolic solution. Subsequent diazotization with isoamylnitrite, followed by hydrolysis with dilute sulfuric acid at elevated temperatures, resulted in 3,6-dichlorogentisic acid whose structure was confirmed by nuclear magnetic resonance and MS analysis. All other chemicals were of the highest purity commercially available. Restriction enzymes were purchased from Amersham, Boehringer (Mannheim, Germany), MBI Fermentas (Vilnius, Lithuania), New England Biolabs, and United States Biochemical. T4 ligase was purchased from New England Biolabs. IPTG was obtained from Roth (Karlsruhe, Germany). Antibiotics were purchased from Sigma (St. Louis, Mo.). X-Gal was obtained from Biomol (Hamburg, Germany). Taq DNA polymerase and calf intestine phosphatase were obtained from Boehringer.
Nucleotide sequence accession number.
The nucleotide sequence presented in this article is deposited in EMBL/GenBank under accession no. AJ224977.
RESULTS AND DISCUSSION
Isolation of strain RW5.
Single strains were isolated from enrichment cultures with 3,6-dichloro-2-methoxybenzoate as the sole energy and carbon source by plating on agar medium containing the target compound. Strain RW5 is an aerobic, gram-negative, nonmotile rod and was identified as a Sphingomonas sp. strain on the basis of 16S ribosomal DNA sequencing.
Purification of gentisate 1,2-dioxygenase from strain RW5.
Cell extracts of strain RW5 bacteria, pregrown on 3,6-dichloro-2-methoxybenzoate, exhibited a GDO with specific activities of 0.426 U/mg with gentisate and 0.023 U/mg with 3,6-dichlorogentisate as the substrates.
GDO from cell extracts of Sphingomonas sp. strain RW5 bacteria, pregrown on the above-mentioned compound, was partially purified according to a two-step purification protocol consisting of the second type of anion-exchange chromatography described in Materials and Methods (ion-exchange chromatography II) and gel filtration chromatography to near homogeneity (>95%). The highly enriched polypeptide exhibited an approximate molecular mass of 40 kDa on SDS-PAGE. Proteins from this gel were blotted onto a ProBlot membrane, and the GDO band was excised for amino-terminal sequencing.
Amino-terminal sequence of gentisate 1,2-dioxygenase.
The 43-residue amino-terminal sequence of purified GDO was determined by automated Edman degradation to be Met-Gln- Pro-Val-Leu-Ala-Asn-Asp-Gln-Gln-Ala-Gln-Leu-Thr-Ala-Leu-Tyr-Asp-Glu-Met-Arg-Pro-Ala-Gly-Leu-Lys-Pro-Leu-Trp-Glu-Val-Leu-His-Ala-Leu-Val-Leu-Ala-Glu-Gln-Pro-Val-Leu-Ala-Asn-Asp-Gln-Gln-Ala-Gln-Leu-Thr-Ala-Leu-Tyr-Asp-Glu-Met-Arg-Pro-Ala-Gly-Leu-Lys-Pro-Leu-Trp-Glu-Val-Leu-His-Ala-Leu-Val-Leu-Ala-Glu-Pro-Ala-Glu- Leu. This amino-terminal sequence showed no homology to the known amino-terminal sequences of the strains Comamonas testosteroni and Comamonas acidovorans (21).
Cloning and sequencing of the gtdA gene encoding gentisate 1,2-dioxygenase.
A gene probe was constructed with degenerate primers based on the amino acid sequences of the N-terminal region and with peptides assumed to be derived from the C-terminal region of GDO. A clone that hybridized strongly with the gene probe was isolated from a partial gene library of strain RW5 DNA constructed in E. coli. The hybrid plasmid containing a 4,103-bp SfuI fragment cloned into pBluescript was termed pJW39. Cell suspensions of E. coli DH5α harboring plasmid pJW39 showed high activities of GDO. Both strands of the 4,103-bp insert of plasmid pJW39 were sequenced, and four open reading frames (ORFs) were identified. The first 43-amino-acid residues deduced from the sequence of ORF 1 were identical to those of the N terminus of the native enzyme purified from strain RW5. This gene of 1,052 bp in length, designated gtdA (Fig. 1), specifies a 350-amino-acid polypeptide with a predicted size of 38.85 kDa. The probable Shine-Dalgarno sequence is located six nucleotides upstream of the initiation codon. No obvious E. coli ς70 promoter consensus sequence upstream of the start codon of gtdA was detected.
