Abstract
Aerobically growing wild-type strains of Saccharomyces cerevisiae are unable to take exogenously supplied sterols from media. This aerobic sterol exclusion is vitiated under anaerobic conditions, in heme-deficient strains, and under some conditions of impaired sterol synthesis. Mutants which can take up sterols aerobically in heme-competent cells have been selected. One of these mutations, designated upc2-1, gives a pleiotropic phenotype in characteristics as diverse as aerobic accumulation of sterols, total lipid storage, sensitivity to metabolic inhibitors, response to altered sterol structures, and cation requirements. During experiments designed to ascertain the effects of various cations on yeast with sterol alterations, it was observed that upc2-1 was hypersensitive to Ca2+. Using resistance to Ca2+ as a screening vehicle, we cloned UPC2 and showed that it is YDR213W, an open reading frame on chromosome IV. This belongs to a fungal regulatory family containing the Zn(II)2Cys6 binuclear cluster DNA binding domain. The single guanine-to-adenine transition in upc2-1 gives a predicted amino acid change from glycine to aspartic acid. The regulatory defect explains the semidominance and pleiotropic effects of upc2-1.
Wild-type cells of Saccharomyces cerevisiae do not accumulate sterol from a growth medium when they are grown aerobically; however, during anaerobic growth, sterol uptake is required, as yeast cannot synthesize ergosterol in the absence of oxygen. This phenomenon, called aerobic sterol exclusion (10), seems counterintuitive as one may expect a selective advantage for aerobic uptake of sterol when it is available in the medium. A yeast that could take up sterol in the presence of oxygen would obviate the need for the very expensive synthesis of the 28-carbon ergosterol, a required membrane component in yeast cells.
Although the mechanism by which yeast cells prohibit uptake of sterol is not known, it is well understood that synthesis of heme is an important determinant in the regulation of sterol uptake (5, 22). Mutations in genes involved in heme biosynthesis prevent the synthesis of heme components, which results in defects in heme-dependent processes such as respiration, unsaturated fatty acid synthesis, sterol synthesis, and aerobic sterol exclusion. Mutations in genes controlling heme synthesis allow the growth of sterol auxotrophs by permitting the uptake of exogenous sterol to complement the block in ergosterol synthesis. This technique has been used to study the structural requirements for sterol in the growth and metabolism of S. cerevisiae (16).
While heme competency is an important factor in regulating sterol uptake, other factors may play significant roles. The level of ergosterol in S. cerevisiae also regulates the uptake of sterol, as treatment of cells with the sterol biosynthesis inhibitor lovastatin reduces cellular levels of sterol and leads to a concomitant increase in the level of sterol uptake (9). In addition, inhibition of ergosterol synthesis with a different inhibitor, fenpropimorph, has a similar effect on sterol accumulation. Fenpropimorph is believed to increase sterol uptake by decreasing the intracellular ergosterol content (11). It should be noted that although heme is required for several steps in ergosterol synthesis, the ability of heme to regulate sterol uptake is in part independent of the regulation of ergosterol synthesis by heme. This independence is demonstrated by the fact that addition of large concentrations of δ-aminolevulenic acid to a strain containing the hem1 erg1 combination is not sufficient to prevent fully sterol uptake (6). δ-Aminolevulenic acid is the product of the enzyme encoded by HEM1. In strains with hem1 and no other heme defect, it permits heme synthesis.
In addition to mutations in heme biosynthesis, other genes that regulate the uptake of sterol in S. cerevisiae have been identified (2, 8, 12). The SUT1 gene was identified as a regulator of sterol uptake in a screen for genes that, when they were overexpressed, caused uptake of sterol (2). Expression of SUT1 on a high-copy-number plasmid leads to a modest 2.6-fold increase in the uptake of cholesterol by yeast, which is much less than the uptake of sterol seen in a hem1 mutant strain (9.6-fold increase). Nevertheless, SUT1 is considered to be a hypoxic gene, expressed highly when cells are cultured anaerobically, the cultural condition under which sterol uptake is greatest. The PDX3 gene, which encodes a pyridoxine (pyridoxamine) phosphate oxidase, is also involved in regulating sterol uptake, presumably through reduced aminolevulinate synthase activity, the first step in heme biosynthesis, which requires pyridoxal phosphate for activity (12).
Yet another sterol uptake control mutant (UPC20; upc2-1) was isolated by Lewis et al. (8) and was found to have multiple alterations in sterol metabolism. Strains containing the upc2-1 allele had increased rates of both sterol synthesis and uptake, although these strains made ergosterol as the principal sterol and were heme competent (8). The upc2-1 mutation was further shown to result in an inability of yeast cells to interconvert steryl ester and free sterol (6); however, this inability is most likely due to increased capacity for sterol synthesis, as inhibition of sterol synthesis in a strain containing a upc2-1 mutation results in conversion of steryl ester to free sterol (7). Since the first isolation of the upc2-1 allele, we have been interested in cloning the wild-type copy of the UPC2 gene. Our initial attempts at cloning the UPC2 gene were unsuccessful due to the absence of an easily scored phenotype for the upc2-1 mutation. We show here that we have successfully cloned the UPC2 gene by using a differential Ca2+ phenotype. We also demonstrate that the upc2-1 allele is semidominant. The pleiotropic effects of upc2-1 arise from an alteration in YDR213W, an open reading frame in the GAL4 transcriptional activator family containing the Zn(II)2Cys6 binuclear cluster DNA binding motif.
