Abstract
Cell-to-cell trafficking of regulatory proteins is a novel mechanism for communication during cell fate specification in plants. Although several developmental proteins traffic cell-to-cell, no signals that are both necessary and sufficient for this function in developmental proteins have been described. We developed a novel trafficking assay using trichome rescue in Arabidopsis. Fusion to KNOTTED1 (KN1) conferred gain-of-trafficking function to the cell-autonomous GLABROUS1 (GL1) protein. We show that the KNOX homeodomain (HD) is necessary and sufficient for intercellular trafficking, identifying a novel function for the HD as the minimal sequence required for trafficking of KN1 and its associated mRNA.
Keywords: KNOX homeodomain, trichome, trafficking signal, Glabrous 1, knotted1
Intercellular trafficking of regulatory proteins and mRNAs through plasmodesmal (PD) channels allows for the non-cell-autonomous regulation of plant development (Lucas et al. 1995; Perbal et al. 1996; Kim et al. 2001, 2003; Nakajima et al. 2001; Wada et al. 2002; Lee et al. 2003; Wu et al. 2003). However, the mechanisms by which macromolecules traffic cell-to-cell are largely unknown. In some cases transport may be nonselective (Wu et al. 2003), whereas in others the developmental regulation of protein or mRNA movement suggests a selective and regulated process (Kim et al. 2001, 2003; Nakajima et al. 2001; Lee et al. 2003; Gallagher et al. 2004; Sena et al. 2004).
If PD trafficking is regulated, one would expect to find specific signals that are both necessary and sufficient for movement in non-cell-autonomous proteins. Such signal sequence(s) should promote trafficking of a cell-autonomous reporter. However no such sequences have been described.
Studies of macromolecular trafficking were pioneered using viral movement proteins (MPs). These proteins traffic through plasmodesmata, increase the size exclusion limit (SEL), and facilitate spread of the viral RNA genome (Wolf et al. 1989; Citovsky et al. 1990; Waigmann et al. 1994). The discovery of a functionally related plant protein, CmPP16, suggests that MPs use an endogenous trafficking mechanism (Xoconostle-Cazares et al. 1999). Microinjection experiments suggested that most PD have an SEL of 1 kDa to a few kilodaltons (Wolf et al. 1989; Waigmann and Zambryski 1995). However, the intercellular trafficking of green fluorescent protein (GFP) suggests that in some cases the PD SEL is larger and is dynamically regulated according to the species, tissue, developmental stage, and environmental conditions (Oparka et al. 1999; Crawford and Zambryski 2000). GFP trafficking is thought to occur by a passive or nonselective diffusion-based mechanism. The developmental transcription factor protein LEAFY may also traffic by a similar nonselective mechanism, raising the question of whether targeting sequences are required for PD transport (Wu et al. 2003).
The maize KNOTTED1 (KN1) homeodomain protein was the first plant protein found to traffic cell-to-cell (Lucas et al. 1995; Kim et al. 2002). This property explains the noncell autonomy of dominant KN1 alleles in maize leaf development (Hake and Freeling 1986), and trafficking may contribute to the normal function of KN1 in stem cell maintenance (Kim et al. 2003).
To ask whether KN1 uses a selective trafficking pathway, a novel gain-of-function trafficking assay in Arabidopsis was developed, and used to identify a cis-signal for intercellular KN1 trafficking in vivo. Here we show that the homeodomain (HD) regions of KNOTTED1 and related KNOX proteins fit the definition of a trafficking signal domain, and can also promote trafficking of the KN1 mRNA.