FIG. 1.
Nucleotide and deduced amino acid sequence of the putative 1,053-bp gene gtdA. The sense DNA strand is shown. The putative Shine-Dalgarno sequence is underlined. A point indicates the stop codon.
The amino acid sequence deduced from gtdA was compared with those of proteins currently available in the National Center for Biotechnology Information and EMBL databases. The sequence of the gtdA gene product showed similarity with very few known polypeptides, including ring-cleaving dioxygenases of the intradiol (<13%) or extradiol class (<13%), except with the 1-hydroxy-2-naphthoate dioxygenase (phdI) of Nocardioides sp. strain KP7 (27%) (25). This dioxygenase, which is involved in phenanthrene degradation, catalyzes the cleavage of 1-hydroxy-2-naphthoate to trans-2′-carboxybenzalpyruvate, a reaction analogous to the cleavage of gentisate to maleylpyruvate. Interestingly, there is a region of 75 amino acids from residue 94 to 167 (GDO sequence numbering) which shows a significantly higher similarity (56% identity). Since this region may contain the active center of the enzyme, it will be of interest to determine whether they are members of the same class of dioxygenases, as the sequence comparison suggests. The sequence comparisons made above suggest that this GDO is a member of a new class of ring-cleaving dioxygenases.
Analysis of sequences downstream of the gentisate 1,2-dioxygenase gene.
Three additional ORFs, all of which are accompanied by upstream sequences exhibiting potential ribosomal binding sites, were found downstream of the gtdA gene. ORF 2, which overlaps the last four nucleotides of the gtdA gene, is predicted to encode a protein of 25 kDa. Its deduced amino acid sequence shows significant homology to a number of ORFs coding for putative isomerases and decarboxylases which may also be associated with genes of degradative pathways, for example, 45 and 34% identity to two ORFs of the dibenzo-p-dioxin- and dibenzofuran-degrading Sphingomonas sp. strain RW1 (3), 29% to an ORF of Methanococcus janashii (8), and 28% to HpcE of Synechocystis sp. (27). The gene product of ORF 2 may thus be involved in an isomerization reaction of the gentisate pathway, such as the isomerization of maleylpyruvate to fumarylpyruvate (10), but the real function remains unknown so far.
ORF 3, whose start codon is located five nucleotides downstream of ORF 2, is predicted to encode a protein with a predicted size of 28 kDa. Its deduced amino acid sequence shows the greatest homology to enzymes of the glutathione S-transferase (GST) superfamily: 32% identity to PcpC of Sphingomonas flava (41), 27% to LigF of Sphingomonas paucimobilis (36), and 25% to ParB of Nicotiana tabacum (52). Even though the highest level of homology found is only 32%, amino acids highly conserved in the alpha, pi, mu, and theta classes of the GST superfamily (53) are present in the predicted amino acid sequence of ORF 3. GST-encoding genes, such as bphK (23), or ORF 3 in the tft operon (12), have been found in numerous operons or gene clusters specifying ring cleavage and further metabolism of aromatic compounds, thus suggesting a role of GST in these pathways. Crawford and Frick reported a glutathione-dependent isomerization of maleylpyruvate to fumarylpyruvate in the gentisate pathway in Moraxella (10). It will therefore be of interest to determine whether the gene product of ORF 3 plays a functional role in gentisate degradation in RW5.
