Abstract
Arginine catabolism produces ammonia without transferring nitrogen to another compound, yet the only known pathway of arginine catabolism in Escherichia coli (through arginine decarboxylase) does not produce ammonia. Our aims were to find the ammonia-producing pathway of arginine catabolism in E. coli and to examine its function. We showed that the only previously described pathway of arginine catabolism, which does not produce ammonia, accounted for only 3% of the arginine consumed. A search for another arginine catabolic pathway led to discovery of the ammonia-producing arginine succinyltransferase (AST) pathway in E. coli. Nitrogen limitation induced this pathway in both E. coli and Klebsiella aerogenes, but the mechanisms of activation clearly differed in these two organisms. We identified the E. coli gene for succinylornithine aminotransferase, the third enzyme of the AST pathway, which appears to be the first of an astCADBE operon. Its disruption prevented arginine catabolism, impaired ornithine utilization, and affected the synthesis of all the enzymes of the AST pathway. Disruption of astB eliminated succinylarginine dihydrolase activity and prevented arginine utilization but did not impair ornithine catabolism. Overproduction of AST enzymes resulted in faster growth with arginine and aspartate. We conclude that the AST pathway is necessary for aerobic arginine catabolism in E. coli and that at least one enzyme of this pathway contributes to ornithine catabolism.
In a defined minimal medium, Escherichia coli and Klebsiella aerogenes can use several amino acids as sole nitrogen sources (reviewed in reference 24). Such catabolism must result in assimilation of nitrogen into glutamate and glutamine, which provide nitrogen for the synthesis of virtually all nitrogen-containing compounds. Because glutamine synthesis requires ammonia, amino acid degradation must produce ammonia, whose concentration and generation limit growth. A low level of ammonia results in a low level of glutamine, which induces proteins that transport, degrade, and assimilate nitrogen-containing compounds. This response, which in effect is a response to ammonia limitation, is called the nitrogen-regulated (Ntr) response (see references 24 and 25 for reviews).
Despite the importance of reactions that produce ammonia, such reactions have not been identified for the catabolism of γ-aminobutyrate, aspartate, arginine, glutamate, proline, putrescine, and other compounds (24). The obvious approach to analyze such catabolism, i.e., to isolate mutants, has not succeeded for aspartate and arginine catabolism. Therefore, we adopted an alternate approach: we characterized 15N-aspartate catabolism by a variety of methods, including nuclear magnetic resonance analysis of cellular extracts (11). When E. coli was incubated with 14N-arginine (in the form of a rapidly metabolized dipeptide) and 15N-aspartate, all of the ammonia was 14N, which implied that arginine catabolism produced ammonia. Alanine, aspartate, and glutamate were heavily labeled, which implied that direct deamination of these amino acids did not produce ammonia. These results were unexpected since the only known pathway of arginine catabolism does not generate ammonia.
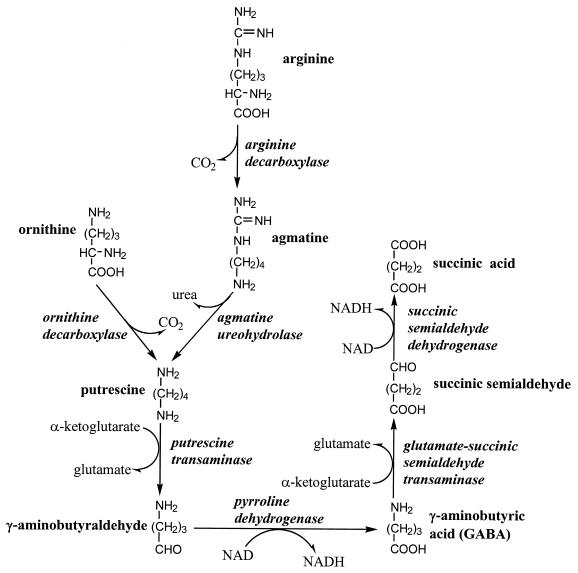
It has been proposed that E. coli degrades arginine via a pathway that is initiated with arginine decarboxylation (30, 31). The agmatine produced from this reaction is metabolized to putrescine, which is eventually degraded to glutamate and succinate (see Fig. 1). A catabolic function for this pathway is consistent with the induction by nitrogen limitation of all the enzymes of this pathway, except for arginine decarboxylase (ADC) itself, which is constitutive (31). Furthermore, mutants deficient in ADC, agmatine ureohydrolase, or putrescine transaminase grow more slowly than the corresponding wild-type strain with arginine as the sole nitrogen source. Nonetheless, we suspected another pathway because this pathway does not produce ammonia and because all mutants with deficiencies in the ADC pathway still grow reasonably well with arginine; for example, a speB (agmatine ureohydrolase-encoding) mutant has a 300-min doubling time, whereas the isogenic wild-type strain has a 160-min doubling time (30). In the present study, we show that the ADC pathway does not significantly contribute to arginine degradation. Instead, we find that E. coli possesses the ammonia-producing arginine succinyltransferase (AST) pathway. We present evidence that the AST pathway is necessary for arginine degradation during nitrogen-limited growth and that it contributes to the degradation of other amino acids.
FIG. 1.
Arginine catabolism by the ADC pathway and the routes of putrescine synthesis.
MATERIALS AND METHODS
Strains and plasmids.
All E. coli K-12 strains used for enzyme assays and growth experiments were derivatives of W3110, which was considered a wild-type strain (29). Strain LR1 also contains glnG10::Tn5, and AKK1 contains a disruption of astB, the fourth gene of the ast operon. AKK1 was constructed by ligating the 2-kb SmaI/NsiI fragment of pLC3-11, which contains the fourth gene of the ast operon, into pUC18, which had been digested with SmaI and PstI. Removing a 300-bp NruI/BssHII fragment from the insert and replacing it with a kanamycin resistance gene resulted in disruption of the cloned gene. A 4.2-kb linear DNA fragment with the disrupted gene was electroporated into strain K4633 (recD), and integrants were selected on kanamycin-containing plates. The disruption was transduced into W3110 with P1 vir. K4633 (recD) was obtained from A. Ninfa (University of Michigan). The K. aerogenes strains were KC2668 (wild type) and KC2738 (glnG2::Tn5-131), which were obtained from R. Bender (University of Michigan). C. Fraley and A. C. Matin (Stanford University) disrupted the astC gene, which codes for succinylornithine transaminase, as part of an independent study. They provided us with a strain containing the disruption, and we transferred the astC::kan into W3110 by P1 transduction. The E. coli Genetic Stock Center provided the Carbon-Clarke plasmid pLC3-11 (3, 26). Plasmid pΔLC3-11 is derived from pLC3-11 by deletion of two adjacent PstI fragments, which were replaced by a kanamycin resistance-encoding cassette from pUC-4-KAPA (Pharmacia). The altered plasmid contains chromosomal DNA from osmE to xthA.
Cell growth.
The minimal growth medium contained W salts (27), 0.4% of the carbon source and 0.2% of each nitrogen source, unless otherwise indicated. Cells were grown and harvested as described previously (27); whole-cell pellets were frozen until needed.
