Abstract
In Anabaena spp., synthesis of the heterocyst envelope polysaccharide, required if the cell is to fix dinitrogen under aerobic conditions, is dependent on the gene hepA. A transcriptional start site of hepA was localized 104 bp 5′ from its translational initiation codon. A 765-bp open reading frame, denoted hepC, was found farther upstream. Inactivation of hepC led to constitutive expression of hepA and prevented the synthesis of heterocyst envelope polysaccharide. However, the glycolipid layer of the heterocyst envelope was synthesized. A hepK mutation blocked both the synthesis of the heterocyst envelope polysaccharide and induction of hepA. The predicted product of hepK resembles a sensory protein-histidine kinase of a two-component regulatory system. Analysis of the region between hepC and hepA indicated that DNA sequences required for the induction of hepA upon nitrogen deprivation are present between bp −574 and −440 and between bp −340 and −169 relative to the transcriptional start site of hepA. Gel mobility shift assays provided evidence that one or more proteins bind specifically to the latter sequence. The Fox box sequence downstream from hepA appeared inessential for the induction of hepA.
Anabaena spp. and related filamentous cyanobacteria can reduce dinitrogen to ammonia, an extremely oxygen-sensitive process. They can do so, despite the fact that they simultaneously produce O2 by photolysis of water, because nitrogen fixation takes place within differentiated cells called heterocysts. These cells maintain a very low internal partial oxygen pressure (pO2) by means of the following mechanisms (60). First, the O2-producing photosystem II that is present in the other, vegetative cells is inactivated in heterocysts. Second, heterocysts are enveloped by a layer of glycolipids that provides a substantial barrier to the entry of O2, and this layer is in turn encompassed by a layer of polysaccharide that protects it from physical damage. Finally, O2 that enters is reduced to water by respiration.
There is increasing evidence that regulation of genetic expression during heterocyst formation occurs primarily at the level of transcription. About 600 to 1,000 genes in the genome of Anabaena variabilis are transcribed exclusively in heterocysts (37). Regulatory and structural genes that are required for heterocyst differentiation in Anabaena sp. strain PCC 7120 have been identified and cloned. Nitrogen control gene ntcA is required for an early response to nitrogen deprivation, for induction of nitrate-assimilatory genes, and for the later response of heterocyst differentiation (26, 54). hanA, not required for expression of the nitrate-assimilatory genes, is required for induction of hetR. hetR, an autoregulatory gene that is induced within 2 h after nitrogen stepdown, may play a key role in determining which cells in the filament become heterocysts (7, 9). hetR strongly represses expression of hetC in vegetative cells; in a hetC mutant, what may be presumptive differentiation is detectable as spaced loci of diminished fluorescence, but morphological differentiation is not observed by bright-field microscopy (34). devA, whose induction at about 5 h after nitrogen stepdown is blocked in a hetR mutant, is required for all but the early morphological stages of heterocyst differentiation. The devA product is similar to the ATP-binding-cassette (ABC) subunit of binding-protein-dependent transport systems and may be involved in the import of nutrients into heterocysts (39) or the deposition of heterocyst envelope glycolipids (25). In particular, devA has been reported to affect the activation of hetM (8, 13), which is in turn required for the synthesis of heterocyst envelope glycolipids (28, 40). hepA (30; see reference 23), whose predicted product is also similar to ABC transporters, is activated at about 5 to 7 h after nitrogen stepdown, nearly exclusively in developing heterocysts (59), and is required (see below) for synthesis of the polysaccharide layer of the heterocyst envelope. Activation of hepA is dependent on hetR (7).
How this cascade of genetic activations is regulated remains obscure. Regions of great nucleotide similarity, 5′-A(G/T)GT ATCTGTPy(C/A)PyATTC(T/A)TTTTTPy(A/C)AATPyG- 3′, each designated a Fox box, are found 3′ from several genes that are required for the fixation of dinitrogen in the presence of oxygen (the Fox+ phenotype [23]), and that are activated between about 5 and 7 h after nitrogen stepdown, and were conjectured to be substrates of a developmental regulatory mechanism (34).
Members of the extensive family of two-component signal transduction systems are composed of a sensory kinase that uses ATP to autophosphorylate an internal histidine residue in response to extracellular or intracellular signals, plus an associated cytoplasmic response regulator that transfers the phosphate from the kinase to an aspartate residue in its own receiver domain (1). Phosphorylation of the response regulator activates either genetic transcription or some other function (e.g., reference 44). The Anabaena sp. patA gene affects the localization of heterocyst formation within filaments (36). The carboxy-terminal domain of its predicted product resembles response regulators that lack a DNA-binding domain (44). Similarly, the predicted product of the developmentally active gene devR of the filamentous cyanobacterium Nostoc sp. strain ATCC 29133 (a cross-hybridizing sequence is present in Anabaena sp. strain PCC 7120) has high similarity to the receiver domains of response regulator proteins, especially CheY and Spo0F (14). Spo0F, a single-domain response regulator, is active in a phosphorelay that regulates the initiation of sporulation of Bacillus subtilis (10, 29, 49). Heretofore identified protein-histidine kinases did not significantly influence heterocyst formation (28).
