Abstract
In this report we present the identification and analysis of two Bacillus subtilis genes, yklA and ykzA, which are homologous to the partially RpoS-controlled osmC gene from Escherichia coli. The yklA gene is expressed at higher levels in minimal medium than in rich medium and is driven by a putative vegetative promoter. Expression of ykzA is not medium dependent but increases dramatically when cells are exposed to stress and starvation. This stress-induced increase in ykzA expression is absolutely dependent on the alternative sigma factor ςB, which controls a large stationary-phase and stress regulon. ykzA is therefore another example of a gene common to the RpoS and ςB stress regulons of E. coli and B. subtilis, respectively. The composite complex expression pattern of the two B. subtilis genes is very similar to the expression profile of osmC in E. coli.
ςB was discovered in 1980 by Haldenwang and Losick and was the first alternative sigma factor of Bacillus subtilis identified (19). However, the function of the regulon controlled by ςB remained a matter for speculation until 1993, when it was shown to be involved in the cellular response to stress. It has subsequently been demonstrated that expression of a large number of genes is induced in a ςB-dependent manner by such different stimuli as heat shock, ethanol, acid, and salt stress, and starvation for oxygen, phosphate, and glucose (6, 7, 10, 11, 21, 22, 40, 43). Since induced expression of more than 50 genes is absolutely dependent on ςB, it was tempting to assume that the gene products perform essential adaptive functions in B. subtilis. However, earlier studies had shown that a null sigB mutant strain was apparently not impaired in sporulation or response to stress compared to the wild type (9, 10, 21, 23, 39). This is unusual, since starving or stressed B. subtilis cells devote a considerable amount of their residual protein-synthesizing capacity to the synthesis of the members of the ςB regulon (8). This apparently anomalous result has prompted a concerted effort to identify the genes of the ςB regulon, to elucidate the function(s) of their gene products, and to establish their contributions to the cellular response to stress and starvation conditions.
Among others, genes encoding a catalase (katE), a nonspecific DNA-binding and protecting protein (dps), and an osmotically activated proline uptake system (opuE) have been shown to be subject to the control of ςB in B. subtilis (5, 13, 44). Interestingly, in Escherichia coli genes like katE, dps, and proP, whose gene products perform similar functions, are subject to a RpoS-dependent regulation (28, 30, 34). RpoS directs the expression of a large group of genes whose expression is induced following starvation and stress (20, 33). Null mutations in rpoS result in a loss of stationary-phase-induced resistance against heat, acid, or oxidative stress and impairment in the ability to survive prolonged periods of starvation (20, 26, 27, 29). Therefore, it was tempting to speculate that ςB-dependent stress proteins may provide the stressed or starved B. subtilis cell with a general multiple-stress resistance similar to that provided by the RpoS-dependent proteins of E. coli. This view is supported by the demonstration that sigB mutants are impaired in stationary-phase-induced resistance to oxidative stress, like E. coli rpoS mutants (4, 12). Recently, the Dps protein of B. subtilis has been shown to play a crucial role in the development of this nonspecific starvation-mediated resistance to oxidative stress (5). However, it is necessary to identify and investigate the physiological roles of additional general stress proteins to further support this hypothesis. Identification of general stress proteins by N-terminal sequencing (3, 8, 40, 41) has greatly benefited from the recent release of the complete sequence of the B. subtilis genome (25). In this paper we present an investigation of two genes, yklA and ykzA, identified in B. subtilis during the genome-sequencing project, which show homology to osmC from E. coli. The expression profile of osmC in E. coli is complex (16, 18). It has two independent promoters, which provide medium-, growth phase-, and stress-dependent expression. One of the promoters is partially controlled by RpoS. Although there are two genes in B. subtilis which have homology to osmC, the composite expression profile of the two genes (medium-, growth phase-, and stress-dependent expression) is very similar to that of the E. coli gene. In addition, expression of the homolog YkzA is directed by ςB, the B. subtilis stress sigma factor.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis wild-type strain 168 and its isogenic sigB mutant strain ML6 (trpC2 sigB::ΔHindIII-EcoRV::cat [21]) were cultivated at 37°C under vigorous agitation in Luria broth (LB) (31) or a synthetic medium described previously (37). The synthetic medium had to be used in order to achieve glucose limitation and in the experiments involving salt stress, in order to avoid the protective effects of osmoprotective substances present in LB. Stresses and starvation were imposed as described previously (40, 42).
