Abstract
Mu DNA transposition proceeds through a series of higher-order nucleoprotein complexes called transpososomes. The structural core of the transpososome is a tetramer of the transposase, Mu A, bound to the two transposon ends. High-resolution structural analysis of the intact transposase and the transpososome has not been successful to date. Here we report the structure of Mu A at 16-Å and the Type 1 transpososome at 34-Å resolution, by 3D reconstruction of images obtained by scanning transmission electron microscopy (STEM) at cryo-temperatures. Electron spectroscopic imaging (ESI) of the DNA-phosphorus was performed in conjunction with the structural investigation to derive the path of the DNA through the transpososome and to define the DNA-binding surface in the transposase. Our model of the transpososome fits well with the accumulated biochemical literature for this intricate transposition system, and lays a structural foundation for biochemical function, including catalysis in trans and the complex circuit of macromolecular interactions underlying Mu DNA transposition.
Keywords: DNA transposition, transpososome, transposase, site-specific recombination, 3D reconstruction, electron spectroscopic imaging, scanning transmission electron microscopy
A variety of DNA processes including replication, recombination, transcription, and transposition make use of higher-order nucleoprotein complexes (Echols 1986). These complexes are built by multiple proteins binding cooperatively at multiple DNA sites with the assistance of accessory proteins, which bend DNA to facilitate interaction of proteins bound at distant sites. The requirement for the assembly of such elaborate structures ensures high levels of specificity and a means of providing elaborate regulatory mechanisms.
Mu DNA was the first mobile element for which an in vitro transposition system (Mizuuchi 1983) was available (for a recent review, see Chaconas and Harshey 2002) and knowledge gained from this system has proven invaluable for more recent analyses of other transposable elements. Interestingly, the Mu system has also turned out to be one of the most complex and elaborate transposition systems studied to date. The Mu strand transfer reaction, the first stage in the transposition process, requires four proteins: Mu A and Mu B encoded by the transposon and HU and IHF from the host cell. Subsequent steps to complete the replicative transposition process of Mu require a variety of host replication proteins.
The Mu A protein active site cleaves at the Mu–host junctions and promotes transesterification of the free 3′OH Mu ends to a target DNA molecule. An acidic, metal coordinating DDE motif is a crucial element of the Mu active site, as for other transposons (Mizuuchi and Baker 2002). The Mu B protein captures the target DNA, recruits it to the active site of Mu A, and allosterically activates Mu A for the transesterification step. The Escherichia coli HU and IHF proteins are accessory factors that bend Mu DNA at specific locations to allow the assembly of a higher-order complex involving three transposase (Mu A)-binding sites at each end of Mu. The two Mu ends are separated by the 38-kb Mu genome. In addition, there is a transpositional enhancer located 1 kb from the left end. The enhancer contains several additional Mu A-binding sites as well as an IHF-binding site. The Mu A-binding sites in the ends and at the enhancer are recognized by separate subdomains of Mu A.
Mu A is inactive as a monomer but becomes catalytically proficient upon assembly into a higher-order nucleoprotein complex or transpososome. The various stages of Mu DNA transposition are mediated by a series of transpososome configurations, as summarized in Figure 1A. The transpososome core is a tetrameric unit of Mu A bound to the two Mu ends, where catalysis occurs in trans (reaction chemistry at one Mu end is promoted by Mu A bound to the other end in the complex [Aldaz et al. 1996; Savilahti and Mizuuchi 1996]). However, the lack of a structural three-dimensional framework for the Mu reaction has hindered a full understanding of the molecular details.
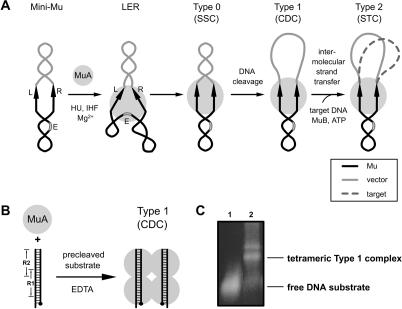
Figure 1.
Mu transpososome assembly. (A) Schematic of the Mu strand-transfer reaction (see Chaconas and Harshey 2002 and Supplementary Legend for Fig. 1 for a detailed legend). (B) Type 1 complex formation under simplified reaction conditions with a pair of identical precleaved linear DNA substrates (50 bp) containing the Mu A protein R1- and R2-binding sites (Savilahti et al. 1995). In the presence of EDTA, Type 1 complex is formed and the reaction cannot proceed beyond this point. Black dots indicate the cleavage point at the Mu termini. This Type 1 transpososome was used for our EM studies. (C) Gel electrophoresis of the Type 1 transpososome used for our EM studies. Electrophoresis was performed on a 2% Metaphor agarose gel, which was subsequently stained with SYBR gold. Lane 1 contains the DNA substrate and lane 2 contains the substrate incubated with Mu A protein. The band above the tetrameric complex contains additional unstably associated Mu A molecules (Wu and Chaconas 1997). Tetrameric complexes were selected for structural analysis based on a mass calculation during image processing of the electron micrographs.
NMR and X-ray crystallography, techniques permitting atomic resolution, have been successful only with isolated domains or subdomains of Mu A and not with the intact transposase or the transpososome. However, with lesser detail, electron microscopy (EM) permits structure determination of macromolecules and complexes that will not crystallize and that are too large for NMR (for reviews and discussions, see Frank 1996; Yang et al. 2003). EM can be coupled with electron spectroscopic imaging (ESI), a technique by which one can map specific atomic elements, such as the phosphorus in DNA or RNA, within a specimen (Egerton 1996). The combination of EM and ESI, and subsequent computer image analysis, has been used by us to determine the three-dimensional structures of a number of large nucleoprotein complexes, such as the ribosome (Beniac et al. 1997) and the nucleosome (Czarnota et al. 1997). To attain sufficient contrast for visibility of even smaller proteins, such as the 75-kDa Mu A monomer in this study, a scanning transmission electron microscope (STEM) equipped with a cryo-stage was used here to lyophilize quick-frozen aqueous specimens in the STEM stage, and then to image the specimens while still in situ in the STEM, by dark-field at cryo-temperatures. Low-electron-dose imaging (∼10 electrons/Å2), equivalent to that of imaging 2D crystals in conventional transmission electron microscopy (Kuhlbrandt et al. 1994), and specimen temperatures of –130°C served to minimize radiation-induced structural alterations.
While dark-field imaging using only scattered electrons is possible for visibility of small protein specimens with any conventional transmission electron microscope (CTEM), the STEM offers several crucial advantages: (1) The instrument has a 10-fold more efficient detection capability for elastically scattered electrons due to very large annular scintillator detectors, resulting in a 10-fold reduction in radiation dose to the specimen for equivalent signal strength. (2) STEM can instantaneously, simultaneously, and without loss in spatial resolution capture the inelastic scatter signal, which for carbonaceous biomolecules is threefold stronger than the elastic signal (Egerton 1996), equating to a further corresponding dose reduction not possible in CTEM. (3) For elemental mapping in STEM ESI (here for DNA-phosphorus) all energy loss images needed for mapping an elemental atomic signal are acquired simultaneously, pixel by pixel, resulting in a lower electron dose than in energy filtered CTEM and requiring no alignment a posteriori of the energy loss images.
Image processing developments in EM technology have led to techniques that can reconstruct a protein or particle in three dimensions from its electron micrographs. For specimens that are oriented randomly on the support film, computational approaches have been developed to calculate the initially unknown relative angular orientation of each 2D molecular image, which can then serve to reconstruct the 3D structure of the molecule by various methods, here by iterative quaternion-assisted angle determination (IQAD) (Farrow and Ottensmeyer 1992, 1993). One crucial advantage of the IQAD procedure is its capability to produce independent 3D reconstructions entirely free of any prior reference model.