MATERIALS AND METHODS
Materials.
The Difco products yeast extract, Bacto Agar, tryptone, peptone, dextrose, and yeast nitrogen base (without amino acids) were obtained from Fisher Scientific. Extraction solvents, potassium hydroxide, pyrogallol, sodium dodecyl sulfate, molecular-biological-grade agarose, phenol, formaldehyde, formamide, and inorganic chemicals were also from Fisher Scientific. Cholesterol, amino acids, nucleic acids, 2-mercaptoethanol, Tergitol Nonidet P-40, tyloxapol, and polyethylene glycol were obtained from Sigma Chemical Co. Restriction DNA endonucleases were purchased from New England Biolabs and Bethesda Research Laboratories, Inc. Expand High Fidelity buffer and Expand High Fidelity polymerase were from Boehringer Mannheim. [14C]cholesterol, used in sterol uptake assays, was obtained from Amersham Life Science Inc.
Strains and culture conditions.
Yeast extract-peptone-dextrose (YPD), selective defined media with yeast nitrogen base, prespore, and sporulation media were made as described previously (17). Selective defined media were supplemented with amino acids and nitrogenous bases for plasmid and strain selection. Luria-Bertani medium was made with ampicillin (50 μg/ml) for plasmid selection according to previously described methods (19). Where Ca2+ ions were added as supplements to growth media, CaCl2 was added from a sterile 5 M stock solution after it was autoclaved. For YPD agar plates containing a gradient of Ca2+ ions, a bottom layer of YPD plus 750 mM CaCl2 was added first to a petri dish and allowed to solidify at an angle prior to the addition of an upper layer of an equivalent amount of YPD agar without additional CaCl2. All yeast cultures were incubated at 30°C; Escherichia coli cultures (DH5α and VQ580) were grown at 37°C.
S. cerevisiae strains used in this study are listed in Table 1. The previously described strain UPC20 (8) was crossed with strain 463-1C to produce SC2-1C and CJ2-A. The resulting strains were subsequently backcrossed with 463-1C. The isogenic strains were then used for most of the experiments in this study. The contribution and derivations of other strains are described below. Plasmid clone libraries were obtained as follows: YpH-1 came from P. Hieter, YCp50 came from the American Type Culture Collection, and YEp13 came from L. Robinson.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| 463-1C | MATa leu2 his3 trp1 ura3 | K.T.b |
| 463-1D | MATα leu2 his3 trp1 ura3 | K.T. |
| UPC20 | MATα upc2-1 | 8 |
| SC2-1C | MATα upc2-1 leu2 his3 trp1 ura3 | This study |
| CJ2-A | MATa upc2-1 leu2 his3 trp1 ura3 | This study |
| CJ625 | MATa upc2::URA3 leu2 ura3 his3 trp1 ura3 | This study |
| CJ296 | a/α leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | This study |
| CJ571 | a/α upc2-1/UPC2 leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | This study |
| CJ581 | a/α upc2::URA3/UPC2 leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | This study |
| CJ573 | a/α upc2::URA3/upc2-1 leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | This study |
| CJ577 | a/α upc2-1/upc2-1 leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | This study |
In this table ydr213::URA3 is designated upc2::URA3.
K.T., Kelly Tatchell, Louisiana State University Medical Center, Shreveport, La.
DNA manipulations and transformations.
All recombinant DNA procedures were carried out according to the methods of Sambrook et al. (19). DNA fragments were recovered from agarose gels by using a GeneClean kit (Bio 101). Primers used in PCRs were synthesized by Integrated DNA Technologies, Inc. All transformations into yeast were performed by the lithium acetate method (4). Transformation into E. coli for plasmid propagation was by the calcium chloride procedure (19). PCRs were performed in a Perkin-Elmer model 2400 thermocycler; conditions for the reaction are described below.
Sequencing YDR213W from the upc2-1 mutant.
Genomic DNA was isolated from SC2-1C (upc2-1) and 463-1C (UPC2). PCR was performed on SC2-1C to amplify the mutant YDR213W open reading frame. This was done with the following components: 2 μg each of the primers YDR-5 (5′-GCACCTTAGCGGGATCGCGT-3′) and YDR-3 (5′-ACCGGCCTCCACTCCAACCA-3′), 0.2 mM each deoxynucleotide, 1× Expand High Fidelity buffer, 2 mM MgCl2, and 3.5 U of Expand High Fidelity polymerase in a final volume of 100 μl. The mutant SC2-1C (upc2-1) DNA template was added at a concentration of 1.5 μg, and the wild type (463-1C) was added at a concentration of 1.3 μg. The reaction was performed under the following conditions: initial denaturation at 94°C for 2 min, followed by 35 cycles consisting of 94°C for 30 s, 60°C for 30 s, and 68°C for 3 min, with a final extension for 7 min at 68°C.
