Abstract
DNA in eukaryotic cells is packaged into nucleosomes, the structural unit of chromatin. Both DNA and bulk histones are extremely long-lived, because old DNA strands and histones are retained when chromatin duplicates. In contrast, we find that the Drosophila HSP70 genes rapidly lose histone H3 and acquire variant H3.3 histones as they are induced. Histone replacement does not occur at artificial HSP70 promoter arrays, demonstrating that transcription is required for H3.3 deposition. The H3.3 histone is enriched in all active chromatin and throughout large transcription units, implying that deposition occurs during transcription elongation. Strikingly, we observed that the stability of chromatin-bound H3.3 differs between loci: H3.3 turns over at continually active rDNA genes, but becomes stable at induced HSP70 genes that have shut down. We conclude that H3.3 deposition is coupled to transcription, and continues while a gene is active. Repeated histone replacement suggests a mechanism to both maintain the structure of chromatin and access to DNA at active genes.
Keywords: H3.3, nucleosome, chromatin, transcription, histone, Drosophila
Transcriptional regulation in eukaryotes requires the coordinated recruitment of RNA polymerase II along with a variety of activators, repressors, chromatin-remodeling factors, and other chromatin-associated proteins. Histone variants have also been implicated in transcriptional regulation. All eukaryotic genomes encode four conserved core histones that package bulk chromatin, but a fraction of chromatin is packaged by alternate histones (Doenecke et al. 1997). Three lines of evidence support the argument that an alternate H3 histone subtype in animals—the variant H3.3—plays a role in transcriptionally active chromatin. First, a pulse of epitope-tagged H3.3 protein localized to gene-rich chromatin in Drosophila cells (Ahmad and Henikoff 2002). Second, H3.3 became enriched at a transgene array after its induction (Janicki et al. 2004). Third, H3.3 is enriched for covalent histone modifications associated with active chromatin, while repressive modifications are enriched on the H3 histone (McKittrick et al. 2004). These observations suggest that H3.3 is a common component of active chromatin.
Studies of the H3.3 variant have also suggested that distinctive nucleosome assembly occurs in active chromatin. Recent biochemical (Altheim and Schultz 1999; Ray-Gallet et al. 2002; Tagami et al. 2004) and in vivo (Ahmad and Henikoff 2001, 2002) assays demonstrate that there are both DNA replication-coupled and replication-independent (RI) nucleosome assembly pathways in eukaryotic nuclei. DNA replication-coupled assembly serves to package newly synthesized DNA into nucleosomes, but the existence of RI assembly pathways implies that previously chromatinized templates are also loading new histones. One RI pathway exclusively deposits newly synthesized histone H4 and the H3.3 variant and operates specifically at active chromatin in interphase nuclei (Ahmad and Henikoff 2002; Tagami et al. 2004). However, the relationship between transcriptional activity and H3.3 has not been defined: It is unknown if H3.3 deposits during or after transcription of a gene, or how frequently RI assembly occurs. Conceivably, occasional RI assembly would have little effect on the physical properties of chromatin and the stability of histone modifications. Alternatively, the structure of active chromatin might be disrupted if RI assembly occurs frequently.
Here, we demonstrate that H3.3 deposition is triggered by gene induction, and deposition occurs while genes are transcribed. We show that H3.3 deposition is also associated with histone removal and degradation that depends on the activity of a locus. Repeated rounds of replacement may function to destabilize active chromatin. The progressive degradation of H3.3 histone subtypes is distinct from the conservative exchange of H2A/H2B subtypes, and explains constitutive synthesis of new H3.3 histone.
Results
Active genes are packaged with the H3.3 histone
To determine the histone composition of transcribed genes, we examined the distribution of GFP-tagged H3 and H3.3 in Drosophila polytene chromosome spreads. Use of the same epitope tag on these two histones eliminates possible differences in detection, and the fusion proteins were expressed throughout development to label all possible deposition sites. Quantitation on Western blots with an antibody to the C terminus of histone H3 showed that each of these fusion proteins constituted <0.5% of the total H3 in cells. We considered these trace amounts that are unlikely to interfere with chromatin functions.
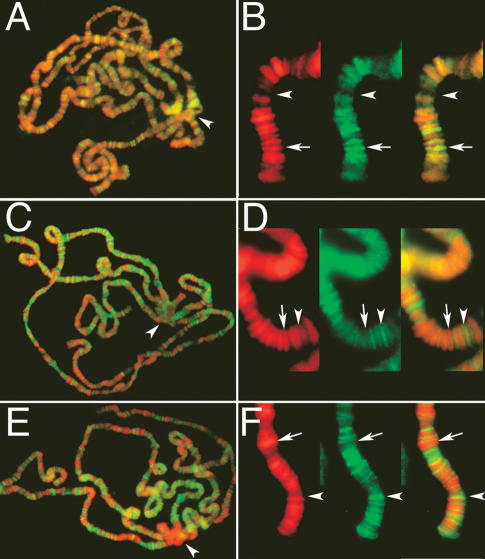
We find that the H3-GFP and H3.3-GFP histones show nearly opposite patterns of distribution in polytene chromosomes. H3-GFP localizes throughout the euchromatic arms and the heterochromatic chromocenter, and entirely overlaps with all DAPI-stained regions (Fig. 1A,B). In contrast, identically expressed H3.3-GFP localizes primarily to the interbands in euchromatin, as well as throughout the chromocenter (Fig. 1C,D). Tagged histone H3 in Drosophila Kc cells only deposits through the replication-coupled pathway (Ahmad and Henikoff 2002), and the band pattern in polytene spreads is consistent with H3 incorporation during DNA replication in the developing salivary gland. H3.3 deposits through both replication-coupled and RI pathways, and its distribution in polytene chromosomes should result from the combination of these two processes.
Figure 1.
RI deposition of H3.3 in active chromatin. Salivary polytene chromosomes from larvae that contain either constitutively expressed H3-GFP (A,B), H3.3-GFP (C,D), or H3.3core-GFP (E,F) fusion genes. (A,C,E) Whole-spread merged images; arrowheads indicate the heterochromatic chromocenters. (B,D,F) Split and merged high-magnification views of chromosome tips with histone-GFP; arrows indicate a condensed DAPI-stained band, and arrowheads indicate an interband. (A,B) H3-GFP localizes to DAPI-stained bands as well as to the chromocenter. (C,D) H3.3-GFP is mostly localized to interbands, although occasional bands are also labeled. H3.3-GFP also stains the chromocenter. (E,F) H3.3core-GFP is specific for interbands and shows little detectable staining of the chromocenter. DAPI-stained DNA is in red, and histone-GFP in green.