Results and Discussion
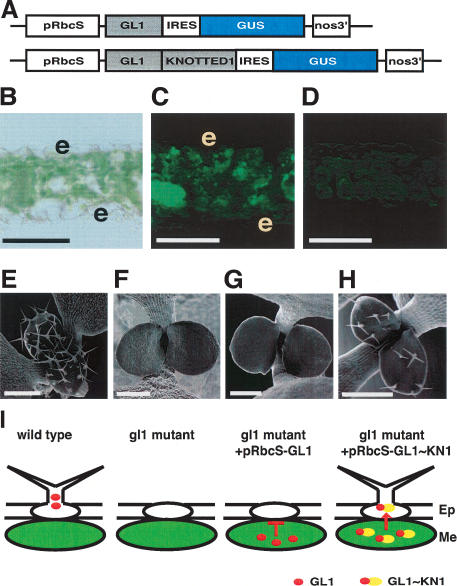
To determine whether KN1 contains specific trafficking signal domains, we designed a novel gain-of-function trafficking assay. Since KN1 and its Arabidopsis homologs are biologically active in the leaf when overexpressed, we developed the assay in the leaf. We also chose to use Arabidopsis, which is more amenable than maize to this kind of analysis. Our assay involves functional complementation of trichome development in glabrous1 (gl1) mutants, mediated by the trafficking of GL1 fusion proteins. GL1, a member of the MYB transcription factor family, is expressed in the epidermis and is required for leaf trichome initiation (Oppenheimer et al. 1991). GL1 probably functions cell autonomously, because sectors lacking GL1 also lack trichomes (M. Hulskamp, pers. comm.). To test the hypothesis that GL1 is cell-autonomous, we expressed it using the Arabidopsis rubisco small subunit 2b (RbcS2b) promoter in a gl1 mutant background. This promoter drives expression specifically in green cells (Kim et al. 2003), including mesophyll cells, which normally do not make trichomes, and epidermal guard cells, which are not relevant here as they are symplasmically isolated (Ding et al. 1997). To confirm that the transgene was expressed tissue-specifically, we included an internal ribosome entry sequence (IRES) followed by the β-glucuronidase (GUS) reporter gene (Fig. 1A). Expression of the cell-autonomous GUS reporter indicated that the bi-cistronic mRNA was expressed as expected in mesophyll cells and not in epidermal pavement cells (Fig. 1C).
Figure 1.
Functional protein trafficking assay using trichome rescue reveals that KN1 fusion to GL1 provides a gain-of-trafficking function. (A) GL1 fusion constructs used for trichome rescue assay. Constructs include the mesophyll-specific RbcS promoter (pRbcS), GL1 or GL1∼KN1 fusions, an internal ribosome entry sequence (IRES) upstream of the GUS coding sequence, and the nopaline synthase (nos) 3′ terminator. (B) Leaf cross-section viewed in bright-field, with epidermal cells labeled “e”. (C) GUS activity was imaged as green fluorescence in mesophyll cells (Immagen Green) in gl1/pRbcS::GL1–IRES–GUS plants. The gl1 background control (D) shows no staining. (E–H) SEM images of wild-type seedling (E), gl1 mutant seedling (F), gl1 seedling carrying the control pRbcS::GL1–IRES–GUS transgene (no trichome rescue) (G), and a gl1 seedling expressing GL1∼KN1 showing trichome rescue (H). (I) Schematic of the functional trafficking assay; in the wild type, GL1 functions in an epidermal precursor cell to initiate trichome formation. GL1 expressed in mesophyll cells is cell-autonomous and cannot rescue trichomes in the gl1 mutant. The KN1 fusion to GL1 can traffic into epidermal cells and rescue trichome formation in the gl1 mutant. (Ep) Epidermal cells; (Me) mesophyll cells. Bars: (B–D), 50 μm; (E–H) 100 μm.
As predicted, the mesophyll-specific expression of GL1 in gl1 plants did not rescue trichome formation, indicating that GL1 is cell-autonomous (data from 329 independent transformed plants) (Fig. 1G). We next asked whether fusion of KN1 to GL1 could promote trafficking of the fusion protein. We fused GL1 at the N terminus of KN1, as previous studies showed that this arrangement preserved the biological function of KN1 (Kim et al. 2002). The GL1∼KN1 fusion rescued trichome formation in 27% (84/316) of independent transgenic plants (Fig. 1H), suggesting that it could traffic from mesophyll to epidermal cells. The fact that rescue was not observed in all lines may be due to variability of transgene expression (Peach and Velten 1991). The rescued plants had a mean of 10.2 trichomes per leaf pair, compared with ∼60 per leaf pair for wild-type siblings. Trichome rescue was stably inherited into the T2 generation (data not shown).