PCR with primers YDR-5 and YDR-3 resulted in a 3.6-kb band when the PCR product was run on a 1% TAE agarose gel and stained with ethidium bromide. This band was cut out of the gel and purified with a GeneClean III kit (Bio 101). The concentration of the purified sample was determined with a DNA Dipstick (Invitrogen). Sequencing primers were constructed with data from the S. cerevisiae Genome Database (Stanford University). All sequencing was performed at the Iowa State University DNA Sequencing Facility. Sequence analysis was done by comparison of SC2-1C sequence data to sequences in the yeast genome database with BLAST(n) searches. The region sequenced was 321 bp upstream from the start codon and 218 bp downstream from the stop codon of YDR213W.
To subclone the wild-type and mutant alleles of YDR213W, the same initial primers used for amplification and sequencing, YDR-5 and YDR-3, were used to amplify the region from −350 to +3227, which encompasses the YDR213W open reading frame (+1 to +2742). The Taq PCR Core Kit (Qiagen) was used in the amplification, and the 3.6-kb products, with 3′-A overhangs, were ligated directly into the EcoRV site of the pGEM-T vector (Promega). The cloned wild-type (UPC2) and mutant (upc2-1) alleles were then excised with SacI and SacII and ligated into the yeast shuttle vector pRS313 (20) to create plasmids pKS100 and pKS101, respectively.
[14C]cholesterol uptake analyses.
Analyses for sterol uptake were performed essentially as described by Lewis et al. (8) with modifications. The growth medium used for the uptake analyses was made by adding [14C]cholesterol to synthetic complete medium lacking leucine, uracil, or histidine in order to maintain selection of plasmids. To make the [14C]cholesterol medium, 6.25 μCi of labeled cholesterol was added to 5 mg of cold cholesterol in toluene and the mixture was dried under nitrogen gas. The cholesterol mixture was then resuspended in 0.4 ml of a 1:1 (vol/vol) mixture of ethanol and tyloxapol and added to 250 ml of growth medium. This routinely yields a specific activity of approximately 2,500 dpm/μg of cholesterol.
Individual 5-ml cultures were grown overnight and were used as inocula for 5-ml cultures of [14C]cholesterol-containing growth media. After an inoculation to a density of 2 × 105 cells/ml, the cultures were allowed to incubate in 15-ml plastic conical tubes for 48 h at 30°C. Following growth, the cultures were centrifuged and the cell pellets were washed twice with 0.5% Tergitol Nonidet P-40 in water. The cell pellets were then lyophilized, weighed, and resuspended in scintillation fluid for counting.
RESULTS
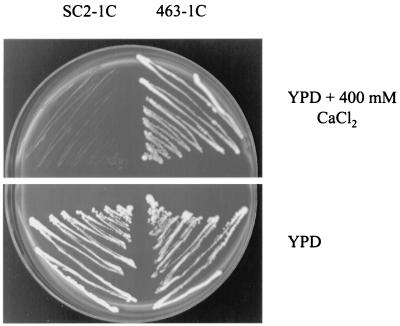
During a study of the effects of sterol alterations on the sensitivity of yeast to cations, it was observed that upc2-1 conferred sensitivity to several salts. It was substantially more sensitive to Ca2+ than the corresponding wild type, with a MIC of 500 versus 800 mM based on YPD-CaCl2 broth analysis. The difference in sensitivities was also evident with cultures on YPD agar plates with 400 mM CaCl2 (Fig. 1).
FIG. 1.
Growth of 463-1C (UPC2) and SC2-1C (upc2-1) on YPD agar medium with and without 400 mM CaCl2.
Transformations of SC2-1C (upc2-1) were performed with the multicopy yeast genomic library contained on YEp13 and the CEN+-based yeast genomic libraries YCp50 (URA3) and YpH-1 (LEU2). After several thousand transformants were screened, one which demonstrated improved resistance to CaCl2 was selected from the YpH-1 library. [14C]cholesterol uptake studies revealed that this transformant also accumulated significantly less exogenously supplied sterol than the mutant. It is important to recall that the wild-type phenotype is a very low level of sterol uptake. The original transformant, PH3, contained two plasmid species, both of which were independently isolated; SC2-1C (upc2-1) was retransformed with each plasmid separately. Of the two plasmids, one resulted in improved CaCl2 tolerance and reduced [14C]cholesterol uptake. This plasmid, named PH36, had an insert of approximately 10 kb (Fig. 2).
FIG. 2.