ORF 4, beginning 31 nucleotides downstream of ORF 3, is predicted to encode a 48-kDa protein. Analysis of its deduced amino acid sequence revealed significant homology to glutathione reductases of many organisms. The highest homologies were 50% to the gor gene of Pseudomonas aeruginosa (43), 49% to TftF of Burkholderia cepacia, involved in the degradation of 2,4,5-trichlorophenoxyacetate (12), and 41% to the gor gene of Streptococcus thermophilus (42). The highly conserved amino acids involved in the binding of NADPH, the substrate glutathione disulphide, and the redox-active cysteins (43), were found in the predicted amino acid sequence of ORF4. The physiological role of the gene product of ORF 4 could involve providing reduced glutathione to the GST encoded by ORF 3.
Expression of recombinant gentisate 1,2-dioxygenase in E. coli.
A pair of synthetic oligonucleotide primers was designed to amplify the gene encoding GDO as described in Materials and Methods. The resulting 1,060-bp DNA fragment was subcloned into the expression vector pT7-7 to construct pJW48. E. coli BL21(DE3) cells containing pJW48 were cultivated in the presence of 0.4 mM IPTG in LB medium at 30°C. Cell extracts of these bacteria showed a specific activity of GDO of 2.83 U/mg of protein. Induced cells of E. coli BL21(DE3) without pJW48 exhibited a specific activity of less than 0.001 U/mg of protein. These results confirm that the 1,052-bp ORF represents the structural gene of GDO.
Purification and characterization of gentisate 1,2-dioxygenase hyperexpressed in E. coli BL21.
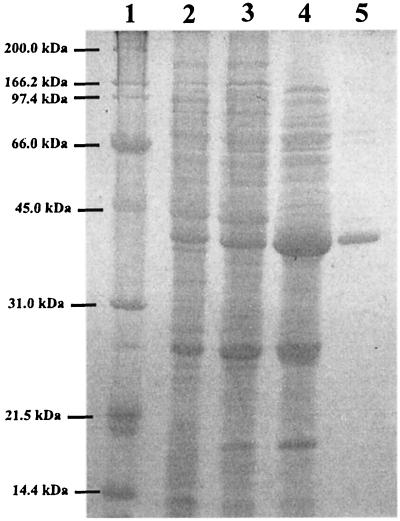
GDO was purified to homogeneity as indicated in Materials and Methods (see also Fig. 2). The results of the purification are summarized in Table 1 and resulted in a 42-fold purification of the specific activity. The molecular mass of GDO estimated by gel filtration chromatography was 178 ± 7.0 kDa (mean ± standard deviation). On SDS-PAGE, GDO migrated as a single band of approximately 39 ± 1.0 kDa (Fig. 2). The molecular mass calculated from the nucleotide sequence is 38.85 kDa. GDO therefore appears to be a homotetramer. Similar results have been reported for the GDOs of Arthrobacter sp. strain GFB 100 (9), C. testosteroni ATCC 49249, C. acidovorans ATCC 17438 (21), and Moraxella osloensis (11). The isoelectric point of our GDO, calculated on the basis of the nucleotide sequence with the GeneWorks software package, was 5.78, very similar to those reported for the GDOs from C. testosteroni ATCC 49249 (21) and Arthrobacter sp. strain GFB100 (9). The pH optimum of the enzyme activity was around 7.0, with a significant decrease in activity below pH 6.0 and above pH 8.0. The enzyme showed a broad activity maximum from pH 6.8 to 8.0 in different buffers such as Tris-HCl or phosphate buffers, as has been reported for other GDOs (11, 21, 29, 49).
FIG. 2.
SDS-PAGE analysis of purified GDO from hyperexpressed gtdA. Protein samples were separated by electrophoresis on a 12% polyacrylamide gel and stained with Coomassie blue. Lane 1, protein standards (in kilodaltons); lane 2, E. coli BL21 (DE3, pJW48) cell extract from induced whole cells; lane 3, GDO fraction from fast-flow DEAE-Sepharose chromatography; lane 4, GDO fraction from Mono Q separation; lane 5, GDO fraction after gel filtration.
TABLE 1.