Using STEM and computer image analysis by IQAD, we determined the 3D structure of the full-length 75-kDa Mu A protein at 16-Å resolution, measured at a Fourier shell correlation (FSC) of 0.5. We also solved the structure of the 367-kDa Mu Type 1 transpososome consisting of a tetramer of Mu A and two identical 50-bp DNA substrates, and determined the path of the DNA through the complex using ESI. The tetrameric transpososome structure, determined with detail at 34 Å, served to locate the constituent Mu A monomers and the separately determined ESI-DNA path within it. We propose a model of the transpososome that fits well with the accumulated biochemical literature for this intricate transposition system, and lays a structural foundation for biochemical function, including catalysis in trans, as well as the complex circuit of macromolecular interactions underlying Mu DNA transposition.
Results and Discussion
3D reconstruction of the Mu A protein monomer
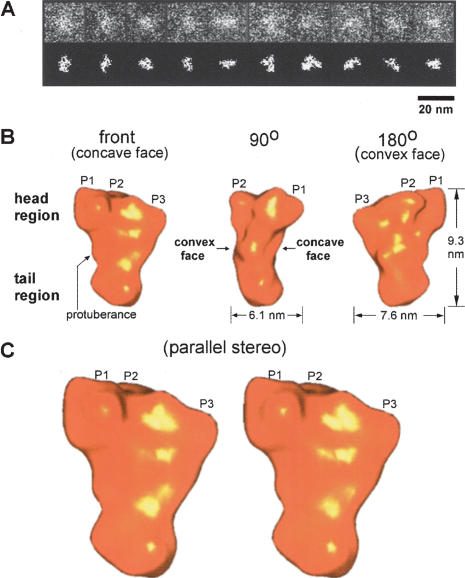
As the initial step we derived a 3D reconstruction of the Mu A monomer, which is the building block for the tetrameric Mu transpososome. Images of the unstained and unshadowed Mu A monomer were collected and processed as described in the Supplemental Material. A representative sampling of the single particle STEM dark-field images of Mu A are shown in the top part of Figure 2A, with the corresponding molecular images computationally segmented from the noisy background signal in the lower panel. The final 3D reconstruction of Mu A shown in Figure 2B and C is derived from 5408 images with a virtually isotropic distribution of orientations (Supplementary Fig. 11).
Figure 2.
3D reconstruction of the Mu A protein monomer. (A) Representative STEM dark-field single-particle images of the Mu A protein monomer. The top row shows the single-particle images cropped from the original STEM micrographs and low-pass filtered to enhance visibility in this photograph. The bottom row shows the corresponding images in the top row contrasted and segmented from the surrounding background signal. (B) 3D reconstruction of the Mu A monomer using 5408 molecular images, filtered to 16-Å resolution. The front view and rotations of 90° and 180° around the vertical axis are shown. The three protrusions in the head region of the structure are designated as P1, P2, and P3. (C) Front view in parallel stereo of the 3D reconstruction of the Mu A monomer.
The resolution of any 3D reconstruction can be determined by FSC (e.g., Cheng et al. 2004), or by phase residual calculations (Frank 1996). For the Mu A reconstruction, the FSC at 0.5 resulted in a measure of 16 Å, with the phase residual calculation yielding 16 Å at 60°, and 14 Å at the information limit of 90°. Since several different resolution cut-off criteria are used currently, full resolution curves are given in Supplementary Figure 10.
Since lyophilization from vitreous was used here as a necessary compromise to attain visibility in dark-field of a protein as small as the 75-kDa Mu A, a concern was the faithfulness of structural preservation during specimen preparation. So small a specimen is not detectable in vitreous ice using bright-field electron microscopy, and even in dark-field an aqueous frozen specimen rapidly loses contrast and visibility with increasing thickness of any support film or any surrounding medium such as vitreous ice. Lyophilization can sometimes, but not always, result in structural collapse of the sample (Yu and Egelman 1992). As a check on structural integrity, we therefore also imaged and reconstructed a uranyl-acetate-stained Mu A monomer structure. Even though the negative stain limits the resolution, and the chemical interaction may have caused some structural changes, the shape and dimensions (9.1 ± 0.7 nm, 7.7 ± 0.8 nm, and 6.7 ± 0.5 nm) of the negatively stained specimen agreed closely with that of the freeze-dried structure, below, suggesting that structural collapse did not occur in our lyophilized samples.
The three-dimensional reconstruction of the unstained lyophilized Mu A monomer structure has the shape of an acute angled triangle (Fig. 2B, front view), with overall dimensions of 9.3 nm, 7.6 nm, and 6.1 nm. This overall shape is in agreement with Mu A behaving like a 110-kDa globular protein on gel filtration chromatography, with a Stokes' radius of 4.2 nm (Kuo et al. 1991). In gross terms, the reconstructed triangular monomer has a concave face and a convex face along the long axis, giving the structure a distinct “head to tail” polarity. The head region has three protrusions (P1–P3). There is also a small protuberance extending from one side of the structure. On the concave face of the structure (Fig. 2C), there is a long continuous shallow groove extending from P1, continuing in front of P2, and then curving to the left to the side protuberance. This groove was identified as the DNA-binding surface for the transposase-binding sites, discussed below. (As an aid to 3D visualization of the structures presented in this paper, a Flash animation and structural files for use with Insight II are available at http://www.med.ucalgary.ca/webs/bprg/Chaconas/DownloadableFiles.html.)
3D reconstruction of the Mu Type 1 transpososome
3D reconstruction using electron micrographs requires that the molecule of interest be imaged in a large number of orientations. This is very difficult to achieve for the Mu Type 1 transpososome built with the usual mini-Mu plasmid, because of the immensity of that DNA substrate. We therefore minimized the size of the DNA substrate by using a 50-bp double-stranded oligonucleotide from the natural cleavage site through the R2 transposase-binding site (Fig. 1B,C; Savilahti et al. 1995). The “noncleaved” DNA strand contained four nucleotides of flanking DNA past the cleavage site. This substrate allowed robust assembly of the Type 1 transpososome in the absence of divalent metal ion, which is required for the subsequent strand transfer step.
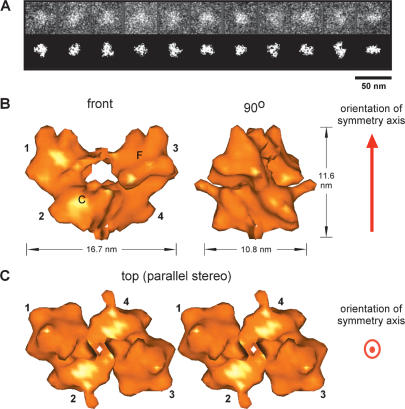
Images of the unstained and unshadowed Type 1 transpososome were collected and processed as described in Supplemental Material. A representative sampling of single particle STEM dark-field images and the corresponding images segmented from the background signal, as for the Mu A monomer, are shown in Figure 3A. The final 3D reconstruction of the Mu Type 1 transpososome, derived from 1912 images, is shown in Figure 3B and C. The resolution of the structure was determined to be 34 Å at an FSC of 0.5, and 36 Å at 60° by phase residual calculations. Orientations of the complex were random except for a disinclination toward two angular directions (see Supplementary Fig. 11).
Figure 3.
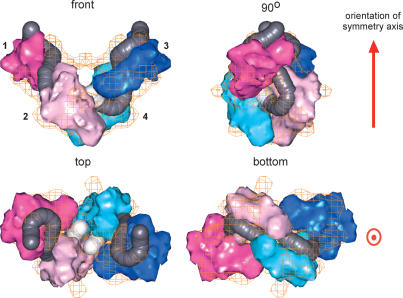
3D reconstruction of the Mu Type 1 transpososome. (A) Representative STEM dark-field single-particle images of the Mu Type 1 transpososome. The top row shows the single-particle images cropped from the original STEM micrographs and low-pass filtered. The bottom row shows the corresponding images contrasted and segmented from the surrounding background signal. (B) 3D reconstruction of the Mu Type 1 transpososome using 1912 molecular images, displayed at the theoretical molecular volume of the transpososome, corrected for DNA and protein densities. The front view and a 90° rotation around the vertical axis are shown. The orientation of the axis about which the structure displays twofold rotational symmetry (C2) is shown in red. Each of the four subunits is designated by a number. In the front view, the flat (slightly concave) face of subunit 3 and the convex face of subunit 2 are labeled F and C, respectively. Images were filtered to 20 Å to facilitate interpretation of fitting of Mu A monomers (Fig. 2) and DNA-phosphorus in subsequent figures. (C) Parallel stereo top view of the 3D reconstruction of the Mu Type 1 transpososome at 90° rotation from the front view around the horizontal axis, at a slightly higher mass-density threshold (smaller volume) than in B, showing a big central cleft in the transpososome structure, which has the appearance of a tunnel in the front view of B.