Cloning, sequencing, and construction of the corresponding mutants.
B. subtilis chromosomal DNA for sequencing was isolated from strain 168 by chromosomal walking with the integrating plasmid pDIA5304 as previously described (24). E. coli TP611 (recBC hsdRM cya610 pcn) was used for cloning large chromosomal DNA fragments (15). E. coli TG1 [K-12 Δ(lac pro) supE thi hsdR F′ traD36 proAB lacI lacZΔM15] was used for subcloning and for preparation of sequencing templates. E. coli strains were routinely grown in LB. The isolated chromosomal DNA from B. subtilis was sequenced by a shotgun strategy and by a directed approach with oligonucleotides. Plasmid DNA (30 μg in 120 μl of Tris-EDTA buffer) was randomly sheared either by sonication (Braun Labsonic sonicator) (seven pulses, 0.22 cycles, 0.25 W) or by using DNase I in the presence of manganese (35). Sequencing reactions were carried out with fluorescent dye primer sequencing kits (GENPAK, Polegate, East Sussex, England) according to the manufacturer’s instructions. Reactions were resolved on an ABI automated sequencer model 373A. Gaps in the sequence were filled with custom-synthesized oligonucleotides (PCR-Mate DNA synthesizer; Applied Biosystems) and the Applied Biosystems dye terminator sequencing kit.
Two oligonucleotide primer pairs were made (Genosys Biotechnologies Ltd., Cambridge, United Kingdom) and used to amplify fragments of DNA located within the yklA (139-bp) and ykzA (152-bp) open reading frames (the positions of the oligonucleotides are given in parentheses): YklA-14F, 5′-AAGCGACAAATCCAGAGC-3′, and YklA-14R, 5′-GCTTCATCCTTTAACAGGC-3′ (33232 to 33371); and YkzA-16F, 5′-CCAAAAAAGAAGGACAAACCGG-3′, and YkzA-16R, 5′-ATCCTTCATGAGGCGACC-3′ (34245 to 34397). The DNA amplified with each pair of primers was isolated, the ends were polished, and the fragments were subcloned in pUC19 to give plasmids pLA004 and pNA005, respectively. The integrity of the cloned fragments was checked by sequencing. Insert DNA was excised from pLA004 and pNA005 with BamHI and HindIII and directionally cloned into the plasmid pMutin4 (an integrating plasmid conferring resistance to erythromycin and containing a promoterless lacZ gene [a gift from V. Vagner and S. D. Ehrlich]) to give plasmids pMutin004 and pMutin005. Plasmids pMutin004 and pMutin005 were integrated into the chromosome of B. subtilis 168 through homology with the yklA and ykzA fragments by a Campbell-type event, which generates strains BFS1816 (carrying a yklA-lacZ transcriptional fusion) and BFS1818 (carrying a ykzA-lacZ transcriptional fusion). The location and structural integrity of the DNA at the integration site was verified by PCR with combinations of oligonucleotides external and internal to the integrated plasmid DNA.
B. subtilis transformation was carried out according to the method of Anagnostopoulos and Spizizen (2). E. coli transformation was carried out according to the method of Sambrook et al. (35).
RNA isolation and analysis of transcription.
RNA was isolated with RNeasy cartridges from Qiagen as described previously (42). Northern blot analysis, hybridization, and quantification of specific hybrids were performed as described by Scharf et al. (36). For the preparation of the digoxigenin-labeled RNA probes, a DNA fragment encompassing the two osmC-homologous genes yklA and ykzA was amplified from chromosomal DNA of the wild-type strain 168 with the synthetic oligonucleotides UV114 (5′-GAGAGGATCCGTGAATAGCGGGGTAATG-3′) and UV115 (5′-GAGAATCGATGTCCGACACCAAAAAACATC-3′). The PCR fragment was digested with BamHI/ClaI and cloned into pBluescript KS(−) digested with the same enzymes. The resulting plasmid, pUV321, was digested with HincII and religated, yielding plasmid pUV521. pUV521 can be used for the production of a digoxigenin-labeled, yklA-specific RNA probe with T3 RNA polymerase after linearization with BamHI. Digestion of pUV321 with BamHI/BglII and religation of the remaining plasmid resulted in pUV520, which is devoid of yklA and can be used for the preparation of digoxigenin-labeled, ykzA-specific RNA probe with T3 RNA polymerase after linearization with SphI.