Enzyme assays.
Cell pellets from a 15-ml culture were resuspended in 1 ml of 50 mM K-PO4 buffer (pH 7.5) (mixture of K2HPO4 and KH2PO4 at pH 7.5) with 1 mM β-mercaptoethanol and sonicated three times for 5 s. Cell debris was removed by centrifugation for 10 min at 4°C in a microfuge.
Syntheses for substrates of enzymes of the AST pathway, i.e., N-succinyl derivatives of arginine, ornithine, and glutamate, have been described (40). The assays for AST pathway enzymes were performed essentially as described previously (14, 40). In brief, AST was assayed by measuring the decrease in succinyl coenzyme A at 232 nm. The activity of succinylarginine dihydrolase was assessed by determination of the amount of ammonia produced, which was coupled enzymatically by glutamate dehydrogenase to the consumption of NADH. Succinylglutamate desuccinylase was analyzed by measuring the liberation of glutamate, which was coupled to the glutamate dehydrogenase-dependent production of NADH, which absorbs at 340 nm. Succinylornithine transaminase was assayed by measuring the product succinylglutamic semialdehyde, which is hydrolyzed to glutamic semialdehyde. Glutamic semialdehyde spontaneously cyclizes to Δ′-pyrroline-5-carboxylic acid, which reacts with o-aminobenzaldehyde to yield a product that absorbs at 440 nm. Succinylglutamic semialdehyde dehydrogenase was measured by monitoring NAD reduction at 340 nm. The substrate for this reaction was synthesized during the reaction by adding succinylornithine and α-ketoglutarate, together with succinylornithine transaminase, which had been purified from E. coli W3110. For all assays and for purification of the transaminase, protein was determined as described earlier (18).
Purification and partial sequencing of succinylornithine transaminase.
The procedure for this enzyme purification differed substantially from that previously described (6). The transaminase was produced from cells of W3110 containing plasmid pΔLC3-11 that had been grown in glucose-arginine minimal medium. The cell extract was prepared by sonication in a pH 7.5 buffer containing 50 mM KPO4, 0.1 mM pyridoxal phosphate, 2.5 mM α-ketoglutarate, 0.1 mM EDTA, and 1 mM dithiothreitol. The protein was precipitated with 60% (NH4)2SO4, redissolved, applied to a DEAE-Sephadex column, and eluted with a KCl gradient (peak activity eluted at 200 mM KCl). The protein was subjected to gel filtration on a G-100 column and applied to a phenyl-Sepharose column, with peak activity eluting at 500 mM (NH4)2SO4. The protein was dialyzed before use. The Harvard Microchemistry Facility sequenced two peptides of the transaminase after proteolytic digestion.
Arginine and urea determination.
Cells were grown at 30°C in media described in Results. Samples of 1.5 ml were centrifuged, and the cell-free culture medium was assayed for arginine and urea. Arginine was converted to urea and ornithine by arginase, and the urea produced was measured. Arginine and urea were added to growth medium and processed as described below to generate standard curves. Arginine reacted with the urea reagent; therefore, it was necessary to physically separate arginine from urea. Separation was achieved by modifying a previously described method (5). Pasteur pipettes containing 2 ml of Dowex 50×4 resin (400 mesh), which had been prepared as described previously (5), were washed with 2 ml of 1 N NaOH and then equilibrated with 2 ml of fresh culture medium, which had been adjusted to pH 2.5 with HCl. A 0.5-ml sample was adjusted to pH 2.5 with HCl, applied to the column, and eluted with 4.5 ml of 0.1 M Na citrate (pH 3.1). The 5 ml of solution that came through was then assayed for urea. Arginine was eluted with 8 ml of a solution that contained 0.5 M Na citrate and 0.35 M acetic acid that had been adjusted to pH 9.5 with NaOH. (The resulting high ion concentration was deliberate.)
Urea was measured essentially as described earlier (22). Samples of 0.35 ml were mixed with 0.25 ml of an acid mixture (H2SO4-H3PO4-H2O, 1:3:1 [vol/vol/vol]) and 0.04 ml of 4% α-isonitrosopropiophenone in 95% ethanol. The mixtures were boiled in the dark for 1 h and then cooled to room temperature; their absorbance at 540 nm was then determined.
For the arginine determination, 0.1 ml of sample was mixed with 0.05 ml of arginase (30 U/ml) in 50 mM maleic acid (pH 7.0) with 50 mM MnSO4 (41) and incubated for 30 min at 37°C; the reaction was stopped with 0.15 ml of the acid mix. A portion of this solution (0.2 ml) was mixed with 0.15 ml of the acid mix, 0.25 ml of H2O, and 0.04 ml of 4% α-isonitrosopropiophenone in 95% ethanol. This mixture was then treated as described above for the urea determination.
RESULTS
Role of ADC in arginine degradation in E. coli.
ADC initiates the only known pathway of arginine degradation in E. coli (Fig. 1; references 24, 30, and 31). The sum of the complete reaction sequence is shown below:
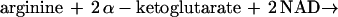 |
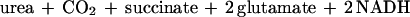 |
To quantify this pathway’s contribution to arginine catabolism, we compared arginine utilization and urea production. E. coli does not metabolize urea, which is rapidly excreted into the medium (21); therefore, we measured urea from the medium. When arginine was used as the sole nitrogen source, less than 3% of the arginine consumed resulted in urea (Table 1). To validate the assay for arginine, we assessed arginine utilization from ammonia-containing medium. Ammonia and the subsequent elevation of the glutamine pool prevent activation of Ntr genes, such as the genes of arginine transport, and should therefore diminish arginine utilization (24). We observed the expected result: there was five times less total arginine consumed despite almost twice the cell growth. Unlike growth with arginine as a nitrogen source, the ADC pathway metabolized 36% of the arginine consumed. It was expected that the ADC pathway would become the major route of putrescine synthesis in ammonia-containing medium, since arginine inhibits the only other route of putrescine synthesis via ornithine decarboxylase and represses the enzymes of ornithine synthesis. To further validate our measurements, we assayed urea production from cells which contain an unusually high level of ADC, i.e., cells grown anaerobically in a complex acidic medium (4, 35, 36). It has been proposed that ADC helps raise the pH by releasing acidic carboxyl groups as CO2 (9). As expected, the urea produced, when normalized to cell mass, was greater than that from any other medium. In summary, these results show that the ADC pathway does not contribute significantly to the degradation of arginine as the sole nitrogen source. Therefore, there must be another pathway for arginine degradation in E. coli.
TABLE 1.
Assessing the role of the ADC pathway in arginine catabolism in E. coli
| Medium | OD600 at saturation | Arg depleted (mM)a | Urea accumulated (mM) | % Arg to urea |
|---|---|---|---|---|
| Minimalb | ||||
| Arginine | 2.66 | 5.0 | 0.15 | 3 |
| NH4+-arginine | 4.74 | 1.1 | 0.40 | 36 |
| NH4+ | 4.74 | NAc | 0.13 | NA |
| Complex acidicd | ||||
| Aerobic growth | 4.12 | NAd | 0.12 | NA |
| Anaerobic growth | 0.95 | NAd | 0.21 | NA |
Calculated as starting concentration (0.2% = 11.5 mM) minus the concentration at the indicated optical density.