The lower spatial resolution of the 3D reconstruction of the complex, compared with that of the monomer (16 Å), is in part due to the lower number of particles used in the reconstruction as well as the coarser digitization (sampling) during imaging. However, another likely cause is a greater flexibility of the complex. Since slightly different conformations cannot be distinguished and separated in the sets of micrographs, the 3D reconstruction is an optimum but slightly blurred average of them all. Such structural flexibility seemed evident as well for 3D mapping of the DNA (below).
The 3D reconstruction represents the total mass of combined protein and DNA components. The transpososome structure displays a twofold rotational symmetry (C2) with overall dimensions of 16.7, 11.6, and 10.8 nm. It comprises four Mu A monomers plus two 50-bp oligomers of DNA, which is in agreement with the Mu transpososome being a tetrameric complex (Lavoie et al. 1991). In our reconstruction, each subunit contacts two others in the complex. Display of the reconstruction at a higher mass-density threshold indicates the extent of those contacts (Fig. 3C), bearing in mind that the DNA and protein mass could not be distinguished at this stage of the analysis. Assigning the structure a “top to bottom” orientation, as shown, the contact between the two bottom subunits (2 and 4) is extensive. The tight connection between them still exists under a very high mass-density threshold. The contact between the two top subunits (1 and 3) is a very tenuous 20-Å bridge. A slightly higher mass threshold completely eliminates the connection between them and leads to the exposure of a big central cleft in the structure (Fig. 3C). Subsequent analysis of individual STEM images with appropriate orientations revealed that the bridge between subunits 1 and 3 is present only in some micrographs, resulting in an averaged weak link between the monomers in the reconstruction. This is discussed further in a later section. Subunits 1 and 2 and subunits 3 and 4 also contact each other. No contact exists between subunits 1 and 4, or subunits 2 and 3.
The top subunits and the bottom subunits appear rotated with respect to each other, and therefore also have a different conformation in relation to their symmetrical opposites. This phenomenon may correspond to the different roles they play during the transposition process. Each monomer in the complex has a convex face along the long axis (Fig. 3B, front view), similar to that of the isolated monomer. However, the opposite face is flat or at most slightly concave in contrast to the very pronounced concave curvature of the Mu A monomer. This difference is due to the presence of Mu DNA in the transpososome as discussed below. Nonetheless, it was possible to assign an initial arrangement of the subunits in the transpososome by their convexity, even before consideration of the DNA component of the complex (see below).
Derivation of the DNA path through the transpososome by ESI
Based on the symmetry of the reconstructed structure, the expectation was that subunits 1 and 2 should bind to one of the substrate oligos, while subunits 3 and 4 should bind to the other. To obtain a measure of the three-dimensional path of the DNA in the transpososome, ESI was used to detect the inelastically scattered electrons interacting with the electronic L-shell of DNA-phosphorus atoms (Egerton 1996).
Obtaining microanalytical elemental information, the phosphorus distribution in this case, of necessity requires a very high electron dose compared with low-dose structural imaging of the complex (Egerton 1996). Nevertheless, the 3D reconstruction of the DNA phosphorus signal still indicated a resolution of 33 Å by phase residual at 60°, and 36 Å at an FSC of 0.5 (full curves are given in Supplementary Fig. 12). For display, the 3D signal was filtered to a cut-off of 30 Å, a level that retained strand continuity and preserved the major structural features (Fig. 4A). The phosphorus signal in the top subunits (1 and 3) is stronger than in the bottom subunits (2 and 4), suggestive of stronger DNA binding or less conformational flexibility in the top subunits. A higher threshold in the top and a lower threshold for the bottom subunits revealed the curvilinear paths formed by the phosphorus signals through subunits 1 and 2, or 3 and 4, respectively. In relation to the transpososome reconstruction, the DNA-phosphorus signal lies in the flat face of both top and bottom subunits, consistent with the initial assignment of the subunits in the complex by their convexity.
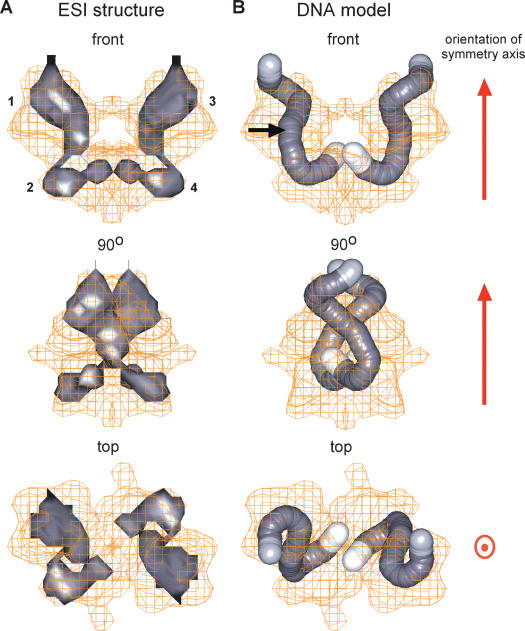
Figure 4.
3D reconstruction of the path of the DNA within the Mu Type 1 transpososome. (A) The transpososome is presented as a wire-mesh contour at the theoretical volume of the combined protein and DNA components. The DNA-phosphorus path (in gray) derived by electron spectroscopic imaging (ESI) is displayed in the complex at the same orientations as in Figure 3B and C, with numbers designating corresponding subunits. (B) Modeled DNA paths within the transpososome represented by a pair of curved rods with a diameter of 20 Å and a length of ∼170 Å. This DNA model was based on the phosphorus distribution from ESI and the modeling of the Mu A monomers into the transpososome structure (see text and Fig. 5). Each of the DNA rods contains the R1- and R2-binding sites and an extension of 5 bp at the R1 end representing the DNA between the R1 site and the Mu-terminal cleavage point (white portion ∼20 Å long at the bottom end of the ESI signal in the front and 90° views). The DNA portion at the top end in the same views is depicted in light gray in recognition of the weak ESI signal, potentially due to the flexible DNA end. The arrow shown in the front view indicates the position of the R1–R2 boundary.
The observed length of each DNA-phosphorus path is ∼150 Å with ∼75 Å in each subunit. In comparison, the expected length was 170 Å, with ∼20 Å more DNA in the bottom subunits, due to the presence of additional DNA in the oligo between the R1-binding site and the site of cleavage. The simplest interpretation of the observed length discrepancy is that the DNA in the area of the cleavage site may be flexible and therefore, similar to flexible protein regions investigated by X-ray crystallography, not seen in the 3D average of the ESI reconstruction. In modeling the DNA path in the transpososome, we therefore tentatively added a length of 5 bp of DNA in the bottom subunit at the R1 end (Fig. 4B). This region might become hyperflexible in the combined absence of divalent metal ion required for cleavage at the Mu–host junction and in the absence of flanking DNA sequences, which provide a large region (∼15 bp) of contact with Mu A in the Type 1 complex (Lavoie et al. 1991; Mizuuchi et al. 1991, 1992). This Mu A–flanking DNA contact is strong enough to destabilize the DNA helix in this region upon transpososome formation (Savilahti et al. 1995; Wang et al. 1996; Mizuuchi and Mizuuchi 2001), and its loss could untether the Mu DNA adjacent to the cleavage site. Although contact-induced DNA structural changes in the complex could also account for a decreased length, it seems unlikely that such changes would be specific to the bottom subunit alone. Similarly, a general weakening of the expected DNA-phosphorus signal strength due to the 33-Å resolution level of the ESI reconstruction might result in an apparent shorter DNA length; but such shortening was not seen in the top subunit, and would not be confined solely to the bottom subunit.