Subclones of the original isolate PH36 and complementation analysis. All growth experiments and [14C]cholesterol uptake assays were performed as described in Materials and Methods. Cholesterol uptake is expressed as micrograms of cholesterol per milligram (dry weight) of cells. SC2-1C (upc2-1) was transformed with each plasmid and tested for CaCl2 tolerance and sterol uptake. Each measurement is representative of the result of an independent assay, and all cultures were grown in duplicate. Controls for these experiments were strains 463-1C (UPC2) and SC2-1C (upc2-1). pFE5k-1 and pFE7k-10 fragments were isolated from the original 10-kb insert. Subsequent subclones were obtained from the 7.2-kb insert of pFE7k-10 (hence the absence of restriction sites from the diagram). N.A., not applicable.
Subsequent subclones of the PH36 insert were ligated into the multicopy YEp352 shuttle vector for restriction analysis and cloning and so that we could use sequencing oligonucleotides previously designed for YEp352. One 7-kb fragment, produced when PH36 was digested with XbaI, was ligated into YEp352, and when it was transformed back into SC2-1C, the result was a reduced sensitivity to CaCl2 (Fig. 2). This plasmid, pFE7k-10, also decreased the level of uptake of [14C]cholesterol by SC2-1C. With the oligonucleotides discussed above which were homologous to the insert-flanking vector sequence, 5′ and 3′ portions of pFE7k-10 were sequenced, yielding approximately 600 bases on both ends of the insert. Once the sequences were obtained, they were subjected to a BLAST(n) search of the S. cerevisiae Genome Database. This search placed the insert on chromosome IV. Its size was 7.2 kb, and three open reading frames, two complete and one partial, were identified. Of the two complete open reading frames present, one was the chaperonin alpha subunit (TCP1) (23), which was centrally located within the insert; the other was an open reading frame of unknown function (YDR213W) located close to the 5′ end of the cloned fragment. The 3′ end of the insert contained the partial sequence of the guanine nucleotide exchange factor (GCD6) (14) (Fig. 2).
We located convenient restriction sites which enabled us to isolate each open reading frame and test for complementation of the upc2-1 mutation. BamHI restriction produced a fragment of approximately 3.7 kb that contained the entire YDR213W open reading frame (2.7 kb). The fragment was ligated into BamHI-digested YEp352, and SC2-1C was transformed with the resulting plasmid (pFE3K-1) to test for complementation. An analysis conducted on YPD solid medium with 400 mM CaCl2 demonstrated that the tolerance of SC2-1C to CaCl2 was enhanced with pFE3k-1 present (Fig. 2). Uptake studies also revealed that the ability of SC2-1C to take up exogenously supplied sterol was diminished, with SC2-1C plus pFE3k-1 taking up eightfold less [14C]cholesterol than SC2-1C untransformed. HindIII-digested pFE7k-10 produced a 3.9-kb fragment which contained the entire TCP1 gene, the promoter for TCP2, and a truncated GCD6 gene. This fragment was subcloned into YEp352, and the resulting plasmid (pFE4k-H) was transformed into SC2-1C. Results showed no increased tolerance to CaCl2, indicating that neither open reading frame present on this insert was responsible for complementation.
Given the partial complementation of the upc2-1 allele by the YDR213W open reading frame, we sought to confirm through gene disruption the identification of YDR213W as UPC2. The YDR213W gene was inactivated by insertion and deletion by the PCR-based technique we refer to as the Belgian knockout (3). The 3.7-kb BamHI fragment containing YDR213W was inserted into pUC18, and this plasmid was used as the template in a PCR amplification with primers UPC2KO-5 (5′-ACAAGCTGATCAACTGAAGACAATAGAATA-3′) and UPC2KO-3 (5′-GATCTTTGATCATTAGGCGATGTCATTAAC-3′), designed to amplify the region flanking the YDR213W gene. PCR with these primers amplified in opposite directions, creating a DNA fragment that consisted of the ends of the YDR213W open reading frame and the entire pUC18 plasmid. After cutting the fragment at BclI sites which had been added at the 5′ end of the primer, the URA3 gene cassette from the YDp-U plasmid (1) was ligated into the BclI site. Although the last 459 bp of YDR213W remain in this construct, the URA3 gene cassette contains translational termination sites in all three reading frames, thus preventing possible construction of chimeric proteins (1).
The BamHI fragment with the null YDR213W open reading frame was used to create the null mutation in vivo by the one-step gene disruption technique (18). Strain CJ296, constructed by crossing 463-1C and 463-1D, was transformed with the null gene construct, and Ura+ colonies were screened by Southern blot analyses to identify null mutants. Of the 17 diploid Ura+ isolates screened, all 17 contained the YDR213W gene disruption ydr213::URA3. Upon sporulation and dissection of tetrads from six diploid transformants, a 2:2 segregation of Ura+/Ura− transformants was observed and all four spores were viable in each tetrad, indicating that disruption of YDR213W is not lethal under these conditions. Southern blot analysis of the segregated haploids confirmed disruption of YDR213W in five of the six diploid strains, where the Ura+ haploid progeny of one of the diploids transformed with the null gene construction demonstrated the presence of both the null construct and a full copy of YDR213W (data not shown). Curiously, the null mutants did not show the sensitivity to calcium ions or sterol uptake seen in strains containing the upc2-1 allele.