Cells were grown in minimal medium with 0.4% glucose and 0.2% of each indicated nitrogen source.
NA, not applicable.
Cells were grown in buffered modified Falkow arginine medium, pH 5.5 (32). This medium contains peptides. The arginine from these peptides might be used before the added arginine. Since the urea produced may result from degradation of the arginine in the peptides, it was not considered possible to compare the urea produced to the arginine consumed.
AST pathway in E. coli.
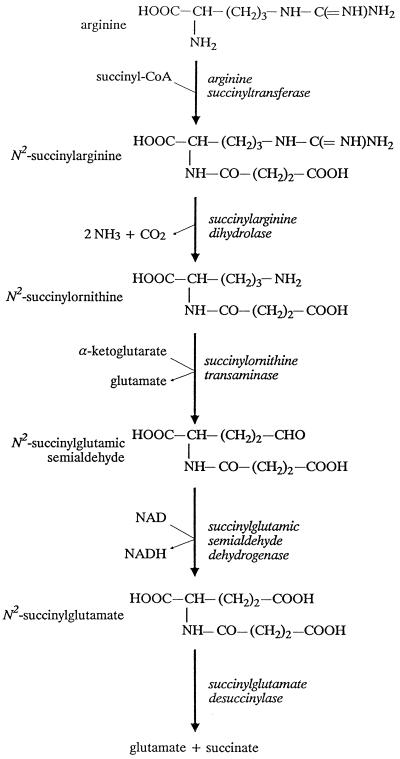
Bacteria contain a surprising variety of pathways for arginine degradation (reviewed in references 4 and 24). We assumed that E. coli contained a pathway that had previously been characterized in another organism. Therefore, we sought a pathway that produces ammonia but not urea. Two well-studied pathways fit these criteria: the arginine deiminase pathway and the AST pathway (4). The arginine deiminase pathway involves three enzymes, arginine deiminase, a catabolic ornithine transcarbamoylase, and carbamate kinase, which catalyze the reactions of arginine to citrulline, of citrulline and phosphate to ornithine and carbamoyl phosphate, and of carbamoyl phosphate and ADP to CO2, NH3, and ATP, respectively. Despite biochemical searches for these enzymes in E. coli and K. aerogenes and careful genetic searches in E. coli, enzymes of this pathway have not been found (8, 15, 16, 20). On the other hand, the AST pathway has been found in K. aerogenes, which is closely related to E. coli (33). Therefore, we focused our attention on the AST pathway. The reactions of this pathway are shown in Fig. 2 and their sum is shown below:
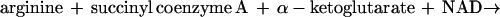 |
 |
As described in more detail below, we successfully assayed all five enzymes of the pathway from wild-type E. coli.
FIG. 2.
AST pathway of arginine catabolism.
Induction of the AST pathway by nitrogen limitation.
To initially analyze the function of the AST pathway, we examined the regulation of its enzymes. The degradation of arginine as the sole nitrogen source limits growth and requires the transcriptional activator nitrogen regulator I (NRI, also called NtrC), the product of glnG (or ntrC) (23). Therefore, we tested whether nitrogen limitation induced enzymes of the AST pathway and, if so, whether such induction required NRI. Nitrogen-limited E. coli had much more succinylarginine dihydrolase and succinylglutamate desuccinylase than cells grown in the corresponding nitrogen-rich (ammonia-containing) medium (compare lines 1 and 3 to lines 2 and 4 in Table 2). Furthermore, the levels of these enzymes were low in glnG mutants (compare lines 3 and 5 in Table 2). By these two criteria, these AST enzymes in E. coli are under nitrogen control.
TABLE 2.
AST pathway enzymes in E. coli and K. aerogenes
| Line | Strain | Mediuma | Enzyme activity, μmol/h/mg of protein (n)b, of:
|
||
|---|---|---|---|---|---|
| Dihydrolase | Desuccinylase | Transaminasec | |||
| E. coli | |||||
| 1 | Wild type | Glucose-arginine | 1.5 ± 0.16 (3) | 2.6 ± 0.10 (3) | 5.4 ± 0.24 (3) |
| 2 | Wild type | Glucose-NH4+-arginine | 0.06 ± 0.033 (2) | 0.09 ± 0.033 (2) | 0.57 ± 0.35 (2) |
| 3 | Wild type | Glucose-glutamine | 0.47 ± 0.04 (4) | 0.84 ± 0.03 (4) | 3.8 ± 0.36 (4) |
| 4 | Wild type | Glucose-NH4+-glutamine | 0.08 ± 0.08 (3) | <0.05 (3) | 2.4 ± 0.14 (3) |
| 5 | glnG | Glucose-glutamine | 0.14 ± 0.03 (4) | 0.22 ± 0.03 (4) | 2.2 ± 0.11 (4) |
| K. aerogenes | |||||
| 6 | Wild type | Arginine | 38 ± 4.0 (2) | 21 ± 0.6 (2) | 64 ± 5.6 (2) |
| 7 | Wild type | Glucose-arginine | 8.0 ± 0.31 (5) | 8.1 ± 0.42 (5) | 24 ± 1.1 (5) |
| 8 | Wild type | Glucose-NH4+-arginine | 3.63 ± 0.20 (3) | 3.32 ± 0.34 (3) | 9.32 ± 0.56 (3) |
| 9 | Wild type | Glucose-glutamine | 0.48 ± 0.058 (2) | 0.44 ± 0.091 (2) | 1.6 ± 0.1 (2) |
| 10 | Wild type | Glucose-NH4+-glutamine | 0.14 ± 0.03 (3) | 0.22 ± 0.033 (3) | 1.2 ± 0.18 (3) |
| 11 | glnG | Glucose-glutamine | 0.17 ± 0.029 (2) | 0.05 ± 0.05 (2) | 7.5 ± 1.1 (2) |
| 12 | glnG | Glucose-arginine | 7.41 ± 0.88 (2) | 6.86 ± 1.3 (2) | 18 ± 1.6 (2) |
Cells were grown in minimal medium with 0.4% glucose and 0.2% of the indicated nitrogen sources. Arginine was at 1% when used as both carbon and nitrogen source.
The variation is expressed as the standard error of the mean, with the number of determinations in parentheses.
Except for cells grown in arginine-containing medium, the assay for this enzyme measures both the arginine-repressible acetylornithine transaminase and the arginine-inducible succinylornithine transaminase.
The results for succinylornithine transaminase, the third enzyme of the pathway, appeared to suggest that this enzyme was not under nitrogen control. However, it is likely that this assay measures the activities of two different enzymes. This suspicion is based on the fact that a mutation in argD, which codes for an arginine-repressible acetylornithine transaminase, an enzyme of arginine synthesis, can be suppressed by a mutation in argM, an arginine-inducible acetylornithine transaminase (10). ArgM was subsequently shown to prefer succinylornithine as a substrate (4, 10). Since both ArgD and ArgM can use acetylornithine as a substrate, we assumed that both can also use succinylornithine. Therefore, to minimize the effects of ArgD, we assayed succinylornithine transaminase activity from cells grown in two different arginine-containing media, one nitrogen limited (glucose-arginine) and the other nitrogen rich (glucose-arginine-ammonia). Succinylornithine transaminase activity was 10 times higher in the nitrogen-limited medium (lines 1 and 2 in Table 2), which indicates that succinylornithine transaminase is also under nitrogen control.