Each DNA trace has two bends, one occurring in the top subunit, the other in the bottom subunit. This corresponds well to the observed 80° bend at R1 and R2 when bound by Mu A (Kuo et al. 1991). Each bend is made up of two kinks measured to occur at positions 6 and 12 from the inside end of each R2 and R1 site in the ESI trace; the first of these kinks occurs at the midpoint between DNase I hypersensitive sites previously reported when these sites are bound by Mu A (Zou et al. 1991). The bends observed in our reconstruction of each oligo are out of plane, but are also out of phase with each other so they do not introduce any net writhe into the structure. Moreover, the two double-stranded oligos are not interwrapped. Recent topological analyses (Pathania et al. 2002, 2003) of the Mu transpososome have revealed a trapping of two negative supercoils between the left and right Mu ends; however, those studies were performed with an intact mini-Mu substrate carrying the intact Mu ends, including the accessory Mu A-binding sites L3 and R3, which are absent from the substrate used in this study.
Docking four Mu A monomers into the transpososome structure
The DNA path derived from the ESI study was used as a starting point for modeling the higher-resolution monomers into the transpososome structure. We assumed that the two Mu A monomers on the same DNA molecule would bind in a similar fashion, since both binding sites, R1 and R2, contain a Mu A consensus sequence (Craigie et al. 1984) in the same orientation. Docking of the four Mu A monomers into the transpososome structure was subject to the following constraints: (1) The paths of DNA within the top and the bottom subunits should be identical. (2) The subunits in the complex should retain the arrangement suggested already by their convexity. (3) The docking should optimize the fitting of the monomer contours with the general contour of the complex structure.
Since the monomer structure contains no DNA, and the transpososome structure is the combined mass of the protein and DNA components, the larger subunit volumes in the complex provided too few geometric constraints for a definitive placement of the Mu A monomers. Therefore, the volume of DNA as determined by ESI was subtracted from the total complex structure in order to help recognize the orientation of each monomeric Mu A subunit in the complex. Different density thresholds were used for the DNA in the top and the bottom subunits of the complex to adjust for the different ESI signal strengths. After DNA subtraction, the Mu A monomer orientations were recognized according to features in general shape, convexity, and protrusions of the remaining mass. In particular, the forward projecting portion of protrusion P1 (Fig. 2C) was now more obvious, as was the apex or tail of the triangular monomer shape, the combination unambiguously defining the location of P3. Minor adjustments in this alignment resulted in additionally matching smaller contour variations at the bottom edge of the complex that were initially overlooked. Small shifts of the monomers inside the contour of the complex then maximized the identity of the path of DNA in the top and the bottom subunits. This led to the model of the Mu Type 1 transpososome (Fig. 5) docked with four Mu A monomers and the DNA substrate comprising two 50-bp oligonucleotides.
Figure 5.
Structural modeling of the Mu Type 1 transpososome. Four Mu A monomer 3D reconstructions filtered to 12.5-Å resolution (solid surface) were docked into the transpososome complex (wire-mesh contour at 20-Å resolution; see Supplemental Material, 3D reconstructions). The two blue monomers bind to one DNA molecule, and the two pink monomers bind to the other. The two monomers with darker color bind to the R2 sites, while the lighter colored ones bind to the R1 sites. The pair of DNA paths within the transpososome is shown in gray and white (see text). Subunit number designation and the symmetry axis orientations are as in Figure 3B and C.
An important point to note is that no major differences in terms of overall shape and convexity were observed between the conformation of the isolated monomer and that of the monomers bound to DNA in the transpososome. The most obvious difference was an elongation of P2 by 13 Å in the bottom subunits of the transpososome (Fig. 5, top and bottom views). This is discussed further in relation to docking of atomic domains below. In addition, a 10% shortening in the apical direction of all subunits hints at a small degree of buckling of the protein around the bound DNA. Finally, a slender 20-Å bridge-like cantilever structure extends bilaterally between the top subunits (Figs. 3B, 5, front view), a structure that was directly visible in only some of suitably oriented molecular images. This structure likely originates from the N-terminal region of Mu A and is discussed later.
In the absence of direct evidence for the orientation of the DNA substrates in the transpososome, it is highly likely that the DNA portion bound by the top subunit (subunit 1 or 3) is the R2 site and that bound by the bottom subunit (subunit 2 or 4) is the R1 site, which is proximal to the Mu end cleavage point (Fig. 4A). Attempts at direct specific labeling of either end of the DNA substrate with nanogold (Nanoprobes) were unsuccessful. Nevertheless, the two top DNA ends are too far apart (116 Å) for DNA cleavage and strand transfer in trans (Aldaz et al. 1996; Savilahti and Mizuuchi 1996) without a massive rearrangement of the structure. The bottom two DNA ends are separated by only 27 Å, suggesting their identification as the R1 ends, which are expected to be near the target DNA splice sites, which are separated by 21 Å (see below). The 5-bp extension in the model (Fig. 4B) resulted in the DNA bound by one bottom subunit extending toward the other bottom subunit (Fig. 5). Given the fact that both the DNA cleavage and strand transfer at each Mu end are catalyzed in trans by the Mu A monomer bound to the partner Mu end (Aldaz et al. 1996; Savilahti and Mizuuchi 1996), such inter-subunit rapprochement makes biological sense.
Docking atomic domain structures into the 3D reconstruction of Mu A
Atomic structures are available for four individual Mu A domains: Iα, Iβ, Iγ, and II (Clubb et al. 1994, 1996, 1997; Rice and Mizuuchi 1995; Schumacher et al. 1997). A logical placement of these domains into the 3D reconstruction of the Mu A monomer needed to be consistent with the structure and function of the transpososome. The starting point for the assembly process was the DNA-binding surface of Mu A; this is delineated by the path of the DNA through the transpososome (Fig. 6B) that coincides with the long continuous groove in the Mu A reconstruction from protrusion 1 to the side protuberance (Fig. 2C). DNA binding here would fill the concave face of Mu A, producing a flat face in the complex (Fig. 6B), as observed in the transpososome reconstruction. This DNA placement, as dictated by the ESI results, is also consistent with hydroxyl radical foot-printing data (Zou et al. 1991); Mu A contacts the DNA helix only on one face in our structure.
Figure 6.
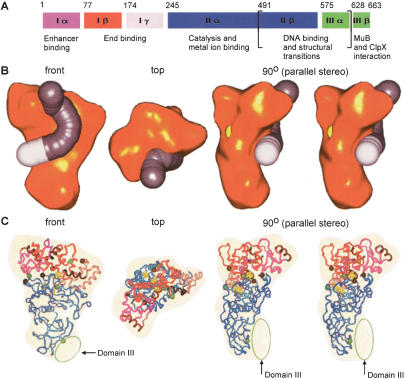
Structural modeling of the Mu A protein monomer. (A) Biochemical domain structure of the 663-amino acid Mu A protein (see Chaconas and Harshey 2002 and Supplementary Legend for Fig. 6 for a detailed legend). Subdomain Iα, Iβ, Iγ, domain II, and domain III are depicted in purple, red, pink, blue, and green, respectively. Numbers above the boxes refer to amino acid positions at the beginning of each functional region. (B) Modeled DNA substrate (curved rod) bound by a Mu A monomer. The DNA rod contains the R1-binding site (in gray with a length of ∼80 Å) and the 5-bp DNA between the R1 site and the Mu-terminal cleavage point (in white with a length of ∼20 Å). The front view, top view, and 90° view in parallel stereo are shown. The DDE motif in the 90° view is represented by a yellow ball structure for D336, the only DDE residue at the surface of the 3D reconstruction as fitted with the aligned atomic structure of domain II (see text and C). The DNA in the R2-binding site follows a similar path on a corresponding Mu A monomer. (C) Docking the known atomic domain structures into the EM 3D reconstruction of Mu A. The color scheme for domains is the same as in A. The views at which the 3D EM structure (transparent yellow) is displayed are the same as in B. The N and C termini of the atomic ribbon structures are represented as black and green spheres, respectively. The extra mass in the fitted EM reconstruction is assigned to domain III (green ellipses). The DNA recognition helices of subdomain Iα, Iβ, and Iγ are depicted in brown. The residues of the DDE motif (D269, D336, and E392) are yellow ball-stick representations.