Purification of recombinant GDO
| Purification step | Vol (ml) | Protein (mg) | Total activity (U) | Sp act (U/mg) | Recovery (%) | Purifi- cation (-fold) |
|---|---|---|---|---|---|---|
| Cell extract | 25 | 674.6 | 1.909 | 0.003 | 100.0 | 1 |
| DEAE-Sepharose | 12 | 120.7 | 1.343 | 0.011 | 70.4 | 3.9 |
| Mono Q | 3.5 | 13.1 | 0.829 | 0.063 | 43.4 | 22.4 |
| Gel filtration | 5.0 | 0.3 | 0.031 | 0.119 | 1.7 | 42.4 |
The purified GDO was highly unstable; it lost 70% of its initial activity when it was stored at −70°C for 2 weeks and 90% of its initial specific activity when it was stored at 4°C for 72 h. The polypeptide could not be stabilized by the addition of reagents such as dithiothreitol (1 mM), ethanol (10% vol/vol), PefaBlock (1 mM), and ferrous sulfate (100 μM). Ammonium sulfate at concentrations of 10% (wt/vol) or more and temperatures of 60°C and above rapidly inactivated the enzyme. Poor stability has also been observed for the GDO purified from Klebsiella pneumoniae M5a1 (49), whereas the GDOs from C. testosteroni and C. acidovorans retained about 85% of their activities when they were stored for 1 year in liquid nitrogen (21). During purification, the GDOs of these Comamonas strains maintained their active form when Fe2+ and cysteine were added to the buffers. Otherwise, activity was lost rapidly and could be restored only partially (20, 21). Previous studies suggested that iron is required for active GDO (11, 49, 51). Harpel and Lipscomb (20, 21) showed, on the basis of EPR studies, that the Fe2+ has no electron-donating ligands such as cysteine or tyrosine in the active center and proposed that the iron binds directly to the organic substrate. It is thus possible that the Fe2+ is a weakly associated cofactor which can be easily abstracted from the enzyme during purification procedures (29). Such irreversible loss of Fe2+ might be the reason for the loss of activity of GDO from strain RW5 during storage.
Identification of maleylpyruvate as the product of cleavage of gentisate.
The ring cleavage product from gentisate formed by GDO has previously been proposed on the basis of a UV spectrum to be maleylpyruvate (30), but so far this has not been confirmed unambiguously by determination of the structure of the product, presumably because maleylpyruvate is labile during extraction with organic solvents from acidified aqueous buffer systems.
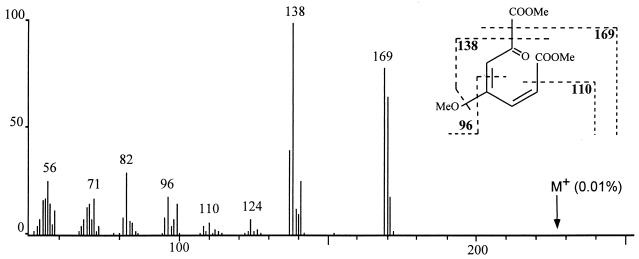
In order to confirm the structure of the reaction product, a 1 mM solution of gentisate was converted stoichiometrically with purified GDO to a single, stable product whose absorption spectrum showed a maximum at 330 nm, as has been described for maleylpyruvate (30). Methylation of the product was performed by direct derivatization of the compound in the aqueous buffer system. One main signal was obtained by GC-MS analysis. The mass spectrum presented in Fig. 3 shows a fragmentation pattern which is in accord with the predicted structure of maleylpyruvate. The molecular ion signal (m/z = 228) was obtained at very low intensity (<0.1%).
FIG. 3.
Mass spectrum (70eV) of trimethylsulfonium hydroxide-derivatized maleylpyruvate.
Turnover characteristics.