To investigate whether KN1 contained specific sequences that conferred this cell-to-cell trafficking function, we made a series of deletions. GL1 fusions to the N-terminal region (KN11–135) or N-terminal/central region (KN11–263) gave no or negligible trichome rescue in >400 independent transgenic plants (two plants out of 159 transformed with the KN11–263 construct made one trichome per leaf pair) (Fig. 2C). In contrast, a GL1 fusion to the C-terminal region (KN1256–359) permitted trichome rescue in 48% of transgenic lines, with a mean of eight trichomes per leaf pair. These data indicate that the C-terminal region contains a sequence that was both necessary and sufficient for rescue. To confirm that the failure to rescue by the KN1 N-terminal/central region fusions was due to an inability to traffic, and not because these regions caused instability or mislocalization, we expressed them ubiquitously using the 35S promoter. These constructs were able to rescue; for example, 68% of the 35S::GL1∼KN11–263 transgenic seedlings showed trichome rescue (n = 108) (Fig. 2C), indicating that this fusion protein is biologically active when expressed in epidermal cells.
Figure 2.
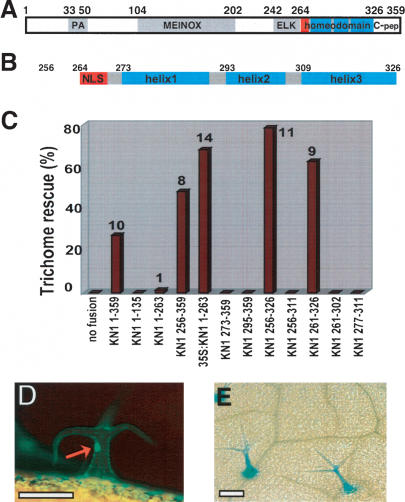
The KN1 homeodomain functions as a trafficking signal domain. (A) Schematic of domains in KN1: peptide antagonist (PA) (Kragler et al. 2000), MEINOX (Bellaoui et al. 2001), ELK (Kerstetter et al. 1994), homeodomain, and C-terminal region (C-pep). Red box indicates nuclear localization signal. (B) The predicted structure of the homeodomain region of KN1. Numbers in A and B indicate amino acids from the ATG. (C) Trichome rescue by different fusions to GL1. Trichome rescue rate (percent of T1 plants showing trichome rescue) is on the Y-axis. The number next to each bar is the average number of trichomes per leaf pair. Note that all constructs are fusions to GL1 driven by the pRbcS promoter, unless labeled “35S”, in which case it is driven by the 35S promoter. (D,E) GFP fluorescence (D) and GUS activity (E) in rescued trichomes of gl1; pGL2::GUS plants containing the pRbcS::GFP∼GL1∼KN1256–359 transgene. Red arrow indicates GFP fluorescence in trichome nucleus. Bars: D,E, 150 μm.
The C-terminal region of KN1 (KN1256–359) that was necessary and sufficient for trafficking included the HD, and we attempted to further delimit trafficking activity. Deletions at the N terminus that removed any part of the HD, for example removal of the NLS region or of a part of the first helix, abolished trafficking activity, as in KN1273–359 or KN1295–359 (Fig. 2C). We also made deletions from the C terminus. We found that removal of the residues C-terminal to the HD (KN1327–359) (C-pep in Fig. 2A) stimulated by ∼1.6-fold the percentage of transgenic plants that showed trichome rescue (construct KN1256–326) (Fig. 2C, cf. KN1256–359). However any further deletion that encroached on the HD, for example removal of part of the third helix in KN1256–311, led to a complete loss of trichome rescue. In summary, sequences within the HD at both the N and C termini were necessary for trichome rescue, and presumably therefore for trafficking. We made a minimal construct that contained only the HD and three amino acids at its N terminus (KN1261–326), and this also gave trichome rescue (Fig. 2C). KN1273–326, composed only of the three predicted HD helices, gave trichome rescue in only 6% of transgenic plants, and the degree of rescue was very low, with an average of only 2.2 trichomes per leaf pair. Consistent with the finding that both N- and C-terminal regions of the HD were necessary for trichome rescue, other constructs that expressed smaller regions of the HD were nonfunctional (e.g., KN1261–302, KN1277–311) (Fig. 2C).