At the atomic level, two independently folded subdomains of Mu A, Iβ and Iγ, are responsible for site-specific binding to a consensus sequence. Subdomain Iβ binds to the more conserved 3′ half and subdomain Iγ binds to the 5′ half at each of the binding sites (Schumacher et al. 1997). Chemical protection and interference studies suggested that Mu A achieves DNA binding specificity primarily through two consecutive major grooves (Zou et al. 1991).
Docking the known atomic domain structures into the 3D reconstruction of Mu A was subject to the following constraints: (1) Subdomain Iβ binds to the 3′ half of each consensus sequence and subdomain Iγ binds to the 5′ half. The DNA recognition helices of subdomain Iβ and Iγ are separated by approximately one helical turn and are positioned such that they can fully contact DNA. (2) The docking of every atomic structure optimizes the fitting with the local contour of the EM structure. (3) Globally, the positioning of the domain structures is in agreement with the order in which they are in the primary amino acid sequence. For example, the N terminus of subdomain Iβ is located in the vicinity of the C terminus of subdomain Iα. The last constraint is relatively flexible because there are 23 disordered amino acids between the atomic structures of subdomain Iα and Iβ, 13 between Iβ and Iγ, and 17 between Iγ and domain II. However, the 3D reconstruction has to accommodate these flexible regions at appropriate locations.
The atomic domain structures were initially docked into the EM structure by manual alignment. The docked subdomain Iα, Iβ, Iγ, and domain II were then combined into a global atomic structure. Using an in-house correlation-based search program, the docking position of the global atomic structure was optimized by translational and rotational searches around the initial docking position. The docking position with the highest correlation coefficient is shown in Figure 6C. Statistically it can be calculated that the accuracy of alignment of the centers of mass of the EM reconstruction and the global atomic structure is better than 1 Å. The rotational accuracy is difficult to quantify, but, as a subjective estimate, deliberate misalignments by as little as 3° from the positions shown in the three views in Figure 6C were visually clearly suboptimal.
In the model the three protrusions at the head region were assigned to subdomain Iα, Iβ, and Iγ, respectively. The large triangular-shaped domain II crystal structure nicely fitted into the contour of the lower portion of the EM structure, including such features as the convex curvature and the side protuberances (Fig. 6C). The remaining space at the tail of the EM structure was designated as domain III, for which no atomic structure is available. In our Mu A model, domain III is not a geometrically separate domain. Its structure closely interlocks with that of domain IIβ. It is known that domain IIβ and domain IIIα belong to a functionally related part of Mu A that has been associated with transpososome assembly and perhaps DNA binding at the Mu–host junction (Wu and Chaconas 1995; Krementsova et al. 1998; Naigamwalla et al. 1998; Namgoong et al. 1998; Mariconda et al. 2000). In our model the C terminus of domain II is suitably located to accommodate a potential domain III centrally in the space near the bottom of the transpososome reconstruction.
Docking of the known domain structures did not fill the entire region of protrusion P2. This region had different conformations in the monomer and transpososome 3D reconstructions as well as in the top and bottom subunits within the complex (see above). This space in the monomer reconstruction faces the C and N termini of docked atomic subdomains Iα and Iβ, respectively, and the C and N termini of Iγ and domain II. NMR and crystallography have shown that the linker regions between these subdomains, of 23 and 17 amino acids, are flexible (Clubb et al. 1994, 1996, 1997; Rice and Mizuuchi 1995; Schumacher et al. 1997). These regions have sufficient mass to fill P2 in the reconstruction, and their flexibility suggests that different conformations may occur in the P2 sites.
The modeling of the atomic subdomains of the Mu A monomer permitted an analysis of the surface electrostatic potential features of the fitted ensemble in relation to the location assigned to the DNA on the basis of ESI phosphorus mapping in the 3D reconstruction. The assigned DNA path on the surface of the fitted monomer was consistent with clear positive charge tracks consisting of eight lysines (18, 41, 44, 95, 148, 154, 461, 479) and 10 arginines (35, 37, 146, 168, 226, 227, 324, 450, 470, 474). Finally, in addition to fulfilling the structural and functional requirements discussed above, it is important to note that in our docked model, the catalytic DDE motif in domain II resides on the convex side of the Mu A monomer (Fig. 6B, stereo view). The position of this important region on the opposite face of the monomer from that used for DNA binding explains why the Mu A monomer is catalytically inactive; the active site residues and the bound substrate are spatially separated, providing a physical barrier to the reaction chemistry.
Structure and function of the Mu transpososome
Docking the available atomic structures of Mu A domains and subdomains into the reconstructed monomers within the complex points to an extensive number of features relating the structure and function of the Mu transpososome model shown in Figure 7. Although these features are derived from the transpososome structure reconstructed from the oligonucleotide system, they nonetheless provide valuable insight into more complex aspects of the reaction when using full-sized plasmid substrates.
Figure 7.
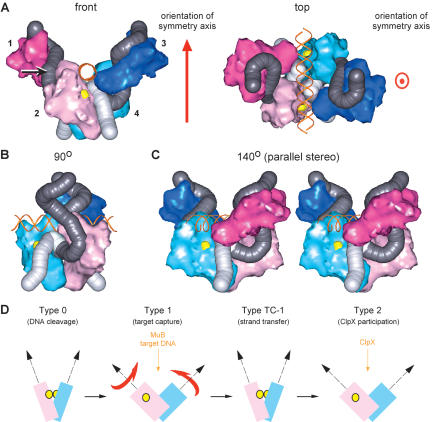
Extended structural modeling of the Mu Type 1 transpososome. (A) The Mu Type 1 transpososome model with target and vector (donor) DNA. A 35-bp target DNA (orange double-stranded helix in standard B form) and the vector DNA beyond the Mu–host junction (in light gray with a length of ∼80 Å) have been added to the structure in Figure 5. The two catalytic DDE motifs are represented as two yellow ball structures. The R1–R2 boundary and point at which the L1–L2 loop would be extruded is indicated by the arrow on the left side of the front view. Note that in relation to this structure and those in B and C, the schematics in Figure 1A are upside down. (B) The 90° rotation of the model is displayed with the darker pink subunit blanked. (C) The 140° rotation of the front view in A around the vertical axis is displayed in parallel stereo. (D) Structural transitions of the Mu transpososome. This model simulates subunits 2 and 4 in the front view of A. The rotational movements of the subunits are indicated by the red arrows when the transpososome transits to the catalytically active configuration. The two catalytic DDE motifs are represented as two yellow balls and are brought into proximity of the scissile phosphates through the combination of the rotational displacement and a scissors-like movement.
Catalysis in trans. As noted in the previous section, catalysis of DNA cleavage or strand transfer cannot occur to DNA bound by a single Mu A monomer. Assembly of the Mu A tetramer in the Type 1 transpososome results in juxtaposition of the DDE motif of subunit 2 to the R1 site bound by subunit 4, and vice versa, thereby allowing catalysis in trans, as expected (Fig. 7A top view, B and C). The DDE motifs in the subunits bound at the R2 sites (subunits 1 and 3) are not in a position to be able to contact the 3′ ends of the Mu DNA. Therefore, only the two DDE motifs in the subunits bound at the R1 sites (subunits 2 and 4) are reasonably disposed for catalysis. This is consistent with biochemical observations (Namgoong and Harshey 1998; Williams et al. 1999).
In our model each 3′-OH end of the Mu DNA is located ∼20 Å away from the in trans catalytic DDE motif. This distance is too large (Lovell et al. 2002) for catalysis and would require a conformational change, as expected from earlier biochemical studies on the Mu transpososome (to be discussed below). The distance between the two 3′-OH ends is ∼30 Å, larger than the 21-Å distance between the two scissile phosphates that are 5 bp apart on the opposite strands of the target DNA across the major groove. Such conformational flexibility, as also suggested by the ESI results, may not be exclusive to this transpososome: In comparison, the 41-Å distance between the two 3′ OH groups in the Tn5 structure is also larger than the expected 35 Å (9 bp) (Davies et al. 2000).