We have sought to elucidate the regulation of hepA and have identified two genes, hepC and hepK, whose mutation is permissive of normal synthesis of the glycolipid layer of the heterocyst envelope but blocks synthesis of the polysaccharide layer of that envelope. The two genes affect the induction of hepA differently.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena sp. strain PCC 7120 and its derivatives (Table 1) were grown at 30°C in the light (ca. 3,500 ergs cm−2 s−1) on a rotary shaker in AA/8 medium (31) supplemented with 5 mM nitrate in 125-ml Erlenmeyer flasks or, for preparation of extracts of protein, in 2-liter batches. Derivative strains were grown in the presence of appropriate antibiotics at the concentrations described by Khudyakov and Wolk (33). Plasmids were introduced by conjugation (19), and double recombinants were selected as described elsewhere (7, 12). Plasmids introduced in this work are listed in Table 1. To induce heterocyst formation, portions of actively growing cultures were washed twice with AA/8, suspended in the original volume of AA/8 without antibiotics, and incubated under growth conditions. Filaments were examined by microscopy 24 to 72 h following nitrogen stepdown. Samples were prepared for electron microscopy (6) and micrographed by S. Burns, MSU Center for Electron Optics.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmida | Derivation and/or relevant characteristicsb |
|---|---|
| Anabaena sp. strainc | |
| PCC 7120 | Wild type, from R. Haselkorn |
| DR1069d (7) | Smr Spr; luxAB-Smr Spr cassette fused at NruI site of hepA |
| DR911 DR1069 | Bmr Nmr Smr Spr; hepC::Tn5-1058 hepA::luxAB-Smr Spr |
| DR1069 DR2053 | Nmr Smr Spr; hepA::luxAB-Smr Spr with cassette C.K2 (53; see reference 20) inserted in the BstXI site of hepC |
| DR1817 | Nmr; cassette C.K3 (20) in the ORF 3′ from hepA |
| DR1818a | Nmr; cassette C.K3 (20) in the ORF 3′ from hepA, and the Fox box 3′ from hepA replaced by a SalI site |
| DR1998a | Smr Spr; bears the omega cassette (45) in the BstXI site of hepC |
| EF116 (57) | hepA Fox− |
| HACb | Bmr Nmr Smr Spr; hepC::Tn5-1058 hepA::luxAB-Smr Spr |
| Y7 (23) | Bmr Nmr Smr; PCC 7120 hepK::Tn5-1058 |
| Plasmids | |
| pRL487 (22) | Cmr Kmr; S.K4/L.XSX1/C.C1 (nomenclature of reference 20) |
| pRL911 | Bmr Cmr Emr Kmr Smr; EcoRV recovery of Tn5-1058 (pRL892) and contiguous DNA from HACb was fused at its EcoRV site to the sacB-bearing EcoRV fragment of pRL1075 (7) |
| pRL1272 (56) | Cmr Emr; RSF1010-based cosmid vector (sequence on request). A region comprised of cos and lox sites, a polylinker that bears unique sites NotI-HindIII-SphI-PstI-SalI-(12 additional sites)-XbaI, and the antibiotic resistance genes is bracketed at both ends by transcriptional stop signals to reduce or eliminate any possible effect of the insert on the copy number of the plasmid |
| pRL1347ab | Cmr Emr Smr Spr; cassette C.CE1 from pRL425 (20) with AccI ends was inserted into the ClaI site of a presumptive helicase gene in a 2.6-kb EcoRV fragment of pRL1756, and that fragment in turn was situated in the XbaI site of pRL277 (7) (joined by short SmaI-XbaI linkers from pRL498 [20]), with the sacB and Emr genes both antiparallel to the presumptive helicase gene |
| pRL1676 | Apr; 3.0-kb NruI-BglII fragment of cosmid 43E7 (57) cloned between the HincII and BamHI sites of pUC19 (62) |
| pRL1706 | Apr; the 1-kb PCR fragment generated by use of the primers 5′-TCCTTCGCGACTTTTACATTACC-3′ (CPW16) and 5′-AGATGTCGACCCCAAAACCATGATTCTGCTC-3′ (CPW17), with pRL351 (30) as template, was cleaved at the italicized NruI and SalI sites and then inserted between the NruI and SalI sites of pIC20R (41) |
| pRL1707 | Apr; the 1.8-kb PCR fragment generated by use of the primers 5′-TTCTGTCGACAAATCGGTATAACTCCCCAAT-3′ (CPW18) and 5′-GTCCGGATCCGGGCCCATTGCTGCTGAAAATAG-3′ (CPW19; ApaI site underlined), with pRL1676 as template, was cleaved at the italicized SalI and BamHI sites and then inserted between the SalI and BamHI sites of pIC20R (41) |
| pRL1708 | Apr; the SalI-BamHI insert-bearing fragment from pRL1707 was transferred between the SalI and BamHI sites of pRL1706 |
| pRL1729 | Apr; 6-kb BglII-ClaI fragment from cosmid 43E7 (57), cloned between the BglII and ClaI sites of pIC20R (41) that had been deleted between the SmaI and NruI sites of its polylinker |
| pRL1730 | Apr; the NruI-ApaI fragment of pRL1708 bearing DNA from PCC 7120 was transferred between the NruI and ApaI sites of pRL1729; differs from pRL1729 by replacement of the Fox box 3′ from hepA by a SalI site |
| pRL1732 | Apr; BamHI fragment bearing Vibrio fischeri luxAB (21) blunted with the Klenow fragment and inserted in the NruI site of pRL1729 |
| pRL1756, pRL1757 | Bmr Kmr Smr; EcoRI and EcoRV recoveries from Y7, respectively |
| pRL1764, pRL1765a,b | Bmr Cmr Emr Nmr Smr; Cmr Emr-oriT(RK2)-sacB-bearing fragment of pRL1075 (7) cut with SstI (and fitted with EcoRI-SstI linkers derived from pIC20R [41]) or (for pRL1765a and pRL1765b) FspI and then ligated to EcoRI-cut pRL1756 or EcoRV-cut pRL1757, respectively |
| pRL1773a | Apr; BamHI fragment bearing V. fischeri luxAB (21) blunted with the Klenow fragment and inserted in the NruI site of pRL1730 |
| pRL1812, pRL1813 | Apr Kmr; cassette C.K3 (20) inserted as a BamHI fragment in the BclI site of the gene 3′ from hepA (30) in pRL1729 and pRL1730, respectively |
| pRL1817, pRL1818a | Cmr Emr Kmr; 7.0-kb EcoRI fragment of pRL1812 and pRL1813, respectively, blunted with the Klenow fragment and inserted in the NruI site of sacB-bearing positive-selection vector pRL271 (7) |
| pRL1826, pRL1827 | Apr Kmr; Kmr cassette C.K3 (20) inserted as a BamHI fragment in the BglII sites of pRL1732 and pRL1773a, respectively |
| pRL1830a, pRL1831a | Cmr Kmr Smr; 8.2-kb EcoRI fragment of pRL1826 and pRL1827, respectively, inserted in the EcoRI site of pKT210 (4) |
| pRL1848 | Apr; BglII-ClaI insert in pRL1729 blunted with the Klenow fragment and inserted in the SmaI site of pIC20R (41) |
| pRL1857 | Apr; 0.4-kb deletion from the ClaI end of the insert of pRL1848 by restriction with SphI and BamHI, followed by digestion with ExoIII and S1 nuclease and religation |
| pRL1858–pRL1861, pRL1882–pRL1884 | Apr; same as pRL1857, but deletions of 1.4, 1.5, 1.8, 1.9, 2.0, 2.1, and 2.3 kb, respectively |
| pRL1902 | Kmr; insert-bearing NruI fragment of pRL1848 transferred between the NruI sites of pRL487 |
| pRL1903–pRL1910 | Kmr; same as pRL1902, but fragments from pRL1857 through pRL1861 and from pRL1882 through pRL1884, respectively |
| pRL1985 | Kmr Smr Spr; the omega interposon (45) as a BamHI fragment and BstXI-cut pRL1902 were blunted by T4 DNA polymerase and ligated |
| pRL1998a | Cmr Emr Smr Spr; 5.0-kb PstI fragment of pRL1985 transferred to the PstI site of pRL271 (7) |
| pRL2078 | Smr Spr; 4.9-kb, hepK-bearing EcoRV fragment from a plaque in a λ-EMBL3 library of wild-type PCC 7120 DNA subcloned between the SmaI sites of pRL1404 (24) |
| pRL2191 | Cmr Emr Kmr; pRL1272 bearing, from PstI to SalI, Kmr cassette C.K2 (53; see reference 20) directed 5′, followed by the c-phycocyanin transcriptional terminator from Agmenellum quadruplicatum (18) directed 3′, and then by E. coli ribosomal terminators T1 and T2 from pKLT6 (43). A cassette (58) bearing V. fischeri luxAB is inserted into the XbaI site of the polylinker |
| pRL2192–pRL2200 | Cmr Emr Kmr; same as pRL2191 but with PCC 7120 inserts shown in Fig. 5 (details of constructions furnished upon request), within the polylinker between the transcriptional terminators and luxAB, oriented for luxAB to report on transcriptional promotion of hepA |
Source or reference: this study, unless otherwise specified (in parentheses).