To ascertain if YDR213W is the UPC2 gene, we performed a series of tests to determine allelism between the ydr213::URA3 and upc2-1 mutant alleles. Strain CJ625 (ydr213::URA3) was crossed with SC2-1C (upc2-1), and the resultant diploid (CJ573) was found to accumulate sterol to a level comparable to that accumulated by the upc2-1 homozygous diploid (CJ577) (Table 2). Diploids from SC2-1C crossed with individual segregants from each of the other diploid null mutants also showed increases in accumulations of sterol compared to the level in the UPC2/upc2-1 diploid strain or to that in the wild-type diploid (data not shown). In addition, upon segregation of tetrads from the CJ625 × SC2-1C diploid, nearly all of the scored tetrads (61 of 67 scored tetrads) were found to be parental ditypes when spores were scored for the Ura+ (ydr213::URA3) and Ca2+-sensitive (upc2-1) phenotypes. These data suggest that the ydr213::URA3 and upc2-1 mutations are allelic.
TABLE 2.
Cholesterol uptake in upc2-1 and upc2::URA3 strains
| Strain | Relevant genotypea | Mean cholesterol uptake (μg of cholesterol/mg [dry weight] of cells) ± SD with plasmidb:
|
|
|---|---|---|---|
| YpH-1 | pUPC2 | ||
| 463-1D | UPC2 | 0.232 ± 0.041 | 0.214 ± 0.044 |
| SC2-1C | upc2-1 | 4.68 ± 1.1 | 0.771 ± 0.059 |
| CJ625 | upc2::URA3 | 0.160 ± 0.018 | 0.178 ± 0.024 |
| CJ296 | UPC2/UPC2 | 0.174 ± 0.018 | 0.178 ± 0.024 |
| CJ571 | upc2-1/UPC2 | 1.01 ± 0.065 | 0.226 ± 0.037 |
| CJ573 | upc2-1/upc2::URA3 | 3.82 ± 0.27 | 0.601 ± 0.21 |
| CJ577 | upc2-1/upc2-1 | 2.56 ± 0.79 | 0.573 ± 0.019 |
| CJ581 | upc2::URA3/upc2::URA3 | 0.279 ± 0.022 | 0.470 ± 0.33 |
In this table ydr213::URA3 is designated upc2::URA3.
Plasmid YpH-1 is the control plasmid lacking the YDR213W open reading frame, and plasmid pUPC2 contains the entire YDR213W coding sequence with the native 5′ untranslated sequence.
As further proof that YDR213W is UPC2, we inserted the 3.7-kb fragment with the YDR213W open reading frame into YpH-1, the low-copy-number LEU2 vector in which the original clone was found. This plasmid, pUPC2, was used to transform a variety of strains in order to determine if YDR213W would complement the upc2-1 phenotypes. The presence of YDR213W in SC2-1C reduced the uptake of sterol from 20-fold over that of the wild type to less than 4-fold over that of the wild type (Table 2). This result shows complementation of the uptake phenotype by YDR213W; however, YDR213W cannot fully complement the uptake phenotype. In our studies with the various diploid strains, we found a very interesting pattern of complementation of the uptake phenotype conferred by the upc2-1 allele. The presence of one copy of the upc2-1 allele in the heterozygous upc2-1/UPC2 diploid (CJ571) resulted in a sixfold increase in sterol uptake over the level found in the wild-type diploid, a result that we have observed consistently throughout our studies (Table 2). This indicates that the upc2-1 allele is semidominant. Cholesterol uptake in the haploid SC2-1C (upc2-1) exceeded the wild-type level by 20-fold. The addition of pUPC2 diminished the uptake of the heterozygous diploid to the wild-type level of sterol uptake (Table 2). As mentioned previously, the upc2-1/ydr213::URA3 diploid (CJ573) accumulated sterol to a high level comparable to the levels accumulated by both the haploid SC2-1C strain and the homozygous upc2-1/upc2-1 diploid strain (CJ577). The addition of pUPC2 also caused a decrease in the uptake of sterol in these strains to levels comparable to that of the SC2-1C strain with the pUPC2 plasmid. As expected, the pUPC2 plasmid had no effect on the uptake of sterol by the wild-type strain or by strains containing only the inactivated YDR213W gene, neither of which showed appreciable levels of sterol uptake.