We could assay the other two enzymes of the AST pathway, arginine succinyltransferase and succinylglutamic semialdehyde dehydrogenase, from wild-type cells grown with arginine as the sole nitrogen source (which induces optimally) but not in any other medium (28). However, in strains with genes of a putative five-gene ast operon on a high-copy-number plasmid, all five enzymes could be measured, and nitrogen limitation induced all of them (see below). In summary, all five enzymes of the AST pathway are nitrogen regulated.
Specific induction of the AST pathway by other amino acids.
All growth-limiting nitrogen sources induce the well-studied glnALG operon equally well (29). However, this was clearly not the case for enzymes of the AST pathway (lines 1 and 3 in Table 2). Therefore, we examined the effect of various amino acids on the induction of the AST pathway (Table 3). Arginine was the best inducer, but other amino acids could also induce, albeit to a lesser extent. For example, aspartate induced to 50% of the maximal induction (Table 3).
TABLE 3.
Activities of AST pathway enzymes in wild-type E. coli grown with different nitrogen sourcesa
| Nitrogen sourceb | Enzyme activity, μmol/h/mg of protein (n)c, of:
|
|
|---|---|---|
| Dihydrolase | Desuccinylase | |
| Arginine | 1.5 ± 0.16 (3) | 2.6 ± 0.10 (3) |
| Glutamine | 0.47 ± 0.037 (4) | 0.84 ± 0.033 (4) |
| Aspartate | 0.83 ± 0.17 (3) | 1.3 ± 0.01 (3) |
| Proline | 0.47 ± 0.04 (2) | 1.3 ± 0.01 (3) |
| Alanine | 0.51 ± 0.03 (2) | 0.72 ± 0.09 (3) |
| Serine | 0.18 ± 0.03 (3) | 0.37 ± 0.08 (3) |
| Arginine-NH4+ | 0.064 ± 0.033 (2) | 0.089 ± 0.033 (2) |
| NH4+ | 0.03 ± 0.03 (2) | 0.01 ± 0.01 (2) |
Only three enzymes of the AST pathway can be measured in wild-type cells grown with ammonia as the sole nitrogen source: succinylarginine dihydrolase, succinylglutamate desuccinylase, and succinylornithine transaminase. Because the assay for the transaminase also measured ArgD, the results for the transaminase assay are not shown.
Cells were grown in minimal medium with 0.4% glucose and 0.2% of each indicated nitrogen source.
See footnote b of Table 2.
One possible explanation for this regulation is that arginine is the true inducer and the other amino acids induce to the extent that they affect arginine levels. Such an explanation can account for induction by aspartate, which is a substrate in the penultimate step of arginine synthesis, i.e., the reaction catalyzed by argininosuccinate synthetase. We tried to test this hypothesis by preventing assimilation of aspartate’s nitrogen into arginine and then examining whether aspartate could still induce. We used an argG strain which is deficient in argininosuccinate synthetase, the enzyme that assimilates aspartate’s nitrogen. Because this strain is an arginine auxotroph, the proposed experiment requires finding a concentration of arginine that satisfies the auxotrophy but does not induce the AST pathway. Unfortunately, such a concentration does not exist: arginine concentrations that limited growth twofold fully induced the AST pathway (28). Further examination of the mechanism of the apparent arginine-dependent induction must await characterization of the transcription of these genes.
Catabolite repression of the AST pathway.
Prior to the results presented here, the AST pathway had been found only in those organisms that degrade arginine as a carbon source, an ability which E. coli lacks (4, 33, 34). Utilization of carbon sources other than glucose frequently involves regulation by catabolite repression. Therefore, we examined whether a residual catabolite repression controlled the AST enzymes in E. coli. With aspartate, succinate, or glycerol as carbon and energy sources, the amounts of succinylarginine dihydrolase were 2- to 10-fold higher and the amounts of succinylglutamate desuccinylase were 9- to 70-fold higher than those with glucose (Table 4).
TABLE 4.
Enzyme activities in wild-type E. coli grown with different carbon sources
| Mediuma | Enzyme activity, μmol/h/mg of protein (n)b, of:
|
|
|---|---|---|
| Dihydrolase | Desuccinylase | |
| Aspartate | 0.33 ± 0.07 (2) | 0.69 ± 0.16 (3) |
| Aspartate-NH4+ | 0.28 ± 0.026 (2) | 0.68 ± 0.15 (3) |
| Succinate-NH4+ | 0.10 ± 0.009 (2) | 0.254 ± 0.008 (3) |
| Glycerol-NH4+ | 0.07 ± 0.010 (2) | 0.090 ± 0.003 (2) |
| Glucose-NH4+ | 0.03 ± 0.03 (2) | 0.01 ± 0.01 (2) |
Cells were grown in minimal medium with 1% aspartate, 0.4% of the other carbon sources, and 0.2% ammonia.
See footnote b of Table 2.
We also examined the expression of succinylarginine dihydrolase and succinylglutamate desuccinylase from cells grown in Luria-Bertani broth and found that these enzymes were induced to 15 to 20% of the level found from cells grown in glucose-arginine minimal medium. This induction was completely abolished by glucose in the Luria-Bertani broth but was not affected by ammonia (28). These results suggest control by catabolite repression.
Nitrogen source utilization and the AST pathway in K. aerogenes.
Unlike E. coli, K. aerogenes can utilize arginine as the sole carbon, energy, and nitrogen source (8, 33). Such growth induced the AST enzymes to much higher levels than those observed in E. coli (line 6 in Table 2). However, like E. coli, K. aerogenes grown in two nitrogen-limited media, glucose-arginine and glucose-glutamine, produced higher levels of succinylarginine dihydrolase and succinylglutamate desuccinylase than cells grown in the corresponding nitrogen-rich (ammonia-containing) medium (compare lines 7 and 9 to lines 8 and 10 in Table 2). Unexpectedly, their induction in K. aerogenes did not require NRI in glucose-arginine medium (lines 7 and 12 in Table 2). This result explains why this glnG mutant of K. aerogenes can still utilize arginine as a nitrogen source (1, 28). However, other results suggest that these two enzymes are under Ntr control in glucose-glutamine minimal medium: their levels were lower in a glnG mutant (lines 9 and 11 in Table 2).
Transaminase activity was unexpectedly higher in the glnG mutant compared to that in the wild-type strain when grown in glucose-glutamine medium (compare lines 9 and 11 in Table 2). Such regulation was not observed in E. coli (see lines 3 and 5 in Table 2). Succinylornithine transaminase is probably regulated in parallel with other enzymes of the AST pathway, which were at very low levels in the glnG mutant (line 11 in Table 2). This implies that another transaminase, probably the biosynthetic acetylornithine transaminase, is repressed by NRI. The function of such control is not apparent.