A gated target DNA-binding pocket. In the transpososome model (Fig. 7A–C) there is an intriguing large central cleft formed by all four monomers. The two catalytic DDE motifs are located at the very bottom of this cleft and the region above them is just large enough to accommodate a target DNA running front to back, through the transpososome. This location for the target DNA provides access to both the DDE motifs in subunits 2 and 4, as well as to the 3′ ends of the Mu DNA. This putative target DNA-binding pocket may also be gated by the slender bridge structure across the top of the cleft (Fig. 3B, front), which was observed in the tetrameric complex. This region may be involved in the conformational changes that accompany transition through the various assembly and reaction steps. Domain III (the tail region) of subunits 1 and 3 surrounds the target DNA-binding cleft. It is therefore ideally positioned for interaction with the Mu B protein on a bound target DNA (Wu and Chaconas 1994).
Enhancer DNA binding. The bridge spanning the cleft and closing the target DNA-binding pocket (Fig. 5, front view) is likely generated by a conformational change in the domain Iα region in subunits 2 and 4, which is the part of the Mu A monomer closest to this region. Since domain Iα is the enhancer-binding domain, its location within the cleft would provide the binding site for yet an additional piece of DNA to be brought into the complex and to establish the required complex set of interactions between the enhancer and the Mu ends (Allison and Chaconas 1992; Jiang et al. 1999). Domain Iα in subunits 1 and 3 would also be easily accessible to enhancer DNA traversing the upper portion of the transpososome. A corollary of this proposal, however, is that enhancer binding would block the access of target DNA into the complex. But this access could be easily accommodated by the transient nature of the bridge (not always present) and the transient nature of the Mu end-enhancer interactions in the LER stage and in the Type 0 and Type 1 complexes (Watson and Chaconas 1996; Kobryn et al. 2002; Pathania et al. 2002, 2003), which would allow windows of opportunity for target entry.
The L1–L2 loop. The usual substrate for Mu transpososome formation contains both a Mu left and a Mu right end, rather than the two right ends used here. During transpososome formation an 80-bp region between L1 and L2 is bound by the E. coli HU protein and is looped out to juxtapose the L1 and L2 sites (Lavoie and Chaconas 1993; Lavoie et al. 1996). Such a loop is compatible with our modeled structure and would exit and re-enter the transpososome in a position that faces the outside of the complex and is clear of any encumbrances that would hinder the extrusion of this DNA loop (arrow, Fig. 7A).
The Mu–Host junction. We modeled 24 bp of host DNA beyond the Mu–host junction in our transpososome model, in light gray (Fig. 7A–C). Approximately 10 bp of vector DNA beyond the Mu–host junction can be bound simultaneously by the domain IIα region of the catalytic subunit (the bottom subunit in trans) and the tail region (domains III and IIβ) of the subunit bound at the R2 site (the top subunit). As noted earlier, subdomains IIβ and IIIα are part of the same complementation group and both display DNA binding activity. Subdomain IIIα has been previously suggested as the region of Mu A interacting with the Mu–host junction (Wu and Chaconas 1995; Naigamwalla et al. 1998). A strong DNA kink put into the model at the Mu–host junction (e.g., Fig. 7B) is consistent with hydroxyl radical hypercleavage at this site (Lavoie et al. 1991), and allows the presence of both host DNA and target DNA in the complex at the same time, without steric clash.
Interactions stabilizing the transpososome. Subdomains IIβ and IIIα of the Mu A protein are associated with transpososome assembly (Baker et al. 1993; Wu and Chaconas 1995; Krementsova et al. 1998; Naigamwalla et al. 1998; Namgoong et al. 1998; Mariconda et al. 2000). In the transpososome model, subunits 2 and 4 interact through their tail regions (subdomain IIβ and domain III). This interaction could be an important part of the transpososome structure scaffold (Supplementary Fig. 8). A small protein–protein contact also exists between the tip of P1 of the bottom subunit and the side protuberance of the top subunit, adjoining the path of the DNA linking these subunits (Supplementary Fig. 9). Except for the above monomer–monomer interactions, all the other interfaces between the subunits (with the exception of the transient bridge) are formed by DNA, consistent with Mu A not being able to tetramerize without a DNA substrate. This is similar to the Tn5 synaptic complex structure in which the C-terminal domain functions as the central point of protein–protein interaction across the molecular twofold axis and protein–DNA contacts provide most of the interactions that stabilize the complex (Davies et al. 2000).
Conformational changes. Formation of the Mu transpososome and progression of the complex through the various reaction stages are accompanied by a series of conformational changes resulting in successive increases in transpososome stability (see Mizuuchi and Mizuuchi 2001; Chaconas and Harshey 2002; Goldhaber-Gordon et al. 2002a,b; Kobryn et al. 2002; Yanagihara and Mizuuchi 2003; Williams and Baker 2004). Some changes in the Mu A monomer structure were apparent upon tetramerization, as noted by comparison (Fig. 5) between the assembled four EM monomer structures and the 3D reconstruction of the actual transpososome (orange cage). However, these changes are small in comparison to the additional conformational changes required for catalysis to occur. In particular, the DDE motif must be appropriately juxtaposed with the cleavage site at the Mu–host junction for the donor cleavage step, and also with the scissile phosphates in target DNA for strand transfer to be effected.
The necessary conformational change in the transpososome structure can be accomplished in our model by rotating and shifting the two dimeric halves (subunits 1 and 2, versus subunits 3 and 4) with respect to each other (Fig. 7D). By a shift of 9.5 Å, and a rotation of the bottom subunit through 41°, subunits 2 and 4 are positioned such that each catalytic DDE motif is juxtaposed in trans with its Mu end, with the distance between the two 3′OH Mu ends small enough for attacking two scissile phosphates that are 5 bp apart on the opposite strands of the target DNA.
This type of conformational change, which is expected to occur during transit into the catalytically competent form of the Mu transpososome at both the donor cleavage and strand transfer steps as depicted in Figure 7D, seems large, but is not unique to the Mu model. Comparison of the Tn5/DNA synaptic complex with the catalytically inactive truncated Tn5 transposase dimer reveals that the relative rotational movement between domain structures accounts for most of the major conformational change that decreases the distance between active sites from 65 to 41 Å, while the separate domain configurations are quite constant with only a few local variations (Davies et al. 2000). Thus, the rotational movement of catalytic subunits (or catalytic domains) proposed above for the Mu transpososome to be catalytically active is of the same order of magnitude as that in the Tn5 synaptic complex. In the Tn5/DNA complex, DNA binding and/or addition of the N-terminal domain cause the catalytic domains to rotate ∼34° from their positions in the truncated dimer. Since in the Mu Type 1 transpososome no dramatic conformational change was discovered between the monomeric subunit and the separate Mu A monomer structure, the rotational movement is possibly triggered by factors other than DNA binding, such as the addition of Mu B or divalent cation.
Conclusion
We have reported the 3D reconstructions from electron micrographs of the Mu transposase and of the tetrameric Type 1 transpososome, including the path of the DNA through the transpososome using ESI. Both of these structures have been intractable to date by high-resolution structural methods. Our model of the transpososome fits well with the accumulated biochemical literature for this intricate transposition system, and lays a structural foundation for biochemical function. It also explains, among other reaction features, catalysis only in trans and the ability of the transpososome to accommodate so many pieces of DNA, including the two Mu ends, the adjacent host DNA, the Mu enhancer, the L1–L2 loop, and the target DNA. Our derived model provides a number of testable features and a framework for future experimentation. The approach described here should be useful for the study of a wide variety of higher-order complexes that are not amenable to study by X-ray crystallographic and NMR approaches alone.
Materials and methods
Protein, substrate, and transpososome formation
See Supplemental Material.
Specimen preparation and image acquisition
Summaries on specimen preparation, image acquisition, image analysis, and reconstruction are given here. Further details are appended in the Supplemental Material.
The electron microscope used in this study was a scanning transmission electron microscope (Vacuum Generators, HB601UX STEM), equipped with a cryo-stage and transfer system, high- and low-angle annular dark-field detectors for elastic low-dose imaging, and a PEELS 666 electron spectrometer (Gatan Inc.). The linear diode array detector of the spectrometer was replaced with a rectangular multichannel photomultiplier tube (Hamamatsu, Model H7260) with four consecutive sets of six channels wired together. This provided simultaneous nanosecond signal acquisition in any or all of four contiguous energy regions of the spectrum for ESI of the phosphorus signal, or in a single energy band in the low energy loss region by combining the four signals for low-dose inelastic imaging.