Since the HD also contains the DNA binding activity, we asked whether trichome rescue was correlated with cell-to-cell trafficking of the KN1 fusion, rather than nonautonomous action of a downstream factor that might be expressed in response to DNA binding by the GL1∼KN1 fusion. We used GLABRA2 (GL2) as a reporter, since its expression in the trichome is dependent on the presence of GL1 (Szymanski et al. 1998). gl1 plants carrying a pGL2::GUS transgene were transformed with a pRbcS::GFP∼GL1∼KN1256–359 construct, which encodes a fusion of GFP, GL1, and the C-terminal domain of KN1. As expected, expression of this construct allowed trichome rescue, and we also detected GFP fluorescence in nuclei of rescued trichomes, indicating that the GFP∼GL1∼KN1256–359 into the protein trafficked epidermal cells (Fig. 2D). Furthermore, we could detect activity of the GL2 promoter as monitored by GUS activity in the rescued trichomes (Fig. 2E). These data strongly suggest that trichome rescue was due to trafficking of the GL1∼KN1 fusion protein into the epidermis, rather than by movement of a nonautonomous downstream factor.
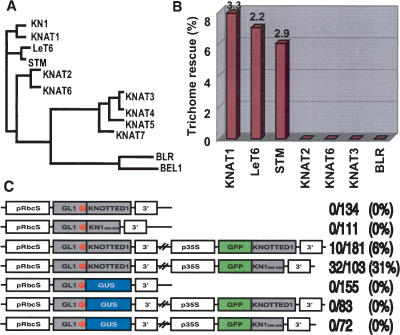
The HD is the defining feature of KNOX family members (Fig. 3A), so we investigated whether the trafficking activity was conserved in other plant HD proteins. The KNOX proteins can be divided into two classes based on sequence conservation and expression patterns (Kerstetter et al. 1994). We found that only the C-terminal regions of closely related class I KNOX proteins (Arabidopsis STM, KNAT1, or tomato LeT6) were functional in trichome rescue (Fig. 3B). The less closely related class I KNOX C-terminal regions from Arabidopsis proteins KNAT2 or KNAT6, or a class II HD (KNAT3) or a BEL-like protein (BELLRINGER) (Byrne et al. 2003) did not show any trichome rescue. Therefore trafficking appeared to be a property only of specific class I HDs closely related to KN1. The trichome rescue rate using these proteins was lower than with KN1, and we cannot at present explain these differences. However the proteins differ in sequences C-terminal to the homeodomain, and this region of KN1 had a negative effect on trafficking. They also differ in the HD itself, and future domain swap experiments should allow the effects of these regions to be distinguished. Our demonstration of the HD region as a minimal trafficking domain that is both necessary and sufficient agrees with earlier mutagenesis studies that found a mutation in the HD that blocked trafficking of microinjected or in vivo expressed proteins (Lucas et al. 1995; Kim et al. 2002). Our results conflict with studies that identified a motif in the N-terminal region of KN1 as a peptide antagonist of trafficking (Kragler et al. 2000), as that region appears not to be sufficient for trafficking in our assay.
Figure 3.
The HD trafficking signal is conserved in closely related class I KNOX homeodomain proteins, and promotes KN1 mRNA movement. (A) Phylogenetic tree of homeodomain sequences: LeT6 (accession no. T04317), BLR (AAP93641), BEL1 (AAM62510), STM (Q38874), KNAT1 (AAM03027), KNAT2 (P46640), KNAT3 (P48000), KNAT4 (P48001), KNAT5 (P48002), KNAT6 (NP_850951), KNAT7 (NP_564805). (B). Trichome rescue assays were performed using HD regions corresponding to KN1256–359. Descriptions of the chart are as in Figure 2C. (C) RNA trafficking assay. Stop codons are shown as a red asterisk after the GL1 coding region. The shorter KN1 regions shown in constructs in lines 2, 4, and 7 correspond to KN1256–359. The number of T1 plants showing trichome rescue/total number of T1 plants observed (rescue rate, percent) is shown on the right side of each construct. (3′)3′ untranslated region (UTR) of KN1 or of nopaline synthase (see Materials and Methods).