GDO activity was assayed at different substrate concentrations in an air-saturated buffer for determination of the Km value (0.5 to 200 μM for gentisate and 30 to 3,000 μM for 3,6-dichlorogentisate). The Km value for gentisate was estimated to be 15 μM, the Vmax was 0.119 U/mg, and the Kcat/Km value was 511 s−1 M−1 × 104. The Km value for 3,6-dichlorogentisate was 754 μM and thus 50-fold higher than for the unchlorinated gentisate. The Vmax with this substrate was 0.204 U/mg, and the Kcat/Km value was 20 s−1 M−1 × 104, i.e., 30-fold lower than with the unchlorinated substrate.
The Km value for gentisate determined here with GDO from RW5 was twofold higher than previously reported for that of M. osloensis (11) but four- to fivefold lower than those described for other strains (9, 21, 49). The turnover numbers of the GDO from RW5 are similar to those reported for GDOs of C. testosteroni and C. acidovorans (21).
From the Kcat/Km values for gentisate and 3,6-dichlorogentisate it can be seen that the chlorosubstituted substrate is well converted by the GDO of RW5. Several types of substituents, including bromine and fluorine at position 3 of gentisate, have been reported to be tolerated by the GDO of C. acidovorans (21).
Sequence analysis of the cloned GDO from cells of strain RW5 grown with 3,6-dichloro-2-methoxybenzoate suggests that it represents a member of a new class of ring-cleaving dioxygenases. The kinetic studies indicated that the cloned GDO tolerates unchlorinated gentisate as well as dichlorinated gentisate as substrates. Although the possibility that strain RW5 harbors another GDO cannot be ruled out, it is likely that our cloned GDO plays an essential role in the degradation of 3,6-dichloro-2-methoxybenzoate.
ACKNOWLEDGMENTS
We thank Edward R. B. Moore and his group for help in sequencing and Jean Armengaud for valuable discussions.
Part of the present work was supported by BMBF grant 0318896C. K. N. Timmis expresses gratitude to the Fonds der Chemischen Industrie for generous support.
REFERENCES
- 1.Arciero D M, Lipscomb J D, Huynh B H, Kent T A, Munck E. EPR and Mössbauer studies of protocatechuate 4,5-dioxygenase. Characterization of a new Fe2+ environment. J Biol Chem. 1983;258:14981–14991. [PubMed] [Google Scholar]
- 2.Armengaud J, Jouanneau Y. Overexpression in Escherichia coli of the FdxA gene encoding Rhodobacter capsulatus ferredoxin II. Protein Expr Purif. 1995;6:176–184. doi: 10.1006/prep.1995.1022. [DOI] [PubMed] [Google Scholar]
- 3.Armengaud J, Timmis K N. Molecular characterization of Fdx1, a putiodaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem. 1997;247:833–842. doi: 10.1111/j.1432-1033.1997.00833.x. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons Inc.; 1994. [Google Scholar]
- 5.Bayly R C, Chapman P J, Dagley S, Di Bernardino D. Purification and some properties of maleylpyruvate hydrolase and fumarylpyruvate hydrolase from Pseudomonas alcaligenes. J Bacteriol. 1980;143:70–77. doi: 10.1128/jb.143.1.70-77.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Bull C, Ballou D P. Purification and properties of protocatechuate 3,4-dioxygenase from Pseudomonas putida. A new iron to subunit stoichiometry. J Biol Chem. 1981;256:12673–12680. [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Chen C-M, Tomasek P H. Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993. Washington, D.C: American Society for Microbiology; 1993. Purification and properties of gentisate 1,2-dioxygenase from Arthrobacter sp. strain GFB 100, abstr. K-102; p. 278. [Google Scholar]