To define sites that result solely from RI deposition, we expressed a truncated H3.3 protein (H3.3core-GFP) that can only participate in the RI pathway (Ahmad and Henikoff 2002). Constitutively expressed H3.3core-GFP localizes to interbands and is absent from the heterochromatic chromocenter (Fig. 1E,F). From comparison of these three patterns, we conclude that the chromocenter and DAPI-banded regions are packaged with histones during DNA replication, while interbands are enriched for RI-deposited H3.3.
Enrichment of H3.3 in interbands is consistent with the idea that this histone variant is enriched at active genes. There is occasional overlap between H3.3core-GFP sites and DAPI bands (Fig. 1F); however, these may be sites where we cannot resolve closely juxtaposed sites and DAPI bands. Alternatively, these sites may have been transcribed earlier in development and still retain tagged H3.3. Apart from these ambiguous sites, it is clear that the vast majority of RI histone deposition occurs in interbands.
Histone replacement occurs during transcription
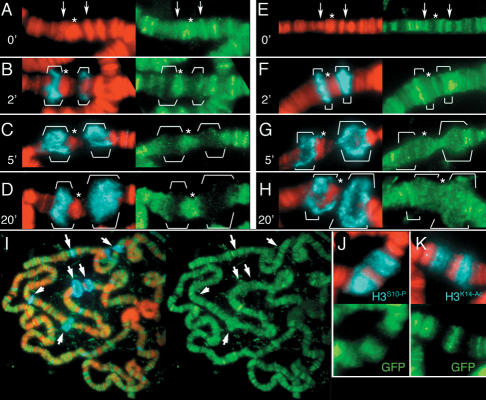
The enrichment of H3.3 in gene-rich interbands suggests that RI deposition occurs during transcription. Previous experiments with rDNA or artificial gene arrays could not distinguish whether RI assembly occurs during or after transcription of individual genes (Ahmad and Henikoff 2002; Janicki et al. 2004). To determine when RI assembly occurs, we looked for changes in histone composition during controlled induction of HSP loci in polytene chromosomes. The HSP70 loci at cytological bands 87A and 87C are compacted before induction, but expand as large puffs during induction and gene expression (Fig. 2A–H; Korge 1975). We measured the summed intensities of histone-GFP signals to compare puffs of different sizes. Strikingly, we find that the fully expanded active puffs show little H3-GFP above background (Fig. 2D; Table 1, 20′ time point). Instead, the puffs are dramatically enriched for the H3.3-GFP histone (Fig. 2H; Table 1). Other heat-shock-inducible loci also show enrichment of H3.3 during induction, indicating that H3.3 enrichment is a common feature of these highly induced genes (Fig. 2I). We conclude that induced loci are predominantly packaged with H3.3 while they are transcribed. Because the H3 subtype is largely absent from puffs, “H3” modifications that are enriched in polytene puffs (Nowak and Corces 2000; Smith et al. 2004) must actually be modifications of the H3.3 subtype. Indeed, immunodetection of H3S10-phospho (Fig. 2J) and H3K14-acetyl epitopes (Fig. 2K) show that these modifications coat puffs that contain no detectable H3 signal.
Figure 2.
Gene induction triggers histone replacement. Heat-shock induction triggers rapid puffing and transcription of the HSP70 genes at polytene bands 87A and 87C. Polytene chromosomes from larvae that contain a constitutively expressed H3-GFP construct (A–D,J,K), or a constitutively expressed H3.3core-GFP construct (E–I) were induced for the indicated times (in minutes) and stained with antibodies against phosphorylated RNA polymerase II (blue), which marks activated puffs. Arrows in A and E indicate the positions of the HSP70 loci at 87A and 87C before induction, while the puffs (B–D,F–H) are bracketed. Asterisks indicate chromosome bands that were used as internal standards for quantifying the summed intensities of histone-GFP signals in puffs. (B,C) Puffs contain some H3 in the first 5 min of induction, but have much less by the time puffs reach their maximal size (D). (F–H) H3.3core-GFP in expanding puffs rapidly increases. (I) After 20 min of heat-shock induction, many active HSP puffs (arrows) have large amounts of H3.3core-GFP, while staining throughout the arms is undiminished. (J,K) Histone H3 modifications that mark heat-shock puffs do not overlap with H3-GFP. DNA is in red, and histone-GFP in green.
Table 1.
Histone-GFP intensities at induced gene puffs
| Gene | Time | H3-GFP | H3.3-GFP |
|---|---|---|---|
| HSP70 (87A and C) | No HS | nd | nd |
| HS + 2′ | 1.2 ± 0.2 | 1.2 ± 0.2 | |
| HS + 5′ | 3.0 ± 0.3 | 5.3 ± 1.6 | |
| HS + 20′ | 2.8 ± 0.6 | 7.4 ± 2.3 | |
| E74 | Before | 3.2 ± 1.6 | 3.5 ± 0.6 |
| Induced | 1.7 ± 0.6 | 7.0 ± 1.5 | |
| E75 | Before | 1.7 ± 0.1 | 1.9 ± 0.3 |
| Induced | 3.1 ± 1.6 | 5.5 ± 1.0 |
Measured area before induction was defined by landmark DAPI-stained bands, and after induction by Pol II staining. Total histone-GFP signals were calculated as the summed signal intensity in the chromosomal area divided by the summed intensity of a nearby control band in the same spread (to standardize for differences in slide preparation). Mean ratio and standard errors were estimated from at least five spreads for each sample.
Quantitative comparison of puffs during induction of the HSP70 genes suggests that changes in histone composition begin in the first few minutes of induction (Table 1). We could not quantitate the amounts of histone-GFP at the HSP70 loci before induction because they are too small to resolve from flanking bands, but intermediate puffs (2–5 min into the induction) show moderate amounts of H3.3, while maximal puffs (20 min) display more H3.3. The rapid gain in H3.3 signals implies that RI deposition is occurring during transcription. Enrichment at the HSP70 genes occurs substantially faster than was observed at multicopy gene arrays in interphase nuclei (Janicki et al. 2004), and may be due to the greater sensitivity of polytene chromosomes. Alternatively, fast gain may be due to the higher rates of transcription at induced HSP70 genes. Such a relationship could explain why the intensities of H3.3-GFP signal vary between different HSP loci (Fig. 2I).