Nonautonomy of KNOX mRNAs has also been reported (Lucas et al. 1995; Kim et al. 2001). To investigate whether trichome rescue was due to trafficking of the KN1 fusion proteins, or of their mRNAs, or both, we introduced a stop codon after the GL1 coding region in the pRbcS::GL1∼KN1 or pRbcS::GL1∼KN1256–359 constructs (Fig. 3C), so that they expressed only the GL1 protein from a GL1STOP–KN1 fusion mRNA. Plants transformed with these constructs did not show trichome rescue (Fig. 3C), suggesting that the GL1∼KN1 fusion protein was required and that the fusion mRNA itself was not sufficient. We also tested whether KN1 could facilitate the trafficking of its own mRNA, as suggested by microinjection studies (Lucas et al. 1995). As class1 KNOX genes are normally not expressed in the Arabidopsis leaf (Lincoln et al. 1994), we coexpressed the pRbcS::GL1STOP–KN1 with GFP∼KN1 or GFP∼KN1256–326 expressed from the 35S promoter. In these plants we observed trichome rescue, suggesting that GFP∼KN1 or GFP∼KN1256–326 could facilitate trafficking of the GL1STOP–KN1 mRNA (Fig. 3C). KN1 mRNA sequences were required for this rescue, because a control construct, pRbcS::GL1STOP∼GUS. did not rescue, even in the presence of GFP∼KN1 or GFP∼KN1256–326 (Fig. 3C). This eliminates the possibility that rescue was due to GL1 protein movement by a nonspecific PD gating effect of GFP∼KN1 overexpression, and suggests that the GL1STOP–KN1 mRNA was specifically transported to the epidermis by GFP∼KN1 or GFP∼KN1256–326.
Our results raise the question of whether trafficking and trichome rescue were due to a signal in the KN1 HD alone, or in the HD in combination with its mRNA. We showed here that KN1 mRNA trafficking is dependent on the presence of the KN1 protein or of the HD. Therefore the trafficking signal is certainly not solely contained within the KN1 mRNA. It remains to be seen whether the KN1 mRNA, in combination with the KN1 HD, is a necessary component of the signal for intercellular trafficking. However, our prior microinjection studies strongly suggest that at least the full-length KN1 protein can traffic without its mRNA (Lucas et al. 1995). Our results also imply a specific interaction between KN1 and its mRNA, because the control GL1–GUS fusion mRNA expressed together with KN1 did not rescue trichomes, and presumably did not traffic. The proposed interaction between KN1 and its mRNA could be mediated at the sequence level through binding of the HD to specific sequences in the KN1 mRNA, similar to the binding of CAUDAL mRNA by the BICOID HD protein (Dubnau and Struhl 1996).
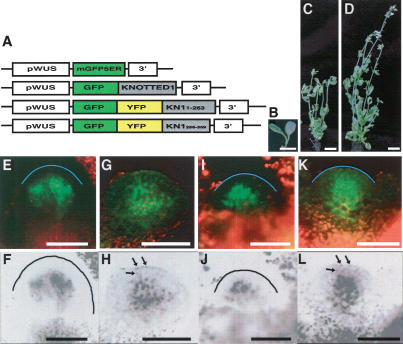
KN1 and other class I KNOX proteins normally function in the shoot apical meristem (Reiser et al. 2000). We therefore investigated whether the trafficking signal region of KN1 was also necessary and sufficient in the shoot apical meristem (SAM). We used WUSCHEL (WUS) regulatory sequences to drive layer-specific expression. WUS is expressed in the L3 layer of the SAM (Mayer et al. 1998), and we found that 1.7 kb of the WUS 5′ region (pWUS) drove expression of a cell-autonomous mGFP5-ER reporter in the SAM, specifically in the L3 layer (Fig. 4E,F). GFP∼KN1 expression using the same promoter showed green fluorescence in all layers, indicating that GFP∼KN1 could traffic from the L3 to the L2/L1 (Fig. 4G,H). This construct was also able to rescue the shootmeristemless mutation (Fig. 4B–D), indicating biological activity of the GFP∼KN1 fusion protein in cells into which it was trafficking. The rescued plants were not completely normal, and this may be because pWUS also drove a low level of expression in developing leaf primordia (data not shown).
Figure 4.