- 10.Crawford R L, Frick T D. Rapid spectrophotometric differentiation between glutathione-dependent and glutathione-independent gentisate and homogentisate pathways. Appl Environ Microbiol. 1977;34:170–174. doi: 10.1128/aem.34.2.170-174.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford R L, Hutton S W, Chapman P J. Purification and properties of gentisate 1,2-dioxygenase from Moraxella osloensis. J Bacteriol. 1975;121:794–799. doi: 10.1128/jb.121.3.794-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fewson C A. Biodegradation of xenobiotic and other persistent compounds: the causes of recalcitrance. Trends Biotechnol. 1988;6:148–153. [Google Scholar]
- 15.Fortnagel P, Harms H, Wittich R-M, Krohn S, Meyer H, Sinnwell V, Wilkes H, Francke W. Metabolism of dibenzofuran by Pseudomonas sp. strain HH69 and the mixed culture HH27. Appl Environ Microbiol. 1990;56:1148–1156. doi: 10.1128/aem.56.4.1148-1156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghisalba O. Chemical wastes and their biodegradation—an overview. Experientia. 1983;398:1247–1263. doi: 10.1007/BF01990362. [DOI] [PubMed] [Google Scholar]
- 17.Gibson D T. Microbial degradation of aromatic compounds. Science. 1968;161:1093–1097. [PubMed] [Google Scholar]
- 18.Grund E, Denecke B, Eichenlaub R. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl Environ Microbiol. 1992;58:1874–1877. doi: 10.1128/aem.58.6.1874-1877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 20.Harpel M R, Lipscomb J D. Gentisate 1,2-dioxygenase from Pseudomonas. Substrate coordination to active site Fe2+ and mechanism of turnover. J Biol Chem. 1990;265:22187–22196. [PubMed] [Google Scholar]
- 21.Harpel M R, Lipscomb J D. Gentisate 1,2-dioxygenase from Pseudomonas. Purification, characterization, and comparison of the enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans. J Biol Chem. 1990;265:6301–6311. [PubMed] [Google Scholar]
- 22.Harwood C S, Parales R E. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 23.Hofer B, Backhaus S, Timmis K N. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene. 1994;144:9–16. doi: 10.1016/0378-1119(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 24.Hopper D J, Chapman P J, Dagley S. Enzymic formation of d-malate. Biochem J. 1968;110:798–800. doi: 10.1042/bj1100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwabuchi T, Harayama S. Biochemical and genetic characterization of 2-carboxybenzaldehyde dehydrogenase, an enzyme involved in phenanthrene degradation by Nocardioides sp. strain KP7. J Bacteriol. 1997;179:6488–6494. doi: 10.1128/jb.179.20.6488-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones D C, Cooper R A. Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch Microbiol. 1990;154:489–495. doi: 10.1007/BF00245233. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 28.Karlson U, Rojo F, van Elsas J D, Moore E. Genetic and serological evidence for the recognition of four pentachlorophenol-degrading bacterial strains as a species of the genus Sphingomonas. Syst Appl Microbiol. 1996;18:539–548. [Google Scholar]
- 29.Kiemer P, Tshisuaka B, Fetzner S, Lingens F. Degradation of benzoate via benzoyl-coenzyme A and gentisate by Bacillus stearothermophilus PK1, and purification of gentisate 1,2-dioxygenase. Biol Fertil Soils. 1996;23:307–313. [Google Scholar]
- 30.Lack L. The enzymic oxidation of gentisic acid. Biochim Biophys Acta. 1959;34:117–123. doi: 10.1016/0006-3002(59)90239-2. [DOI] [PubMed] [Google Scholar]
- 31.Lack L. Enzymic cis-trans isomerization of maleylpyruvic acid. J Biol Chem. 1961;236:2835–2840. [PubMed] [Google Scholar]
- 32.Ladd J N. Oxidation of anthranilic acid by a species of Achromobacter isolated from soil. Nature. 1962;194:1099–1100. doi: 10.1038/1941099b0. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Leisinger T. Microorganism and xenobiotic compounds. Experientia. 1983;39:1183–1191. doi: 10.1007/BF01990355. [DOI] [PubMed] [Google Scholar]