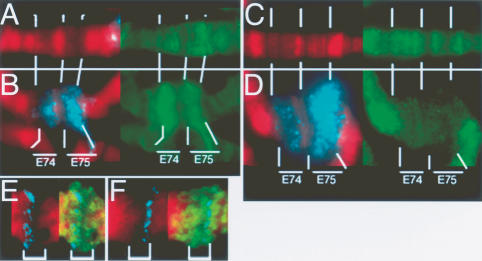
We expected that since induced loci acquire H3.3 as a puff expands, H3-GFP signals should reciprocally decrease. However, the small size of the HSP70 genes (∼2 kb) hampered measuring how much tagged histone was present before induction. Instead, we examined the much larger ecdysone-responsive (EcR) genes E74 and E75, which each encompasses 80–100 kb across characteristic sets of DAPI-stained bands (Segraves and Hogness 1990). Measurable amounts of both H3 and H3.3 are present in these intervals before induction (Fig. 3A,C; Table 1). The E74 and E75 genes also generate chromosomal puffs when they are induced. Just as we observed for heat-shock loci, the transcribing EcR puffs are heavily labeled with H3.3 (Fig. 3B). At these larger genes, the H3-subtype histones show reciprocal changes after induction: H3 amounts decrease in the puff, while H3.3 amounts increase (Fig. 3D; Table 1). These experiments support our observations with the heat-shock-inducible genes, and confirm that induced genes become depleted for H3 and enriched for H3.3 during transcription.
Figure 3.
Ecdysone-induced genes are broadly labeled with H3.3. (A,B) Chromosome divisions spanning the large ecdysone-responsive genes E74 and E75 from larvae constitutively expressing H3.3core-GFP before (A) and after (B) induction. (C,D) The same chromosomal region before and after induction from larvae constitutively expressing H3-GFP. Both regions show moderate amounts of H3.3core-GFP and H3-GFP before induction, but H3.3core is increased while H3 is depleted during transcription. Lines indicate the position of landmark DAPI bands that flank the E74 and E75 genes. DNA is in red, histone-GFP in green, and RNA polymerase II is in blue. (E,F) Fluorescence in situ hybridization of DNA probes (blue) to the 3′-end (E) and 5′-end (F) of the induced E75 gene. Brackets delineate the gene. H3.3core-GFP (green) is enriched in the intervening body of the gene.
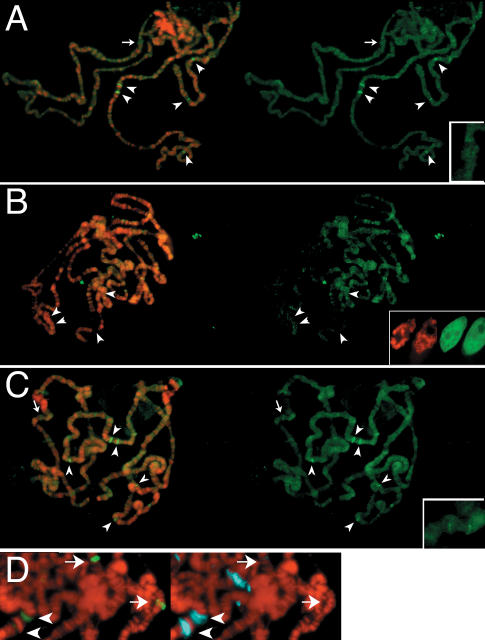
Our observations of loss of H3 and gain of H3.3 at expanding puffs are not due to epitope differences or accessibility as chromatin decompacts, because we used the same epitope (GFP) and see opposite results in puffs with each histone. However, to demonstrate that quantitation of chromosomal puffs correctly reported changes in histone composition, we used a heat-shock-inducible construct to produce a pulse of new tagged H3.3 at the same time as the endogenous HSP70 loci are induced. We predicted that producing new H3.3 and inducing the HSP70 genes would result in deposition only at those loci. Indeed, we find that the newly produced H3.3-GFP localized exclusively to the HSP loci (cf. Figs. 4A and 1B), demonstrating that new nucleosomes are being assembled from new histones at transcribed sites. A similar pulse of H3 protein does not incorporate into chromatin at this time, confirming that the new nucleosomes only use H3.3 (Fig. 4B). We conclude that RI histone deposition is initiated by gene induction and occurs as the gene is being transcribed. The consequence of replacement is that activated loci lose H3 and become enriched for H3.3.
Figure 4.
Histone replacement uses newly produced H3.3. Late-third-instar larvae containing a heat-shock-inducible histone-GFP construct were heat-shocked for 1 h to induce the construct and endogenous HSP genes, and then allowed to recover for 4 h. (A–C) DNA (red) and histone-GFP (green) images are shown as merges on the left, and the histone-GFP signal alone are shown on the right. (A) Full-length H3.3-GFP has deposited specifically at the heat-shock loci (arrowheads indicate the 87A, 87C, 95D, 63E, and 67F loci). (B) H3-GFP is not incorporated into chromatin. H3-GFP fills the interchromatin space within unfixed salivary gland nuclei after induction, but remains diffuse (inset). (C) Spreads with H3.3-GFP and two HSP70 promoter transgene arrays show intense H3-GFP signals at HSP loci but little signal at the arrays. Arrows in A and C indicate the 43E region without an array (A) and with an array (C). (Insets) Magnified images of 43E are shown. (D) Chromosomes from larvae carrying P[H3.3-GFP]B6 and the HSP70 promoter transgene arrays at positions 30A and 43E during a 20-min heat-shock induction. The endogenous HSP70 loci (arrowheads) bind moderate amounts of HSF activator (green), while arrays (arrows) bind large amounts. Conversely, HSP70 loci have large amounts of engaged RNA polymerase II (blue), while arrays have little.
The overall behavior of H3.3 differs from that of many markers of active transcription. While RNA polymerase II components are enriched at HSP loci and depleted from most other loci during the heat-shock response (Jamrich et al. 1977), H3.3-GFP that was deposited earlier in development remains distributed throughout chromosomes (Fig. 2I). As a heat-shock response terminates, active RNA polymerase II rapidly leaves these loci and transcription of the HSP70 genes shuts off. In contrast, tagged H3.3 remains stably incorporated at the HSP70 genes hours after a transient induction (Fig. 4A). Signals at HSP70 genes are undiminished even 48 h after the induction (data not shown). We conclude that H3.3 remains incorporated even after transcription has ceased. In differentiated cells where the cessation of DNA replication precludes expression of the H3 histone and replication-coupled deposition, H3.3 enrichment will be permanent.
RI deposition is associated with transcriptional elongation
In budding yeast, specific nucleosomes within the promoters of some genes are dissembled during transcriptional induction, and then reassembled when repressed (Adkins et al. 2004). In contrast, we observe increases in H3.3 signals at induced genes during active transcription, suggesting that at least some H3.3 deposition is distinct from promoter nucleosome changes. Indeed, H3.3-GFP signals are distributed throughout induced gene puffs, supporting the argument that new nucleosome assembly is not limited to promoters. This is especially clear at the E74 and E75 puffs, which encompass 80 and 100 kb of transcribed DNA, respectively (Fig. 3). At the E75 gene, probes to the 5′- and 3′-ends of the transcription unit label the ends of the puff, and the bulk of the H3.3 signal lies between these probes (Fig. 3E,F), confirming that H3.3 has been enriched in transcribed chromatin.