The KN1 trafficking signal domain functions in the meristem, and L3-specific expression of KN1 is sufficient to complement shootmeristemless-11 (stm-11) mutants. (A) GFP fusion constructs used for protein trafficking assays in the SAM. Constructs include the L3-specific WUS promoter (pWUS), mGFP5-ER, GFP, or GFP∼YFP fusions to KN1 or to KN1 deletions constructs, and the nopaline synthase terminator (nos 3′) or the KN1 3′UTR (3′). The third construct contains KN11–263 and the fourth contains KN1256–359. (B) stm-11 mutants show fused cotyledons and no true leaves. pWUS::KN1 expression (C) rescued 14 plants out of 46 stm-11 T1 homozygotes, and pWUS::GFP∼KN1 expression (D) rescued four plants out of 10 stm-11 T1 homozygotes. Thus pWUS-driven KN1 or GFP∼KN1 expression can rescue the function of STM, which is normally expressed in all three layers of the SAM (Long et al. 1996). (E–L) Confocal microscope images of meristems expressing these GFP constructs; mGFP5-ER is restricted to the L3 (E,F). GFP∼KN11–359 shows movement into L2 and L1 cells, seen as green nuclear fluorescence (G) or in grayscale with labeled nuclei indicated by arrows (H). GFP∼YFP∼KN11–263 is restricted to the L3 (I,J), and GFP∼YFP∼KN1256–359 shows movement into L2 and L1 cells (K,L), indicating that the trafficking signal region identified in KN1256–359 also functions in the meristem. Note that in the inverted grayscale images, the fluorescence of nuclei in the L1 and L2 layers is more pronounced. Labeled nuclei in the L1 and L2 layers are indicated by arrows in H,L. The outer surface of the L1 is marked by a blue line in E,I,K and by a black line in F,J. Bars: E–L, 50 μm.
Because the SEL in the meristem is larger than in the leaf (Wu et al. 2003), we next used a GFP∼YFP fusion as a reporter. Expression of GFP∼YFP∼KN11–265 using pWUS resulted in fluorescence restricted to the L3 layer, suggesting that, as in the leaf, the N-terminal/middle regions of KN1 were not sufficient for trafficking in the SAM (Fig. 4I,J). In contrast, the GFP∼YFP∼KN1256–359 fusion could traffic from the L3 to the L2/L1 layers (Fig. 4K,L). Each pattern was confirmed in a minimum of four independent transgenic plants. Therefore the KN1 C-terminal trafficking signal identified using trichome rescue was also necessary and sufficient for trafficking in the SAM.
The trafficking signal that we report here is the first report of one that is both necessary and sufficient for selective intercellular trafficking in the context of a heterologous cell-autonomous reporter. During plant evolution, HD proteins may have gained a selective trafficking function to regulate cell fate through non-cell-autonomous signaling. The differential regulation of KNOX protein trafficking may provide a mechanism to differentiate homologous, potentially redundant KNOX gene functions when they are expressed in the same tissues. Indeed, closely related KNOX proteins have some degree of redundant function; for example, KNAT1 expression can partially complement the stm phenotype (Byrne et al. 2002), and KNAT1 may also regulate epidermal cell fate nonautonomously (Venglat et al. 2002).
Our functional trafficking assay could be used to determine whether diverse proteins use a selective trafficking pathway in vivo. Owing to the requirement for gain-of-trafficking function, only proteins carrying a positive trafficking signal will promote trichome rescue. The LEAFY transcription factor was recently suggested to move by a nontargeted mechanism (Wu et al. 2003); however, it is possible that it contains redundant selective signals, and it will be interesting to test the ability of LEAFY and other plant proteins to promote trafficking in our system. Trafficking signals identified in these studies may be useful tools to selectively control the misexpression of other cell-autonomous proteins. However, our results do not imply that the KN1 HD can confer protein trafficking in any context, because its own trafficking is developmentally regulated (Kim et al. 2003). In conclusion, using a functional trafficking assay we found a novel role for the KNOX HD. In addition to its function as a DNA-binding domain, this domain also acts as an intercellular protein trafficking signal, and as a mediator for selective cell-to-cell mRNA trafficking.
Materials and methods
Trichome rescue assay
The gl1-1 mutant line originally isolated in Landsberg erecta (Ler) and introgressed to Wassilewskija (Ws) was transformed using Agrobacteria-containing constructs to express GL1 fusion proteins, according to Kim et al. (2003). Transgenic seedlings were screened on MS medium containing 50 μg/mL hygromycin. We scored trichome rescue from the first four leaves of 2-wk-old seedlings using a stereo microscope.