Identification of the E. coli gene for succinylornithine transaminase.
To identify the genes of the AST pathway, we tried to isolate mutants that could not utilize arginine but which could use ammonium sulfate as the sole nitrogen source. We assayed just over 100 mutants, and none of them were defective in AST enzymes. (The reason for this failure is not obvious since, as shown below, AST mutants cannot utilize arginine as a nitrogen source.) Therefore, we tried a reverse genetic strategy to isolate mutants deficient in AST enzymes. We focused on analyzing succinylornithine transaminase because its purification was required for assay of the fourth enzyme of the AST pathway, succinylglutamic semialdehyde dehydrogenase. This transaminase, called the product of argM, had been previously characterized because high levels of this enzyme suppressed a mutation in argD, which codes for the biosynthetic acetylornithine transaminase (26). It was subsequently shown to have a higher affinity for succinylornithine than for acetylornithine (4, 10).
Sequencing of two internal peptides gave the following se- quences: VLELINTPEMLNGVK and (Q)(P)ITRENF-(E) (W)MIPVYAP(A). (The parentheses indicate probable assignments, while the dash indicates that an assignment was not possible.) These peptides show a perfect match with the deduced product of a gene (called b1748 in the latest annotation of the genome) that starts at bp 1830006 of the E. coli genome (about minute 39.3) (Fig. 3). The gene predicts a 406-residue protein and a subunit mass of 43,665 Da, which agrees with the subunit size of the purified transaminase. (This protein was previously reported to have a subunit mass of 30 kDa [see reference 6]. The discrepancy results from proteolysis, which readily occurs if precautions against cleavage are not taken [28].) The gene for the catabolic succinylornithine transaminase is 60% identical to argD, which codes for the arginine-repressible acetylornithine transaminase. Because this enzyme codes for the third enzyme of the AST pathway, we propose that its gene should be designated as astC instead of argM.
FIG. 3.
Deduced amino acid sequence of succinylornithine transaminase. The underlined residues correspond to those determined by direct protein sequencing.
Preliminary characterization of an astCADBE operon.
Computer analysis of the regulatory region and four open reading frames (ORFs) downstream from astC suggests that it is the first of a five-gene, nitrogen-regulated astCADBE operon. (The complete sequence can be accessed in GenBank under ECAE000269.) Binding sites for ς54-RNA polymerase and NRI would be expected for Ntr genes, and such sites are readily identifiable. A probable ς54-RNA polymerase-binding site, TGGCACN5CTGCA, is located 72 bases upstream from the initiating methionine codon. It differs from the consensus binding site, TGGCACN5TTGC(A/T), by only one base (19). Two possible NRI-binding sites are located between 195 and 241 bases upstream from the upstream edge of the RNA polymerase site. While these sites are farther from the RNA polymerase site than is normal for activators of ς54-dependent promoters, there is precedent for such spacing (12). The four genes downstream from astC are transcribed in the same direction, and an apparent rho-independent terminator follows the last gene. The end of each gene overlaps with the beginning of the next; there is a 4-bp overlap for the first three junctions and an 8-bp overlap for the fourth.
Homology searches suggest that the four downstream genes code for enzymes of the AST pathway. The second ORF (numeric identifier b1747), which we designate as astA, codes for a 344-residue protein that appears to be AST. This enzyme, the first of the AST pathway, has been purified from Pseudomonas aeruginosa and has been shown to have two different subunits (38). Only the first few amino acids of each subunit have been determined, but these residues are homologous to each other and to the deduced sequence of the N terminus coded by the second ORF (Table 5). The third ORF (numeric identifier b1746) apparently codes for a protein of 492 residues. It is 32% identical to gabD, which specifies succinic semialdehyde dehydrogenase. This gene probably codes for the fourth enzyme of the AST pathway, succinylglutamic semialdehyde dehydrogenase; hence, we designate this gene as astD. The fourth ORF (numeric identifier b1745) codes for a 447-residue protein that is homologous to nothing in the databases. Such a result would be expected for succinylarginine dihydrolase, the second enzyme of the AST pathway, because it catalyzes an unusual reaction, the complete degradation of the guanidino group of succinylarginine. Disruption of this ORF results in selective loss of dihydrolase activity (described below), therefore, we designate this gene as astB. The fifth ORF (numeric identifier b1744) appears to code for a 322-residue protein. The gene product is homologous to human aspartoacylase, which deacetylates N-acetylaspartate. Aspartoacylase catalyzes a reaction very similar to that of succinylglutamate desuccinylase, the last enzyme of the AST pathway; therefore, we designate the gene as astE.
TABLE 5.
Comparison of the N termini of AST from P. aeruginosa and the protein from the second gene of the putative ast operon from E. coli
| Protein | Sequence |
|---|---|
| P. aeruginosa peptide 1 | MLVMRPAQAAD |
| P. aeruginosa peptide 2 | MIV RPVTSADLPALI |
| E. coli | MMVIRPVERSDVSALM |
Phenotypes of mutants with disruption of ast genes.
Table 6 shows growth rates for strains with disruptions of astB and astC. These disruptions had no effect on the utilization of ammonium sulfate, glutamine, alanine, proline, and aspartate as nitrogen sources. However, either of these disruptions completely prevented arginine utilization (there was no growth after 1 week). Curiously, a disruption of astC, but not of astB, severely impaired ornithine catabolism. (In the Discussion, we will propose that succinylornithine transaminase, but not other enzymes of the AST pathway, contributes to ornithine degradation.)
TABLE 6.
Generation times of wild-type E. coli and mutants with disruptions of astB and astC
| Nitrogen sourcea | Generation time, min ± SEM (n)b, for:
|
||
|---|---|---|---|
| W3110 | astB | astC | |
| NH3 | 77.8 ± 3.6 (6) | 83.0 ± 1.7 (3) | 71.3 ± 0.5 (3) |
| Glutamine | 120 ± 7.0 (6) | 129 ± 2.9 (3) | 105 ± 1.2 (3) |
| Alanine | 120 ± 2.1 (6) | 126 ± 4.1 (3) | 106 ± 3.5 (3) |
| Proline | 265 ± 7.6 (3) | 288 ± 4.9 (3) | 310 ± 1.7 (3) |
| Aspartate | 329 ± 13 (6) | 331 ± 13 (2) | 326 ± 3.6 (3) |
| Ornithine | 301 ± 7.1 (9) | 303 ± 6.5 (6) | 655 ± 24 (6) |
| Aspartate-ornithinec | 237 ± 12 (6) | 249 ± 15 (6) | 236 ± 11 (3) |
| Arginine | 540 ± 32 (9) | NDd | ND |
| Arginine-aspartatec | 105 ± 0.8 (3) | ND | ND |
| Aspartate-argininec | 105 ± 5.0 (6) | 178 ± 4.8 (3) | 271 ± 26 (3) |
Cells were grown in minimal medium with 0.4% glucose and 0.2% of the indicated nitrogen source.
The generation times are expressed as minutes ± standard error of the mean, with the number of determinations given in parentheses.