Ap, ampicillin; Bm, bleomycin; Cm, chloramphenicol; Em, erythromycin; Km, kanamycin; Nm, neomycin; Sm, streptomycin; Sp, spectinomycin.
Single recombinants portrayed in Fig. 4 and PCC 7120 bearing replicating plasmids are omitted.
The notation DRx refers to a product of double-reciprocal recombination of pRLx with the genome of PCC 7120, as confirmed by Southern hybridization.
DNA manipulation.
Recombinant DNA procedures were performed in the standard manner (46). Enzymes were purchased from New England BioLabs, Beverly, Mass. (and occasionally from other suppliers), and used as recommended. Clones bearing transposon Tn5-1058 (erroneously reported as Tn5-1065 in reference 23) were recovered from mutant Y7 (23) by excision with EcoRI or EcoRV, ligation, and transfer to Escherichia coli and were then subcloned. One subclone was used to identify λ-EMBL3 clones bearing corresponding wild-type DNA (8). Unidirectional, nested deletions of subclones and of the region 5′ from hepA (via pRL1848) were generated by exonuclease III (ExoIII) digestion (46). Automated sequencing (Applied Biosystems Inc., Foster City, Calif.) was performed on both strands of the DNA by use of universal primers. Database comparisons and alignments of the translated sequences were performed by using the default settings of the algorithm developed by Altschul et al. (2), using the BLAST network service at the National Center for Biotechnology Information. DNA fragments containing nested deletions were placed individually between the NruI sites of pRL487 (yielding plasmids pRL1902 to pRL1910) and transferred to hepA::luxAB strain DR1069, and recombinants were isolated.
T. A. Black isolated transposon-induced mutant HACb, in which hepA is transcribed constitutively, by mutagenizing DR1069 with Tn5-1058 and imaging the resulting colonies as described by Wolk et al. (58). The transposon and contiguous DNA were recovered by use of EcoRV as described for mutant Y7, and the mutation was reconstructed upon ligation of the resulting plasmid to pRL1075, conjugal transfer to PCC 7120 (wild type) and DR1069, and selection for double recombinants (7).
To delimit one or more cis-acting elements in the region between hepC (5′ from hepA) and hepA, a series of overlapping DNA deletions (windows [5]) of the intergenic region was constructed either by ExoIII digestion or by PCR amplification. DNA fragments bearing these deletions upstream from a hepA::luxAB fusion were placed individually in pRL2091, a plasmid capable of replicating in PCC 7120 because it is based on plasmid RSF1010 (48, 51). The resulting plasmids were introduced into wild-type PCC 7120.
Plasmid pRL1730 contains an extended sequence of cloned PCC 7120 DNA in which a sequence denoted a Fox box was replaced with a SalI site and was used to introduce the same replacement into strain DR1818a and into autonomously replicating plasmid pRL1831a. The degenerate oligonucleotide 5′-C(A/G)ATT(G/T)(A/G)AAAAA(A/T)GAAT(A/G)(G/T)(A/G)ACAGATAC(A/C)T-3′ was used to probe for genomic copies of the Fox box.
Primer extension assay.
Total RNA was isolated from Anabaena sp. strain PCC 7120 10 h after nitrogen stepdown by extraction with glass beads and phenol (27). Primer extension analysis was performed as described by Ausubel et al. (3), using the 29-nucleotide (nt) primer 5′-TGTATATGGGGGGAATCGGCCAAGCATCA-3′. This primer was labeled at its 5′ terminus with [γ-32P]ATP (6,000 Ci/mmol; Amersham, Arlington Heights, Ill.) by T4 polynucleotide kinase. The primer extension reaction was carried out in the presence of 100 ng of primer and 50 μg of purified total RNA. A transcriptional start site (TSS) was determined by electrophoresis of the products on sodium dodecyl sulfate (SDS)–6% polyacrylamide sequencing gels, run in parallel with DNA sequencing reactions that were generated with the same primer.
Luciferase assays.
Luciferase activity of suspensions was measured with an ATP photometer (Turner Designs, Sunnyvale, Calif.) (21) and was normalized to the concentration of chlorophyll in the sample, which was measured in methanolic extracts (38). For tests of the response of strains DR1069 and DR1069 DR2053 to NH4+, 3-μl portions of a suspension of cells (3 μg of chlorophyll ml−1) were transferred to small pieces of filter which were placed for 48 h on petri dishes of nitrogen-free solidified medium (31), with or without supplementation with 2 mM NH4Cl and 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer (pH 7.5), with a change of petri dish after 0.5 h. At 48 h, the pieces of filter were placed together on the nitrogen-free medium and the luminescence of the cells was measured with a Hamamatsu Photonics system (model C1966-20) (7). Measurements were corrected for instrumental background.
Preparation of protein extracts from Anabaena sp. strain PCC 7120.
Two-liter cultures of Anabaena sp. in AA/8 medium with nitrate were harvested by centrifugation, washed, and resuspended in fresh AA/8 medium, half with and half without nitrate. After 10 h, protein was extracted from these cultures and fractionated as described by Schmidt-Goff and Federspiel (47) except that (NH4)2SO4 was added to the supernatant solutions to 30, 50, 70, and 100% saturation.
Southwestern hybridization and DNA mobility shift assays.
Southwestern hybridization and mobility shift assays were performed as described previously (16, 32). The probes used were generated by PCR amplification with pRL1848 as the template and primers 5′-GCTCTAGAATTAGGTTTATCC-3′ (CPW63) and 5′-ACACTAGTAAAAATAATGGAAT-3′ (CPW64) for fragment A and primers 5′-GCGGTACCCACCCTATACTTA-3′ (CPW65) and 5′-CCGTCGACAACCTAATTTTT-3′ (CPW66) for fragment B.
Nucleotide sequence accession numbers.
The nucleotide sequences reported have been submitted to GenBank under accession no. AF031959 (hepA 5′ sequence) and U68034 (hepK).
RESULTS
Functional analysis of the region upstream from hepA.