We used artificial promoter arrays to test if H3.3 enrichment at the HSP70 genes could be accounted for by replacement of promoter nucleosomes. These arrays contain ∼40 tandem copies of HSP70 promoter elements, but do not support RNA polymerase firing (Park et al. 2001). Accordingly, the arrays bind extremely large amounts of transcriptional activator, but show little active RNA polymerase II (compared to that at the endogenous HSP70 genes in the same spreads) (Fig. 4D; Park et al. 2001). While the endogenous genes are heavily labeled after a pulse of H3.3-GFP, we find that promoter arrays show little or no labeling above background (Fig. 4C). We conclude that transcription factor binding and promoter activation are insufficient to stimulate H3.3 deposition. These results suggest that transcriptional elongation is required, and our observation of H3.3 throughout the very long transcription units of EcR genes is consistent with this idea.
H3.3 is a short-lived histone
If transcription triggers RI nucleosome assembly by first displacing histones from the template, we wondered what happens to these released histones. Recent experiments have characterized an activity that performs a histone H2A/H2B dimer exchange reaction between nucleosomes and histone chaperones (Mizuguchi et al. 2004). In vivo, H2A/H2B dimers release from chromatin and rebind to other sites, indicating that they can exchange and be reused (Jackson 1987; Kimura and Cook 2001). However, these experiments detected little or no exchange of the H3 or H4 histones. Our experiments here show that RI assembly can use newly produced H3.3, suggesting that old, displaced H3 subtype histones might be destroyed. Indeed, when we compared histone-GFP signals in whole-mount polytene nuclei from larvae induced 0.5 to 3 d earlier, we observed that H3.3 signals decreased approximately four times faster than H3 signals (data not shown). However, in this time period polytene cells continue to duplicate their chromatin by replication-coupled nucleosome assembly, which complicates measures of histone stability. We directly tested the possibility that H3.3 is unstable by measuring chromatin-bound histones after a pulse of production. To measure only RI-deposited histones, we produced the pulse in adult flies. The vast majority of cells in the adult are post-mitotic, and these cells should not synthesize H3 or perform replication-coupled nucleosome assembly. The main exception is the germline lineages, and we therefore discarded the abdomens of adult flies before analysis.
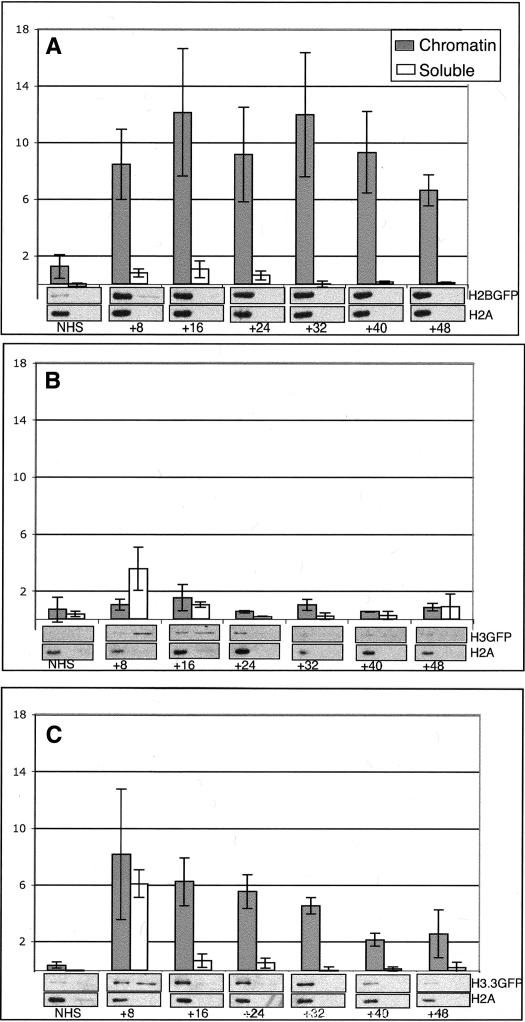
We distinguished chromatin-bound histones from soluble pools by their resistance to extraction with 400 mM salt (Kimura and Cook 2001). Induction of histone-GFP constructs produces soluble pools of protein that are available for deposition for several hours (Fig. 5). As expected for differentiated cells, a pulse of H3-GFP in adults is not incorporated (Fig. 5B). In contrast, a pulse of H3.3-GFP rapidly enters chromatin (Fig. 5C): This must be due to RI assembly. While the amounts of chromatin-bound H3.3 greatly increase within 8 h after production, this enrichment is transient, and after 4 d the amounts of chromatin-bound H3.3 have returned to a low level. We estimate that the bulk chromatin-bound H3.3 has a half-life of ∼24 h. This is in contrast to the stability of chromatin-bound H3, where a half-life of hundreds of days has been estimated (Pina and Suau 1987). The decay of H3.3-GFP is not due to a defect in the fusion protein, because an identical pulse of H2B-GFP becomes chromatin-bound in adults but does not significantly decrease over the chase period (Fig. 5A), and H3-GFP incorporated during development is also stable (data not shown). Instead, a short half-life appears to be a general feature of replacement histones, because the replacement H3 subtype in plants also disappears rapidly (Waterborg 1993). The progressive degradation of H3.3 suggests that some step in RI replacement consumes the histone, and distinguishes RI replacement from a simple exchange process.
Figure 5.
Chromatin-bound H3.3 is unstable in post-mitotic cells. (A–C) Adult flies carrying a heat-shock-inducible H2B-GFP (A), H3-GFP (B), or H3.3-GFP (C) construct were frozen without heat shock or at the indicated times (in hours) after induction. Samples were extracted with 400 mM NaCl and centrifuged to separate soluble from pelleted chromatin-bound histones. Ten percent of the supernatant and 20% of the pellet fractions were separated by SDS-PAGE, and GFP-tagged histones were detected with anti-GFP antibody. Anti-H2A antibody was used as an internal control for the total nucleosome content at each time point (hours after induction). The chromatin-bound (black bars) and soluble forms (empty bars) of each histone-GFP protein are graphed as the ratio of GFP signal to H2A signal (arbitrary units). (A) A pulse of H2B-GFP rapidly enters chromatin, and almost all soluble H2B-GFP is used up within 8 h of production. The bulk of H2B-GFP is stable in chromatin. (B) Soluble H3-GFP is produced, but there is no detectable incorporation into chromatin. (C) A pulse of H3.3-GFP incorporates into chromatin, and reaches its maximum 8 h after production. Soluble H3.3-GFP has disappeared by 16 h, and the bulk of chromatin-bound tagged protein is gone by 48 h.