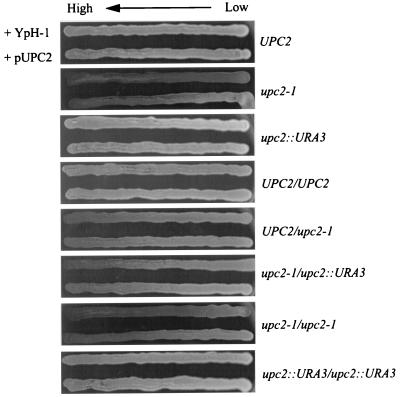
In addition to complementation of the uptake phenotype by pUPC2, we found that the presence of this plasmid could also complement the sensitivity to calcium ions conferred by the upc2-1 allele. We examined the sensitivity to calcium ions using YPD agar plates containing a gradient of calcium ions. In general, the effect of the pUPC2 plasmid in strains containing the upc2-1 allele closely mirrored the results of the sterol uptake analyses. The pUPC2 plasmid increased the resistance of SC2-1C to calcium ions when results were compared to the results with the strain containing the control plasmid (Fig. 3). The heterozygous UPC2/upc2-1 diploid, CJ571, showed slight sensitivity to calcium ions, which is in agreement with the semidominant nature of the upc2-1 allele. The addition of pUPC2 increased the resistance of both the upc2-1/ydr213::URA3 (CJ573) and the upc2-1/upc2-1 (CJ577) diploid; however, complementation of the Ca2+-sensitive phenotype in the UPC2/upc2-1 diploid was less obvious. These complementation data in combination with the finding of allelism between the upc2-1 and ydr213::URA3 alleles provide compelling evidence that the YDR213W open reading frame is the UPC2 gene.
FIG. 3.
Growth on CaCl2 gradient plates of strains of yeast with combinations of UPC2, upc2-1, and upc2::URA3 (ydr213::URA3) as indicated. In each panel the yeast in the upper streak has been transformed with the empty vector (YpH-1) while the yeast in the lower streak has the vector containing UPC2 (YDR213W).
When the sequence data for the mutant YDR213W open reading frame from SC2-1C (upc2-1) was compared to sequences in the yeast genome database, it was observed that only one nucleotide was altered. This point mutation, which was a transition from guanine to adenine, occurred at nucleotide 2663. The predicted protein sequence showed a change at residue 888 from glycine (GGT) to aspartate (GAT). To verify that this mutation was not a variation in the wild-type strain being used, 260 nucleotides around nucleotide 2663 in YDR213W of the wild-type strain (463-1C; UPC2) were sequenced. Figure 4 shows the comparison of the wild type to the mutant at the nucleotide and amino acid levels.
FIG. 4.
Determined nucleotide sequence and predicted amino acid residues of the YDR213W open reading frame of the wild type, 463-1C (UPC2), showing the difference in the mutant strain, SC2-1C (upc2-1). The single difference at nucleotide 2665 (G→A) and the predicted amino acid alteration at residue 888 (G→D) are indicated above and below the wild-type sequence, respectively. Only the carboxy terminus of the predicted protein is presented, as the remaining sequence of the open reading frame of the mutant was identical to that of the wild-type strain. The entire sequence of YDR213W has been reported (GenBank accession no. Z68194) and is not shown here for brevity.
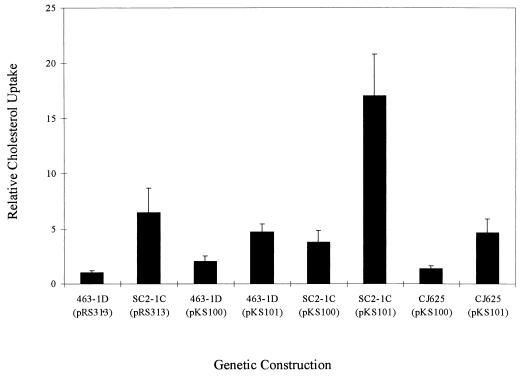
In order to confirm that the point mutation in YDR213W confers the uptake phenotype, we subcloned both wild-type and mutant copies of the YDR213W open reading frame into the low-copy-number yeast shuttle vector pRS313 (20). The region from −350 to +3227 encompassing YDR213W was amplified by PCR from strains 463-1C (UPC2) and SC2-1C (upc2-1) and subcloned into E. coli pGEM-T. The region containing the guanine-to-adenine transition (nucleotide 2663) was sequenced to confirm the presence of this nucleotide base change in the mutant form (upc2-1). Wild-type (UPC2) and mutant (upc2-1) forms of YDR213 were then subcloned into pRS313 to create pKS100 and pKS101, respectively. The ability of strains with the upc2-1 allele to take up sterol was then determined by transforming strains containing various genomic forms of the UPC2 gene (Fig. 5). Most critical in this analysis was that pKS101 (upc2-1) increased uptake fivefold in CJ625 (upc2::URA3) compared to the level of uptake by CJ625 with pKS100 (UPC2) or wild-type 463-1D with the vector only. In addition, the presence of upc2-1 results in a significant increase in uptake over that in the presence of UPC2 alone in the wild type (P < 0.001) in accordance with the previously observed semidominance of this mutant form of the UPC2 gene.
FIG. 5.
Effects of pKS100 (UPC2) and pKS101 (upc2-1) on sterol uptake in S. cerevisiae. Strains 463-1D (UPC2), SC2-1C (upc2-1), and CJ625 (upc2::URA3) were transformed with pKS100 and pKS101, and sterol uptake was performed as described in Materials and Methods. As controls, 463-1D and SC2-1C were transformed with the empty vector (pRS313). Standard deviations are based on six observations for each condition.