The first nitrogen source was at 0.2%; the second nitrogen source was at 0.01%.
ND, no detectable growth after 1 week.
Experiments to analyze aspartate catabolism in an argG mutant, which is an arginine auxotroph, led to the unexpected observation that certain mixtures of amino acids support significantly faster growth than that with either amino acid alone (28). Furthermore, a trace amount of the second amino acid, i.e., 0.01%, instead of the 0.2% used for amino acids as sole nitrogen source, was sufficient for this effect. For example, 0.2% aspartate and 0.2% arginine supported generation times of about 330 and 540 min, respectively, while a mixture of 0.2% aspartate with 0.01% arginine or, conversely, of 0.2% arginine with 0.01% aspartate both sustained generation times of 105 min. We tested whether this synergism requires the AST pathway when arginine is one component of this mixture. When arginine is the major nitrogen source, the low level of aspartate does not restore growth in either mutant (Table 6). However, when aspartate is the major nitrogen source and arginine is a trace nitrogen source, the effect is not as dramatic but growth is still impaired (Table 6). Therefore, arginine catabolism requires the AST pathway when arginine is one component of a synergistic binary mixture.
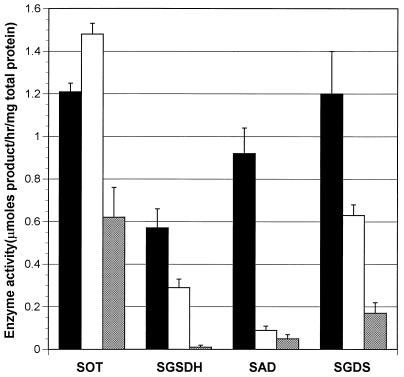
We also examined the disruptions’ effects on AST pathway enzymes. Because these mutants could not utilize arginine, cells were grown in medium with 0.2% of glutamine (which induces somewhat) and 0.1% arginine (which induces best). Despite the presence of arginine, the level of induction in the wild-type strain was virtually the same as that found in wild-type cells grown without the arginine. As a result, we could not detect AST, but we could detect the other four AST pathway enzymes from a wild-type strain (Fig. 4). Disruption of astC, the first gene of the operon, severely affected the levels of the remaining enzymes, except for transaminase activity, which was reduced only twofold (Fig. 4). This latter result presumably reflects assay of the biosynthetic acetylornithine aminotransferase, which may not be completely repressed by the arginine in the medium. Nevertheless, these results provide strong evidence for an ast operon. Disruption of the fourth gene most severely affected succinylarginine dihydrolase, had no effect on transaminase activity, and reduced dehydrogenase and desuccinylase activity twofold (Fig. 4). The effects on transaminase and dehydrogenase activities were expected, since these enzymes appear to be encoded by genes upstream from the insertion. The twofold reduction in desuccinylase activity was unexpected, since the gene for this enzyme is downstream from the insertion. For reasons that are not clear, this disruption appears to be less polar than the first. Nonetheless, the effects of this disruption shows that the fourth gene codes for succinylarginine dihydrolase.
FIG. 4.
AST enzyme activities in mutants with disruptions in astC and astB. Activity in W3110 (wild type) (black bars), in the astB derivative (white bars), and in the astC derivative (cross-hatched bars) is shown. The values given are the averages from at least five cultures. Activities are given in micromoles of product per hour per milligram of total protein; the error bars show the standard error of the mean. SOT, succinylornithine transaminase; SGSDH, succinylglutamic semialdehyde dehydrogenase; SAD, succinylarginine dihydrolase; SGDS, succinylglutamate desuccinylase.
Plasmid pLC3-11 and AST enzyme overproduction in E. coli.
Plasmid pLC3-11 contains chromosomal DNA from min 39 of the E. coli chromosome. A map of restriction endonuclease sites verified that this plasmid contained astC (28). It had been proposed that pLC3-11 contained astC (previously called argM), because Riley and Glansdorff showed that it was one of two plasmids that could complement a mutation in argD (26). (The other complementing plasmid, pLC2-28, contained argD itself.)
Plasmid pLC3-11 caused an approximately fivefold increase in all of the enzymes of the AST pathway (28). We deleted 8 kb from the 19-kb chromosomal insert in pLC3-11, which produced plasmid pΔLC3-11. The remaining insert contained the five genes of the putative ast operon and very little else. Cells with this plasmid had a 6- to 12-fold increase in all five AST pathway enzymes (Table 7). Such cells also grew faster with arginine and aspartate as nitrogen sources, reducing the doubling time from 296 to 166 min with arginine and from 418 to 245 min with aspartate (Table 7). Deletion into the most upstream or downstream region of these five genes on the plasmid resulted in loss of expression of all five gene on the plasmid, i.e., we could detect only activities that resulted from expression of the chromosomal genes.
TABLE 7.
Effect of overproduction of the enzymes of the AST pathway
| Strain | Nitrogen sourcea | Generation time (min) | Enzyme activity, μmol/h/mg of protein (n)b, of:
|
||||
|---|---|---|---|---|---|---|---|
| Dihydrolase | Desuccinylase | Transaminase | Dehydrogenase | Transferase | |||
| W3110 | Arginine | 296 | 1.5 ± 0.16 (3) | 2.6 ± 0.10 (3) | 5.4 ± 0.24 (3)c | 0.48 ± 0.12 (2) | 2.4 ± 0.4 (2) |
| W3110(pΔLC3-11) | Arginine | 166 | 19.2 ± 0.39 (2) | 16.6 ± 0.89 (2) | 48.22 ± 0.28 (2)c | 5.6 ± 0.6 (2) | 20 ± 1.2 (2) |
| W3110 | Aspartate | 418 | 0.83 ± 0.17 (3) | 1.3 ± 0.01 (3) | 4.0 ± 0.3 (3)d | NDe | ND |
| W3110(pΔLC3-11) | Aspartate | 245 | 6.16 ± 0.82 (2) | 4.16 ± 0.38 (2) | 11.2 ± 0.6 (2)d | ND | ND |
| W3110 | Ammonia | 103 | 0.03 ± 0.03 (2) | 0.01 ± 0.01 (2) | 2.6 ± 0.30 (3)d | ND | ND |
| W3110(pΔLC3-11) | Ammonia | 97 | 0.09 ± 0.09 (2) | 0.84 ± 0.12 (2) | 3.8 ± 0.56 (2)d | 0.24 ± 0.033 (2) | <0.8 (2) |
Cells were grown in minimal medium with 0.4% glucose and 0.2% of the indicated nitrogen source.
See footnote b of Table 2.
This measurement assessed only succinylornithine transaminase because arginine should repress acetylornithine transaminase.
This measurement may reflect both succinylornithine and acetylornithine transaminases because arginine is not present to repress the latter.
ND, not determined.