Histone H3.3 is rapidly removed from active chromatin
Our estimate of H3.3 turnover is based on bulk measurements of all the tagged histone in a cell. However, since transcriptional rates vary between sites in the nucleus, histone half-lives at each site might differ. Indeed, a pulse of H3.3-GFP shows labeling with a wide range of intensities in the nucleus (Ahmad and Henikoff 2002). These intensity differences might indicate densities of H3.3 in chromatin, but an alternative explanation is that intense sites are where the pulse of tagged H3.3 rapidly enters chromatin.
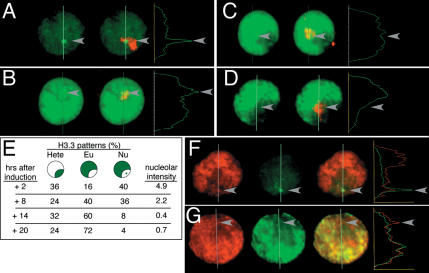
Overall, the summed intensity of H3.3-GFP within nuclei decreases over a time course (data not shown). This is similar to what we observed in polytene nuclei, and is consistent with turnover of the histone variant in interphase cells. However the deposition rate hypothesis predicts that the dimming of individual sites will differ, with the brightest foci of H3.3-GFP being the first sites to lose the tagged histone. To detect turnover rates at particular loci, we tracked the subnuclear distribution of H3.3-GFP immediately after a pulse of production and subsequent time points (Fig. 6A–E). We classified labeled nuclei into “nucleolar” (Nu), “euchromatic” (Eu), and “heterochromatic” (Hete) patterns. Previous experiments have shown that nucleolar patterns only result from RI deposition, and heterochromatic ones from replication-coupled deposition. Both early S-phase replication and RI deposition produce euchromatic labeling. Two hours after producing tagged H3.3 in an asynchronous cell culture, 40% of labeled cells show focal staining within the nucleolus (Fig. 6A,E). These rDNA foci are approximately five times brighter than the next brightest sites in euchromatin. The rest of the labeled nuclei show replication-coupled H3.3 deposition patterns. However, over the next few hours both the nucleolar pattern becomes rare and the intensity of GFP within nucleoli diminishes (Fig. 6B–E). By 20 h after the pulse there are no obvious labeled rDNA foci, and the brightest site within nucleoli averages ∼0.8 the intensity of euchromatic sites in the same nucleus. This change in nucleolar intensities is statistically significant (P < 0.001, two-tailed comparison of ratio of means). Thus, the amounts of H3.3-GFP at active rDNA genes drop at least sixfold over the 20-h chase. We conclude that the rDNA foci do gain and then lose the tagged histone more rapidly than other sites in the nucleus. Our estimate of the nucleolar turnover rate must be an underestimate, because soluble H3.3 is available for at least the first 8 h of the time course. In spite of this, the turnover rate at the highly transcribed rDNA genes appears to be much faster than the bulk rate, suggesting that this rate corresponds to transcriptional activity.
Figure 6.
H3.3 is rapidly displaced from highly active chromatin. (A–D) Kc cells transfected with the HS-H3.3-GFP construct were heat-shocked to induce the construct. Samples were collected 2 h (A), 8 h (B), 14 h (C), and 20 h (D) later and lightly extracted with Triton X-100 to remove soluble proteins before fixation. The location of the nucleolus is marked by antibody detection of the nucleolar protein fibrillarin (red). Arrowheads indicate the position of H3.3-GFP nucleolar foci. Line profiles show the pixel intensities of the GFP signal (arbitrary units) through the nucleus. The ratio of H3.3-GFP spot intensities within the nucleolus to that of the brightest site in euchromatin was used as a single-cell measure of the amount of histone deposited at the active rDNA genes. (A) Two hours after induction many cells have intensely labeled rDNA foci. (B–D) H3.3-GFP signals in nucleoli become dimmer over time, and by 20 h nucleolar foci are rare. Euchromatic signals reach their maximum 8 h after production, and then diminish slowly. (E) Quantitation of H3.3-GFP nuclear patterns and intensities over time. Twenty-five GFP-positive cells were counted for each sample. Both the frequency of nuclei with nucleolar foci and the intensity of those foci drop over time. The decrease in focal intensity is statistically significant (P < 0.001, comparison of ratio of means). (F) Constitutive expression of H3.3-RFP (red) in Kc cells gives similar intensities in nucleolar foci and in euchromatin. In the same cells, a pulse of H3.3-GFP (green) 2 h earlier intensely labels the same nucleolar foci. (G) Twenty hours later, nucleolar H3.3-GFP signal is dimmer than the constitutive H3.3-RFP signal.
If the early intense signals at nucleolar foci are due to the kinetics of RI assembly in the nucleus, continual production of H3.3-RFP should be deposited throughout the nucleus. Indeed, we find that euchromatin and nucleolar foci are labeled with roughly uniform intensities, while a pulse of H3.3-GFP in the same cells intensely labels nucleolar foci (Fig. 6F). Thus, with continual production of the labeled histone, it appears that all sites within the nucleus reach a steady state. Notably, at later times the H3.3-GFP signals within the nucleolus are always dimmer than the constitutive H3.3-RFP signals, as if the pulse of protein does not freely exchange within the nucleus (Fig. 6G). We conclude that the early intense labeling after pulsed expression results from the more rapid deposition of the histone at these sites. This suggests that the bulk measurements of histone turnover are the average of many sites undergoing replacement, with some sites rapidly replacing histones while many sites do so more slowly.
Discussion
H3.3 becomes enriched in active chromatin
Cells use two pathways for the assembly of nucleosomes. The replication-coupled pathway suffices to package the bulk genome as DNA is replicated, using histones synthesized from the major canonical genes. The RI pathway acts on transcriptionally active loci and specifically uses the H3.3 histone variant (Ahmad and Henikoff 2002; Janicki et al. 2004; Tagami et al. 2004). Here, we have shown that RI assembly begins within minutes after gene induction and operates while genes are active. The binding of transcription factors to artificial promoter arrays is insufficient to trigger H3.3 deposition, implying that RI assembly is tied to a late step in gene induction. Our findings that H3.3 becomes distributed throughout long transcribed genes suggest that RNA polymerase progression itself is involved in histone displacement and replacement. Finally, the stability of H3.3 in chromatin depends on transcriptional status: At continually active loci, H3.3 deposits and then leaves, but when genes become repressed, incorporated H3.3 becomes stable. This dependence is explained if transcription causes the displacement of chromatin-bound histones.