A 5-μL drop of the transpososome reaction mixture at 50 μg/mL was spread on a 30-Å-thin carbon film supported on a holey-plastic-coated copper grid. After incubation and wash, the wet specimen grids were freeze-plunged, freeze-dried to achieve sufficient image contrast, and imaged under cryo conditions. Dark-field electron micrographs were acquired at 500,000× magnification (3.3 Å/pixel) with a dose at 14.7 e/Å2, to image the Mu A protein monomer, whereas the transpososomes were imaged at 252,000× magnification (6.54 Å/pixel) with a dose at 7.4 e/Å2.
2D image analysis
All calculations and processing were performed using the SPIDER and WEB software package (Wadsworth Laboratories) unless otherwise stated. Single molecule images were extracted from the initial micrographs, selected based on calculated molecular mass, normalized to correct for changes in beam intensity over the period of image acquisition, low-pass filtered to reduce the background noise for initial processing, and then segmented from the surrounding carbon background signal. After molecular orientation calculations were made, 3D reconstructions used the unfiltered original images at those orientations.
3D reconstructions
Thirty-two 3D reconstructions for the Mu A protein monomer were initially computed from 32 independent image sets via IQAD using randomly chosen starting images (Farrow and Ottensmeyer 1992, 1993). The initial 32 3D reconstructions were then refined to a final structure using an approach based on projection matching (Penczek et al. 1994).
Twenty-four 3D reconstructions were initially computed for the Mu Type 1 transpososome. No symmetry was imposed at this stage. The initial 3D reconstructions of the transpososome complex manifested a twofold, but not fourfold, rotational symmetry. Subsequent refinement of the transpososome structure was processed in a similar manner as the Mu A monomer, with the exception that twofold rotational symmetry was now imposed in the data.
Resolution of the final 3D reconstructions was determined by comparison of half, third, and quarter subaverages by Fourier shell correlation and by phase residual calculations (Frank 1996).
The 3D reconstructions were visualized using InsightII (Accelrys). For surface representations, threshold values were selected using the molecular volume as a guide. Docking of the monomer protein structure into the tetramer nucleoprotein structure was assisted by details in the structures filtered variously to levels between 12.5 and 34 Å.
Electron spectroscopic imaging (ESI)
In addition to low-dose imaging for molecular reconstruction, the transpososomes were also imaged at higher doses for phosphorus microanalysis using ESI in the STEM at 252,000× magnification. Each ESI image set comprised three images taken simultaneously in three 12.5-eV energy bands centered at 112.5, 125, and 150 eV energy loss. For phosphorus mapping the detector-sensitivity-corrected intensities (I) of the images at energy losses (E) of 112.5 and 125 eV were used to obtain an extrapolated background signal at 150 eV using a pixel-by-pixel fit to the canonical curve of I = A*E–R (Egerton 1996). An image of the characteristic net phosphorus signal was obtained by subtracting the extrapolated 150 eV background signal from the corresponding experimental 150 eV image. Image selection was based on the strength of the net phosphorus signal, with 747 of 1100 three image sets with the highest net phosphorus signal being retained for reconstruction. A sum image, adding each of the 112.5-, 125-, and 150-eV energy loss sets, was calculated as well to maximize the total structural information content of the transpososome protein and DNA mass images. These sum images were used for orientation determination by projection matching alignment with the low-dose reconstruction. For 3D reconstruction, the images of the corresponding net phosphorus signal at the same orientations were then back-projected into a 3D structure.
Acknowledgments
We thank Ilana Goldhaber-Gordon and Tania Baker for help with the Mu oligo transposition system. We also thank Michael Surette, Kerri Kobryn, Colin Coros, Marie Fraser, and Ed Egelman for a critical reading of the manuscript. This research was undertaken, in part, thanks to funding from the Canadian Institutes of Health Research (CIHR) to G.C. and F.P.O., the National Science and Engineering Research Council (NSERC) to F.P.O., the National Cancer Institute of Canada to F.P.O., and the Canada Research Chairs Program to G.C. J.F.Y. was supported by a CIHR Studentship and G.C. was supported by a Distinguished Scientist Award from the CIHR, a Scientist Award from the Alberta Heritage Fund for Medical Research, and a Canada Research Chair in the Molecular Biology of Lyme Disease.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1291405.
References
- Aldaz H., Schuster, E., and Baker, T.A. 1996. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85: 257–269. [DOI] [PubMed] [Google Scholar]
- Allison R.G. and Chaconas, G. 1992. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. A complex circuit of interactions involving the Mu ends and the transpositional enhancer. J. Biol. Chem. 267: 19963–19970. [PubMed] [Google Scholar]
- Baker T.A., Mizuuchi, M., Savilahti, H., and Mizuuchi, K. 1993. Division of labor among monomers within the Mu transposase tetramer. Cell 74: 723–733. [DOI] [PubMed] [Google Scholar]
- Beniac D.R., Czarnota, G.J., Rutherford, B.L., Ottensmeyer, F.P., and Harauz, G. 1997. The in situ architecture of Escherichia coli ribosomal RNA derived by electron spectroscopic imaging and three-dimensional reconstruction. J. Microsc. 188: 24–35. [DOI] [PubMed] [Google Scholar]
- Chaconas G. and Harshey, R.M. 2002. Transposition of phage Mu DNA. In Mobile DNA II (eds. N.L. Craig et al.), pp. 384–402. ASM Press, Washington, DC.
- Cheng Y., Zak, O., Aisen, P., Harrison, S.C., and Walz, T. 2004. Structure of the human transferrin receptor–transferrin complex. Cell 116: 565–576. [DOI] [PubMed] [Google Scholar]
- Clubb R.T., Omichinski, J.G., Savilahti, H., Mizuuchi, K., Gronenborn, A.M., and Clore, G.M. 1994. A novel class of winged helix–turn–helix protein: The DNA-binding domain of Mu transposase. Structure 2: 1041–1048. [DOI] [PubMed] [Google Scholar]
- Clubb R.T., Mizuuchi, M., Huth, J.R., Omichinski, J.G., Savilahti, H., Mizuuchi, K., Clore, G.M., and Gronenborn, A.M. 1996. The wing of the enhancer-binding domain of Mu phage transposase is flexible and is essential for efficient transposition. Proc. Natl. Acad. Sci. 93: 1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clubb R.T., Schumacher, S., Mizuuchi, K., Gronenborn, A.M., and Clore, G.M. 1997. Solution structure of the Iγ subdomain of the Mu end DNA-binding domain of phage Mu transposase. J. Mol. Biol. 273: 19–25. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi, M., and Mizuuchi, K. 1984. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell 39: 387–394. [DOI] [PubMed] [Google Scholar]
- Czarnota G.J., Bazett-Jones, D.P., Mendez, E., Allfrey, V.G., and Ottensmeyer, F.P. 1997. High resolution microanalysis and three-dimensional nucleosome structure associated with transcribing chromatin. Micron 28: 419–431. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Goryshin, I.Y., Reznikoff, W.S., and Rayment, I. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85. [DOI] [PubMed] [Google Scholar]
- Echols H. 1986. Multiple DNA–protein interactions governing high-precision DNA transactions. Science 233: 1050–1056. [DOI] [PubMed] [Google Scholar]
- Egerton R.F. 1996. Electron energy-loss spectroscopy in the electron microscope. Plenum Press, New York.
- Farrow N.A. and Ottensmeyer, F.P. 1992. A posterori determination of relative projection directions of arbitrarily oriented macromolecules. J. Opt. Soc. Am. A9: 1749–1760. [Google Scholar]
- ____. 1993. Automatic 3D alignment of projection images of randomly oriented objects. Ultramicroscopy 52: 141–156. [Google Scholar]
- Frank J. 1996. Three-dimensional electron microscopy of macromolecular assemblies. Academic Press, San Diego.