DISCUSSION
Although ergosterol has been recognized as a major constituent of fungi for over a century, little is known about its functions in the cell. It is peculiar that wild-type yeast cells cannot accumulate sterols from growth media aerobically but that they can easily do so anaerobically. In order to conduct a variety of experiments using altered sterols that could be fed to the organism, we isolated a strain (UPC20) that could take up free sterols from a growth medium aerobically. The basis for the selection was the identification by radioautography of colonies that took up [14C]cholesterol from agar plates. The utility of such a yeast mutant has generated an enormous amount of interest, and the culture has been widely distributed to research and industrial laboratories. In addition to its use in probing the mechanism and functions of sterols in the organism, the study of the uptake of sterols is particularly valuable for bioengineered organisms that were designed to modify sterols exogenously supplied to high-value products. An intensive effort was undertaken to define the upc2-1 mutation that was involved and to determine the physiological consequences of that mutation. However, an assay based on diminished uptake of labeled sterols was far too time-consuming and expensive to use as a screening tool for isolation of the UPC2 gene. Therefore, other selective criteria were attempted. Differential sensitivity to inhibitors of sterol synthesis and function, growth differences in cultures accumulating less acceptable sterols than endogenously synthesized ergosterol, fluorescence-activated cell sorting based on accumulated lipids, and sensitivity to a large assortment of metabolic inhibitors were tested as potential diagnostic criteria. While each technique was based on an observed physiological difference between upc2-1 and its isogenic wild type, none of these procedures was successful.
Several studies have shown that alterations in sterols cause a change in the sensitivities of the organisms to various cations (15). During such experiments in our laboratory, it was observed that the strains containing the upc2-1 allele were substantially more sensitive to calcium than was an isogenic wild type. Attempts to clone the UPC2 gene were predicated on the complementation of calcium sensitivity in the mutant to wild-type levels of resistance to Ca2+. This screen has allowed us to identify the YDR213W open reading frame as UPC2. Specifically, strains with upc2-1, when they were transformed with YDR213W, developed resistance to CaCl2 and reduced levels of cholesterol uptake that were comparable to those of strains with UPC2. A null mutation of YDR213W (ydr213::URA3) was constructed and transformed into a UPC2/UPC2 diploid. Following sporulation, the URA3 marker of the null mutation segregated 2:2. When strain CJ625 (ydr213::URA3) was crossed with SC2-1C (upc2-1), 61 of the 67 scored tetrads were parental ditypes for the Ura+ and Ca2+-sensitive phenotypes. This strongly supports the allelism of ydr213::URA3 and upc2-1. By sequencing the YDR213W region from upc2-1 and comparing it to the YDR213W region of UPC2, a single base transition was observed between the two genes. The upc2-1 gene had a guanine-to-adenine change at nucleotide 2663. This difference predicted an amino acid change from glycine to aspartic acid at amino acid 888 in the product of upc2-1. Upon cloning of this upc2-1 allele from strain SC2-1C (upc2-1), it was observed that a plasmid-borne copy of upc2-1 caused increased accumulation of exogenous sterol but that the wild-type UPC2 did not.
Based on these results it is now possible to offer an explanation for the pleiotropic phenotype and semidominant characteristic of upc2-1 over UPC2. Sequence analysis and BLAST search results revealed that YDR213W is a predicted member of the Zn(II)2Cys6 binuclear cluster of DNA binding proteins. For a recent comprehensive review of these structures, see Todd and Andrianopoulos (21). These proteins are unique to fungi. GAL4, LEU3, and HAP1 are among the approximately 80 examples of predicted DNA binding proteins in this group. Almost all of the proteins with known functions have been shown to have diverse regulatory roles, generally as transcription activators. In S. cerevisiae, several of the proteins act both as repressors and as activators. This may account for the pleiotrophy attributed to upc2-1, if indeed its encoded protein is a member of this DNA binding group. As mentioned above, differential sensitivities to inhibitors, cations, and unusual sterols were observed (14a). Large accumulations of neutral fat and steryl esters have been seen (8). If, as predicted, upc2-1 has an alteration in a multifunctional regulatory protein, varied phenotypes with upc2-1 are understandable.