GUS staining, confocal and scanning electron microscopy
GUS staining using X-glucuronide was performed as described previously (Kim et al. 2003). The stained tissues were cleared in 70% ethanol for 3 h at 37°C, and photographed. Staining using the ImmagenGreen kit was performed according to the manufacturer (Molecular Probes). In brief, leaf cross-sections were incubated in 100 mM phosphate buffer (pH 7.5) containing 50 μM C12FDGluU at 37°C for 2 h. Confocal and scanning electron microscopy were performed as described (Taguchi-Shiobara et al. 2001; Kim et al. 2003).
DNA constructs
The cloning of GFP∼KN1 and pRbcS was described in Kim et al. (2002, 2003); the pRbcS was upstream of GFP∼KN1 in pCambia1300. pRbcS::GL1∼KN1 was made by replacing GFP with GL1. The IRES sequence (Kim et al. 2002) was inserted in pCambia2301 to make the IRES–GUS construct; pRbcS::GL1∼KN1 was then inserted upstream to make pRbcS::GL1∼KN1–IRES–GUS, and pRbcS::GL1–IRES–GUS was produced by deleting KN1 from this clone. To make deletion constructs of KN1 or KNOX proteins, specific regions were amplified by PCR and inserted to replace KN1 in pRbcS::GL1∼KN1. For constructs used in the RNA trafficking assay, an in-frame stop codon was inserted after the GL1 coding region to produce pRbcS::GL1STOP–KN1 or pRbcS::GL1STOP–GUS. The pRbcS::GL1STOP–KN1 construct carries the 3′ untranslated region (UTR) of KN1, and pRbcS::GL1STOP–GUS carries the 3′UTR of nopaline synthase. To engineer RNA trafficking constructs to overexpress KN1 or KN1266–326, a HindIII fragment of 35S::GFP∼KN1 (Kim et al. 2002) or of 35S::GFP∼KN1256–326 was end-filled by Klenow treatment and ligated into the PmlI site of pRbcS::GL1STOP–KN1 or the SmaI site of pRbcS::GL1STOP–GUS. pWUS was produced by PCR using primers GGGGAATTCTGGATGGAATCCAAAAGTTTAT and GGGGGTAC CCCGGGTGTGTTTGATTCGACTTTTGT and subcloned upstream of “double GFP” reporters. The pWUS constructs carrying KN1 or its fragments (KN11–263, KN1256–359) have the 3′UTR of KN1. All PCRs were carried out using high-fidelity DNA polymerase (Pfu, Stratagene) and were confirmed by sequencing.
In the primers listed below, f indicates forward, r denotes reverse, and numbers indicate locations of first or last amino acid of each amplified fragment. Lowercase letters include restriction enzyme sites added for cloning.
The following primers were used: KN1-1f, ggggagatctTATGGAGGA GATCACCCAACA; KN1–135r, ggggggtaccTTACGCCGACACCTCCG GTG; KN1-263r, ggggggtaccTTATGACAGTTCTTGCTTGAGCG; KN1-261f, ggggagatctGTCAAAGAAGAAGAAGAAAGG; KN1-302r, ggggggta ccTTAAGCCAGTGCCACCTTCTG; KN1-277f, ggggagatctCCTTAGC TGGTGGGATCAG; KN1-311r, ggggggtaccTTAGAACCAGTTGTTGA TCTGCT; KN1-295f, ggggagatctGAAGGTGGCACTGGCTGA; KN1-360r, ggggggtaccCATCCATCATCAGGTGGT; KNAT1-f, ggggagatctAA GCAGTTTGAAGCAAGAAC; KNAT1-r, ggggggtaccTTATGGACCGA GACGATAAG; KNAT1-f, ggggagatctCAGTTCATTGAAACTCGAGTT; KNAT1-r, ggggggtaccTTACTCGGTAAAGAATGTTTCA; KNAT1-f, gg ggagatctAGTAGACATAAGAGAGGAGATATTA; KNAT1-r, ggggggtac cCTACGCGAACCGCTCTCT; KNAT1-f, ggggagatctTAGTACTTTAA AGCTTGAGTTCTC; KNAT1-r, ggggggtaccTCATTCCTCGGTAAAG AATG; STM-f, ggggagatctAGGGAGCCTCAAGCAAGA; STM-r, gggggg taccTCAAAGCATGGTGGAGGAG; BLR-f, ggggagatctATTTCCTGA TCATCATGCTCCT; BLR-r, ggggggtaccGTCGAGTCTCCAGCATGT GAA; LeT6-f, ggggagatctGGGAAGCCTTAAGCAGGAGT; LeT6-r, ggg gggtaccTAGCAAGTACAAACTATTTAATTCCATAATTA.