- Goldhaber-Gordon I., Early, M.H., Gray, M.K., and Baker, T.A. 2002a. Sequence and positional requirements for DNA sites in a mu transpososome. J. Biol. Chem. 277: 7703–7712. [DOI] [PubMed] [Google Scholar]
- Goldhaber-Gordon I., Williams, T.L., and Baker, T.A. 2002b. DNA recognition sites activate MuA transposase to perform transposition of non-Mu DNA. J. Biol. Chem. 277: 7694–7702. [DOI] [PubMed] [Google Scholar]
- Jiang H., Yang, J.Y., and Harshey, R.M. 1999. Criss-crossed interactions between the enhancer and the att sites of phage Mu during DNA transposition. EMBO J. 18: 3845–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobryn K., Watson, M.A., Allison, R.G., and Chaconas, G. 2002. The Mu three-site synapse: A strained assembly platform in which delivery of the L1 transposase binding site triggers catalytic commitment. Mol. Cell 10: 659–669. [DOI] [PubMed] [Google Scholar]
- Krementsova E., Giffin, M.J., Pincus, D., and Baker, T.A. 1998. Mutational analysis of the Mu transposase. Contributions of two distinct regions of domain II to recombination. J. Biol. Chem. 273: 31358–31365. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W., Wang, D.N., and Fujiyoshi, Y. 1994. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367: 614–621. [DOI] [PubMed] [Google Scholar]
- Kuo C.F., Zou, A.H., Jayaram, M., Getzoff, E., and Harshey, R. 1991. DNA–protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 10: 1585–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D. and Chaconas, G. 1993. Site-specific HU binding in the Mu transpososome: Conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes & Dev. 7: 2510–2519. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Chan, B.S., Allison, R.G., and Chaconas, G. 1991. Structural aspects of a higher order nucleoprotein complex: Induction of an altered DNA structure at the Mu–host junction of the Mu type 1 transpososome. EMBO J. 10: 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Shaw, G.S., Millner, A., and Chaconas, G. 1996. Anatomy of a flexer–DNA complex inside a higher-order transposition intermediate. Cell 85: 761–771. [DOI] [PubMed] [Google Scholar]
- Lovell S., Goryshin, I.Y., Reznikoff, W.R., and Rayment, I. 2002. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat. Struct. Biol. 9: 278–281. [DOI] [PubMed] [Google Scholar]
- Mariconda S., Namgoong, S.Y., Yoon, K.H., Jiang, H., and Harshey, R.M. 2000. Domain III function of Mu transposase analysed by directed placement of subunits within the transpososome. J. Biosci. 25: 347–360. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. 1983. In vitro transposition of bacteriophage Mu: A biochemical approach to a novel replication reaction. Cell 35: 785–794. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. and Baker, T.A. 2002. Chemical mechanisms for mobilizing DNA. In Mobile DNA II (eds. N.L. Craig et al.), pp. 12–23. ASM Press, Washington, D.C.
- Mizuuchi M. and Mizuuchi, K. 2001. Conformational isomerization in phage Mu transpososome assembly: Effects of the transpositional enhancer and of MuB. EMBO J. 20: 6927–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi M., Baker, T.A., and Mizuuchi, K. 1991. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc. Natl. Acad. Sci. 88: 9031–9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1992. Assembly of the active form of the transposase–Mu DNA complex: A critical control point in Mu transposition. Cell 70: 303–311. [DOI] [PubMed] [Google Scholar]
- Naigamwalla D.Z., Coros, C.J., Wu, Z., and Chaconas, G. 1998. Mutations in domain IIIα of the Mu transposase: Evidence suggesting an active site component which interacts with the Mu–host junction. J. Mol. Biol. 282: 265–274. [DOI] [PubMed] [Google Scholar]
- Namgoong S.Y. and Harshey, R.M. 1998. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 17: 3775–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S.Y., Kim, K., Saxena, P., Yang, J.Y., Jayaram, M., Giedroc, D.P., and Harshey, R.M. 1998. Mutational analysis of domain IIβ of bacteriophage Mu transposase: Domains IIα and IIβ belong to different catalytic complementation groups. J. Mol. Biol. 275: 221–232. [DOI] [PubMed] [Google Scholar]
- Pathania S., Jayaram, M., and Harshey, R.M. 2002. Path of DNA within the Mu transpososome: Transposase interactions bridging two Mu ends and the transposition enhancer trap five DNA supercoils. Cell 109: 425–436. [DOI] [PubMed] [Google Scholar]
- ____. 2003. A unique right end-enhancer complex precedes synapsis of Mu ends: The enhancer is sequestered within the transpososome throughout transposition. EMBO J. 22: 3725–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P., Grassuci, R.A., and Frank, J. 1994. The ribosome at increased resolution: New techniques for merging and orientation refinement in 3D cryoelectron microscopy of biological particles. Ultramicroscopy 53: 251–270. [DOI] [PubMed] [Google Scholar]
- Rice P. and Mizuuchi, K. 1995. Structure of the bacteriophage Mu transposase core: A common structural motif for DNA transposition and retroviral integration. Cell 82: 209–220. [DOI] [PubMed] [Google Scholar]
- Savilahti H. and Mizuuchi, K. 1996. Mu transpositional recombination—Donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell 85: 271–280. [DOI] [PubMed] [Google Scholar]
- Savilahti H., Rice, P.A., and Mizuuchi, K. 1995. The phage Mu transpososome core—DNA requirements for assembly and function. EMBO J. 14: 4893–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher S., Clubb, R.T., Cai, M., Mizuuchi, K., Clore, G.M., and Gronenborn, A.M. 1997. Solution structure of the Mu end DNA-binding Iβ subdomain of phage Mu transposase: Modular DNA recognition by two tethered domains. EMBO J. 16: 7532–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Namgoong, S.-Y., Zhang, X., and Harshey, R.M. 1996. Kinetic and structural probing of the pre-cleavage synaptic complex (type 0) formed during phage Mu transposition: Action of metal ions and reagents specific to single stranded DNA. J. Biol. Chem. 271: 9619–9626. [DOI] [PubMed] [Google Scholar]
- Watson M.A. and Chaconas, G. 1996. Three-site synapsis during Mu DNA transposition: A critical intermediate preceding engagement of the active site. Cell 85: 435–445. [DOI] [PubMed] [Google Scholar]
- Williams T.L. and Baker, T.A. 2004. Reorganization of the Mu transpososome active sites during a cooperative transition between DNA cleavage and joining. J. Biol. Chem. 279: 5135–5145. [DOI] [PubMed] [Google Scholar]
- Williams T.L., Jackson, E.L., Carritte, A., and Baker, T.A. 1999. Organization and dynamics of the Mu transpososome: Recombination by communication between two active sites. Genes & Dev. 13: 2725–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. and Chaconas, G. 1994. Characterization of a region in phage Mu transposase that is involved in interaction with the Mu B protein. J. Biol. Chem. 269: 28829–28833. [PubMed] [Google Scholar]
- ____. 1995. A novel DNA binding and nuclease activity in domain III of Mu transposase: Evidence for a catalytic region involved in donor cleavage. EMBO J. 14: 3835–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997. The Mu transposase tetramer is inactive in unassisted strand transfer: An auto-allosteric effect of Mu A promotes the reaction in the absence of Mu B. J. Mol. Biol. 267: 132–141. [DOI] [PubMed] [Google Scholar]
- Yanagihara K. and Mizuuchi, K. 2003. Progressive structural transitions within Mu transpositional complexes. Mol. Cell 11: 215–224. [DOI] [PubMed] [Google Scholar]
- Yang S., Yu, X., Galkin, V.E., and Egelman, E.H. 2003. Issues of resolution and polymorphism in single-particle reconstruction. J. Struct. Biol. 144: 162–171. [DOI] [PubMed] [Google Scholar]
- Yu X. and Egelman, E.H. 1992. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 227: 334–346. [DOI] [PubMed] [Google Scholar]
- Zou A.H., Leung, P.C., and Harshey, R.M. 1991. Transposase contacts with Mu DNA ends. J. Biol. Chem. 266: 20476–20482. [PubMed] [Google Scholar]