Primer extension experiments were performed with synthetic oligonucleotides UV117 (5′-GACATCAAGCTCAAGAAC-3′) and UV116 (5′-CATTTGGCATGAAATATC-3′), complementary to the regions encoding the N termini of yklA and ykzA, as described previously (45). A DNA-sequencing ladder prepared with the same primers and pUV320 plasmid DNA as a template was used to assign the 5′ end of the mRNAs.
Two-dimensional protein gel electrophoresis.
Protein extracts were prepared by passage through a French press after cells had been harvested on ice. Equal amounts of protein (300 μg) were loaded. The proteins were separated with immobiline dry strips, pH 4 to 8, in the first dimension on the multiphor apparatus supplied by Pharmacia, equilibrated, loaded onto 12.5% polyacrylamide gels, and separated according to their molecular masses with the InvestigatorTM electrophoresis system of ESA Inc. (Chelmsford, Mass.).
Enzyme assays.
Expression of lacZ was measured as described by Ferrari et al. (14) with the following modifications: activity units are expressed in nanomoles per minute per microgram of protein and the cells were lysed for 25 min in Z buffer containing 10 μg of lysozyme per ml, 1 mM dithiothreitol, 0.00025% Triton X-100, and 1 μg of DNase I per ml.
Computer sequence analysis.
Sequence alignment and editing were performed with the XBAP program of the STADEN package. Conceptual translation of the sequence and other sequence analyses were performed with the NIP program of the STADEN package. The GenBank database was accessed with ACNUC (17), and homology searches of the database were performed with the TBLASTN program (1). Multiple sequence alignments were performed with CLUSTAL W (38).
RESULTS
Cloning and sequencing of the chromosomal region containing yklA and ykzA.
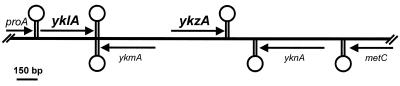
Two homologs of osmC from E. coli were identified during the B. subtilis genome-sequencing project (25). They are called yklA and ykzA and are arranged as shown in Fig. 1 at approximately 105° on the chromosome. Their sequence can be found in GenBank entry AJ002571. Both genes are transcribed in the direction of chromosomal replication and are separated by ykmA, which is transcribed in the opposite direction. There is a putative ςA promoter positioned upstream of yklA and a putative ςB promoter located upstream of the ykzA gene. The paralogs YklA and YkzA differ in size by five amino acids (141 and 136, respectively) and are 49% identical (67% similar) to each other. Both YklA and YkzA are approximately 28% identical (42% similar) to OsmC from E. coli.
FIG. 1.
Schematic diagram of chromosomal organization in the region containing the yklA and ykzA genes. Open reading frames are indicated by arrows, and their positions indicate whether they are encoded on the top (above) or the bottom (below) DNA strand. Terminators are indicated by “lollipops” on the top (above) or the bottom (below) strand. The terminator between yklA and ykmA is bidirectional.
Transcriptional regulation of yklA and ykzA.
Expression of the osmC gene from E. coli is subject to osmotic and growth phase-dependent regulation, which are partially dependent on the presence of the stress and stationary-phase sigma factor RpoS. Therefore, we wanted to determine if one or both of the B. subtilis homologous genes yklA and ykzA are similarly regulated by osmotic stress or starvation. Total RNA was prepared from exponentially growing cells and from bacteria which had been treated with sodium chloride or which entered the stationary phase as a result of the exhaustion of glucose. An analysis of the ykzA mRNA level revealed that the expression of this gene was strongly and rapidly induced by salt stress (Fig. 2). The induction was transient, reaching a maximum between 9 and 12 min after the imposition of stress. ykzA was also induced during the exhaustion of glucose, but clearly the level of induction was less pronounced than during salt stress. Induction by stress was not confined to salt stress. A similar very strong and transient induction of ykzA was also measured following heat shock and ethanol stress (Fig. 2). Therefore, ykzA belongs to the group of general stress genes induced by multiple stimuli in B. subtilis. When the same RNA preparations were probed with a yklA-specific probe, we failed to detect significant changes in the expression of yklA in response to any of the stimuli examined (Fig. 2).