We identified a 765-bp open reading frame (ORF), denoted hepC, that terminates 787 bp 5′ from hepA (Fig. 1). Its predicted translation product, HepC (molecular weight, 29,110; pI 9.73 [calculated by the ExPASy web server, Geneva, Switzerland]), shows greatest similarity (score = 56.2 bits, expect = 2e-07, identities = 39/138 [28%], positives = 66/138 [47%], gaps = 31/138 [22%]) to a galactosyltransferase from Actinobacillus actinomycetemcomitans and (expect = 2e-07 also) a UDP-galactose-lipid carrier transferase from Erwinia amylovora; similarity (score = 53.1 bits, expect = 2e-06, identities = 60/245 [24%], positives = 101/245 [40%], gaps = 41/245 [16%]) over nearly its entire length to a predicted glycosyltransferase from Synechocystis sp.; and lesser similarity (score = 46.1 bits, expect = 2e-04, identities = 34/136 [25%], positives = 58/136 [42%], gaps = 30/136 [22%]) to a predicted enzyme, involved in synthesis of lipopolysaccharide, from the same strain of Anabaena sp. (2, 61).
FIG. 1.
Nucleotide sequence of the region 5′ from the hepA gene. The transcriptional start site of hepA is underlined and labeled +1. Presumptive translational initiation codons are shown in boldface, and the BstXI site is indicated in bold italics. The 9 bp duplicated upon transposition of Tn5-1058 in the HACb mutant are double underlined. Underlined leucines 118, 125, 132, and 139, properly spaced to form a leucine zipper (11), overlap a potential membrane-spanning region (italicized; L-121 through M-145; analysis by TMPRED via Institut Suisse de Recherche Expérimentale sur le Cancer web server at Epalinges s/Lausanne, Switzerland). Regions identified in Fig. 5 as bearing cis-acting elements are indicated in lowercase.
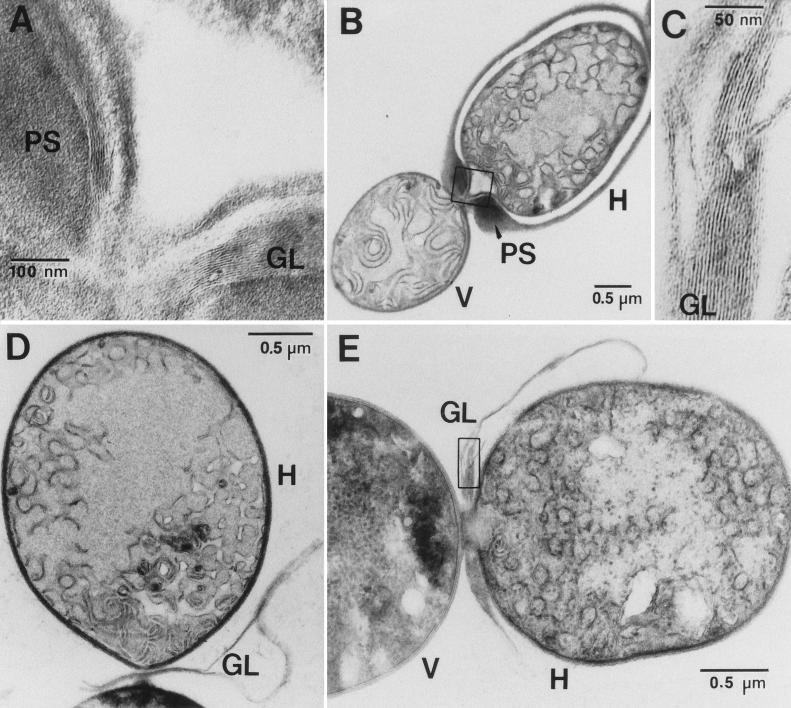
In transposon mutant HACb, hepA is expressed constitutively. Consistent with the mapping results of Kuritz et al. (35), sequencing showed that the locus of transposition of Tn5-1058 in HACb lies within hepC. A reconstruction of the HACb mutant as strain DR911 DR1069 and the similar strain DR1069 DR2053 showed the same hepA-constitutive phenotype as HACb, indicating that insertion of the transposon is responsible for the phenotype of the HACb mutant. For example, in a representative experiment and in units of photons detected by the Hamamatsu Photonics System (per nanogram of chlorophyll per minute) (mean ± standard error of the mean; n = 4), strain DR1069 showed luminescence of 0.2 ± 0.8 after 48 h on NH4+ and 238 ± 53 after 48 h on N2, whereas strain DR1069 DR2053 showed luminescence of 57 ± 6 and 232 ± 33 after 48 h on NH4+ and N2, respectively. In addition, hepC was insertionally inactivated by recombination with plasmid pRL1998a bearing, in the unique BstXI site of hepC, the omega cassette (45) (this cassette confers resistance to streptomycin and spectinomycin). The resulting mutant was Fox−, and as shown by electron microscopy, the immature-appearing heterocysts that differentiated upon nitrogen stepdown formed a laminated layer of glycolipids (55) but no envelope polysaccharide layer (Fig. 2). High-magnification electron micrographs of hepA mutant DR1069 showed that, in contrast to our earlier interpretations of lower-magnification images (42, 57) but like the results for hepC and hepK, the remaining envelope material is laminated (55) and therefore glycolipid, not polysaccharide.
FIG. 2.
hepC and hepK mutants form a laminated layer of heterocyst envelope glycolipids but no heterocyst envelope polysaccharide layer. (A; see box in panel B) In a heterocyst of wild-type PCC 7120, the laminated layer of glycolipids (55) is enveloped by a layer of polysaccharide. In contrast, the only envelope layer seen in heterocysts of hepC mutant DR1998 (D) or hepK mutant Y7 (E) is the laminated layer of glycolipids (C; see box in panel E; similar images of laminations were obtained for DR1998). H, heterocyst; V, vegetative cell; GL, glycolipid layer; PS, polysaccharide layer.
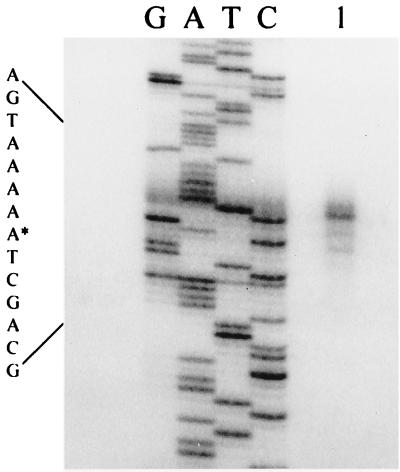
Consistent with the S1 mapping results of Holland and Wolk (30), primer extension analysis indicated the presence of a TSS 104 bp 5′ from the translational initiation codon of hepA (Fig. 3). We proceeded in two ways to identify cis-acting elements that influence the transcription of hepA. First, by making nested deletions of a fragment that extended 2,103 bp 5′ from the TSS, we generated a series of sequences whose Anabaena sp. portion extended for various distances 5′ from hepA. The resulting fragments were placed into pRL487, a plasmid that cannot replicate in Anabaena, and the constructs were transferred to Anabaena sp. hepA::luxAB derivative DR1069. The resulting single recombinants, which were validated by Southern analysis (data not shown), bore different lengths of DNA 5′ from hepA between the vector portion of the added constructs and the TSS.