In vitro experiments show that RNA polymerases often pause when they run into a nucleosome (Polach and Widom 1996). Transcription through a nucleosome requires the transient disruption of histone–DNA contacts so that polymerase can progress. A key question is whether the loss of these contacts releases histones from DNA as polymerase passes, or if the histones remain associated. The current view is that accessory factors assist polymerases by moving histone octamers along the template, or by returning any histones that are displaced (Narlikar et al. 2002). However, the rapid stimulation of H3.3 replacement by gene induction implies that at least sometimes displaced histones are not restored. The enrichment of H3.3 at all active chromatin in Drosophila indicates that the loss of displaced histones is a general feature of transcription from chromatin templates.
There are two processes that might link histone variant replacement to transcription. First, the polymerase may displace histones, thus producing a naked template suitable for RI assembly machinery. Such a dependence on prior nucleosome disassembly would effectively restrict H3.3 replacement to active genes. Second, factors that accompany elongating RNA polymerase may catalyze histone replacement. Support for this idea comes from the detection of transcriptional regulators in isolated H3.3 predeposition complexes (Tagami et al. 2004). Tagami et al. (2004) suggested that these regulators might serve as protein interfaces to recruit H3.3 to sites of active transcription. Indeed, a protein interaction between the histone chaperone CAF large subunit and the DNA replication fork protein PCNA serves to enhance replication-coupled deposition of histone H3 (Shibahara and Stillman 1999). An analogous role might be performed by transcription factors that interact with the HIRA chaperone in H3.3 predeposition complexes. Additionally, the histone-binding proteins SPT5 and SPT6 are known to localize to transiently induced HSP70 genes, and are thought to manipulate nucleosomes for transcription (Andrulis et al. 2000). Such factors might drive histone replacement to clear DNA for polymerase passage. SPT5 and SPT6 are not known to localize to sites of RNA Pol I transcription, so there may also be analogous polymerase-specific factors that clear DNA. Since SPT5 and SPT6 leave HSP genes after transient induction, replacement would also cease and the genes would retain high levels of H3.3. Thus, H3.3 replacement leaves behind an extremely stable marker of past transcriptional activity.
Degradation of histones from active chromatin
Our observation that chromatin-bound H3.3 has a short half-life (∼24 h) is in stark contrast to the half-life of histones from bulk chromatin (Pina and Suau 1987). Measurements of in vivo histone exchange have demonstrated recycling of H2A and H2B between sites within a nucleus (Kimura and Cook 2001), and at least one complex is capable of exchanging these histones between chromatin and histone chaperones (Mizuguchi et al. 2004). However, recycling of H3 and H4 was not detected, and in our experiments bulk H2B-GFP does not degrade. The fate of H3.3 when it is displaced from chromatin distinguishes H3.3 replacement from other kinds of chromatin exchange and remodeling, and suggests that RI nucleosome assembly and disassembly is not an exchange reaction. This appears to be a general property of replacement H3 histones, because alfalfa replacement H3 also has a fast rate of turnover (Waterborg 1993). The rapid turnover of replacement H3 histones explains why all eukaryotes have constitutively expressed H3.3-type genes (Thatcher et al. 1994; Malik and Henikoff 2003), because a continuous supply will be required to replenish active chromatin. Perhaps the purpose of specific histone degradation is to prevent exchange between chromatin sites. Chromatin-bound H3 and H4 are heavily modified (Zhang et al. 2002), and we suggest that H3/H4 subparticles are degraded to prevent moving modified histones to new sites. In contrast, the relative paucity of modifications on the H2A and H2B histones means that their exchange between sites has no detrimental effect.
Two other instances of histone degradation have been well-characterized. Chromatin-bound centromeric histones in budding yeast (CSE4) are subject to proteasome-mediated degradation that may limit this histone variant to centromeres (Collins et al. 2004). A separate system that degrades excess soluble histones has also been described (Gunjan and Verreault 2003). Our experiments do not distinguish whether H3.3 is degraded as it is displaced from chromatin or once it is soluble. Degradation of the histone may occur by a proteosome-catalyzed mechanism, or by a general response to damaged proteins. However, histone degradation in differentiated cells appears to discriminate between histone subtypes, because H2B-GFP is not similarly degraded. As RI histone deposition uses specialized chromatin assembly factors (Tagami et al. 2004), our results raise the possibility that there are histone disassembly factors that specifically degrade H3.3. This could be tested with drugs that block known degradation pathways.
Dynamics of chromatin states
The H3 and H3.3 histones are extremely similar, differing by only four amino acid residues in Drosophila. Three of these specify whether the histone can be used for replication-coupled or RI nucleosome assembly, but these residues are not accessible in the complete nucleosome (Ahmad and Henikoff 2002). This extreme similarity raises a paradox: What can the function of RI assembly be, since new assembly results in a virtually identical nucleosome? In fact, the similarity between H3 and H3.3 distinguishes it from other histone variants like H2A.Z, which do introduce structural differences into the nucleosome (Redon et al. 2002). A possible resolution to this paradox comes from our observations of the dynamics of RI assembly. Replication-coupled histone deposition only occurs once every cell cycle at a site, but RI deposition is a continual process at active genes. Thus, inactive chromatin retains the same histones for long periods of time, but active chromatin repeatedly shed its histones. Rapid turnover of histones at active genes has three implications for chromatin structure.
First, the maintenance of any modification state in active chromatin will require ongoing activity of histone-modifying enzymes to modify newly arriving histones. Acetyl modifications do rapidly turn over in vivo (Waterborg 1998), and can be removed enzymatically, but histone replacement will also contribute to turnover rates. Replacement may enhance the specificity of histone-modifying enzymes by continually removing modifications from active chromatin. Histone replacement also provides a mechanism to remove histone modifications that may not be metabolized. One example of this is the activation of heterochromatic gene arrays (Ahmad and Henikoff 2002; Janicki et al. 2004), where chromatin is marked by methylation at Lys 9 of the H3 tail (H3K9me). Activation of these gene arrays results in the coincident disappearance of H3K9me and deposition of H3.3, as if removal of the methylation mark occurs by histone replacement. In contrast, some histone modifications that are set during gene expression remain present after transcription and histone replacement cease (Kouskouti and Talianidis 2005). Indeed, the retention of H3.3 at previously active chromatin may require that the variant maintain its extreme similarity with H3 so that they both can be repressed by the same modification systems.