FIG. 2.
Levels of yklA- and ykzA-specific mRNA before and after the imposition of different stresses. RNA was prepared from the wild-type strain 168 at the times indicated on the x axes. Specific RNAs were detected with digoxigenin-labeled RNA probes, and the intensities of the signals were quantified with a laser densitometer as described previously (36). The amount of RNA present during exponential growth was set to one. The induction ratios of yklA (solid bars) and ykzA (shaded bars) at the different time points are displayed. All stresses were applied at time zero with the exception of glucose limitation, where zero indicates the point at which the culture ceased to grow.
In view of its response to multiple stimuli, transcription of ykzA was analyzed in more detail by Northern blot analysis. A signal of 0.5 kb was observed, which is the expected size of a monocistronic transcript encoding only ykzA. However, two additional, but less intense, signals corresponding to transcripts of 0.8 and 1.4 kb were also detected (Fig. 3). Only the 0.5-kb transcript was detected during growth, but the intensities of all three transcripts increased upon stress or starvation. Northern analysis experiments with probes spanning the regions upstream and downstream of ykzA indicated that the signals corresponding to the two larger transcripts result from readthrough at the terminator downstream of ykzA (data not shown).
FIG. 3.
Northern blot analysis of ykzA. Equal amounts of total RNA (10 μg) prepared from the wild-type 168 or the isogenic sigB mutant (ML6) before (co) and at different times (in minutes) after exposure to stress were separated on denaturing gels, transferred onto a positively charged nylon membrane, and hybridized with digoxigenin-labeled RNA probe specific for ykzA.
In B. subtilis, expression of most of the general stress genes induced by stress or starvation depends on the sigma factor ςB. Since YkzA belongs to this family of general stress proteins and there is a putative ςB-dependent promoter upstream of ykzA, it was decided to examine expression of ykzA in a sigB mutant strain. Total RNA isolated from sigB mutant strain ML6 after exposure to heat, ethanol, and salt was probed with a ykzA-specific probe by Northern analysis. No ykzA-specific signals were detected in this strain after exposure to any of the stresses (Fig. 3).
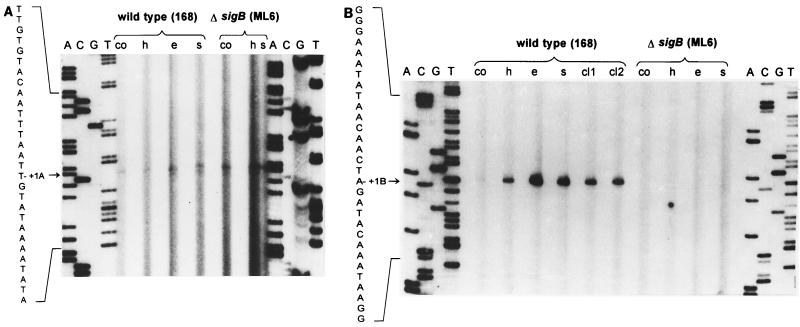
The promoters of yklA and ykzA were mapped by primer extension analysis. Primers were designed complementary to the DNA regions encoding the N termini of yklA and ykzA as outlined in Materials and Methods. A very weak signal was detected for yklA, which did not significantly increase upon stress. A signal of similar intensity was also obtained with RNA isolated from a sigB mutant (Fig. 4A). The size of this reverse transcript is consistent with expression of yklA being driven by the ςA-type promoter which was identified by sequence analysis (TTGACA-17 nucleotides-TACAAT). Primer extension analysis with the ykzA-specific primer revealed a reverse transcript (Fig. 4B) which was barely detectable with RNA from exponentially growing bacteria, but its intensity increased dramatically with RNA isolated from cells which had been exposed to stress (Fig. 4B). No transcript was detected with RNA isolated from a similarly stressed sigB mutant strain (Fig. 4B). The point of initiation of transcription for ykzA is consistent with transcription being driven from the ςB-type promoter (GTTTAA-12 nucleotides-GGGAAA) identified in the sequence analysis.
FIG. 4.