- 35.Lipscomb J D, Orville A M. Mechanistic aspects of dihydroxybenzoate dioxygenases. In: Sigel H, Sigel A, editors. Metal ions in biological systems. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 243–298. [Google Scholar]
- 36.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound beta-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 37.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 38.Neidle E L, Hartnett C, Bonitz S, Ornston L N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol. 1988;170:4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilson A H, Allard A-S, Remberger M. Biodegradation and transformation of recalcitrant compounds. In: Hutzinger O, editor. The handbook of environmental chemistry. Berlin, Germany: Springer-Verlag KG; 1985. pp. 29–86. [Google Scholar]
- 40.Ohmoto T, Sakai K, Hamada N, Ohe T. Salicylic acid metabolism through a gentisate pathway by Pseudomonas sp. TA-2. Agric Biol Chem. 1991;55:1733–1737. [Google Scholar]
- 41.Orser C S, Dutton J, Lange C, Jablonski P, Xun L, Hargis M. Characterization of a Flavobacterium glutathione S-transferase gene involved in reductive dechlorination. J Bacteriol. 1993;175:2640–2644. doi: 10.1128/jb.175.9.2640-2644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pebay M, Holl A C, Simonet J M, Decaris B. Characterization of the gor gene of the lactic acid bacterium Streptococcus thermophilus CNRZ368. Res Microbiol. 1995;146:371–383. doi: 10.1016/0923-2508(96)80283-x. [DOI] [PubMed] [Google Scholar]
- 43.Perry A C, Ni Bhriain N, Brown N L, Rouch D A. Molecular characterization of the gor gene encoding glutathione reductase from Pseudomonas aeruginosa: determinants of substrate specificity among pyridine nucleotide-disulphide oxidoreductases. Mol Microbiol. 1991;5:163–171. [PubMed] [Google Scholar]
- 44.Pfennig N, Lippert K D. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol. 1966;55:245–256. [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Singleton I. Microbial metabolism of xenobiotics: fundamental and applied research. J Chem Technol Biotechnol. 1994;59:9–23. [Google Scholar]
- 47.Smith M R. The biodegradation of aromatic compounds by bacteria. In: Ratledge C, editor. Physiology of biodegradative microorganisms. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1990. pp. 191–206. [Google Scholar]
- 48.Starovoitov I I, Nefedova G I, Yakovlev A M, Zyakun A M, Adanin V M. Gentisic acid as a microbial oxidation product of naphthalene. Izv Akad Nauk SSSR Ser Khim. 1975;9:2091–2092. [Google Scholar]
- 49.Suarez M, Ferrer E, Martin M. Purification and biochemical characterization of gentisate 1,2-dioxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol Lett. 1996;143:89–95. doi: 10.1111/j.1574-6968.1996.tb08466.x. [DOI] [PubMed] [Google Scholar]
- 50.Suemori A, Kurane R, Tomizuka N. Purification and properties of gentisate 1,2-dioxygenase from Rhodococcus erythropolis S-1. Biosci Biotechnol Biochem. 1993;57:1781–1783. [Google Scholar]
- 51.Sugiyama S I, Yano K, Tanaka H, Komagata K, Arima K. Metabolism of aromatic compounds by bacteria, I. Gentisic acid oxidase and protocatechuic acid oxidases of Pseudomonas ovalis S-5. J Gen Appl Microbiol. 1958;4:223–240. [Google Scholar]
- 52.Takahashi Y, Nagata T. parB, an auxin-regulated gene encoding glutathione S-transferase. Proc Natl Acad Sci USA. 1992;89:56–59. doi: 10.1073/pnas.89.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheelis M L, Palleroni N J, Stanier R Y. The metabolism of aromatic acids by Pseudomonas testosteroni and P. acidovorans. Arch Mikrobiol. 1967;59:302–314. doi: 10.1007/BF00406344. [DOI] [PubMed] [Google Scholar]
- 55.Yano K, Arima K. Metabolism of aromatic compounds by bacteria, II. m-hydroxybenzoic acid hydroxylase A and B; 5-dehydroshikimic acid, a precursor of protocatechuic acid: a new pathway from salicylic acid to gentisic acid. J Gen Appl Microbiol. 1958;4:241–258. [Google Scholar]