DISCUSSION
E. coli has the AST pathway.
We examined arginine catabolism in E. coli because an analysis of amino acid catabolism suggested that arginine catabolism produces ammonia but that direct deamination of alanine, aspartate, and glutamate does not (11). The previously described ADC pathway does not produce ammonia (Fig. 1), and we showed that it could account for only about 3% of the arginine consumed (Table 1). Friedrich and Magasanik, based on similar evidence, also concluded that the ADC pathway does not degrade arginine in K. aerogenes (8). We sought an ammonia-generating pathway of arginine catabolism and found activities for all five enzymes of the AST pathway, which is initiated by the succinylation of arginine’s amino group.
We identified an astCADBE operon, which codes for the five enzymes of the AST pathway. The gene for succinylornithine transaminase was found based on the identity of the protein sequence and the deduced gene product. We have designated this gene astC, and it appears to be the first gene of the operon. Homology searches of the second, third, and fifth ORFs of this operon suggest that they code for AST, succinylglutamic semialdehyde dehydrogenase, and succinylglutamate desuccinylase, the first, fourth, and fifth enzymes of the AST pathway, respectively. A homology search did not suggest a function for the fourth ORF, but its disruption specifically eliminated the activity of the second enzyme of the pathway, succinylarginine dihydrolase. The observation that each gene overlaps with the one that follows it is consistent with an operon organization and suggests that translation of a polycistronic mRNA may require just one ribosome. Disruption of the first gene impairs synthesis of all the AST enzymes, a finding which is also consistent with an operon structure. Finally, these observations, together with the fact that cells with plasmid pΔLC3-11, which contains this putative operon, have elevated levels of all five enzymes, provide further evidence for an astCADBE operon.
Functions of the AST pathway.
Mutants deficient in the AST pathway fail to utilize arginine, which shows that the AST pathway is necessary for catabolism of arginine as the sole nitrogen source during aerobic exponential growth. Other evidence also suggests a catabolic function. The presence of the astCADBE operon on plasmid pΔLC3-11 resulted in high levels of all of the AST enzymes and caused faster growth with arginine. Furthermore, the regulation of AST enzymes is consistent with a catabolic function, i.e., the levels are highest when arginine is the sole nitrogen source.
The AST pathway also appears to contribute to ornithine degradation. Disruption of astC, which codes for succinylornithine aminotransferase, significantly impaired ornithine degradation. It has previously been proposed that ornithine is degraded via putrescine and γ-aminobutyrate as indicated in Fig. 1 (30). However, the phenotype of the astC::kan mutant suggests that the AST pathway contributes to ornithine catabolism. There is precedent for involvement of the AST pathway in ornithine catabolism (39). In P. aeruginosa, succinylation of ornithine by a multispecific heteromeric succinyltransferase initiates such catabolism, and the resulting succinylornithine is then degraded by the last three enzymes of the AST pathway (38, 39). However, such a pathway may not function in E. coli, since disruption of astB, which reduces the expression of astE twofold (Fig. 4), does not impair ornithine catabolism. Furthermore, the E. coli succinyltransferase appears to be homomeric (more accurately, there is no evidence for a second homologous subunit) and may not have the capacity to succinylate ornithine. It is possible that the only enzyme of the AST pathway that contributes to ornithine catabolism is the transaminase, the product of astC, which has been shown to utilize ornithine as a substrate (2). Such a reaction would produce glutamic semialdehyde, which cyclyzes to form Δ′-pyrroline-5-carboxylate, the immediate precursor for proline synthesis. It could be hypothesized that Δ′-pyrroline-5-carboxylate is directly metabolized to glutamate. However, the PutA protein catalyzes this reaction, and its induction requires proline. Therefore, we propose that ornithine degradation requires its conversion to proline. A similar pathway has been proposed for ornithine catabolism in Pseudomonas putida (37).
Some evidence also suggests that the AST pathway contributes to the degradation of aspartate. First, cells with pΔLC3-11 grew faster with aspartate as the nitrogen source (Table 7). Second, aspartate induces the AST pathway (Table 3). Third, glutamate and aspartate accumulate in an argG mutant when aspartate is the sole nitrogen source (11). If arginine were an intermediate in aspartate degradation, then this result would be expected because the last two enzymes of arginine synthesis, argininosuccinate synthetase and argininosuccinase (products of argG and argH, respectively), would then be required for aspartate degradation. However, mutants with low levels of AST enzymes grew normally with aspartate as nitrogen source. There is an unidentified NRI-independent pathway of aspartate degradation that also contributes to aspartate catabolism (23). It is possible that this pathway compensates for loss of the AST pathway. Despite its potential to contribute to aspartate catabolism (as suggested by the faster growth that accompanies elevated levels of AST pathway enzymes), the evidence is inadequate to demonstrate that the AST pathway contributes to aspartate catabolism.
The regulation of the AST pathway by conditions other than nitrogen limitation suggests additional functions. The AST pathway is induced by growth in broth (28). The AST pathway also appears to be induced upon entry into stationary-phase growth (7). The function of the AST pathway under such conditions is not clear. Unlike all other organisms that contain the AST pathway, E. coli does not degrade arginine as a carbon source (33, 34). E. coli may fail to utilize arginine as a carbon source because the ast genes may lack an appropriately controlled promoter or because of inadequate transport during carbon-limited growth.
Functions of ADC in E. coli.
If the AST pathway degrades arginine, then the question arises as to what is the function of ADC. E. coli has two ADC isozymes. The first, encoded by speA, is constitutively synthesized and its product is referred to as the biosynthetic ADC. It probably synthesizes putrescine when the major route of putrescine synthesis via ornithine decarboxylase is blocked by arginine, which represses the enzymes of ornithine synthesis, i.e., the first five enzymes of arginine synthesis (36) (see Fig. 1). Tabor and Tabor have suggested that a biosynthetic ADC is present in E. coli only because of the absence of a mechanism to degrade arginine to ornithine (36). A role for the biosynthetic ADC in putrescine synthesis accounts for why mutants deficient in this isozyme grew slowly with arginine as the sole nitrogen source (30, 31). It also explains why 36% of arginine utilized is metabolized via ADC when ammonia is in the medium (Table 1). In this situation, ammonia supplies the cell with nitrogen, as indicated by the fact that arginine is not efficiently catabolized (Table 1), and ADC must initiate the synthesis of putrescine to the extent that arginine represses the enzymes of ornithine synthesis.
The second ADC isozyme is specified by the adi gene and is generally referred to as the biodegradative ADC (17, 35). This enzyme is required for arginine-dependent acid resistance for E. coli grown anaerobically in complex medium (17). During such growth, this ADC can become a few percent cellular protein (reference 35 and references therein), and there is significant urea production (Table 1). However, it is not clear how arginine degradation promotes acid resistance, since it is known that ammonia generation does not contribute to survival under these conditions (17).
In summary, arginine is an energy-rich amino acid that various bacteria can use for nitrogen, carbon, or energy and for other functions, such as survival in an acidic environment. E. coli appears to contain only one pathway of arginine catabolism. In contrast, P. aeruginosa has at least four pathways of arginine catabolism with overlapping functions (13, 37). The limited number of such pathways in E. coli undoubtedly reflects its ability to grow in a much narrower ecological niche.
ACKNOWLEDGMENTS
This work was supported by National Institute of General Medical Sciences grant GM47965 and by National Science Foundation grant MCB-9723003.