Mapping of the 5′ ends of the yklA (A) and ykzA (B) mRNA during growth and after exposure to stress. Total RNA was isolated from the wild-type strain and its isogenic sigB mutant during exponential growth (co), 10 min after the imposition of the different stresses (h, heat shock; e, ethanol stress; s, salt stress), and 30 or 40 min after the limitation of glucose (cl1 and cl2, respectively). The primer extension analysis was performed as described in Materials and Methods. The 5′ ends of the transcripts were determined by comparison with a DNA-sequencing ladder generated with the same primer and run in parallel on the same gel (lanes A, C, G, and T).
Expression of yklA and ykzA in minimal and rich medium during the growth cycle and after induction by salt.
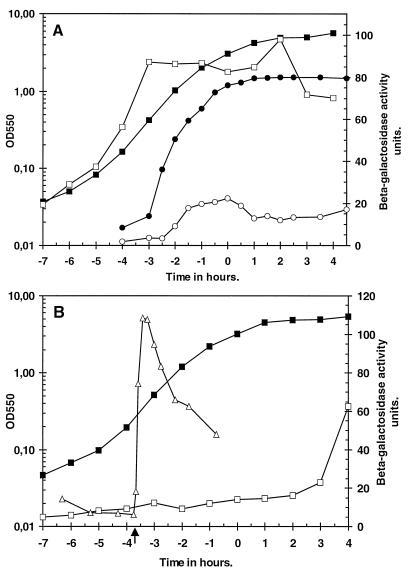
Two strains (BFS1816 and BFS1818), mutated in yklA and ykzA, respectively, were constructed by integration of pMutin004 and pMutin005 into the chromosome of B. subtilis. The pMutin-derived plasmids contained an internal fragment of the yklA and ykzA open reading frames, respectively, cloned immediately upstream of a promoterless lacZ gene, allowing the expression of each gene to be examined. The strains grew and sporulated normally both on minimal medium and on medium containing 0.3 M NaCl. Expression of yklA-lacZ and ykzA-lacZ was examined in nutrient broth and minimal medium throughout the growth cycle. When cells harboring yklA-lacZ were grown in nutrient medium, β-galactosidase activity reached approximately 20 U during exponential growth and decreased slightly at the onset of the stationary phase (Fig. 5A). When these cells were grown in minimal medium, β-galactosidase levels rose during the early period of the growth cycle. This accumulation reached a plateau of approximately 90 U by the midpoint of the growth cycle (T−3), and this level was maintained for the remainder of the growth cycle (Fig. 5A). Addition of 0.3 M NaCl to exponentially growing cells containing yklA-lacZ did not affect the β-galactosidase activity (data not shown), confirming the results of the slot blot analysis, which showed that expression of yklA is not responsive to osmotic stress (Fig. 2).
FIG. 5.
Expression of the yklA-lacZ (A) and ykzA-lacZ (B) transcriptional fusions during the growth cycle in minimal medium or nutrient broth and after exposure to salt stress. Cells were grown as described in Materials and Methods, and samples were taken every 30 or 60 min as indicated. (A) solid symbols indicate growth of B. subtilis BFS1816, and open symbols indicate β-galactosidase activity. Circles, nutrient medium; squares, minimal medium. (B) Solid squares represent growth of B. subtilis BFS1818 in minimal medium, and open squares indicate β-galactosidase activity. The influence of the addition of salt during exponential growth on the accumulation of β-galactosidase is also indicated (open triangles). The point of salt addition is indicated by an arrow. OD550, optical density at 550 nm.
When cells harboring ykzA-lacZ were grown in nutrient medium, the level of β-galactosidase remained at ≤20 U throughout the exponential and stationary phases of the growth cycle (data not shown). When these cells were grown in minimal medium the level of β-galactosidase also remained at ≤20 U until approximately 2 to 3 h into the stationary phase, when an increase in activity was discernible (Fig. 5B). When salt was added to exponentially growing cells containing ykzA-lacZ in minimal medium (to a final concentration of 0.3 M NaCl), there was a rapid 10-fold increase in β-galactosidase activity, which peaked approximately 20 min after the addition of salt. The β-galactosidase level declined during the subsequent 2.5 h of growth but still remained approximately three- to fivefold higher than that in unstressed cells at the end of this time interval (Fig. 5B). These results demonstrate that expression of these two paralogs is complex but complementary: expression of yklA is medium dependent but is not responsive to stress. In contrast, expression of ykzA is medium independent but is responsive to osmotic and other stresses (as shown by transcription analysis).
Identification of YkzA on two-dimensional protein gels; level of YkzA during exponential growth and after imposition of stress.