FIG. 3.
5′ TSS of the hepA gene. Primer extension was performed with a 29-nucleotide (nt) oligonucleotide (see Materials and Methods) complementary to a sequence that starts 14 nt 3′ from the 5′ end of the hepA coding region. Lanes G, A, T, and C show sequencing reactions generated with the same primer. Lane 1 shows the extension product of total RNA from cells 10 h after nitrogen stepdown. The 5′ end of the transcript is marked by an asterisk in the DNA sequence shown at the left.
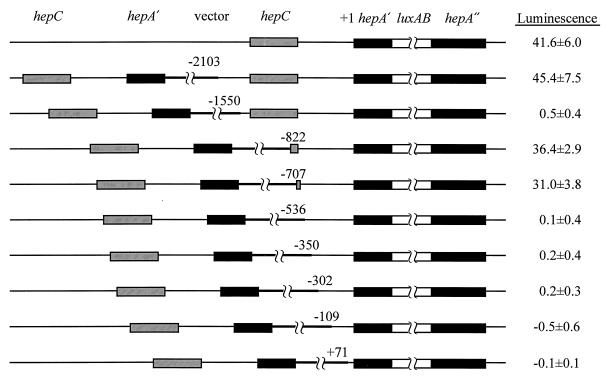
Measurements of the luminescence of these single recombinants showed the following (Fig. 4). (i) High induction of the hepA::luxAB fusion required a sequence of DNA between bp −707 and −536. (ii) Induction was greatly reduced if the luxAB fusion was under the direct control of a DNA fragment that extended to bp −1550, which includes the entire coding region of hepC together with about 100 bp upstream from hepC. (iii) Extension of this DNA fragment to bp −2103 led to increased induction of hepA::luxAB.
FIG. 4.
Luminescence, in response to nitrogen stepdown, of hepA::luxAB fused to different lengths of hepA-5′ DNA. Left, structures of single recombinants; right, corresponding values of luminescence, expressed as ATP photometer units per microgram of 0-h chlorophyll, at 10 h after nitrogen stepdown, corrected for initial values (maximally 1.9 ± 0.5 ATP photometer units per μg of chlorophyll). Gray bars, hepC or its 3′ end; black bars, 5′ end (hepA′) and 3′ end (hepA") of hepA; white bars, luxAB-bearing cassette (4.38 kb; not shown to scale); thin lines, other chromosomally derived DNA; thick lines, vector (2.72 kb; not shown to scale). Numbers represent base pairs relative to the transcriptional start site of hepA, at the junction of the vector and PCC 7120 DNA. Results from top to bottom correspond to strain DR1069 and to single-crossover recombinants of PCC 7120 with pRL1902 through pRL1910.
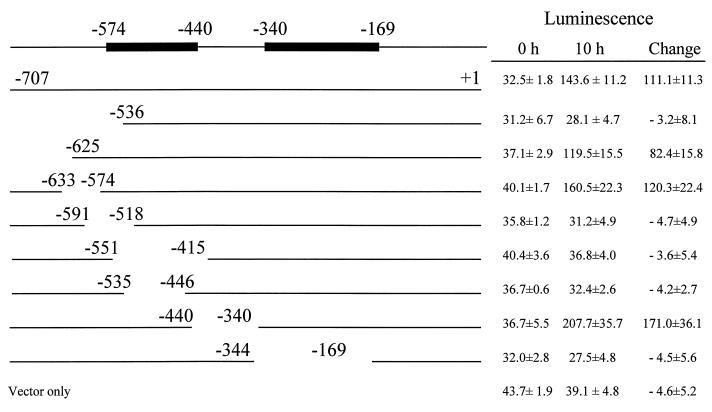
For further delimitation of cis-acting elements 5′ from hepA, we constructed a series of clones with overlapping DNA deletions from the hepC-hepA intergenic region (Fig. 5). These deletions, which averaged about 110 bp in length, extended from bp −707 to −169. DNA fragments bearing these deletions were fused to hepA::luxAB within pRL2191 and introduced into wild-type PCC 7120. Regions extending from bp −574 to −440 and from bp −340 to −169 were required for the induction of hepA::luxAB in response to nitrogen stepdown (Fig. 5).
FIG. 5.
Delimitation of cis-acting elements in the region between hepC and hepA. RSF1010-based plasmids bearing hepA::luxAB fusions downstream from intact or fenestrated intergenic sequences were introduced into wild-type PCC 7120, and the luminescence of the resulting strains was measured (as ATP photometer units per microgram of 0-h chlorophyll) at 0 and 10 h after nitrogen stepdown. Results from top to bottom correspond to transfer to PCC 7120 of pRL2199, pRL2200, pRL2192 through pRL2198, and pRL2191. Positions shown are relative to the transcriptional start site of hepA at bp +1. Bars at the top represent regions of cis-acting elements that were required for high induction of hepA::luxAB in response to nitrogen stepdown.
Southwestern and gel mobility shift assays of the cis-acting elements.
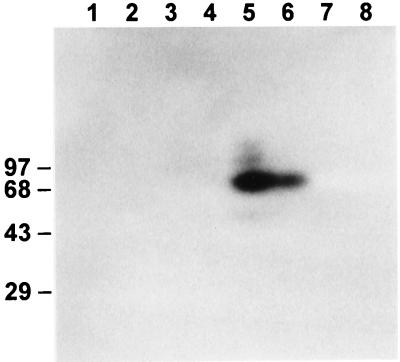
Upon Southwestern hybridization with labeled fragment B (bp −349 to −168 [Fig. 6]), the only proteins labeled were those in fractions F70+ and F70−, i.e., those precipitating between 50 and 70% saturation with ammonium sulfate and prepared from cells grown in the presence and absence, respectively, of fixed nitrogen. The strongest signal corresponded to a protein with a mass of ca. 73 to 83 kDa, with faint signals observed also for larger and smaller proteins. No signals were seen consistently with labeled fragment A (bp −580 to −445 [data not shown]).
FIG. 6.
Characterization of a DNA-binding factor by SDS-polyacrylamide gel electrophoresis and Southwestern analysis. Proteinaceous extracts from cells grown without (lanes 1, 3, 5, and 7) or with (lanes 2, 4, 6, and 8) fixed nitrogen were precipitated successively with 30, 50, 70, and 100% saturated ammonium sulfate (lanes 1 and 2, 3, and 4, 5 and 6, and 7 and 8, respectively) and separated by electrophoresis on an SDS–10% polyacrylamide gel. The proteins were then electroblotted to a nitrocellulose membrane, which was incubated with labeled fragment B and exposed to X-ray film. Sizes are indicated in kilodaltons.