Acknowledgments
We thank Myeong-Min Lee (YonSei University) for gl1 and gl1 (GL2::GUS) mutant seeds, and M. David Marks (University of Minnesota) for GL1 cDNA. D.J. was supported by the National Science Foundation Integrative Plant Biology Grant 9727959 and Cold Spring Harbor Laboratory institutional funds. J.Y.K was funded by a grant from KOSEF/MOST to the Environmental Biotechnology National Core Research Center (R15-2003-012-01003-0) and a grant from the BioGreen 21 Program, Rural Development Administration, Republic of Korea. This work was supported by Korea Research Foundation Grant KRF-2003-042-C00124.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.332805.
Corresponding authors.
References
- Bellaoui M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. 2001. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13: 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski, J., and Martienssen, R.A. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. 2003. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130: 3941–3950. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Knorr, D., Schuster, G., and Zambryski, P. 1990. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647. [DOI] [PubMed] [Google Scholar]
- Crawford K.M. and Zambryski, P.C. 2000. Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10: 1032–1040. [DOI] [PubMed] [Google Scholar]
- Ding B., Kwon, M.O., Hammond, R., and Owens, R. 1997. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12: 931–936. [DOI] [PubMed] [Google Scholar]
- Dubnau J. and Struhl G. 1996. RNA recognition and translational regulation by a homeodomain protein. Nature 379: 694–699. [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette, A.J., Nakajima, K., and Benfey, P.N. 2004. Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14: 1847–1851. [DOI] [PubMed] [Google Scholar]
- Hake S. and Freeling, M. 1986. Analysis of genetic mosaics shows that the extraepidermal cell divisions in Knotted1 mutant maize plants are induced by adjacent mesophyll cells. Nature 320: 621–623. [Google Scholar]
- Kerstetter R., Vollbrecht, E., Lowe, B., Veit, B., Yamaguchi, J., and Hake, S. 1994. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6: 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Canio, W., Kessler, S., and Sinha, N. 2001. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Yuan, Z., Cilia, M., Khalfan-Jagani, Z., and Jackson, D. 2002. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl. Acad. Sci. 99: 4103–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Yuan, Z., and Jackson, D. 2003. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362. [DOI] [PubMed] [Google Scholar]
- Kragler F., Monzer, J., Xoconostle-Cazares, B., and Lucas, W.J. 2000. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 19: 2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. 2003. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299: 392–396. [DOI] [PubMed] [Google Scholar]
- Lincoln C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan, E.I., Medford, J.I., and Barton, M.K. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983. [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena, G., Nawy, T., and Benfey, P.N. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Oparka K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. 1999. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754. [DOI] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493. [DOI] [PubMed] [Google Scholar]
- Peach C. and Velten, J. 1991. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol. Biol. 17: 49–60. [DOI] [PubMed] [Google Scholar]
- Perbal M.C., Haughn, G., Saedler, H., and Schwarz-Sommer, Z. 1996. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122: 3433–3441. [DOI] [PubMed] [Google Scholar]
- Reiser L., Sanchez-Baracaldo, P., and Hake, S. 2000. Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42: 151–166. [PubMed] [Google Scholar]
- Sena G., Jung, J.W., and Benfey, P.N. 2004. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. 1998. Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F., Yuan, Z., Hake, S., and Jackson, D. 2001. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes & Dev. 15: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. 99: 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M.D., Shimura, Y., and Okada, K. 2002. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419. [DOI] [PubMed] [Google Scholar]
- Waigmann E. and Zambryski, P. 1995. Tobacco mosaic virus movement protein-mediated protein transport between trichome cells. Plant Cell 7: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Lucas, W.J., Citovsky, V., and Zambryski, P. 1994. Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc. Natl. Acad. Sci. 91: 1433–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom, C.M., and Beachy, R.N. 1989. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379. [DOI] [PubMed] [Google Scholar]
- Wu X., Dinneny, J.D., Crawford, K.M., Rhee, Y., Citovsky, V., Zambryski, P., and Weigel, D. 2003. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. 1999. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283: 94–98. [DOI] [PubMed] [Google Scholar]