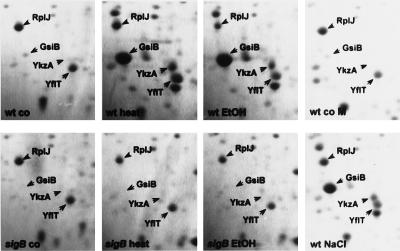
We have determined the N-terminal sequences of general stress proteins of B. subtilis by microsequencing (3, 8, 40). When comparing these sequences with the sequences of YklA and YkzA, we discovered that the N-terminal sequence of the general stress protein Gsp17o (ALFTAKVTAR GGRAAHITSD D) matched the amino acid sequence deduced from the ykzA DNA sequence (with the exception of the alanine residue at position 15). Therefore, the ATG codon at position 34145 of the DNA sequence is indeed the start codon of ykzA, and the N-terminal formyl-methionine is subsequently removed. Two-dimensional protein gel electrophoresis of crude protein extracts from cells harvested during exponential growth or after imposition of stress was used to show that levels of YkzA significantly increased following heat, salt, and ethanol stress and that the stress sigma factor ςB was required for this increase to occur (Fig. 6). The intensity of a reference spot corresponding to the ribosomal protein RplJ did not increase during the same time interval (Fig. 6).
FIG. 6.
Level of YkzA in the wild type (wt) and the sigB mutant (sigB) before (co) and after the imposition of stresses (heat, ethanol [EtOH], and NaCl). Bacteria were grown in LB (co, heat, and EtOH) or minimal medium (co M and NaCl) and harvested during exponential growth, 60 min after the imposition of heat shock or ethanol, or 90 min after exposure to NaCl. Crude protein extract (300 μg) was separated by two-dimensional gel electrophoresis and stained with Coomassie brilliant blue R-350. Besides YkzA, GsiB is indicated as an additional ςB-dependent stress protein and RplJ is labeled as a vegetative marker protein. The identities of the labeled proteins were verified by microsequencing or mass spectrometry.
DISCUSSION
The expression profiles of two B. subtilis genes, yklA and ykzA, whose products are highly similar to the general stress protein OsmC first identified in E. coli, have been presented. The profiles are complex, with expression of the genes being growth phase, medium, and stress dependent. Expression of yklA is directed by a ςA-type promoter, and it is maximally expressed during exponential growth. The expression of yklA is four- to fivefold higher in minimal medium than in a rich medium, and it is not induced by stress.
Expression of ykzA, in contrast, is directed by a promoter which requires ςB, the so-called stress sigma factor of B. subtilis, for initiation of transcription. Expression of ykzA is not medium dependent and is very low throughout exponential growth. However, expression of ykzA is rapidly induced by salt and ethanol stress, heat shock, and starvation. Therefore, the expression patterns, although complex, appear complementary, with yklA being medium responsive whereas ykzA is stress and starvation responsive.
It is instructive to compare the organization and expression of osmC from E. coli with those of yklA and ykzA from B. subtilis. There is only one osmC gene in E. coli, whose expression is directed by two overlapping but apparently independent promoters (16, 18). In contrast, B. subtilis has two osmC homologs, each expressed from a single promoter. The osmCp1 from E. coli is recognized by the housekeeping sigma factor Sigma70. Similarly, the yklA gene from B. subtilis seems to be recognized by the housekeeping sigma factor ςA. Expression of osmC from the osmCp2 promoter and expression of ykzA are directed by the stress sigma factors RpoS and ςB, respectively. Induction of these genes, which can be effected by a variety of stresses, is dependent on these sigma factors. Expression directed by osmCp2 also increases when the cells enter stationary phase, as does expression of ykzA in B. subtilis. Although only the ςB-dependent promoter of ykzA is induced following salt stress in B. subtilis, both promoters of osmC of E. coli are salt responsive.
Nevertheless, despite the overt differences in gene organization between the two bacteria, the similarity of the composite expression profiles is striking. It is evident that both bacteria must regulate the level of the general stress protein with great precision, and expression must be responsive to growth, medium, and stress conditions. In E. coli this is achieved by the expression of one gene being directed by two promoters, whereas in B. subtilis it is achieved by having two genes each directed by a single promoter.