Gel mobility shift assays were also performed with the same fragments A (data not shown) and B (Fig. 7). The fragments were again incubated with fractions prepared from cells grown with or without fixed nitrogen, and again it was only the fractions F70+ and F70− that shifted the electrophoretic mobility of labeled fragment B. No alteration of the mobility of fragment A was observed with any of the fractions, nor was any complexed form of fragment B seen in the absence of added protein (data not shown).
FIG. 7.
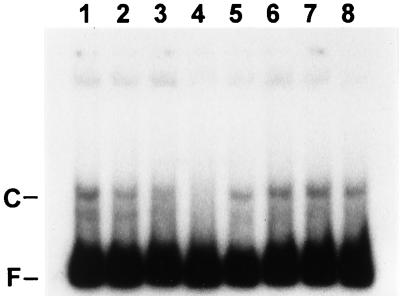
Competition assays. Protein extract fraction F70− was incubated for 20 min with labeled fragment B after 15 min of preincubation without competitor DNA (lanes 1 and 5) or in the presence of specific (fragment B; lanes 2 to 4) or nonspecific (fragment A; lanes 6 to 8) competitor DNA at 10-fold (lanes 2 and 6)-, 50-fold (lanes 3 and 7)-, or 100-fold (lanes 4 and 8)-higher molar concentration. The samples were subjected to electrophoresis on a native 5% polyacrylamide gel, which was dried and exposed to X-ray film at −80°C. C and F, complexed and free fragment B, respectively.
The specificity of the mobility-shifting interaction between a protein or proteins and fragment B was tested by means of competition experiments in which unlabeled competitor DNA was added to the protein fractions prior to incubation with labeled fragment B. A 100-fold molar excess of unlabeled fragment B greatly reduced the intensity of the band (C) of mobility-shifted, labeled fragment B (Fig. 7, lanes 1 to 4), whereas poly(dI-dC) · poly(dI-dC) (Pharmacia) (data not shown) and unlabeled fragment A (Fig. 7, lanes 5 to 8) did not reduce the intensity of that band derived from labeled fragment B. The data shown were obtained by using the protein fraction isolated from a culture grown without fixed nitrogen; very similar results were obtained with the protein fraction from a culture grown with fixed nitrogen (data not shown).
Dependence on hepK of the induction of hepA.
Upon nitrogen stepdown, transposon mutant Y7 synthesizes heterocyst envelope glycolipid but no heterocyst envelope polysaccharide (Fig. 2). Reconstruction of the mutation by sacB selection with pRL1764, pRL1765a, and pRL1765b led to the same phenotype (data not shown). The transposon is present within an ORF which we designate hepK. Its predicted translation product resembles sensory protein-histidine kinases of two-component regulatory systems (Fig. 8). The H, N, D/F, and G boxes that are characteristic of such proteins, and the H-box histidine residue that is presumptively phosphorylated (50), are all observed. Plasmid pRL1830a, which bears a hepA::luxAB fusion in RSF1010-based plasmid pKT210 (4), was introduced into wild-type PCC 7120 and into mutant Y7. The luminescence of the wild-type strain containing the fusion increased from 1.4 ± 0.1 to 53.4 ± 10.9 relative light units (μg of chlorophyll a)−1 min−1 between 0 and 10 h after nitrogen stepdown, whereas the luminescence of Y7 bearing the fusion changed only from 0.1 to 0.3 relative light units (μg of chlorophyll a)−1 min−1 during the same time interval.
FIG. 8.
HepK. Predicted Anabaena sp. strain PCC 7120 HepK (An) sequence compared with the sequences of typical protein-histidine kinases of two-component regulatory systems: Sy, a hypothetical such protein from Synechocystis sp. strain PCC 6803 (DDBJ accession no. D90912, locus 1653308); Cc, such a protein from Caulobacter crescentus (GenBank accession no. M91449); Ec, ArcB from E. coli (GenBank accession no. X53315; amino acids 77 to 502 out of 778; score = 107 bits, expect = 2e-22, identities = 117/453 [25%], positives = 188/453 [40%; indicated with •], gaps = 46/453 [10%]). Bold italics, residues that the predicted protein from PCC 7120 shares with the other sequences shown; ∗, presumptively phosphorylated histidine residue. Conserved H, N, D/F, and G boxes of such kinases (50) are doubly overlined, singly underlined, singly overlined, and doubly underlined, respectively. Lowercase, possible membrane-spanning helical regions of HepK (analysis by TMPRED).
Starting 259 bp 3′ from the termination codon of hepK is an ORF whose predicted product shows 61% amino acid sequence identity (75% sequence similarity) over its ca. 768 amino acids (predicted partially by sequencing of a single strand of DNA) to a predicted 793-amino-acid DNA helicase II [locus D64002, PID|d1011041] from Synechocystis sp. strain PCC 6803. Insertional inactivation of the presumptive helicase gene by double-reciprocal recombination with plasmid pRL1347ab led to no evident developmental phenotype upon nitrogen stepdown (data not shown). Moreover, the mutation in strain Y7 was complemented in all of 20 exconjugants to which had been transferred pRL2078, a pDU1-based plasmid that contains a 4.9-kb DNA fragment bearing wild-type hepK, an upstream region, and the first 389 bp of the presumptive helicase II coding region.
The Fox box.
Hybridization with a degenerate probe corresponding to the Fox box indicated the presence of ca. 12 hybridizing sites in the genome of PCC 7120 (data not shown). We sought in several ways to determine whether the Fox box 3′ from hepA has regulatory significance. This sequence was replaced by a SalI restriction site by synthesis of the DNA fragments flanking the Fox box via PCR and ligation of the resulting products. The fidelity of the PCR products was established by sequencing (data not shown). Cells bearing plasmid pRL1830a, which contains a Fox box downstream from its hepA::luxAB fusion, showed luminescence of 2.3 ± 0.6 and 87.9 ± 2.4 relative light units (μg of chlorophyll a)−1 min−1 0 and 10 h after nitrogen stepdown, respectively, whereas cells bearing plasmid pRL1831a, in which the downstream Fox box is replaced by a SalI site, showed luminescence of 2.4 ± 0.6 and 114.1 ± 38.4 relative light units (μg of chlorophyll a)−1 min−1, respectively, at those same times. Thus, no significant decrease of induction of hepA::luxAB was observed upon nitrogen stepdown in the absence of a Fox box. pDU1-based plasmid pRL1729, which bears a 6.0-kb ClaI-BglII DNA fragment that contains the hepA gene, complemented hepA mutant EF116, restoring a Fox+ phenotype. However, so did plasmid pRL1730, equivalent to pRL1729 but with the Fox box 3′ from hepA substituted by a SalI site. Similarly, replacement of the chromosomal copy of the Fox box 3′ from hepA in wild-type PCC 7120 by a SalI site by double-reciprocal recombination with plasmid pRL1818a, producing strain DR1818, led to no obvious effect on the development of the mutant or its capacity for aerobic fixation of N2. (DR1818 can be considered a variant of DR1817, which also showed no evident developmental or N2 fixation phenotype compared to wild-type PCC 7120.) These experiments suggested that neither the transcription of hepA nor the biological effect of that gene on aerobic nitrogen fixation is significantly affected by the presence of the Fox box 3′ from that gene.