Second, continual histone replacement means that active regions have little time to “mature” their chromatin between rounds of histone deposition. Nascent chromatin produced by replication-coupled assembly matures into a nuclease-resistant form within 1 h, and this delay is thought to reflect the rates for removing predeposition modifications, establishing new modifications, and condensing chromatin (Cusick et al. 1983; Wolffe 1998). If the rates for modification and condensation after RI assembly are similar to replicated chromatin, rapid histone turnover will continually reset chromatin to an “immature” uncondensed state.
Third, high DNA accessibility is a characteristic feature of active chromatin (Weintraub and Groudine 1976), and this feature may simply result from ongoing histone replacement. This idea is supported by recent experiments in budding yeast demonstrating that the inducible accessibility of the PHO5 promoter is due to loss of nucleosomes (Boeger et al. 2003; Reinke and Horz 2003). Earlier studies observed a slight decrease in the density of histone H4 at activated HSP70 genes, and concluded that transcription occurs without displacing histones from chromatin (Solomon et al. 1988). However, a small decrease could result if displaced histones are rapidly replaced with new ones. Indeed, in budding yeast the density of histones appears to drop at very high transcription rates (Schwabish and Struhl 2004). Such transient structural instability of active chromatin would allow efficient polymerase elongation.
Materials and methods
DNA constructs
Constructs carrying the H2B-GFP, H3-GFP, H3.3-GFP, and H3.3core-GFP fusion genes were previously described (Henikoff et al. 2000; Ahmad and Henikoff 2002). The H3.3core-GFP protein is deleted for amino acid residues 4–36 of the histone N-terminal tail, and can only undergo RI assembly. The constitutive H3.3-RFP construct was generated by replacing the UAS and HS promoters from pPUASHSH3.3GFP with the Copia LTR promoter (Rio and Rubin 1985), and replacing GFP with mRFP (Campbell et al. 2002). This produced the construct pPCoH3.3-RFP.
Cell culture
Drosophila Kc cells were maintained, electroporated, and prepared for cytology as previously described (Ahmad and Henikoff 2002). Twenty micrograms of plasmid DNA was used for each transfection. For experiments with both H3.3-RFP and H3.3-GFP, we transfected cells with equal amounts of each plasmid and then cultured them for 48 h before inducing H3.3-GFP production. The location of the nucleolus in Kc cells was identified using anti-Fibrillarin antiserum (Abcam; 1:100 dilution), with secondary detection using goat anti-rabbit IgG-Texas Red antibodies (JacksonImmunoResearch; 1:100).
Fly lines
All flies were raised at 25°C on standard media. Germline transformants of P-element constructs carrying the H2B-GFP, H3-GFP, H3.3-GFP, and H3.3core-GFP fusion genes were generated using standard procedures (Rubin and Spradling 1983). The lines P[H2B-GFP]A4 (X-chromosome insertion), P[H3-GFP]C4 (chromosome 2), P[H3.3-GFP]B6 (chromosome 2), and P[H3.3 core-GFP]G5E (chromosome 3) were used here. The fusion genes in each of these constructs carry a heat-shock-inducible HSP70 promoter and a GAL4-dependent promoter. A pulse of tagged histone was produced by incubating larvae or flies in a waterbath at 37°C as previously described (Schwartz et al. 2004), for the time specified in the text. Tagged histone in the salivary glands of larvae was constitutively produced in animals carrying the histone-GFP gene and a dppGAL4 driver (Staehling-Hampton et al. 1994) in a separate P element. The fly line CXM40-4—carrying arrays of HSP70 promoter elements (Park et al. 2001)—was a gift from Dr. John Lis (Cornell University, Ithaca, NY).
Polytene chromosome analysis
Chromosome spreads for protein detection were prepared and fixed as previously described (Schwartz et al. 2004). Late-third-instar larvae with everted anterior spiracles were chosen to obtain polytene chromosomes with induced E74 and E75 genes. Primary antibodies were the JL-8 anti-GFP (Clontech; 1:100 dilution), H14 anti-RNA polymerase II (specific for the Ser 5-phosphorylated form of Pol II CTD, BabCo; 1:50 dilution), anti-histone H3S10-phospho (Upstate; 1:100 dilution), and anti-histone H3K14Ac (Upstate; 1:25 dilution). Rabbit antiserum to Drosophila HSF was a gift from Dr. John Lis. We used conjugated goat anti-mouse IgG-FITC, goat anti-mouse IgM-Texas Red, goat anti-rabbit IgG-Texas Red, and goat anti-rabbit IgG-FITC antibodies (JacksonImmunoResearch; 1:100) for secondary detection. DNA–protein FISH experiments were performed as described in the Roche Applied Science Application Manual. Briefly, chromosomes were spread in 40% acetic acid and hybridized at 55°C with probes to the 5′-end of the E75 “B” promoter (coordinates in Drosophila Genome Sequence Release 4.0, 3L:18026253–18025011) or to the 3′-end of the coding sequence (coordinates 3L:17921033–17920872). Probes were synthesized by PCR, labeled with the Digoxygenin (Dig) Hi-Prime labeling kit (Roche), and detected with rhodamine-conjugated anti-Dig sheep antibody. The histone-GFP fusion proteins remained fluorescent under these conditions. All preparations were stained with 0.05 μg/mL DAPI to detect DNA.
Microscopy and image analysis
Salivary gland and cell culture nuclei were examined on a Nikon TE2000E epifluorescence microscope using 360/40, 457/17 (DAPI), 484/15, 517/30 (FITC), and 555/25, 605/40 (TRITC) filter sets (Chroma Corp.). Single-channel digital images were collected with a Cooke 12-bit SensiCam CCD driven by IPLab 3.9 software. Quantitative image analysis and whole-image adjustments were performed using 12-bit files in IPLab, and exported as 8-bit files to Adobe Photoshop for formatting.
Protein analysis
Samples of five adult flies carrying histone-GFP constructs were frozen in liquid nitrogen. Heads and thoraces were dounced on ice with a glass homogenizer in 10 mM Tris buffer containing protease inhibitors. Supernatants from these samples were adjusted to 400 mM NaCl to release soluble histones, and centrifuged to pellet chromatin. Proteins from the soluble and chromatin fractions were separated in 12% SDS-PAGE gels and transferred to nitrocellulose. Histone-GFP protein was detected using JL-8 antibody (1:1000 dilution), and anti-Drosophila H2A rabbit antisera (Leach et al. 2000) was used to detect H2A in each sample as a loading control (1:4000 dilution). Conjugated goat anti-mouse or anti-rabbit IgG-HRP antibodies (JacksonImmunoResearch; 1:1250 dilution) were used for secondary detection. Western blots were developed with an ECL kit (Amersham) and exposed to film. The integrated intensities of each band were measured using Image Gauge V3.45 software, and the background-corrected ratio between histone-GFP and H2A signals is a standardized measure of the amount of histone-GFP in adult animals.