Our data clearly demonstrate that there are three RNA transcripts produced upon induction of ykzA. All three transcripts begin at the ykzA promoter. The major transcript is 0.5 kb in length, which is consistent with transcription ceasing at the putative terminator located immediately downstream of ykzA. The lengths of the other two transcripts are consistent with transcription proceeding through the ykzA terminator and ending at the putative terminators for the ykoA and metC genes, respectively, which are expressed from the strand opposite to ykzA. Therefore, it is evident from our data that there is a disparity between the strength of the ykzA promoter and that of the terminator.
Despite our extensive knowledge of the organization and control of expression of osmC, yklA, and ykzA, the functions of the proteins are unknown. It is evident that they play roles in cellular response to a variety of stressful conditions, but their precise functions remain to be established. No obvious phenotype is observed, even under stressful conditions, when osmC is inactivated in E. coli or when either gene is inactivated in B. subtilis. The occurrence of osmC-homologous genes among bacteria does not correlate with any bacterial group or ecological niche and so does not shed any light on its function. There are now 11 members of this gene family distributed among the following bacteria: E. coli (one copy), B. subtilis (two copies), Mycoplasma pneumoniae (one copy), Mycoplasma genitalium (one copy), Acinetobacter calcoaceticus (one copy), Xanthomonas campestris (one copy), Pseudomonas aeruginosa (two copies), and Deinococcus radiodurans (two copies). However, no member of this gene family has been identified in the complete genome sequences of Haemophilus influenzae, Helicobacter pylori, or Synechocystis sp. An alignment of the 11 proteins reveals four regions which are absolutely conserved (data not shown): (i) a glycine residue near the amino terminus, (ii) an NPEQ/EXL motif, (iii) a CF motif, and (iv) an AXXXCPXS motif. These motifs do not show similarity to any other motif in the database. However, conservation of the two cysteine residues is interesting, suggesting that perhaps the protein contains a disulfide bond which is required for activity. Alternatively, it may bind a metal ion or may participate in maintaining disulfide bonds in other proteins, i.e., a type of disulfide bond chaperone. At least the ohr gene of X. campestris, which is a member of the osmC family, is required for protection against organic hydroperoxides (32). A phylogenetic analysis of the eleven proteins (Fig. 7) shows that they can be grouped into three families: (i) the E. coli family, which includes, besides osmC, one each of the D. radiodurans and P. aeruginosa genes; (ii) the Mycoplasma family; and (iii) a family containing yklA and ykzA from B. subtilis, one each of the D. radiodurans and P. aeruginosa genes, and the genes of A. calcoaceticus and X. campestris. The interesting feature of this tree is that yklA and ykzA are more closely related to each other than to any other member of the family. In contrast to P. aeruginosa and D. radiodurans, which also have two copies of osmC, the paralogs fall into distinct phylogenetic groups. This suggests that the duplicated genes in B. subtilis have not evolved to fulfill separate functions within the cell. Instead we propose that the duplication provides a mechanism for the Bacillus cell to regulate OsmC levels in response to a wide range of environmental and nutritional stimuli by placing each copy of the gene under the control of different but complementary expression signals. In E. coli, the environmental and nutritional conditions under which osmC is expressed are very similar to those in Bacillus. However, the mechanism through which this is achieved differs in that expression of a single gene is directed by two different but independent promoters.
FIG. 7.
Unrooted phylogenetic tree of the relationships between the 11 members of the osmC gene family produced from a multiple alignment by using the CLUSTAL W program. The data were bootstrapped 1,000 times, and the values are indicated on the horizontal axes. Drad, D. radiodurans; Paer, P. aeruginosa; Ecol, E. coli; Xant; X. campestris; Acal, A. calcoaceticus; Bsub, B. subtilis; Mpne, M. pneumoniae; Mgen, M. genitalium.
The complexity of osmC gene expression in E. coli and B. subtilis suggests that it plays an important role in the response of the cells to stress. Since protection of stressed or starving cells from oxidative stress seems to be a premier function of the ςB regulon, we are currently investigating whether YkzA and/or YklA is involved in establishing a protective resistance, as does Ohr of X. campestris (32).
ACKNOWLEDGMENTS
U. Völker and K. K. Andersen have contributed equally to this study.
We are grateful to R. Schmid for determining the N-terminal sequence of Gsp17o and to A. Harang and R. Gloger for excellent technical assistance.