DISCUSSION
Testing of a series of nested deletions 5′ from the transcriptional start site of hepA showed that upon deletion of the DNA sequence between 707 and 536 bp upstream from that start site, extensive induction of hepA::luxAB upon nitrogen stepdown is lost (Fig. 4). That sequence may overlap the binding site(s) for one or more transcriptional regulators. Despite the presence of an intact copy of hepC in all single recombinants illustrated in Fig. 4, induction of hepA in a recombinant with pRL1903 was greatly reduced by the presence of a sequence that included a second copy of hepC, a result consistent with the observation that insertion of a transposon or a cassette in hepC (Fig. 1) led to constitutive activity of hepA. The possible interpretation of these results that HepC directly regulates hepA might seem to receive support from the presence in HepC of a potential leucine zipper, a motif by means of which certain DNA-binding proteins are known to dimerize (11). However, that interpretation encounters the difficulties that HepC (i) lacks a known DNA-binding motif and (ii) has what appears to be a transmembrane helix that overlaps the region of its possible leucine zipper. The protein that binds to sequence B (see below), upstream from hepA, appears more than twice as large as HepC. A second possibility, that a portion of hepC acts in cis to down-regulate hepA, cannot account for the divergent effect of the sequence 5′ from hepC in the recombinant with pRL1902. The extensive similarity (2) of the predicted HepC to UDP-galactose-lipid carrier transferases (and similar enzymes; a different but catalytically similar enzyme is involved in synthesis of the lipopolysaccharide of vegetative cells [61]) and the absence of heterocyst envelope polysaccharide in hepC mutants (Fig. 2) lead us to favor a third interpretation. We suggest that HepC plays a role in the synthesis of the repeating subunit of that polysaccharide, perhaps by involvement in the addition of galactosyl side branches such as are present in all known heterocyst envelope polysaccharides (15), and that HepC may affect transcription of hepA indirectly, by generating a metabolite that helps to repress hepA or by utilizing a metabolite that helps to induce hepA. It may be that in the recombinant of PCC 7120 with pRL1903 (Fig. 4), sequences 5′ from hepC that limit its expression are absent, so that hepC is overexpressed, whereupon hepA is strongly repressed, whereas in the recombinant with pRL1902 (next-to-top construct in Fig. 4), in which additional sequence 5′ from hepC is present, hepC may be normally regulated.
Further analysis (Fig. 5) showed that deletion from bp −707 to −625 or from bp −633 to −574 had little effect on transcription of hepA::luxAB, whereas deletion of the sequence from bp −707 to −536 resulted in extensive loss of transcription. We conclude that a sequence between bp −574 and −536 is required for transcription. Similar analysis indicated that a region from bp −535 to −440 and a region from bp −340 to −169 are required for induction of hepA after nitrogen deprivation. Perhaps regulatory proteins bind to the regions −574 to −440 (sequence A) and −340 to −169 (sequence B) and, together with RNA polymerase, lead to initiation of transcription. Despite the hepA-constitutive phenotype of a hepC mutation, none of the deletions upstream from hepA in the experiments represented by Fig. 5 led to high, nitrogen-independent expression of hepA::luxAB. Perhaps a corresponding regulatory region lies 3′ from bp −169 or perhaps a window or windows that overlapped such a region overlapped also a region that negated its effect. Southwestern analysis confirmed that certain proteins bound to fragment B but gave no information about the specificity of the interaction except that the same proteins did not appear to bind to fragment A.
Gel mobility shift assays also indicated that a certain proteinaceous factor(s), derived from extracts of cultures grown with or without fixed nitrogen, bound specifically to fragment B (bp −349 to −168). Available data cannot distinguish whether the binding factor(s) represses or activates transcription. Possible repression might be expressed only in vegetative cells. Then, because only about 10% of vegetative cells differentiate into heterocysts, a culture grown without fixed nitrogen may contain almost as much of such a putative repressor as would an uninduced culture. If the binding factor activates transcription, it may always bind to fragment B but become active only in developing heterocysts.
That no alteration of the mobility of fragment A was observed may mean that fragment A is not, or is only part of, a protein-binding site or that proteins that bind to that site are present in extracts of whole filaments at a concentration too low to permit detection by gel mobility shift assays. The latter might be the case if, for example, a binding protein were present only in the small percentage of cells that differentiates.
Unlike inactivation of hepC, inactivation of hepK blocks expression of hepA. The following facts suggest that a protein that receives a phosphate group from HepK may regulate, directly or indirectly, the hepA promoter. (i) The putative product of hepK resembles sensory protein-histidine kinases of two-component signal transduction systems; (ii) a mutation in hepK (like a mutation in hepC) blocks the synthesis of heterocyst envelope polysaccharide while not qualitatively affecting synthesis of heterocyst envelope glycolipid; (iii) the phenotype of a hepK mutation is not a polar effect on the gene 3′ from it, because insertional inactivation of the latter ORF leads to no comparable phenotype and the hepK mutation is complemented by a sequence that includes little of the 3′ ORF; (iv) hepA is also required for synthesis of heterocyst envelope polysaccharide; and (v) induction of hepA in response to nitrogen deprivation requires an intact hepK gene.
It is interesting that according to the results of BLAST searching, HepK shows the most protracted similarity to ArcB (Fig. 8). This E. coli protein-histidine kinase is involved in the repression of genes of aerobic metabolism under anaerobic conditions (17). It is also involved in the activation of expression, as O2 becomes limiting, of the cydAB operon, which encodes a cytochrome d oxidase complex whose affinity for oxygen is greater than that of the cytochrome o oxidase complex (52). Perhaps HepK is involved in sensing, directly or indirectly, the decreasing pO2 within immature heterocysts. However, whether HepK is involved in the regulation of genes other than hepA, including possible pO2-responsive genes, in developing heterocysts remains to be determined.
ACKNOWLEDGMENTS
We thank Xudong Xu for helpful discussions and Kelly Zarka for expert assistance.
This work was supported by the U.S. Department of Energy under grant DE-FG02-91ER20021 and by NSF grant MCB 9723193.