Acknowledgments
We thank R. Glaser for antisera to Drosophila H2A, John Lis for CXM40-4 flies and antiserum to Drosophila HSF, Steve Henikoff for assistance generating the H2B-GFP fly lines, and Akiko Sakai for helpful comments. This work was supported by the NIHGMS.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1259805.
References
- Adkins M.W., Howar, S.R., and Tyler, J.K. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Ahmad K. and Henikoff, S. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Altheim B.A. and Schultz, M.C. 1999. Histone modification governs the cell cycle regulation of a replication-independent chromatin assembly pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman, E., Doring, P., Werner, J., and Lis, J.T. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 14: 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H., Griesenbeck, J., Strattan, J.S., and Kornberg, R.D. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.A., Furuyama, S., and Biggins, S. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. [DOI] [PubMed] [Google Scholar]
- Cusick M.E., Lee, K.S., DePamphilis, M.L., and Wassarman, P.M. 1983. Structure of chromatin at deoxyribonucleic acid replication forks: Nuclease hypersensitivity results from both prenucleosomal deoxyribonucleic acid and an immature chromatin structure. Biochemistry 22: 3873–3884. [DOI] [PubMed] [Google Scholar]
- Doenecke D., Albig, W., Bode, C., Drabent, B., Franke, K., Gavenis, K., and Witt, O. 1997. Histones: Genetic diversity and tissue-specific gene expression. Histochem. Cell Biol. 107: 1–10. [DOI] [PubMed] [Google Scholar]
- Gunjan A. and Verreault, A. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537–549. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad, K., Platero, J.S., and van Steensel, B. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. 97: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. 1987. Deposition of newly synthesized histones: New histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry 26: 2315–2325. [DOI] [PubMed] [Google Scholar]
- Jamrich M., Greenleaf, A.L., and Bautz, E.K. 1977. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc. Natl. Acad. Sci. 74: 2079–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto, T., Salghetti, S.E., Tansey, W.P., Sachidanandam, R., Prasanth, K.V., Ried, T., Shav-Tal, Y., Bertrand, E., Singer, R.H., et al. 2004. From silencing to gene expression: Real-time analysis in single cells. Cell 116: 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. and Cook, P.R. 2001. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korge G. 1975. Chromosome puff activity and protein synthesis in larval salivary glands of Drosophila melanogaster. Proc. Natl. Acad. Sci. 72: 4550–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A. and Talianidis, I. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach T.J., Mazzeo, M., Chotkowski, H.L., Madigan, J.P., Wotring, M.G., and Glaser, R.L. 2000. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275: 23267–23272. [DOI] [PubMed] [Google Scholar]
- Malik H.S. and Henikoff, S. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10: 882–891. [DOI] [PubMed] [Google Scholar]
- McKittrick E., Gafken, P.R., Ahmad, K., and Henikoff, S. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen, X., Landry, J., Wu, W., Sen, S., and Wu, C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan, H.Y., and Kingston, R.E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487. [DOI] [PubMed] [Google Scholar]
- Nowak S.J. and Corces, V.G. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes & Dev. 14: 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Werner, J., Kim, J.M., Lis, J.T., and Kim, Y.J. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8: 9–19. [DOI] [PubMed] [Google Scholar]
- Pina B. and Suau, P. 1987. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 123: 51–58. [DOI] [PubMed] [Google Scholar]
- Polach K.J. and Widom, J. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258: 800–812. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy, J.P., Scamps, C., Martini, E.M., Lipinski, M., and Almouzni, G. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9: 1091–1100. [DOI] [PubMed] [Google Scholar]
- Redon C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12: 162–169. [DOI] [PubMed] [Google Scholar]
- Reinke H. and Horz, W. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11: 1599–1607. [DOI] [PubMed] [Google Scholar]
- Rio D.C. and Rubin, G.M. 1985. Transformation of cultured Drosophila melanogaster cells with a dominant selectable marker. Mol. Cell. Biol. 5: 1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling, A.C. 1983. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 11: 6341–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish M.A. and Struhl, K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B.E., Werner, J.K., and Lis, J.T. 2004. Indirect immunofluorescent labeling of Drosophila polytene chromosomes: Visualizing protein interactions with chromatin in vivo. Methods Enzymol. 376: 393–404. [DOI] [PubMed] [Google Scholar]
- Segraves W.A. and Hogness, D.S. 1990. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes & Dev. 4: 204–219. [DOI] [PubMed] [Google Scholar]
- Shibahara K. and Stillman, B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585. [DOI] [PubMed] [Google Scholar]
- Smith S.T., Petruk, S., Sedkov, Y., Cho, E., Tillib, S., Canaani, E., and Mazo, A. 2004. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat. Cell Biol. 6: 162–167. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Larsen, P.L., and Varshavsky, A. 1988. Mapping protein–DNA interactions in vivo with formaldehyde: Evidence that histone H4 is retained on a highly transcribed gene. Cell 53: 937–947. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Jackson, P.D., Clark, M.J., Brand, A.H., and Hoffmann, F.M. 1994. Specificity of bone morphogenetic protein-related factors: Cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5: 585–593. [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Thatcher T.H., MacGaffey, J., Bowen, J., Horowitz, S., Shapiro, D.L., and Gorovsky, M.A. 1994. Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena. Nucleic Acids Res. 22: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg J.H. 1993. Histone synthesis and turnover in alfalfa: Fast loss of highly acetylated replacement histone variant H3.2. J. Biol. Chem. 268: 4912–4917. [PubMed] [Google Scholar]
- ____. 1998. Dynamics of histone acetylation in Chlamydomonas reinhardtii. J. Biol. Chem. 273: 27602–27609. [DOI] [PubMed] [Google Scholar]
- Weintraub H. and Groudine, M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193: 848–856. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. 1998. Chromatin: Structure and function, 3rd ed. Academic Press, New York.
- Zhang K., Tang, H., Huang, L., Blankenship, J.W., Jones, P.R., Xiang, F., Yau, P.M., and Burlingame, A.L. 2002. Identification of acetylation and methylation sites of histone H3 from chicken erythrocytes by high-accuracy matrix-assisted laser desorption ionization-time-of-flight, matrix-assisted laser desorption ionization-postsource decay, and nanoelectro-spray ionization tandem mass spectrometry. Anal. Biochem. 306: 259–269. [DOI] [PubMed] [Google Scholar]