The work of M. Hecker and U. Völker was supported by grants from the Fonds der Chemischen Industrie and the Deutsche Forschungsgemeinschaft (He 1887/2-4 and Vö 629/2-2). Work in the laboratory of K. M. Devine was supported by the EU Biotechnology Programme (BIO2-CT93-9272 and BIO2-CT95-0278) and by a grant from the Danish Research Academy to Kasper Krogh Andersen.
REFERENCES
- 1.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 4.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antelmann H, Engelmann S, Schmid R, Sorokin A, Lapidus A, Hecker M. Expression of a stress- and starvation-induced dps/pexB homologous gene is controlled by the alternative sigma factor ςB in Bacillus subtilis. J Bacteriol. 1997;179:7251–7256. doi: 10.1128/jb.179.23.7251-7256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson A K, Haldenwang W G. The ςB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 9.Binnie C, Lampe M, Losick R. Gene encoding the ς37 species of RNA polymerase ς factor from Bacillus subtilis. Proc Natl Acad Sci USA. 1986;83:5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςB by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelmann S, Hecker M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol Lett. 1996;145:63–69. doi: 10.1111/j.1574-6968.1996.tb08557.x. [DOI] [PubMed] [Google Scholar]
- 13.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a ςB-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari E, Howard S, Hoch J. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986;166:173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser P, Kunst F, Arnaud M, Coudart M P, Gonzales W, Hullo M F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilis genome project—cloning and sequencing of the 97kb region from 325 degrees to 333 degrees. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 16.Gordia S, Gutierrez C. Growth-phase-dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol Microbiol. 1996;19:729–736. doi: 10.1046/j.1365-2958.1996.418945.x. [DOI] [PubMed] [Google Scholar]
- 17.Gouy M, Gautier C, Attimonelli M, Lanave C, diPaola G. ACNUC—a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. CABIOS. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez C, Devedjian J C. Osmotic induction of gene osmC expression in Escherichia coli K12. J Mol Biol. 1991;220:959–973. doi: 10.1016/0022-2836(91)90366-e. [DOI] [PubMed] [Google Scholar]
- 19.Haldenwang W, Losick R. Novel RNA polymerase ς factor from Bacillus subtilis. Proc Natl Acad Sci USA. 1980;77:7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengge-Aronis R. Back to log phase: ςS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 21.Igo M, Lampe M, Ray C, Schafer W, Moran C P, Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 23.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogh S, Oreilly M, Nolan N, Devine K M. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology. 1996;142:2031–2040. doi: 10.1099/13500872-142-8-2031. [DOI] [PubMed] [Google Scholar]
- 25.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Düsterhöft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Hénaut A, Hilbert H, Holsappel S, Hosono S, Hullo M-F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S-M, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Moszer I, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S-H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schroeter R, Schleich S, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B-S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H-F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 26.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 27.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 28.Lomovskaya O L, Kidwell J P, Matin A. Characterization of the ς38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol. 1994;176:3928–3935. doi: 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCann M P, Kidwell J P, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellies J, Wise A, Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol. 1995;177:144–151. doi: 10.1128/jb.177.1.144-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of ςS turnover: a role for DnaK and relationship between stress responses mediated by ςS and ς32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Scharf C, Riethdorf S, Ernst H, Engelmann S, Völker U, Hecker M. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J Bacteriol. 1998;180:1869–1877. doi: 10.1128/jb.180.7.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stülke J, Hanschke R, Hecker M. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truitt C L, Weaver E A, Haldenwang W G. Effects on growth and sporulation of inactivation of a Bacillus subtilis gene (ctc) transcribed in vitro by minor vegetative cell RNA polymerases (E-ς37, E-ς32) Mol Gen Genet. 1988;212:166–171. doi: 10.1007/BF00322460. [DOI] [PubMed] [Google Scholar]
- 40.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 41.Völker U, Mach H, Schmid R, Hecker H. Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J Gen Microbiol. 1992;138:2125–2135. doi: 10.1099/00221287-138-10-2125. [DOI] [PubMed] [Google Scholar]
- 42.Völker U, Völker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Völker U, Völker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VonBlohn C, Kempf B, Kappes R M, Bremer E. Osmostress response in Bacillus subtilis—characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor Sigma B. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 45.Wetzstein M, Völker U, Dedio J, Lobau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]