REFERENCES
- 1.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1987. pp. 4.8.1–4.8.3. [Google Scholar]
- 4.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 5.Bergh K T, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black K, Buikema W, Haselkorn R. The hglK gene is required for localization of heterocyst-specific glycolipids in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1995;177:6440–6448. doi: 10.1128/jb.177.22.6440-6448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 8.Black T A, Wolk C P. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J Bacteriol. 1994;176:2282–2292. doi: 10.1128/jb.176.8.2282-2292.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buikema W J, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 10.Burbulys D, Trach K A, Hoch J A. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 11.Busch S J, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, Wolk C P. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J Bacteriol. 1997;179:267–271. doi: 10.1128/jb.179.1.267-271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell E L, Hagen K D, Cohen M F, Summers M L, Meeks J C. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1996;178:2037–2043. doi: 10.1128/jb.178.7.2037-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardemil L, Wolk C P. The polysaccharides from the envelopes of heterocysts and spores of the blue-green algae Anabaena variabilis and Cylindrospermum licheniforme. J Phycol. 1981;17:234–240. [Google Scholar]
- 16.Chastain C J, Brusca J S, Ramasubramanian T S, Wei T-F, Golden J W. A sequence-specific DNA-binding factor (VF1) from Anabaena sp. strain PCC 7120 vegetative cells binds to three adjacent sites in the xisA upstream region. J Bacteriol. 1990;172:5044–5051. doi: 10.1128/jb.172.9.5044-5051.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotter P A, Melville S B, Albrecht J A, Gunsalus R P. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 18.de Lorimier R, Bryant D A, Porter R D, Liu W-Y, Jay E, Stevens S E. Genes for the α and β subunits of phycocyanin. Proc Natl Acad Sci USA. 1984;81:7946–7950. doi: 10.1073/pnas.81.24.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 20.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 21.Elhai J, Wolk C P. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 1990;9:3379–3388. doi: 10.1002/j.1460-2075.1990.tb07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elhai, J., and C. P. Wolk. Unpublished data.
- 23.Ernst A, Black T, Cai Y, Panoff J-M, Tiwari D N, Wolk C P. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J Bacteriol. 1992;174:6025–6032. doi: 10.1128/jb.174.19.6025-6032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Piñas F, Leganés F, Wolk C P. A third genetic locus required for the formation of heterocysts in Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176:5277–5283. doi: 10.1128/jb.176.17.5277-5283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiedler G, Arnold M, Hannus S, Maldener I. An ABC-exporter is essential for the localisation of envelope material in heterocysts of cyanobacteria, abstr. 69A. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. Abstracts of the IX International Symposium on Phototrophic Prokaryotes. 1997. [Google Scholar]
- 26.Frías J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 27.Golden S S, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 28.Haselkorn R. Molecular genetics of nitrogen fixation in photosynthetic prokaryotes. In: Tikhonovich I A, Provogov N A, Romanov V I, Newton W E, editors. Nitrogen fixation: fundamentals and applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 29–36. [Google Scholar]
- 29.Hoch J A. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction system. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 129–144. [Google Scholar]
- 30.Holland D, Wolk C P. Identification and characterization of hetA, a gene that acts early in the process of morphological differentiation of heterocysts. J Bacteriol. 1990;172:3131–3137. doi: 10.1128/jb.172.6.3131-3137.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu N-T, Thiel T, Giddings T H, Wolk C P. Anabaena and Nostoc cyanophages from sewage settling ponds. Virology. 1982;114:236–246. doi: 10.1016/0042-6822(81)90269-5. [DOI] [PubMed] [Google Scholar]
- 32.Keller A D, Maniatis T. Selection of sequences recognized by a DNA binding protein using a preparative southwestern blot. Nucleic Acids Res. 1991;19:4675–4680. doi: 10.1093/nar/19.17.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khudyakov I, Wolk C P. Evidence that the hanA gene coding for HU protein is essential for heterocyst differentiation in, and cyanophage A-4(L) sensitivity of, Anabaena sp. strain PCC 7120. J Bacteriol. 1996;178:3572–3577. doi: 10.1128/jb.178.12.3572-3577.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khudyakov I, Wolk C P. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:6971–6978. doi: 10.1128/jb.179.22.6971-6978.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuritz T, Ernst A, Black T A, Wolk C P. High-resolution mapping of genetic loci of Anabaena PCC 7120 required for photosynthesis and nitrogen fixation. Mol Microbiol. 1993;8:101–110. doi: 10.1111/j.1365-2958.1993.tb01207.x. [DOI] [PubMed] [Google Scholar]
- 36.Liang J, Scappino L, Haselkorn R. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc Natl Acad Sci USA. 1992;89:5655–5659. doi: 10.1073/pnas.89.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynn M E, Bantle J A, Ownby J D. Estimation of gene expression in heterocysts of Anabaena variabilis by using DNA-RNA hybridization. J Bacteriol. 1986;167:940–946. doi: 10.1128/jb.167.3.940-946.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- 39.Maldener I, Ernst A, Fernández-Piñas F, Wolk C P. Characterization of devA, a gene whose mutation by a transposon prevents maturation of proheterocysts in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176:7543–7549. doi: 10.1128/jb.176.24.7543-7549.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maldener I, Arnold M, Englmeier L, Fiedler G, Hannus S. Characterization of cyanobacterial mutants that are impaired in heterocyst differentiation, abstr. 152A. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. Abstracts of the IX International Symposium on Phototrophic Prokaryotes. 1997. [Google Scholar]
- 41.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 42.Murry M A, Wolk C P. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch Microbiol. 1989;151:469–474. [Google Scholar]
- 43.Orosz A, Boros I, Venetianer P. Analysis of the complex transcription termination region of the Escherichia coli rrnB gene. Eur J Biochem. 1991;201:653–659. doi: 10.1111/j.1432-1033.1991.tb16326.x. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 45.Prentki P, Binda A, Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991;103:17–23. doi: 10.1016/0378-1119(91)90385-o. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Schmidt-Goff C M, Federspiel N A. In vivo and in vitro footprinting of a light-regulated promoter in the cyanobacterium Fremyella diplosiphon. J Bacteriol. 1993;175:1806–1813. doi: 10.1128/jb.175.6.1806-1813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 49.Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 50.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 51.Thiel T. Genetic analysis of cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 581–611. [Google Scholar]
- 52.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 53.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 54.Wei T-F, Ramasubramanian T S, Golden J W. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkenbach F, Wolk C P, Jost M. Lipids of membranes and of the cell envelope in heterocysts of a blue-green alga. Planta. 1972;107:69–80. doi: 10.1007/BF00398015. [DOI] [PubMed] [Google Scholar]
- 56.Wolk, C. P. Unpublished data.
- 57.Wolk C P, Cai Y, Cardemil L, Flores E, Hohn B, Murry M, Schmetterer G, Schrautemeier B, Wilson R. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J Bacteriol. 1988;170:1239–1244. doi: 10.1128/jb.170.3.1239-1244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolk C P, Cai Y, Panoff J-M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolk C P, Elhai J, Kuritz T, Holland D. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol Microbiol. 1993;7:441–445. doi: 10.1111/j.1365-2958.1993.tb01135.x. [DOI] [PubMed] [Google Scholar]
- 60.Wolk C P, Ernst A, Elhai J. Heterocyst metabolism and development. In: Bryant D, editor. Molecular genetics of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- 61.Xu X, Khudyakov I, Wolk C P. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:2884–2891. doi: 10.1128/jb.179.9.2884-2891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]