By direct analysis and genetic evidence, several of the Zn(II)2Cys6 DNA binding proteins have been shown to be homodimers (21). Others are predicted to be so. Dimerization of the protein encoded by YDR213W (UPC2) has not been determined. In the likelihood that it does function as a homodimer, it is easy to understand how a dimer consisting of the wild-type gene (UPC2) and its mutant allelic pair (upc2-1) could give intermediate results in the Ca2+ sensitivity and [14C]cholesterol uptake analyses. An interesting curiosity of our results was the observation that the ydr213::URA3 null mutants did not show sensitivity to calcium ions or have the high levels of sterol uptake seen in strains carrying the upc2-1 allele. It is anticipated that ydr213::URA3 does not code for a protein and is without function. However, the single amino acid substitution in the predicted protein from upc2-1, particularly the neutral (glycine) replacement by aspartate, may substantially alter the controlling activities of the modified regulator. Such precedents have been set with other members of the GAL4 family of activators. Substitution or truncation mutations in the last 80 amino acids of the PUT3 transcriptional activator can lead to constitutive activity, as the carboxy termini of these activators are often involved in interaction with other proteins (13). The location of the mutation only 26 amino acids from the end in the upc2-1 allele described here suggests a similar activity. The altered protein may behave in an activation mode to sustain the synthesis of proteins that contribute to sterol uptake. Such an activator role is consistent with the known functions of other members of the Zn(II)2Cys6 group of DNA binding proteins. To speculate specifically how this single amino acid alteration could mediate the observed results in upc2-1 would be premature. However, intriguing possibilities arise that suggest avenues of further investigation.
ACKNOWLEDGMENTS
Frank W. Leak, Jr., James H. Crowley, and Kevin V. Shianna contributed equally to the data presented in this report.
This research was supported by the North Carolina Agricultural Research Service, by a grant from the U.S. Army Research Office (DAAAH04-97-003), by an AASERTS award (DAAAH04-94-G0179), and by a generous gift of funds from the NutraSweet Kelco Company.
We thank P. Hieter and L. Robinson for the YpH-1 and YEp13 libraries. Over the past 10 years, virtually every member of the Microbial Physiology Research Group has contributed to discussions on the cloning of UPC2. L. W. Parks gratefully acknowledges the many people who have contributed cumulatively to this intriguing problem.
REFERENCES
- 1.Berben G, Dumont J, Gilliquet V, Bolle P A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 2.Bourot S, Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165:97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- 3.Eberhardt I, Hohmann S. Strategy for deletion of complete open reading frames in Saccharomyces cerevisiae. Curr Genet. 1995;27:306–308. doi: 10.1007/BF00352097. [DOI] [PubMed] [Google Scholar]
- 4.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gollub E G, Liu K P, Dayan J, Adelsberg M, Sprinson D B. Yeast mutations deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3 oxidosqualene. J Biol Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- 6.Keesler G A, Casey W M, Parks L W. Stimulation by heme of steryl ester synthase and aerobic sterol exclusion in the yeast Saccharomyces cerevisiae. Arch Biochem Biophys. 1992;296:474–481. doi: 10.1016/0003-9861(92)90600-2. [DOI] [PubMed] [Google Scholar]
- 7.Leber R, Zinser E, Hrastnik C, Paltauf F, Daum G. Export of steryl esters from lipid particles and release of free sterols in the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1234:119–126. doi: 10.1016/0005-2736(94)00270-y. [DOI] [PubMed] [Google Scholar]
- 8.Lewis T L, Keesler G A, Fenner G P, Parks L W. Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast. 1988;4:93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 9.Lorenz R T, Parks L W. Effects of lovastatin (mevinolin) on sterol levels and on activity of azoles in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1990;34:1660–1665. doi: 10.1128/aac.34.9.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz R T, Parks L W. Involvement of heme components in sterol metabolism of Saccharomyces cerevisiae. Lipids. 1991;26:598–603. doi: 10.1007/BF02536423. [DOI] [PubMed] [Google Scholar]
- 11.Lorenz R T, Parks L W. Physiological effects of fenpropimorph on wild-type Saccharomyces cerevisiae and fenpropimorph-resistant mutants. Antimicrob Agents Chemother. 1991;35:1532–1537. doi: 10.1128/aac.35.8.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loubbardi A, Marcireau C, Karst F, Guilloton M. Sterol uptake induced by an impairment of pyridoxal phosphate synthesis in Saccharomyces cerevisiae: cloning and sequencing of the PDX3 gene encoding pyridoxine (pyridoxamine) phosphate oxidase. J Bacteriol. 1995;177:1817–1823. doi: 10.1128/jb.177.7.1817-1823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marczak J E, Brandiss M C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991;11:2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niederberger P, Aebi M, Hutter R. Identification and characterization of four new GCD genes in Saccharomyces cerevisiae. Curr Genet. 1986;10:657–664. doi: 10.1007/BF00410913. [DOI] [PubMed] [Google Scholar]
- 14a.Parks, L. W. Unpublished results.
- 15.Parks L W, Casey W M. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez R J, Low C, Bottema C D K, Parks L W. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim Biophys Acta. 1985;837:336–343. doi: 10.1016/0005-2760(85)90057-8. [DOI] [PubMed] [Google Scholar]
- 17.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 18.Rothstein R. Targeting, description, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Todd R B, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 22.Trocha P J, Sprinson D B. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch Biochem Biophys. 1976;174:45–51. doi: 10.1016/0003-9861(76)90322-2. [DOI] [PubMed] [Google Scholar]
- 23.Ursic D, Culbertson M R. The yeast homology of mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991;11:2629–2640. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]