We are grateful to Catherine Bailey for the preparation of figures, to Warren Goux (Department of Chemistry, University of Texas at Dallas) for assistance in synthesis of the N-succinylated substrates required for the enzyme assays, to Cres Fraley and A. C. Matin (Stanford University) for providing a strain with a disruption of astC, and to A. Ninfa and R. Bender (University of Michigan) for the strains. This work is presented as partial fulfillment for the requirements for the Ph.D. degree at the University of Texas at Dallas for B. L. Schneider and A. K. Kiupakis.
REFERENCES
- 1.Bender, R. Personal communication.
- 2.Billheimer J T, Carnevale H N, Leisinger T, Eckhardt T, Jones E E. Ornithine δ-transaminase activity in Escherichia coli: its identity with acetylornithine δ-transaminase. J Bacteriol. 1976;127:1315–1323. doi: 10.1128/jb.127.3.1315-1323.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke L, Carbon J. A colony bank containing synthetic Col E1 hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 4.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dugan E L. Measurement of amino acids by column chromatography. Methods Enzymol. 1957;3:492–504. [Google Scholar]
- 6.Forsyth G W, Theil E C, Jones E E, Vogel H J. Isolation and characterization of arginine-inducible acetylornithine δ-transaminase from Escherichia coli. J Biol Chem. 1970;245:5354–5359. [PubMed] [Google Scholar]
- 7.Fraley C D, Kim J H, McCann M P, Matin A. The Escherichia coli starvation gene cstC is involved in amino acid catabolism. J Bacteriol. 1998;180:4287–4290. doi: 10.1128/jb.180.16.4287-4290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedrich B, Magasanik B. Utilization of arginine by Klebsiella aerogenes. J Bacteriol. 1978;133:680–685. doi: 10.1128/jb.133.2.680-685.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale E F. The bacterial amino acid decarboxylases. Adv Enzymol. 1946;6:1–32. [Google Scholar]
- 10.Glansdorff N. Arginine and polyamine biosynthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 408–434. [Google Scholar]
- 11.Goux W J, Strong A A D, Schneider B L, Lee W-N P, Reitzer L J. Utilization of aspartate as a nitrogen source in Escherichia coli: analysis of nitrogen flow and characterization of the products of aspartate catabolism. J Biol Chem. 1995;270:638–646. doi: 10.1074/jbc.270.2.638. [DOI] [PubMed] [Google Scholar]
- 12.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 13.Jann A, Matsumoto H, Haas D. The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J Gen Microbiol. 1988;134:1043–1053. doi: 10.1099/00221287-134-4-1043. [DOI] [PubMed] [Google Scholar]
- 14.Jann A, Stalon V, Vander Wauven C, Leisinger T, Haas D. N2-succinylated intermediates in an arginine catabolic pathway of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1986;83:4937–4941. doi: 10.1073/pnas.83.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jessop A P, Clugston C. Amplification of the argF region in strain HfrP4X of E. coli K-12. Mol Gen Genet. 1985;201:347–350. doi: 10.1007/BF00425683. [DOI] [PubMed] [Google Scholar]
- 16.Legrain C, Stalon V, Glansdorff N, Gigot D, Pierard A, Crabeel M. Structural and regulatory mutations allowing utilization of citrulline or carbamoylaspartate as a source of carbamoylphosphate in Escherichia coli K-12. J Bacteriol. 1976;128:39–48. doi: 10.1128/jb.128.1.39-48.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 20.Morris D R, Pardee A B. Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem. 1966;241:3129–3135. [PubMed] [Google Scholar]
- 21.Morris D R, Koffron K L. Urea production and putrescine biosynthesis by Escherichia coli. J Bacteriol. 1967;94:1516–1519. doi: 10.1128/jb.94.5.1516-1519.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oginsky E L. Isolation and determination of arginine and citrulline. Methods Enzymol. 1957;3:639–643. [Google Scholar]
- 23.Pahel G, Tyler B. A new glnA-linked regulatory gene for glutamine synthetase in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:4544–4548. doi: 10.1073/pnas.76.9.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 380–390. [Google Scholar]
- 25.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 26.Riley M, Glansdorff N. Cloning the Escherichia coli K-12 argD gene specifying acetylornithine δ-transaminase. Gene. 1983;24:335–339. doi: 10.1016/0378-1119(83)90095-1. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein D M, Pahel G, Tyler B, Magasanik B. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc Natl Acad Sci USA. 1980;77:7372–7376. doi: 10.1073/pnas.77.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider, B. L., and L. J. Reitzer. Unpublished results.
- 29.Schneider B L, Shiau S-P, Reitzer L J. Role of multiple environmental stimuli in control of transcription from a nitrogen-regulated promoter in Escherichia coli with weak or no activator-binding sites. J Bacteriol. 1991;173:6355–6363. doi: 10.1128/jb.173.20.6355-6363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaibe E, Metzer E, Halpern Y S. Metabolic pathway for the utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol. 1985;163:933–937. doi: 10.1128/jb.163.3.933-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaibe E, Metzer E, Halpern Y S. Control of utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol. 1985;163:938–942. doi: 10.1128/jb.163.3.938-942.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Bennett G N. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J Bacteriol. 1994;176:7017–7023. doi: 10.1128/jb.176.22.7017-7023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stalon V. Evolution of arginine metabolism. In: Schleifer K H, Stackebrandt E, editors. Evolution of prokaryotes. New York, N.Y: Academic Press; 1985. pp. 277–308. [Google Scholar]
- 34.Stalon V, Vander Wauven C, Momin P, Legrain C. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol. 1987;133:2487–2495. doi: 10.1099/00221287-133-9-2487. [DOI] [PubMed] [Google Scholar]
- 35.Stim K P, Bennett G N. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabor C W, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tricot C, Stalon V, Legrain C. Isolation and characterization of Pseudomonas putida mutants affected in arginine, ornithine and citrulline catabolism: function of the arginine oxidase and arginine succinyltransferase pathways. J Gen Microbiol. 1991;137:2911–2918. doi: 10.1099/00221287-137-12-2911. [DOI] [PubMed] [Google Scholar]
- 38.Tricot C, Vander Wauven C, Wattiez R, Falmagne P, Stalon V. Purification and properties of a succinyltransferase from Pseudomonas aeruginosa specific for both arginine and ornithine. Eur J Biochem. 1994;224:853–861. doi: 10.1111/j.1432-1033.1994.00853.x. [DOI] [PubMed] [Google Scholar]
- 39.Vander Wauven C, Jann A, Haas D, Leisinger T, Stalon V. N2-succinylornithine in ornithine catabolism of Pseudomonas aeruginosa. Arch Microbiol. 1988;150:400–404. doi: 10.1007/BF00408314. [DOI] [PubMed] [Google Scholar]
- 40.Vander Wauven C, Stalon V. Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J Bacteriol. 1985;164:882–886. doi: 10.1128/jb.164.2.882-886.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worthington Biochemical Corporation. Worthington enzyme manual. Freehold, N.J: Worthington Biochemical Corporation; 1993. Arginase; pp. 45–47. [Google Scholar]